Abstract
We sought to identify genome-wide variants influencing antihypertensive drug response and adverse cardiovascular outcomes, utilizing data from four randomized controlled trials in the International Consortium for Antihypertensive Pharmacogenomics Studies (ICAPS). Genome-wide antihypertensive drug-single nucleotide polymorphism (SNP) interaction tests for four drug classes (β-blockers, n = 9,195; calcium channel blockers (CCBs), n = 10,511; thiazide/thiazide-like diuretics, n = 3,516; ACE-inhibitors/ARBs, n = 2,559) and cardiovascular outcomes (incident myocardial infarction, stroke, or death) were analyzed among patients with hypertension of European ancestry. Top SNPs from the meta-analyses were tested for replication of cardiovascular outcomes in an independent Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) study (n = 21,267), blood pressure (BP) response in independent ICAPS studies (n = 1,552), and ethnic validation in African Americans from the Genetics of Hypertension Associated Treatment study (GenHAT; n = 5,115). One signal reached genome-wide significance in the β-blocker-SNP interaction analysis (rs139945292, Interaction P = 1.56 × 10−8). rs139945292 was validated through BP response to β-blockers, with the T-allele associated with less BP reduction (systolic BP response P = 6 × 10−4, Beta = 3.09, diastolic BP response P = 5 × 10−3, Beta = 1.53). The T-allele was also associated with increased adverse cardiovascular risk within the β-blocker treated patients’ subgroup (P = 2.35 × 10−4, odds ratio = 1.57, 95% confidence interval = 1.23–1.99). The locus showed nominal replication in CHARGE, and consistent directional trends in β-blocker treated African Americans. rs139945292 is an expression quantitative trait locus for the 50 kb upstream gene NTM (neurotrimin). No SNPs attained genome-wide significance for any other drugs classes. Top SNPs were located near CALB1 (CCB), FLJ367777 (ACE-inhibitor), and CES5AP1 (thiazide). The NTM region is associated with increased risk for adverse cardiovascular outcomes and less BP reduction in β-blocker treated patients. Further investigation into this region is warranted.
Hypertension (HTN) affects ~ 31% (1.39 billion) of adults worldwide and ~ 46% (166.4 million) of adults in the United States.1,2 Additionally, HTN is the leading cause of cardiovascular disease globally.2 Numerous antihypertensive drug classes are considered appropriate first-line therapy to lower blood pressure (BP), including thiazide diuretics, calcium channel blockers (CCBs), angiotensin-converting enzyme (ACE) inhibitors, and angiotensin receptor blockers (ARBs).3 Although β-blockers were previously considered appropriate first-line therapy in uncomplicated HTN,4 they are currently considered secondary agents in uncomplicated HTN and first-line agents in some patients with complicated HTN (e.g., patients with stable ischemic heart disease or post-myocardial infarction (MI)).3 Additionally, β-blockers are first-line therapy in patients with heart failure.5 All antihypertensive drugs are ultimately prescribed to reduce or prevent the long-term cardiovascular complications of HTN.3 However, there exists great interpatient variability in antihypertensive drug response, with only about 50% of patients achieving an adequate BP response to any one drug, and limited data available to guide treatment selection.3 Why patients respond differently to the same drug and why some patients experience adverse cardiovascular outcomes despite BP control while others do not remains poorly understood.
Multiple landmark randomized controlled trials (RCTs) have been conducted investigating different antihypertensive therapies and adverse cardiovascular outcomes.6–10 Through these studies, clinical factors, such as age, sex, race, smoking, history of MI, history of diabetes, prior stroke, and history of heart failure, have been associated with increased risk for adverse cardiovascular outcomes during antihypertensive treatment.11 Additionally, pharmacogenomic studies have identified several candidate genes associated with adverse cardiovascular outcomes and various antihypertensive drugs.12–17 However, these candidate genes often fail to replicate between studies. One prior genome-wide association study (GWAS) meta-analysis studying antihypertensive treatment and adverse cardiovascular outcomes within the observational study Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium has been published.18 Although the study included ~ 15,000 individuals and investigated antihypertensive drug-SNP (single nucleotide polymorphism) interaction associations with adverse cardiovascular outcomes for four different antihypertensive drug classes (ACE-inhibitors, β-blockers, CCBs, and diuretics), no statistically significant interactions were identified.18
The International Consortium for Antihypertensive Pharmacogenomic Studies (ICAPS), aimed to further prior research by conducting this genome-wide meta-analyses to identify single nucleotide polymorphisms (SNPs) that influence adverse cardiovascular outcomes during antihypertensive drug treatment, using data obtained from RCTs, as opposed to the observational nature of the data from the CHARGE studies. We were interested to see whether pharmacogenetics analyses within more appropriately designed RCTs enable improved detection of genetic variants influencing adverse cardiovascular risk. Herein, we report the genome-wide meta-analysis antihypertensive drug class-SNP interaction results for adverse cardiovascular outcomes in participants of European ancestry utilizing data from four RCTs within ICAPS and testing over 6.4 million SNPs.
METHODS
Study participants and inclusion criteria
Within the discovery phase, we conducted drug × SNP interaction GWAS meta-analyses for four antihypertensive drug classes including (i) β-blockers, (ii) CCBs, (iii) ACE inhibitors/ARBs, and (iv) thiazide/thiazide-like diuretics. The discovery phase only included patients with HTN of European/European-American ancestry. The GWAS meta-analyses for β-blockers and CCBs included participants from four HTN RCTs, which consisted of five study cohorts: Anglo-Scandinavian Cardiac Outcomes Trial–United Kingdom and Ireland (ASCOT-UK), 19 Anglo-Scandinavian Cardiac Outcomes Trial-Scandinavians (ASCOT-SC), 19 Action to Control Cardiovascular Risk in Diabetes (ACCORD),20 International Verapamil SR-Trandolapril Study (INVEST),21 and Secondary Prevention of Small Subcortical Strokes (SPS3).22 Whereas the GWAS meta-analyses for ACE-inhibitors/ARBs and thiazide/thiazide-like diuretics only included three RCTs: ACCORD, INVEST, and SPS3. Details of the individual studies are available in the Supplementary Methods. Briefly, each study was a large, adverse cardiovascular outcome RCT comparing antihypertensive drug strategies in HTN participants (ASCOT and INVEST),19,21 or with a BP-lowering arm comparing target BP control in diabetic participants (ACCORD)20 or lacunar-stroke participants (SPS3).22 All participants included in the analyses were hypertensive.
For independent replication, the prior results from the GWAS meta-analysis studying antihypertensive drug and adverse cardiovascular outcomes within the CHARGE consortium were used for lookups.18 Details of the individual studies included in CHARGE are available in the Supplementary Methods and have been previously published.18
The BP response validation utilized the results from the discovery phase of the ICAPS β-blocker response GWAS meta-analysis. 23 Data from African American participants from the Genetics of Hypertension-Associated Treatment (GenHAT) were included for ethnic validation.24 Details of the individual studies included in the validation phases are available in the Supplementary Methods.
All studies were approved by local institutional review boards or ethics committees. All studies were conducted in accordance with the regulations set forth by the Declaration of Helsinki. The data from ACCORD are publicly available on the Biological Specimen and Data Repository Information Coordinating Center (BioLINCC): https://biolincc.nhlbi.nih.gov/studies/accord/. The data from INVEST are available on the database of Genotypes and Phenotypes (dbGaP; dbGaP Study Accession: phs002319.v1.p1). The data from SPS3 are included within the NINDS Stroke Genetics Network (SiGN) upload on dbGaP (dbGaP Study Accession: phs000625.v1.p1).
Definition of drug exposure
We examined four broad antihypertensive drug class: (i) β-blockers, (ii) CCBs, (iii) ACE inhibitors/ARBs, and (iv) thiazide/thiazide-like diuretics. A study participant was considered to be exposed to a particular drug class if there was a documented prescription for the drug at two or more study visits. Study participants meeting this exposure criteria were included in the exposed group, and study participants who were users of other drug classes were included in the reference group. Thus, if participants were exposed to multiple antihypertensive drug classes, these individuals appeared in multiple analyses, but only within either the exposed or reference group.
Definition of adverse cardiovascular outcome
The primary phenotype of the study was adverse cardiovascular outcomes. An adverse cardiovascular outcome was defined as the first occurrence of nonfatal stroke, nonfatal MI, and all-cause death. Adverse cardiovascular outcomes included in the meta-analyses were outcomes adjudicated during the defined follow-up period from each RCT, similar to an intent-to-treat design.6–9,19,21,22,25
Genotyping, quality control, and imputation
Genome-wide SNP genotyping was conducted within each study using an Affymetrix or Illumina genotyping panel. Additional details on genotyping in each study are available in the Supplementary Methods. Each study underwent standard quality control (QC) procedures including exclusion of participants based on sex mismatch or duplication, exclusions of participants based on heterogeneity, exclusion of samples with genotyping call rates < 95%, and exclusion of SNPs with genotyping call rates < 95 to < 99%. Principal components for ancestry were identified using the EIGENSTRAT method.26 Genotype imputation for the discovery phase was conducted by study to the 1000G Phase I reference panel (ASCOT) and 1000G Phase III reference panel (ACCORD, INVEST, and SPS3). For the replication phase and validation phase, CHARGE was imputed to the HapMap Phase II reference panel, ICAPS β-blocker response was imputed to the 1000G Phase III reference panel, and GenHAT was imputed to the 1000G Phase III reference panel.
Statistical analysis
The demographics and characteristics for each study were described as numbers and percentages (categorical variables) and as mean ± SD (continuous variables).
We conducted a multistage GWAS meta-analysis (Figure 1). The first stage (Discovery) was a discovery GWAS meta-analysis for each of the four drug classes, followed by testing the top signals from each drug class in the CHARGE replication cohort (Replication). The validation stage consisted of a BP response validation (BP Response Validation) for genome-wide significant signals from our discovery β-blocker drug class GWAS interaction analysis, and a directional validation and ethnic validation (Directional and Ethnic Validation) for SNPs passing the replication or validation thresholds in the replication stage or BP response validation stage.
Figure 1.
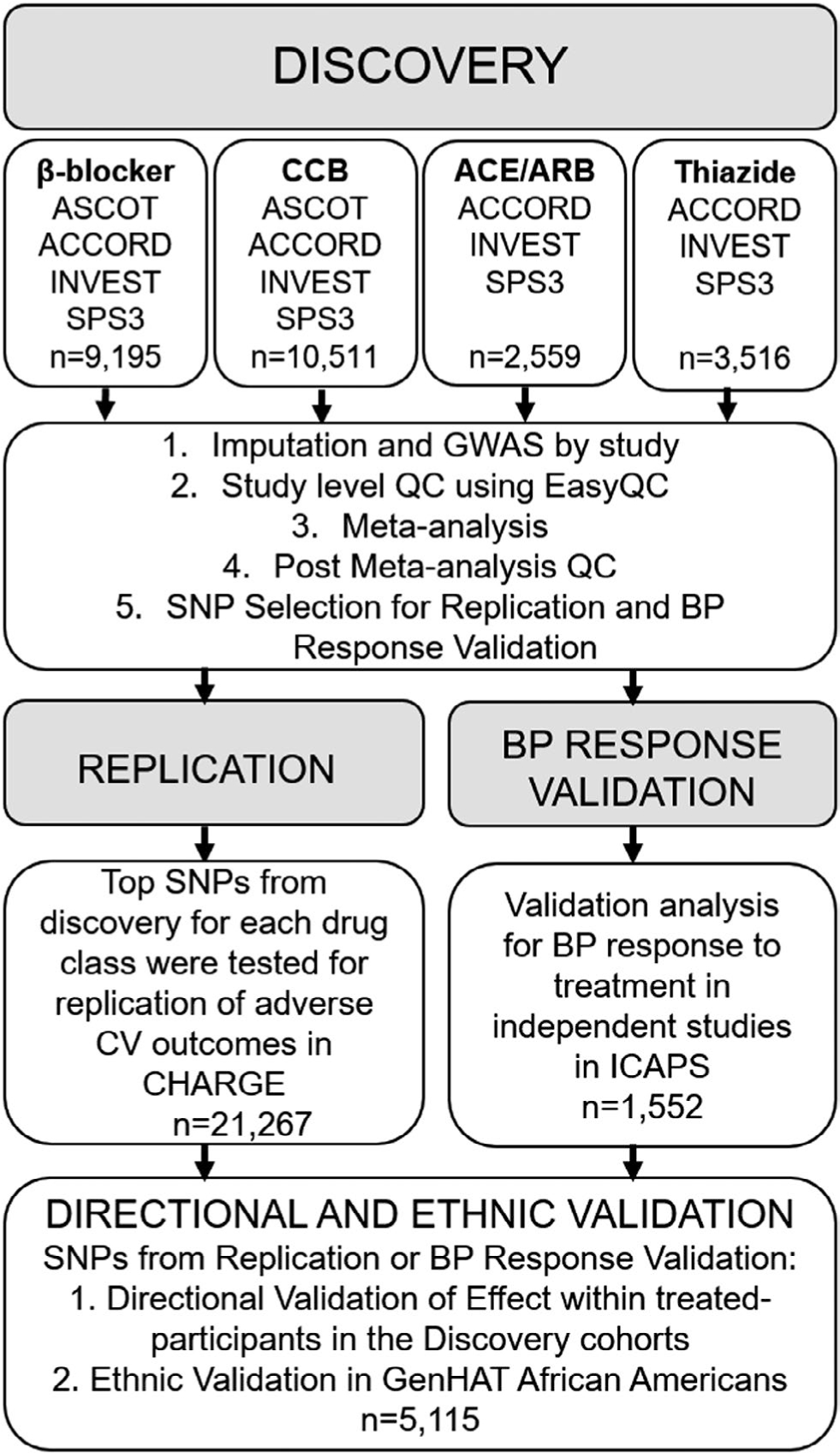
Study framework for the discovery meta-analysis, replication analysis and validation analyses. ACCORD, Action to Control Cardiovascular Risk in Diabetes; ASCOT, Anglo-Scandinavian Cardiac Outcomes Trial; BP, blood pressure; CCB, calcium channel blocker; CV, cardiovascular; GenHAT, Genetics of Hypertension Associated Treatment; GWAS, genome-wide association study; ICAPS, International Consortium for Antihypertensive Pharmacogenomics Studies; INVEST, International Verapamil SR-Trandolapril Study; QC, quality control; SNP, single nucleotide polymorphism; SPS3, Secondary Prevention of Small Subcortical Strokes.
Discovery analysis.
Each study conducted two (ASCOT) or four (ACCORD, INVEST, and SPS3) antihypertensive drug × SNP GWAS for adverse cardiovascular outcomes using logistic regression across at least 6.4 million imputed autosomal SNPs. The antihypertensive drug × SNP interaction model allows for the identification of SNPs where there is a difference in odds for adverse cardiovascular outcomes by SNP genotype and antihypertensive drug treatment. All regression models included an antihypertensive drug × additive SNP interaction term and were adjusted for age, sex, principal components for ancestry, and other antihypertensive drug classes (exposure > 1 visit). Additionally, other variables that were previously associated with adverse cardiovascular outcomes in the individual studies were included as covariates.8,9,11 ACCORD included adjustment for the glycemic arm and cardiovascular disease history; INVEST included adjustment for prior MI, history of diabetes, and history of heart failure; and SPS3 included adjustment for the antiplatelet arm, body mass index, prior MI, diabetes, and history of heart failure. Regression analyses were conducted using ProbABEL or PLINK.27,28 SNP filtering was conducted by study at a minor allele frequency cutoff of > 5% and an imputation QC (Rsq) of > 0.30.
Results from each antihypertensive drug × SNP GWAS by study underwent the central QC protocol outlined by Winkler et al. using the EasyQC package.29 Summary data were analyzed for heterozygosity, allele mismatches, and missingness. The QC results were visualized through quantile-quantile plots, P-Z plots, and allele frequency mismatch plots. The post-QC GWAS results from each cohort were combined by antihypertensive drug class in a fixed effects inverse variance weighted meta-analysis using METAL.30
After meta-analysis, SNPs were filtered based on presence in at least two cohorts and with heterogeneity P > 0.05. The genome-wide significance threshold for antihypertensive drug × SNP interaction was P < 5.0 × 10−8. The suggestive threshold for association, used for replication look-ups, was set at Interaction P < 1.0 × 10−4.
Replication analysis.
SNPs that met the suggestive threshold for association were compared to the results from the previously published CHARGE analyses.18 If the exact SNP was not available in the CHARGE data, the closest proxy (r2 ≥ 0.70) was used. The number of independent signals (linkage disequilibrium (LD) pruned at r2 > 0.20) by antihypertensive drug were used to calculate a Bonferroni-corrected α level for declaring significant replication. If there was at least one genome-wide significant signal, the α level was split so that the genome-wide significant signals were tested at 0.025/# of independent genome-wide signals, and then the suggestive signals were tested at 0.025/# of independent sub-genome-wide suggestive signals. If there were no genome-wide significant signals, the α level was calculated as 0.05/# of independent sub-genome-wide suggestive signals. Nominal significance was defined as P < 0.05.
BP response validation analysis.
SNPs that met the genome-wide significance threshold from the discovery interaction meta-analyses were tested for validation through a BP response analysis. SNPs were looked up in the results from the previously published ICAPS β-blocker discovery GWAS meta-analysis. 23 This analysis consisted of independent studies and participants from those included in the discovery phase of this study. The BP response validation significance threshold was set at 0.05/# of independent genome-wide significant signals tested.
Directional and ethnic validation analysis.
SNPs that passed the replication analysis or the BP response validation analysis were tested for directional validation and ethnic validation. The directional validation was conducted in the discovery studies (ASCOT, ACCORD, INVEST, and SPS3). SNPs were tested for association with adverse cardiovascular outcomes by study within the subgroup of only β-blocker treated participants, and within the subgroup of participants not exposed to β-blockers. Logistic regression models were adjusted for the same covariates as in the discovery interaction analyses. Results from each study were combined by meta-analysis to confirm consistent directionality across studies. Second, to examine associations in other ancestries, SNPs and proxies (r2 ≥ 0.70), were tested for association with adverse cardiovascular outcomes in African American participants in GenHAT who were exposed to β-blockers, and in African American participants in GenHAT who were not exposed to β-blockers.
RESULTS
The demographics and characteristics of the participants included in the discovery analysis are shown in Table 1. All participants were of European/European-American ancestry. The sample size included from ACCORD varied by drug and the participants included in the β-blocker analysis are shown in Table 1. The demographics of the participants included in the CCB, diuretic, and ACE/ARB analyses from ACCORD are shown in Table S1. Overall, there were 9,195 participants included in the β-blocker analysis, 10,511 in the CCB analysis, 3,516 in the diuretic analysis, and 2,559 in the ACE/ARB analysis. Men were more represented in four of the five cohorts, and, on average, participants were middle-aged (mean age = 61–71 years; Table 1). The demographics and characteristics of the participants included in the replication and validation analyses are shown in Tables S2–S4.
Table 1.
Demographics and characteristics of the participants included in the discovery meta-analysis
| ASCOT-UK |
ASCOT-SC |
ACCORDa |
INVEST |
SPS3 |
|
|---|---|---|---|---|---|
| Characteristic | (n = 3,804) | (n = 2,462) | (n = 2,015) | (n = 568) | (n = 346) |
|
| |||||
| Age, years, mean ± SD | 63.7 ± 8.1 | 61.0 ± 8.7 | 62.7 ± 6.6 | 70.8 ± 9.5 | 63.0 ± 10.6 |
|
| |||||
| Sex, male | 3,131 (82%) | 1,833 (74%) | 920 (46%) | 300 (53%) | 236 (64%) |
|
| |||||
| BMI, kg/m2 (mean ± SD) | 29.0 ± 4.8 | 28.5 ± 4.6 | 32.6 ± 5.4 | 27.2 ± 5.2 | 29.1 ± 5.9 |
|
| |||||
| SBP, mmHg (mean ± SD) | 161.6 ± 17.3 | 164.6 ± 17.3 | 140.9 ± 16.0 | 149.9 ± 18.4 | 145.3 ± 18.0 |
|
| |||||
| DBP, mmHg (mean ± SD) | 92.4 ± 9.9 | 96.4 ± 10.0 | 76.1 ± 10.4 | 81.3 ± 10.7 | 79.5 ± 10.5 |
|
| |||||
| History of | |||||
|
| |||||
| Myocardial infarction | 114 (3%) | 591 (24%) | -- | 231 (41%) | 16 (5%) |
|
| |||||
| Heart failure | 0 | 0 | -- | 38 (7%) | 3 (1%) |
|
| |||||
| Diabetes | 841 (22%) | 517 (21%) | 2,015 (100%) | 112 (20%) | 115 (31%) |
|
| |||||
| CV Outcomes caseb | 396 (10%) | 232 (9%) | 220 (11%) | 142 (25%) | 19 (5%) |
|
| |||||
| Randomized antihypertensive drug strategies | CCB vs. β-Blocker | CCB vs. β-Blocker | BP control target | CCB vs. β-Blocker | BP control target |
Values are presented as number (percentage) unless otherwise noted.
ACCORD, Action to Control Cardiovascular Risk in Diabetes; ASCOT-UK, Anglo-Scandinavian Cardiac Outcomes Trial-United Kingdom & Ireland; ASCOT-SC, Anglo-Scandinavian Cardiac Outcomes Trial-Scandinavians; BMI, body mass index; BP, blood pressure; CCB, calcium channel blocker; CV, cardiovascular; DBP, diastolic blood pressure; INVEST, International Verapamil SR-Trandolapril Study; SBP, systolic blood pressure; SPS3, Secondary Prevention of Small Subcortical Strokes.
Data presented from ACCORD are from patients included in the β-Blocker analysis, additional demographics from ACCORD are presented in Table S1.
CV Outcomes case defined as myocardial infarction (MI), stroke, or death.
Discovery analysis
The Manhattan plots and corresponding QQ plots for the association results from each of the four drug classes are shown in Figure 2 and Figure S1, respectively. Three SNPs (rs2511774, rs139945292, and rs80175218) near NTM on chromosome 11 reached genome-wide significance in the β-blocker discovery interaction meta-analysis (Table 2, Figure S2). After filtering by presence in at least 2 studies and heterogeneity P > 0.05, there were 608 SNPs that met the suggestive level of significance (Interaction P < 1 × 10−4) in the β-blocker analysis, 628 SNPs in the CCB analysis, 447 SNPs in the ACE-inhibitor/ARB analysis, and 651 in the thiazide/thiazide-like analysis. The top SNPs from each drug analysis (Interaction P < 1 × 10−6) are shown in Tables S5–S8.
Figure 2.
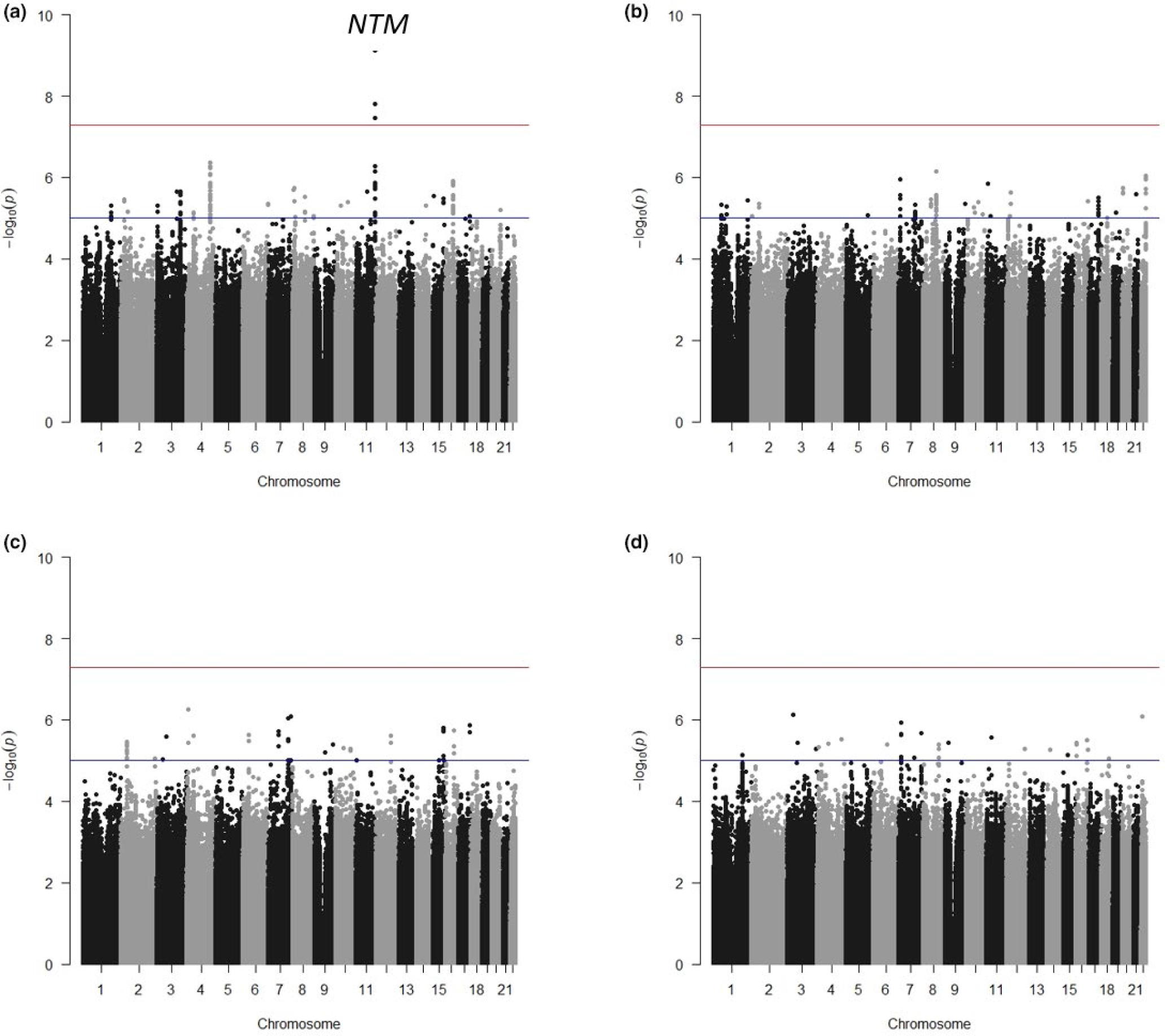
Antihypertensive drug × SNP interaction and cardiovascular outcomes genome-wide association study (GWAS) results by drug. (a) β-blocker. (b) Calcium channel blocker (CCB). (c) ACE-inhibitor/ARB. (d) Thiazide/Thiazide-like diuretic. Cardiovascular outcomes defined as myocardial infarction, stroke, or death.
Table 2.
Genome-wide significant β-blocker treatment × SNP interaction discovery meta-analysis results and adverse CV outcomes association results in β-blocker treated and non-β-blocker participants
| βB Treated meta-analysis |
Non βB treated meta-analysis |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Chr | Position | Nearest gene | EA | EAF | Interaction meta-analysis P value | Odds | P value | Direction | Odds | P value | Direction |
|
| ||||||||||||
| rs2511774 | 11 | 131,204,524 | NTM | C | 0.844 | 7.58E-10 | 0.66 | 2.53E-05 | −−−−+ | 1.16 | 0.0761 | ++++− |
|
| ||||||||||||
| rsl39945292 | 11 | 131,190,603 | NTM | T | 0.089 | 1.56E-08 | 1.57 | 2.35E-04 | +++++ | 0.76 | 0.0098 | −−−−+ |
|
| ||||||||||||
| rs80175218 | 11 | 131,167,910 | NTM | A | 0.884 | 3.52E-08 | 0.71 | 0.0012 | −−−−+ | 1.15 | 0.1228 | ++−++ |
Order of Direction: ASCOT-UK, ASCOT-SC, ACCORD, INVEST, SPS3.
ACCORD, Action to Control Cardiovascular Risk in Diabetes; ASCOT-UK, Anglo-Scandinavian Cardiac Outcomes Trial-United Kingdom & ireland; ASCOT-SC, Anglo-Scandinavian Cardiac Outcomes Trial-Scandinavians; βB, β-Blocker; CV, cardiovascular; EA, effect allele; EAF, effect allele frequency; INVEST, International Verapamil SR-Trandolapril Study; SNP, single nucleotide polymorphism; SPS3, Secondary Prevention of Small Subcortical Strokes.
Replication analysis
There were three independent signals (LD r2 < 0.20) within the genome-wide significant region in the discovery β-blocker interaction analysis. None of the three SNPs (rs2511774, rs139945292, and rs80175218) were available in the CHARGE results. However, proxies were identified for rs139945292 and rs80175218. Neither of the proxies met the Bonferroni-corrected replication threshold (Interaction P < 0.0083) in CHARGE (Table S9); however, rs7107440, the proxy for rs80175218, did show evidence of nominal significance (Interaction P = 0.0435; Table S9).
After LD pruning the suggestive SNPs from each drug analysis, there were 131 independent SNPs from the β-blocker analysis (replication threshold Interaction P < 1.9 × 10−4), 183 independent SNPs from the CCB analysis (replication threshold Interaction P < 2.7 × 10−4), 126 independent SNPs from the ACE-inhibitor/ ARB analysis (replication threshold Interaction P < 4.0 × 10−4), and 139 SNPs from the thiazide/thiazide-like diuretic analysis (replication threshold Interaction P < 3.6 × 10−4). No SNPs met the replication significance threshold for any drug in CHARGE. However, five SNPs from the β-blocker analysis showed nominal evidence of replication in CHARGE (Table S10). Additionally, two SNPs from the CCB analysis, nine SNPs from the ACE-inhibitor/ARB analysis, and three SNPs from the thiazide/thiazide-like diuretic analysis showed nominal evidence of replication in CHARGE (Tables S11–S13). As no SNPs met the replication significance thresholds, no SNPs from the replication analysis were moved forward to the directional and ethnic validation analysis.
BP response validation analysis
The 3 independent genome-wide significant SNPs (rs2511774, rs139945292, and rs80175218) near NTM on chromosome 11 were tested for BP response validation (validation threshold P < 0.0167). One SNP, rs139945292, passed the validation threshold for both systolic blood pressure (SBP) response (P = 0.0006, Beta = 3.09), and diastolic blood pressure (DBP) response (P = 0.0054, Beta = 1.53) in the independent studies included from the ICAPS β-blocker BP response meta-analysis (Table 3). In addition, rs80175218 showed nominal significance with SBP response (P = 0.0174, Beta = -1.59; Table 3). rs139945292 was moved forward for directional and ethnic validation analysis.
Table 3.
Association of the chromosome 11 region with BP response in independent studies from ICAPS β-blocker meta-analysis
| SNP | Chr | Position | Nearest gene | EA | ICAPS BP response | |||
|---|---|---|---|---|---|---|---|---|
| SBP | DBP | |||||||
| Beta | P value | Beta | P value | |||||
| rs2511774 | 11 | 131,204,524 | NTM | C | −1.32 | 0.0810 | −0.46 | 0.3351 |
| rs139945292 | 11 | 131,190,603 | NTM | T | 3.09 | 0.0006 | 1.53 | 0.0054 |
| rs80175218 | 11 | 131,167,910 | NTM | A | −1.59 | 0.0174 | −0.76 | 0.0677 |
BP, blood pressure; DBP, diastolic blood pressure; EA, effect allele; ICAPS, International Consortium for Antihypertensive Pharmacogenomics Studies; SBP, systolic blood pressure; SNP, single nucleotide polymorphism.
Directional and ethnic validation analysis
The T allele of rs139945292 was associated with an increased risk of adverse cardiovascular outcomes in β-blocker treated participants (P = 2.35 × 10−4, odds ratio (OR) = 1.57, 95% confidence interval (CI) = 1.23–1.99; Table 2), whereas in non β-blocker treated participants the T allele was associated with a decreased risk of adverse cardiovascular outcomes (P = 0.0098, OR = 0.76, 95% CI = 0.61–0.94; Table 2). This is consistent with the weaker BP reduction that was observed in the ICAPS β-blocker response analysis (SBP response Beta = 3.09 mmHg, DBP response Beta = 1.53 mmHg, per T allele; Table 3). The association of rs139945292 and adverse cardiovascular outcomes by β-blocker treatment in each study included in the discovery analysis is shown in Figure 3. Overall, there were consistent associations and trends across all five cohorts for an increased risk of adverse cardiovascular outcomes in β-blocker treated participants.
Figure 3.
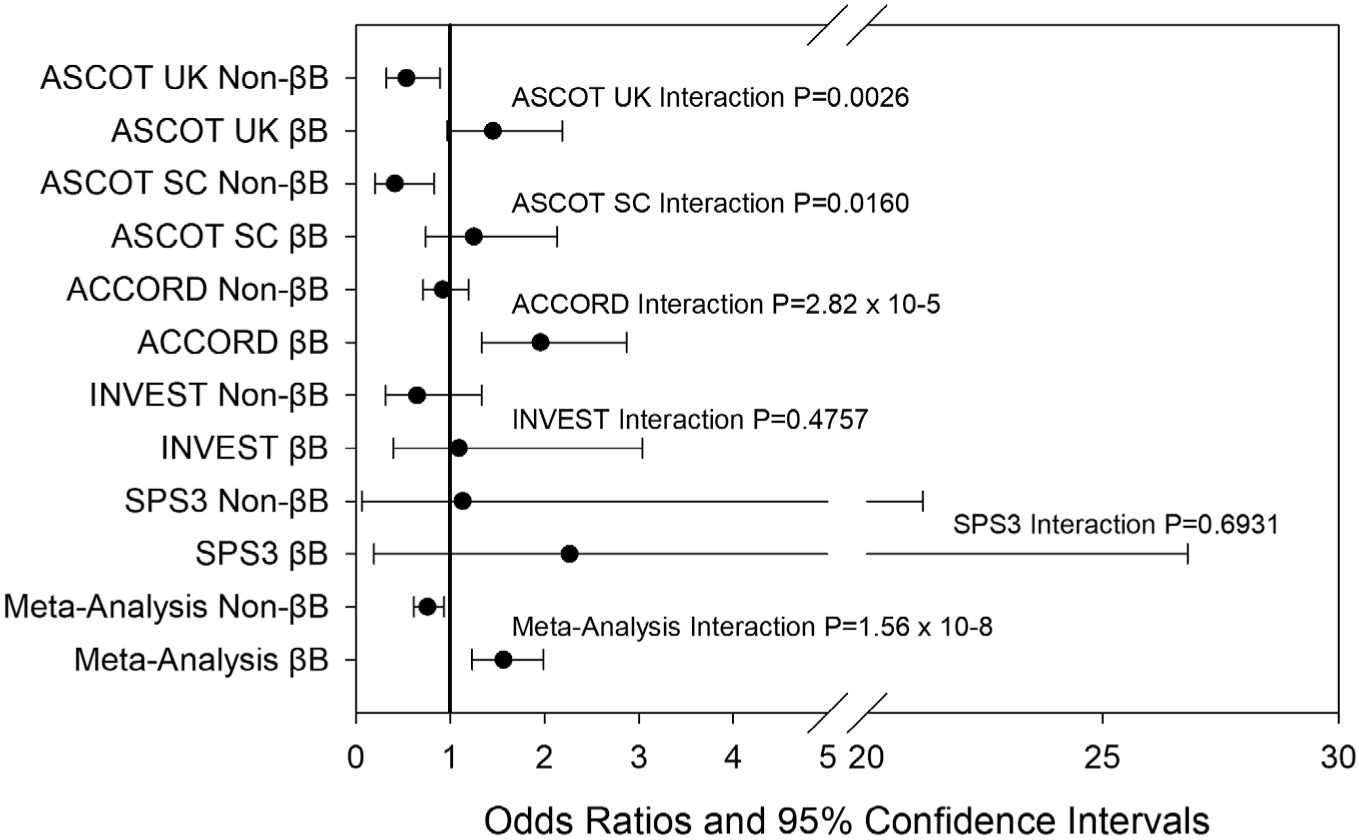
Forest plot for each discovery study and meta-analysis results of association with rs139945292 and cardiovascular outcomes in β-blocker (βB) and non-β-blocker (non-βB). Cardiovascular outcomes defined as myocardial infarction, stroke, or death. Odds ratios and 95% confidence intervals for adverse cardiovascular outcomes per T allele of rs139945292. ACCORD, Action to Control Cardiovascular Risk in Diabetes; ASCOT, Anglo-Scandinavian Cardiac Outcomes Trial; INVEST, International Verapamil SR-Trandolapril Study; SPS3, Secondary Prevention of Small Subcortical Strokes.
Proxies for rs139945292 were also examined in African American participants from GenHAT. Although there was no evidence of significance (P < 0.0083), there were similar trends observed for rs76944577, a proxy for rs139945292 in African populations (r2 = 1.0). A nonsignificant increased risk was observed with the A allele of rs76944577 in β-blocker treated participants (P = 0.1206, OR = 1.63, 95% CI = 0.88–3.04; Table S14).
DISCUSSION
To our knowledge, this is the first genome-wide meta-analysis for the association of antihypertensive drug class-SNP interactions and adverse cardiovascular outcomes to utilize data obtained from RCTs. By using the high-quality data from RCTs, we examined SNP-antihypertensive drug interactions for four antihypertensive drug classes: (i) β-blockers, (ii) CCBs, (iii) ACE-inhibitors/ARBs, and (iv) thiazide/thiazide-like diuretics. Through our multistage analysis plan, we identified a genome-wide significant association with β-blocker treatment × SNP interaction and adverse cardiovascular outcomes at rs139945292 near the NTM locus. We were able to validate the association with rs139945292 through BP response to β-blockers. We showed a consistent direction with the T allele of rs139945292 associated with a weaker BP reduction after β-blocker treatment and an increased risk for adverse cardiovascular outcomes with β-blocker treatment. We also observed nominal β-blocker treatment × SNP interaction associations at the NTM locus in CHARGE, and similar trends of increased risk for adverse cardiovascular outcomes in β-blocker treated African American participants from GenHAT. Meta-analyses of over 60 prospective observation studies of BP and mortality have shown a strong and direct relationship between BP level and vascular and overall mortality. 31 This direct link between BP level and adverse cardiovascular outcomes strengthens our findings and validation approach.
NTM is located ~ 50 kb upstream (5′) of rs139945292. NTM encodes an immunoglobulin (Ig) domain-containing GPI-anchored cell adhesion molecule that belongs to the IgLON family of Igs. NTM is highly expressed in the brain, but also shows evidence of expression in tibial nerves, lungs, and the atrial appendage of the heart.32 In lung tissue, rs139945292 is an expression quantitative trait locus for NTM with the T allele associated with increased expression of NTM.32 A gene-based association analysis for blood lipid levels in the Genetic Epidemiology Network of Salt-Sensitivity (GenSalt) study found a significant association with NTM and triglyceride levels.33 Other studies have also found evidence of increased NTM protein levels in plasma serum of patients with heart failure,34 and a balanced translocation in NTM in a family with thoracic aortic aneurysm.35 Many have hypothesized that NTM plays a role in neurite extension and neurite outgrowth; however, the role of NTM in cardiovascular function and development is still unclear.36,37 Our results suggest that the higher expression of NTM associated with the T allele at rs139945292 may weaken BP response to β-blockers and increase risk of adverse cardiovascular outcomes during β-blocker treatment.
Although none of the suggestive SNPs from the CCB, ACE inhibitor/ARB, or thiazide/thiazide-like discovery analyses passed replication thresholds in CHARGE, we did observe some noteworthy nominal associations in CHARGE. In the CCB replication, there was a nominal association observed with rs1938577, an intronic SNP in LRRC7, a gene with prior associations with HTN.38 In both the ACE-inhibitor/ARB replication, and the thiazide/thiazide-like replication there were nominal associations observed in BEND4, a gene previously associated with lung cancer. 39 Finally, in the β-blocker replication, there were nominal associations observed with SNPs in or near ADAMTSL1 (ADAMTS like 1) and DDAH1. SNPs in ADAMTSL1 have been previously associated with QT interval prolongation after thiazide diuretic treatment,40 whereas SNPs in DDAH1 have been previously associated with pre-eclampsia. 41
Our study is not without limitations. Although data collection and outcome determination may be more accurate within an RCT, by conducting post hoc analyses of RCTs, we have potentially biased our results. Overall, the populations included in RCTs may be biased and not represent the general population. Specifically, the RCTs included in our study are heterogeneous in regard to inclusion criteria, BP interventions, antihypertensive drugs, and primary outcomes.6–9 However, our study design included study-specific covariate adjustment within each RCT, analyses based on drug exposure as opposed to BP intervention arm, analyses by antihypertensive drug class, and standardization of adverse cardiovascular outcomes that were included in the analysis. We were unable to formally replicate any of our genome-wide significant SNPs or suggestive SNPs in CHARGE. This may be, in part, due to the observational nature of the data included in the CHARGE study.18 Medication information was collected through medication inventories or prescription drug records. These methods may not capture an accurate medication exposure for each study participant.18 The prior antihypertensive drug-SNP interaction study from CHARGE was unable to identify any genome-wide significant signals for adverse cardiovascular outcomes, also possibly highlighting that there could have been some misclassification in the medication exposure variable.18 By using data from RCTs, our medication inventories were likely more accurate. However, it is possible we still had some misclassification of medication exposure due to participant nonadherence. Additionally, we were able to validate our genome-wide significant SNPs through BP-response validation. This analysis approach also relied on the careful phenotyping and BP measurement collection that occurs within clinical trials.23 The availability of appropriate replication cohorts in pharmacogenomic studies is often challenging due to the need for access to a large cohort, study, or RCT with similar eligibility criteria, drug exposure, phenotypic or outcome data, and genetic samples or existing genetic data.42 However, approaches similar to our validation strategy provide alternative methods to confirm top signals.43
We only observed significant associations within the β-blocker interaction analyses. Whereas many of the prior candidate gene and genome-spanning studies that investigate SNP-antihypertensive drug interactions with adverse cardiovascular outcomes have included β-blockers, 13,15 there have also been prior significant findings with CCBs,12,16,44 ACE inhibitors,16 and thiazide diuretics.14,16 Although this is the largest analysis to date using data from RCTs for these drugs classes, we remain underpowered for interaction analyses for ACE inhibitors and thiazide diuretics (power ~ 43% for an interaction OR = 3.0 and a minor allele frequency = 25%). However, we were adequately powered (power ≥ 80%) to detect significant interactions at minor allele frequencies ≥ 10% for β-blockers and CCBs. Yet, in order to increase our sample size, we combined dihydropyridine and non-dihydropyridine CCBs into the same class (CCB), thiazide and thiazide-like diuretics into the same class (thiazide/thiazide-like diuretics), and ACE inhibitors and ARBs into the same class (ACE-inhibitor/ARB). The subtle differences in mechanisms of action or structure between these subclasses could possibly play a larger role in the contribution to adverse cardiovascular outcomes. Further study in larger sample sizes is warranted to investigate these differences. Whereas pharmacogenomic signals may be difficult to replicate, and interaction analyses require large sample sizes; pharmacogenomic studies can provide insight into both disease biology and mechanisms of drug response.45,46 These types of discoveries, as novel drug targets or new biomarkers, are needed as we continue to strive toward precision medicine.43,47
In conclusion, we conducted genome-wide association antihypertensive drug-SNP interaction analyses for β-blockers, CCBs, ACE inhibitor/ARBs, and thiazide/thiazide-like diuretics. We identified one locus on chromosome 11, near the NTM gene, with genome-wide significant β-blocker treatment × SNP interactions. We found that the T allele of rs139945292 was associated with increased risk of adverse cardiovascular outcomes during β-blocker treatment and weaker SBP and DBP response after treatment with a β-blocker. Additionally, prior functional studies showed that the T allele of rs139945292 was associated with higher expression of NTM in lung tissue. These data suggest that this locus and the NTM gene may mediate BP response to β-blockers and impact risk of adverse cardiovascular outcomes during β-blocker treatment. Future studies are needed to understand this mechanism, and further validate these findings.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
There is great interpatient variability in antihypertensive drug response, and limited data available to guide treatment selection. Clinical factors, such as age, sex, race, smoking, history of cardiovascular disease, and several candidate genes, have been associated with adverse cardiovascular outcomes and various antihypertensive drugs.
WHAT QUESTION DID THIS STUDY ADDRESS?
We sought to identify genome-wide significant variants influencing antihypertensive drug response and adverse cardiovascular outcomes.
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
We identified one locus on chromosome 11, near the NTM gene, with genome-wide significant β-blocker treatment × single nucleotide polymorphism interactions. We found that the T allele of rs139945292 was associated with increased risk of adverse cardiovascular outcomes during β-blocker treatment and weaker systolic and diastolic blood pressure response after treatment with a β-blocker.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
Our results suggest that the higher expression of NTM associated with the T allele at rs139945292 may weaken blood pressure response to β-blockers and increase risk of adverse cardiovascular outcomes during β-blocker treatment.
FUNDING
C.W.M. is supported by National Institutes of Health (NIH) grant K01 HL141690. ASCOT was funded by the National Institutes for Health Research (NIHR) as part of the portfolio of translational research of the NIHR Barts Biomedical Research Unit and the NIHR Biomedical Research Centre at Imperial College, the International Centre for Circulatory Health Charity, and the Medical Research Council through G952010. ACCORD research was supported by NIH grants R01 HL110380 and R01 HL110400, and intramural funds from the National Institute of Environmental Health Sciences. ACCORD cohort was supported by Contract Nos. N01-HC-95178, N01-HC-95179, N01-HC-95180, N01-HC-95181, N01-HC-95182, N01-HC-95183, N01-HC-95184, IAA #Y1-HC-9035, and IAA #Y1-HC-1010 from the National Heart, Lung, and Blood Institute (NHLBI), with additional support from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), the National Eye Institute (NEI), the National Institute on Aging (NIA), and the Centers for Disease Control and Prevention (CDC). INVEST was supported by the NIH grants R01 HL074730, U01 GM074492, and NIH CTSA grant UL1 RR092890, as well as grants from the University of Florida Opportunity Fund and Abbott Pharmaceuticals. The genome-wide genotyping for INVEST-GENES was provided by RIKEN. SPS3 was supported by NIH grants R01 NS073346, U01 GM074492-05S109, and U01 NS038529. Infrastructure for the CHARGE Consortium and the CHARGE Pharmacogenetics Working Group is supported in part by the NHLBI grants R01 HL105756, R01 HL103612, and R01 HL085251, and CHS contract number 75N92021D00006. Study descriptions and acknowledgements for the participating CHARGE cohorts are available in https://doi.org/10.1371/journal.pone.0140496. Both the PEAR and PEAR-2 studies were supported by NIH grants U01-GM074492 and UL1 TR000064 (University of Florida); UL1 TR000454 (Emory University), and UL1 TR000135 (Mayo Clinic). PEAR was also supported by funds from the Mayo Foundation. LIFE-Fin and GENRES were supported by the Sigrid Juselius Foundation, The Finnish Foundation for Cardiovascular Research, and the Helsinki University Central Hospital. BB-SS studies were supported by the HYPERGENES project (FP7-HEALTHF4-2007-201550), InterOmics (PB05 MIUR-CNR Italian Flagship Project), and the “Associazione per lo sviluppo della ricerca sull’ipertensione arteriosa e sulle malattie cardiovascolari—ONLUS.” GenHAT was supported by NIH grants R01 HL123782 and R01 HL063082, and N.A. was also supported by NIH grant T32 HL007457.
Footnotes
CONFLICT OF INTEREST
M.J.C. is Chief Scientist for Genomics England, a UK Government company. All other authors declared no competing interests for this work.
SUPPORTING INFORMATION
Supplementary information accompanies this paper on the Clinical Pharmacology & Therapeutics website (www.cpt-journal.com).
Supplementary Material
References
- 1.Virani SS et al. Tsao CW and Subcommittee AHACoEaPSCaSS. Heart disease and stroke statistics-2020 Update: a report from the American Heart Association. Circulation 141, e139–e596 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Mills KT, Stefanescu A & He J The global epidemiology of hypertension. Nat. Rev. Nephrol. 16, 223–237 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whelton PK et al. 2017 ACC/AHA/AAPA/ABC/ACPM/AGS/ APhA/ASH/ASPC/NMA/PCNA guideline for the prevention, detection, evaluation, and management of high blood pressure in adults: executive summary: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Hypertension 71, 1269–1324 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Chobanian AV et al. The Seventh Report of the Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure: the JNC 7 report. JAMA 289, 2560–2572 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Maddox TM et al. 2021. Update to the 2017 ACC expert consensus decision pathway for optimization of heart failure treatment: answers to 10 pivotal issues about heart failure with reduced ejection fraction: a report of the American College of Cardiology Solution Set Oversight Committee. J. Am. Coll. Cardiol. 77, 772–810 (2021). [DOI] [PubMed] [Google Scholar]
- 6.Pepine CJ et al. A calcium antagonist vs a non-calcium antagonist hypertension treatment strategy for patients with coronary artery disease. The International Verapamil-Trandolapril Study (INVEST): a randomized controlled trial. JAMA 290, 2805–2816. (2003). [DOI] [PubMed] [Google Scholar]
- 7.Dahlαf. B et al. Prevention of cardiovascular events with an antihypertensive regimen of amlodipine adding perindopril as required versus atenolol adding bendroflumethiazide as required, in the Anglo-Scandinavian Cardiac Outcomes Trial-Blood Pressure Lowering Arm (ASCOT-BPLA): a multicentre randomised controlled trial. Lancet 366, 895–906 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Cushman WC et al. Effects of intensive blood-pressure control in type 2 diabetes mellitus. N. Engl. J. Med. 362, 1575–1585 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benavente OR et al. Blood-pressure targets in patients with recent lacunar stroke: the SPS3 randomised trial. Lancet 382, 507–515 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.The ALLHAT Officers and Coordinators for the ALLHAT Collaborative Research Group. Major outcomes in high-risk hypertensive patients randomized to angiotensin-converting enzyme inhibitor or calcium channel blocker vs diuretic: the Antihypertensive and Lipid-Lowering Treatment to Prevent Heart Attack Trial (ALLHAT). JAMA 288, 2981–2997 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Pepine CJ et al. Predictors of adverse outcome among patients with hypertension and coronary artery disease. J. Am. Coll. Cardiol. 47, 547–551 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Beitelshees AL et al. KCNMB1 genotype influences response to verapamil SR and adverse outcomes in the INternational VErapamil SR/Trandolapril STudy (INVEST). Pharmacogenet. Genomics 17, 719–729 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McDonough CW et al. Pharmacogenomic Association of Nonsynonymous SNPs in SIGLEC12, A1BG, and the selectin region and cardiovascular outcomes. Hypertension 62(1), 48–54 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McDonough CW et al. Association of variants in NEDD4L with blood pressure response and adverse cardiovascular outcomes in hypertensive patients treated with thiazide diuretics. J. Hypertens. 31, 698–704 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pacanowski MA et al. , beta-adrenergic receptor gene polymorphisms and beta-blocker treatment outcomes in hypertension. Clin. Pharmacol. Ther. 84, 715–721 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lynch AI et al. Gene panels to help identify subgroups at high and low risk of coronary heart disease among those randomized to antihypertensive treatment: the GenHAT study. Pharmacogenet. Genomics 22, 355–366 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lynch AL et al. Antihypertensive pharmacogenetic effect of fibrinogen-beta variant -455G>A on cardiovascular disease, end-stage renal disease, and mortality: the GenHAT study. Pharmacogenet. Genomics 19, 415–421 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bis JC et al. Drug-gene interactions of antihypertensive medications and risk of incident cardiovascular disease: a pharmacogenomics study from the CHARGE Consortium. PLoS One 10, e0140496 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sever PS et al. Rationale, design, methods and baseline demography of participants of the Anglo-Scandinavian cardiac outcomes trial. ASCOT investigators. J. Hypertens. 19, 1139–1147. (2001). [DOI] [PubMed] [Google Scholar]
- 20.Williamson JD et al. The Action to Control Cardiovascular Risk in Diabetes Memory in Diabetes Study (ACCORD-MIND): rationale, design, and methods. Am. J. Cardiol 99, 112i–122i (2007). [DOI] [PubMed] [Google Scholar]
- 21.Pepine CJ et al. Rationale and design of the International Verapamil SR/Trandolapril Study (INVEST): an internet-based randomized trial in coronary artery disease patients with hypertension. J. Am. Coll. Cardiol. 32, 1228–1237 (1998). [DOI] [PubMed] [Google Scholar]
- 22.Benavente OR et al. The Secondary Prevention of Small Subcortical Strokes (SPS3) study. Int. J. Stroke 6, 164–175 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh S et al. Genome-wide meta-analysis of blood pressure response to β. J. Am. Heart Assoc. 8, e013115 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arnett DK et al. Pharmacogenetic association of the angiotensin-converting enzyme insertion/deletion polymorphism on blood pressure and cardiovascular risk in relation to antihypertensive treatment: the Genetics of Hypertension-Associated Treatment (GenHAT) study. Circulation 111, 3374–3383. (2005). [DOI] [PubMed] [Google Scholar]
- 25.Buse JB et ai. Action to Control Cardiovascular Risk in Diabetes (ACCORD) trial: design and methods. Am. J. Cardiol 99, 21i–33i (2007). [DOI] [PubMed] [Google Scholar]
- 26.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA & Reich D Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Purcell S et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aulchenko YS, Struchalin MV & van Duijn CM ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics 11, 134 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winkler TW et al. Quality control and conduct of genome-wide association meta-analyses. Nat. Protoc. 9, 1192–1212 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilier CJ, Li Y & Abecasis GR METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewington S, Clarke R, Qizilbash N, Peto R, Collins R & Prospective Studies Collaboration. Age-specific relevance of usual blood pressure to vascular mortality: a meta-analysis of individual data for one million adults in 61 prospective studies. Lancet 360, 1903–1913 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Consortium. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li C et al. Genome-wide linkage and positional association analyses identify associations of novel AFF3 and NTM genes with triglycerides: the GenSalt study. J. Genet. Genomics 42, 107–117 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao TH et al. Identification of novel biomarkers in plasma for prediction of treatment response in patients with heart failure. Lancet 385(Suppl 1), S26 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Luukkonen TM et al. A balanced translocation truncates Neurotrimin in a family with intracranial and thoracic aortic aneurysm. J. Med. Genet. 49, 621–629 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanz R, Ferraro GB & Fournier AE IgLON cell adhesion molecules are shed from the cell surface of cortical neurons to promote neuronal growth. J. Biol. Chem. 290, 4330–4342 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh K et al. The combined impact of IgLON family proteins Lsamp and Neurotrimin on developing neurons and behavioral profiles in mouse. Brain Res. Bull. 140, 5–18 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Kim SJ et al. Genetic association of short sleep duration with hypertension incidence-a 6-year follow-up in the Korean genome and epidemiology study. Circ. J 76, 907–913 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Kettunen E et al. Asbestos-associated genome-wide DNA methylation changes in lung cancer. Int. J. Cancer 141, 2014–2029. (2017). [DOI] [PubMed] [Google Scholar]
- 40.Avery CL et al. Drug-gene interactions and the search for missing heritability: a cross-sectional pharmacogenomics study of the QT interval. Pharmacogenomics J. 14, 6–13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Akbar F, Heinonen S, Pirskanen M, Uimari P, Tuomainen TP & Salonen JT Haplotypic association of DDAH1 with susceptibility to pre-eclampsia. Mol. Hum. Reprod. 11, 73–77 (2005). [DOI] [PubMed] [Google Scholar]
- 42.loannidis JP To replicate or not to replicate: the case of pharmacogenetic studies: Have pharmacogenomics failed, or do they just need larger-scale evidence and more replication? Circ. Cardiovasc. Genet 6, 413–418 (2013); discussion 418. [DOI] [PubMed] [Google Scholar]
- 43.Aslibekyan S, Claas SA & Arnett DK To replicate or not to replicate: the case of pharmacogenetic studies: Establishing validity of pharmacogenomic findings: from replication to triangulation. Circ. Cardiovasc. Genet. 6, 409–412 (2013); discussion 412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beitelshees AL et al. CACNA1C gene polymorphisms, cardiovascular disease outcomes, and treatment response. Circ. Cardiovasc. Genet 2, 362–370 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meyer UA Pharmacogenetics -five decades of therapeutic lessons from genetic diversity. Nat. Rev. Genet. 5, 669–676 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Kraus VB Biomarkers as drug development tools: discovery, validation, qualification and use. Nat. Rev. Rheumatol 14, 354–362. (2018). [DOI] [PubMed] [Google Scholar]
- 47.Aronson SJ & Rehm HL Building the foundation for genomics in precision medicine. Nature 526, 336–342 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


