Abstract
The formation of the biomolecule corona on the surface of nanoparticles upon exposure to biological fluids critically influences nanocarrier performance in drug delivery. It has been shown that in some cases corona proteins can mediate specific nanoparticle interactions with cell receptors. Within this context, in order to identify corona proteins affecting nanoparticle uptake, in this work, correlation analysis is performed between the corona composition of a panel of silica nanoparticles of different sizes and surface functionalities and their uptake in four endothelial cell types derived from different organs. In this way, proteins that correlate with increased or decreased uptake were identified, and their effects were validated by studying the uptake of nanoparticles coated with a single protein corona and competition studies in brain and liver endothelium. The results showed that precoating nanoparticles with histidine-rich glycoprotein (HRG) alone strongly decreased uptake in both liver and brain endothelium. Furthermore, our results suggested the involvement of the transferrin receptor in nanoparticle uptake in liver endothelium and redirection of the nanoparticles to other receptors with higher uptake efficiency when the transferrin receptor was blocked by free transferrin. These data suggested that changes in the cell microenvironment can also affect nanoparticle uptake and may lead to a different interaction site with nanoparticles, affecting their uptake efficiency. Overall, correlating the composition of the protein corona and nanoparticle uptake by cells allows for the identification of corona molecules that can be used to increase as well as to reduce nanoparticle uptake by cells.
Keywords: protein corona, correlation analysis, targeted drug delivery, endothelial cells, nanoparticle uptake
Introduction
Nanomaterials have shown tremendous potential for biomedical applications, such as drug delivery and diagnosis.1−3 However, in order to properly apply them in this context, the fundamental interactions that govern biological processes once nanomaterials come into contact with living systems should be thoroughly investigated.4 It is now widely known that when applied in biological fluids, nanomaterials adsorb proteins and biomolecules on their surface, forming a layer which is known as the biomolecule corona.5−7 In the development of targeted nanocarriers, this rapidly forming protein corona has been considered a crucial element, since the interactions of nanomaterials with cells can be greatly affected by this acquired biomolecular layer.8−15 In fact, the protein corona can direct nanocarriers to specific receptors resulting in enhanced uptake by certain cell types, but in some cases, it can also inhibit targeted drug delivery by masking the targeting ligands on nanocarriers.14,16,17 Furthermore, the protein corona can also influence other biological processes, such as biodistribution, immune cell activation,18 and cytotoxicity.19 More importantly, the formation of this protein layer cannot be fully prevented by coating of the nanocarriers with antifouling agents, such as, for instance, polyethylene glycol (PEG). Actually, it has been shown that PEGylated surfaces acquire the ability to reduce clearance by immune cells thanks to the adsorption of specific proteins in their corona.20
Given the many effects of the protein corona on the interactions of nanoparticles with cells, understanding which corona components and cellular receptors can affect the biological fate of nanoparticles is important for improving the design of nanocarriers. Many reports have shown that the protein corona can interact with specific cell receptors.11−13 This opens up new ways for the improvement of nanocarrier targeting by exploiting interactions mediated by their corona, as for instance, it was demonstrated for approved nanomedicines currently in the clinic.21 Moreover, it is known that the corona composition varies with nanoparticle properties, such as size, shape, and surface charge–among many others.22−25 Thus, nanoparticles may be tailored to adsorb specific biomolecules in their corona and, in this way, be directed to specific cell receptors.16,17 Recent studies have exploited this concept to predict cellular association using a library of different nanoparticles and by characterizing their protein corona fingerprints.8,26−28 Similarly, by correlating the composition of the corona formed on different nanoparticles and their cell uptake efficiency, corona proteins that can promote or reduce uptake by cells have been identified.28,29 For instance, a previous study in which this approach was applied discovered that apolipoproteins (ApoH, ApoA4, and ApoC3) were responsible for regulating nanoparticle uptake in human mesenchymal stem cells.29
Since different cell types may have different interactions with the protein corona, by comparing the association in multiple target cells new strategies for targeting specific cells may be discovered. For this, endothelial cells represent an important target cell model, given the fact that they are one of the major barriers nanomedicine encounters in vivo after administration.
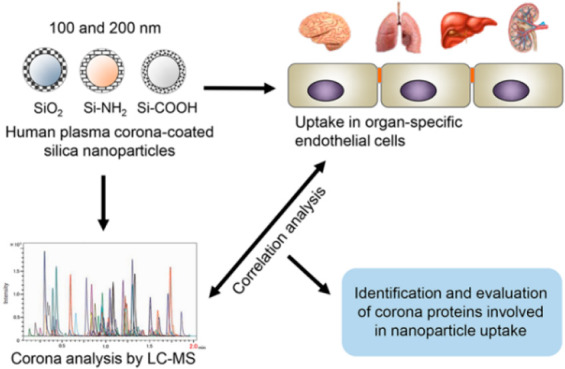
Within this context, in this work, we used endothelial cells from brain, lungs, liver, and kidneys as target cell models for different organs. It was recently reported that endothelial cells from different organs, because of their heterogeneity, show differences in nanoparticle uptake efficiency.30 Thus, a panel of six nanoparticles consisting of plain, carboxylated, and amino-modified silica in two different sizes (100 and 200 nm) was used to form different protein coronas in full human plasma. Their uptake efficiency in the different endothelial cells was determined, and the composition of the protein corona formed on each nanoparticle type was analyzed by mass spectrometry. Then, correlation analysis between the corona composition and uptake levels in the different endothelial cells was applied to identify key candidate corona proteins affecting nanoparticle uptake efficiency. In order to validate the correlation results, the uptake of nanoparticles with “artificial” coronas composed of the candidate proteins was compared to that of nanoparticles with a “natural” corona. Additionally, RNA interference and competition experiments were used to block the corresponding receptors in order to determine their role in nanoparticle uptake.
Materials and Methods
Cell Culture
Each cell line was cultured using media of different composition and coating flasks in different ways to improve cell adhesion as previously described.30−34 Briefly, immortalized hCMEC/D3 cells were used as a model for human brain endothelium.31 Cells were cultured in EBM-2 endothelial basal medium (LONZA, Allendale, NJ, USA) to which 10 mM HEPES (ThermoFisher Scientific), 1 μg mL–1 hydrocortisone (Sigma-Aldrich, St. Louis, USA), 200 ng mL–1 bFGF (Peprotech, London, United Kingdom), and 1% chemically defined lipid concentrate (ThermoFisher Scientific) were added, together with 5% fetal bovine serum (FBS, Gibco ThermoFisher Scientific, Landsmeer, Netherlands). In order to improve cell adhesion, flasks were precoated with 0.1 mg mL–1 rat-tail collagen type-1 (Corning, NY, USA). Cells were cultured at 37 °C and 5% CO2 and used between passages 29–38, refreshing the medium every 2–3 days.35
Human HPMEC-ST1.6R immortalized cells were used as a model for lung microsovascular endothelium.32 Cells were cultured in EBM-2 basal medium supplemented with the EGM-2 bullet kit (LONZA) at 37 °C and 5% CO2. In order to improve cell adhesion, a cold solution of 0.2% gelatin (Sigma-Aldrich) was used to precoat the cell flasks. Every 2–3 days, the cell culture medium was refreshed.
Human TRP3 immortalized cells were used as a model for liver endothelial sinusoidal cells.33 Cells were cultured in MCDB 131 medium (Gibco ThermoFisher Scientific) to which 50 μg mL–1 endothelial cell growth supplement (ECGS, Corning), 250 μg mL–1 cAMP (Sigma-Aldrich), and 1 μg mL–1 hydrocortisone (Sigma-Aldrich) were added, together with 10 mM glutamine (ThermoFisher Scientific) and 20% FBS (Gibco ThermoFisher Scientific). Cells were seeded in flasks precoated with a cold solution of gelatin at 0.1% and cultured at 37 °C and 5% CO2, refreshing the cell culture medium every 2–3 days.
Conditionally immortalized CiGENC cells were used as a model for kidney glomerular endothelial cells.34 Cells were cultured in EBM-2MV medium to which all components of the EGM-2MV bullet kit (LONZA) except the growth factor VEGF were added. Fibronectin (Corning) at 1 μg cm–2 was used to coat cell flasks to improve cell adhesion. The cells were cultured in 5% CO2 at a temperature of 33 °C until 90% confluency and then as the temperature was increased to 37 °C for a further 3 days in order for the cells to develop kidney glomerular endothelial phenotypes. The medium was exchanged with fresh cell culture medium every 2–3 days.
Physicochemical Characterization of Nanoparticles
Green fluorescently labeled silica nanoparticles (maximum excitation 485 nm and emission 510 nm) of 100 and 200 nm diameter, and with a plain, amino-modified or carboxylated surface (SiO2, SiO2–NH2, and SiO2–COOH, respectively), were purchased from Micromod Partikeltechnologie GmbH (Rostock, Germany). All amino and carboxylated nanoparticles had a surface charge density of 1 μmol g–1, except for 200 nm SiO2–NH2, which had a surface charge density of 4 μmol g–1. The same batch of nanoparticles was used for all studies, with the exception of the 200 nm SiO2–NH2 results shown in Supporting Figures S5 and S6 and one of the 3 experiment repeats shown in Figure 5a,b. A Malvern Zetasizer Nano ZS (Malvern Instrument Ltd., Worcestershire, UK) was used for nanoparticle characterization by dynamic light scattering (DLS) to obtain the nanoparticle size distribution and for zeta potential (ζ-potential) measurements. Briefly, dispersions of the nanoparticles or of the corona-coated nanoparticles (50 μg mL–1 and 30 μg mL–1, for 100 and 200 nm nanoparticles, respectively) in PBS were measured at 20 °C in disposable capillary cells (Malvern) just after preparation. For each sample, measurements were repeated at least 3 times with 5 runs each.
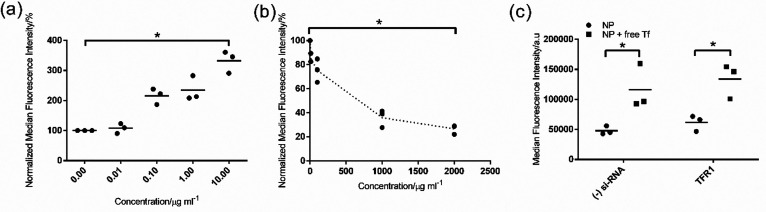
Figure 5.
Effect of free transferrin on nanoparticle uptake in TRP3 liver endothelium cells. (a) Uptake of 30 μg mL–1 corona-coated nanoparticle complexes formed on 200 nm SiO2–NH2 in full human plasma in the presence of increasing concentrations of human transferrin in serum-free medium after a 4 h exposure. (b) Uptake of 10 μg mL–1 Alexa Fluor 546 fluorescently labeled transferrin in the presence of increasing concentrations of the isolated corona-coated nanoparticle complexes in serum-free medium. (c) Uptake of corona-coated nanoparticle complexes in TRP3 cells after silencing the expression of transferrin receptor 1 (TFR1). TFR1 expression was silenced as described in the Materials and Methods section, then cells were exposed for 4 h to 30 μg mL–1 nanoparticle-corona complexes in serum-free medium or in the presence of 1 mg mL–1 human transferrin. The results of three independent experiments are shown, together with their average indicated with a line. One of the 3 repeated experiments of panels a and b was performed using a different batch of nanoparticles (see Figure S3, Supporting Information for more details). Nevertheless, as shown in these panels, the results were highly reproducible. The competition experiments showed that free transferrin increased nanoparticle uptake instead of competing with it, while the uptake of transferrin decreased when corona-coated nanoparticle complexes were added. A Mann–Whitney test was applied to compare the uptake level in serum-free conditions (0 μg mL–1 competitor) and when (a) transferrin or (b) the corona-coated nanoparticles were added at the highest concentration. For the results in part c, a Mann–Whitney test was applied to compare uptake levels after addition of transferrin. p ≤ 0.05 was considered significant (indicated with an asterisk).
Nanoparticle-Corona Preparation and Characterization
Prior to physicochemical characterization and incubation on cells, nanoparticle-corona complexes were prepared and isolated. All 6 nanoparticle types were dispersed at 1 mg mL–1 in full human plasma (human plasma from pooled donors, prepared using citrate as anticoagulant, total protein concentration 86 mg mL–1, from TCS BioSciences Ltd., Botolph Claydon, Buckingham, UK) at 37 °C for 1 h under continuous shaking at 300 rpm. Then, the nanoparticle-corona complexes were separated from the excess unbound proteins by centrifugation at 16,000 g for 1 h at 15 °C. The nanoparticle-corona complexes in the pellet were resuspended in PBS. For incubation on cells, the dispersion of nanoparticle-corona complexes was diluted in serum-free medium to 50 or 30 μg mL–1 nanoparticles for the 100 or 200 nm silica, respectively. In order to isolate clean hard corona-coated nanoparticles for SDS PAGE and mass spectrometry analysis, three more centrifugations were performed using the same settings (each for 1 h at 16,000 g and 15 °C). After the last wash, nanoparticle fluorescence was measured with a spectrofluorometer in order to determine nanoparticle concentration in the final samples. Then, hard corona-coated nanoparticles corresponding to 200 or 300 μg nanoparticles, for the 100 or 200 nm silica, respectively, were resuspended in gel loading buffer. The samples were incubated for 5 min at 95 °C and then loaded onto a 10% polyacrylamide gel for SDS-PAGE. The proteins in the gel were stained by incubation for 1 h in 0.1% w/v Coomassie blue R-250 in a solution containing water, methanol, and glacial acetic acid (5:4:1). Then, the gel was washed with Milli-Q-grade water. Images of the gel were acquired using a ChemiDoc XRS (Biorad, USA).
Mass Spectrometry and Correlation Analysis
The samples for mass spectrometry were prepared and measured as previously described.13 Briefly, the protein concentration in the isolated nanoparticle-corona complexes prepared as described above was determined with a BCA Protein Assay Kit (Pierce, ThermoFisher Scientific). Then, for each sample, equal amounts of proteins were loaded on a gel (10% polyacrylamide) and separated from the nanoparticles by SDS-PAGE for 5 min. The proteins in the gel were stained by incubation for 1 h in an InstantBlue solution (Sigma-Aldrich) and washed with Milli-Q-grade water. The gel containing corona proteins was then reduced and alkylated by incubation at 56 °C in 10 mM DTT for 30 min and, after that, at room temperature in 55 mM chloroacetamide in the dark for other 30 min. Then, the gel was washed with Milli-Q water and cut to separate it from the nanoparticles and from the other samples. Each piece of cut gels was further washed to remove the remaining InstantBlue solution in 100 mM ammonium bicarbonate:acetonitrile (70:30) at room temperature for 30 min, then in ammonium bicarbonate:acetonitrile (50:50), and finally in acetonitrile. Then, the samples were dried at 60 °C for 5 min. Afterward, the proteins in the gels were digested by overnight incubation at 37 °C in sequencing grade modified trypsin (Promega Corporation, Madison, WI, USA) (1:100) in 100 mM ammonium bicarbonate. In order to stop protein digestion, 75% v/v acetonitrile and 25% of a solution of 5% v/v formic acid in water were added. The digested peptides extracted out of the gels were diluted in 0.1% v/v formic acid in water and loaded onto SPE (Solid Phase Extraction) GracePure columns (W. R. Grace & Co., Columbia, MD, USA) which were conditioned with 0.1% v/v formic acid in acetonitrile two times and then with 0.1% v/v formic acid in water an additional two times. Next, the loaded samples were washed with 0.1% v/v formic acid in water two times, and peptides were eluted with 0.1% v/v formic acid in 50% v/v acetonitrile two times. The recovered peptides were dried for 2–3 h in a speed vacuum, then 0.1% v/v formic acid in water was added, and Acclaim PepMap 100 C18 LC Columns (ThermoFisher Scientific) were used to load the samples into a Q Exactive Plus Hybrid Quadrupole-Orbitrap Mass Spectrometer (ThermoFisher Scientific). PEAKS 10 software (Bioinformatics Solutions Inc., Waterloo, ON, Canada) was used to identify the proteins in the samples using the human proteome database from UniProtKB/Swissprot. A 10-ppm parent mass error tolerance was used to search the experimental data, and one missed cleavage was allowed, setting fixed carbamidomethylation and variable oxidation as modifications. Spectral counts (Spectra) were normalized by the molecular weight of the identified proteins, and for each protein, the relative protein abundance (RPA, Spectrax) was calculated as follows:
| 1 |
Then, for each protein identified, the Pearson Product-Moment Correlation Coefficient was calculated using the following equation as previously described29
| 2 |
where x is the RPA of each protein, and y is the median cell fluorescence intensity measured by flow cytometry. This correlation coefficient was used as a statistical measurement for the correlation between the abundance (RPA) of each protein in the corona formed on the nanoparticles (as obtained by mass spectrometry) and the cell uptake efficiency in the different cell types (as measured by flow cytometry). An r value close to +1 indicated strong positive correlation (proteins associated with higher uptake), while an r value close to −1 indicated a negative correlation (proteins associated with lower uptake).
Nanoparticle Uptake and Flow Cytometry Analysis
Cellular uptake of nanoparticles was measured by flow cytometry. Briefly, 25,000 or 50,000 cells cm–2 for the HPMEC-ST1.6R and the other cell lines, respectively, were seeded in a 24-well plate (Greiner Bio-One BV, A. Alphen on den Rijn, Netherlands) precoated with an extracellular matrix as described above for each cell type. CiGENC and TRP3 cells were grown for 3 days, and the hCMEC/D3 and HPMEC-ST1.6R were grown for 4 days in order to form endothelial cell barriers.30 Then, cells were exposed to 50 or 30 μg mL–1 of corona-coated 100 or 200 nm silica nanoparticles (plain, amino-modified, and carboxylated), respectively, prepared as described above just before addition to cells. After exposure, in order to wash away the extracellular nanoparticles and reduce those adhering on the cell membrane, cell culture medium supplemented with 10% FBS was used to wash the cells, followed by two washes with PBS. Afterward, cells were harvested using 0.05% trypsin-EDTA for 5 min, and after centrifugation at 300 g for 3 min to discard the medium with trypsin, cells were resuspended in PBS. Then, nanoparticle uptake was determined by measuring the green cell fluorescence by flow cytometry using a Cytoflex S (Beckman Coulter, Woerden, The Netherlands). A 488 nm laser was used for nanoparticle excitation. Flowjo data analysis software (Flowjo, LLC) was used to analyze the results. Cell debris and cell doublets were excluded from the analysis by setting gates in the double scatter forward and side scattering plot. For each sample, the fluorescence of at least 15,000 individual cells was measured in order to obtain a cell fluorescence distribution, and for each condition, two replicate samples were made. Experiments were repeated 3 times to confirm reproducibility (unless specified). For each condition, the median cell fluorescence intensity averaged over two replicate samples was calculated, and the results obtained in each individual experiment are shown, together with a line which indicates their average (unless specified).
Uptake of Single Protein Corona-Coated Nanoparticles
In order to coat nanoparticles with a single protein corona, green fluorescently labeled 200 nm SiO2–NH2 were incubated with histidine-rich glycoprotein (HRG, Peprotech, London, United Kingdom), transferrin, human serum albumin (HSA), or alpha-1 antitrypsin (all from Sigma-Aldrich) in serum-free medium. In order to coat the entire nanoparticle surface area, 30 μg mL–1 nanoparticles were added to a 15 μg mL–1 solution of each single protein. As additional controls, 30 μg mL–1 green fluorescently labeled 200 nm SiO2–NH2 nanoparticles were also coated with 15 μg mL–1 human plasma (as in the conditions used for single protein coronas, referred to as “human plasma” in Figure 4) or 1 mg mL–1 nanoparticles with 86 mg mL–1 human plasma (as in the conditions used for mass spectrometry and nanoparticle uptake studies, referred to as “full human plasma” in Figure 4). Then, after incubation at 37 °C for 1 h with shaking at 300 rpm, the corona-coated nanoparticles were pelleted by centrifugation for 1 h at 16,000 g and resuspended to a concentration of 30 μg mL–1 nanoparticles in serum-free cell culture medium. In order to determine their uptake, 50,000 cells cm–2 hCMEC/D3 and TRP3 cells were seeded in a 24-well plate (Greiner) precoated with an extracellular matrix as described above. Three or 4 days after seeding, for TRP3 or hCMEC/D3, respectively, cells were exposed to 30 μg mL–1 single protein-coated or human plasma-coated SiO2–NH2 (200 nm) in serum-free medium, freshly prepared just prior to addition to cells. After 4 h of exposure, cells were washed, and samples were prepared for flow cytometry analysis as described above.
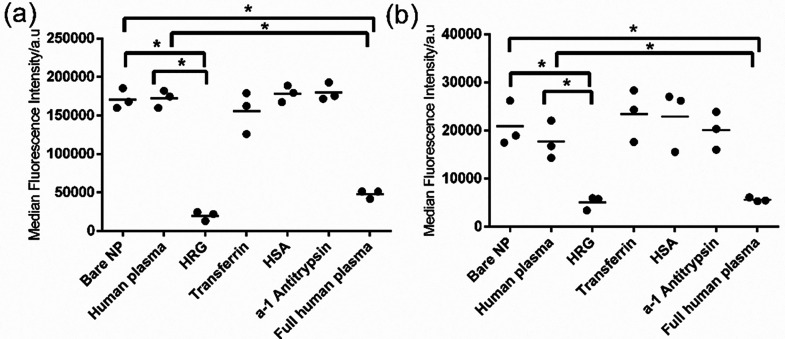
Figure 4.
Uptake of single protein corona-coated nanoparticles in liver (a) and brain endothelium (b). The 200 nm SiO2–NH2 were coated with 15 μg mL–1 human plasma, HRG, transferrin, HSA, or alpha-1 antitrypsin as described in the Materials and Methods section, and 30 μg mL–1 corona-coated nanoparticles were added to cells for 4 h in serum-free medium. As additional controls, the uptake in serum-free medium of 30 μg mL–1 bare nanoparticles (bare NP) and nanoparticles coated with full human plasma corona prepared as described in the Materials and Methods section (full human plasma) was also measured. The results of three independent experiments are shown, together with their average indicated with a line. A Kruskal–Wallis test was used to compare the different groups and indicated significant differences in both panels. A Mann–Whitney test with Bonferroni correction for multiple testing was applied to compare the uptake level of single protein corona-coated nanoparticles to the uptake of bare nanoparticles (bare NP) or nanoparticles coated with 15 μg mL–1 human plasma (human plasma). p ≤ 0.05 was considered significant (indicated with an asterisk).
Competition Study of Nanoparticles with Individual Proteins
For the competition study, 50,000 cells cm–2 hCMEC/D3 and TRP3 cells were seeded in a 24-well plate (Greiner) precoated with extracellular matrix as described above. Three or 4 days after seeding, for TRP3 or hCMEC/D3, respectively, cells were exposed to human plasma corona-coated SiO2–NH2 (200 nm) (30 μg mL–1 nanoparticles in serum-free medium, prepared as described above just prior addition to cells) in the presence of 1 (or increasing concentrations) unlabeled transferrin or 5 mg mL–1 unlabeled HSA. Alternatively, cells were exposed to fluorescently labeled Alexa Fluor 546 transferrin (Life Technologies, NY, USA) at a concentration of 10 μg mL–1 in serum-free medium in the presence of increasing concentrations of human plasma corona-coated SiO2–NH2 (200 nm). Then, cells were washed and collected for flow cytometry analysis as described above. A 561 nm laser was used for Alexa Fluor 546 transferrin excitation.
TFR1 Silencing Using RNA interference
In order to silence the expression of the transferrin receptor TFR1 on TRP3, 13,000 cells cm–2 were seeded in a 24-well plate (Greiner) precoated with 0.1% cold gelatin as described above. Twenty-four h after seeding, the expression of TFR1 was silenced as previously described.36 Briefly, cells were incubated in serum-free medium for 20 min, and then the medium was replaced with 250 μL of a siRNA (small interfering RNA) mix made with 2 μL of oligofectamine (ThermoFisher Scientific), 20 pmol siRNA directed toward TFR1 (Silencer Select, ThermoFisher Scientific), and Opti-MEM (ThermoFisher Scientific), following the manufacturer’s instructions. A scrambled siRNA was used as a negative control. After 4 h, 125 μL of TRP3 growth medium containing 60% v/v FBS was added, and cells were grown for a further 72 h in standard conditions (37 °C, 5% CO2). Then, cells were exposed for 4 h to 30 μg mL–1 freshly prepared hard corona-coated nanoparticles or for 10 min to Alexa Fluor 546 transferrin (10 μg mL–1) in serum-free medium. Finally, samples were collected and prepared as described above for flow cytometry analysis.
Quantification of TFR1 mRNA Expression
The expression level of silenced TFR1 in TRP3 cells was determined by quantitative real time PCR (RT-PCR) using forward (left) TGAAGAGAAAGTTGTCGGAGAAA and reverse (right) CAGCCTCACGAGGGACATA primers. After 72 h of silencing, the content of 3 wells was merged, and a Maxwell 16 LEV simplyRNA Cells Kit (Promega, Madison, WI, USA) was used to isolate the total mRNA in a Maxwell instrument according to the instructions provided by the manufacturer. cDNA was prepared by reverse transcription of mRNA using a Reverse Transcription System (Promega, Leiden, The Netherlands). The full procedure and cycle details are described in Aliyandi et al.30 An ABI7900HT sequence detection system (Applied Biosystems, Foster City, CA, USA) was used to determine the transcription levels by quantitative RT-PCR using a SensiMix SYBR kit (Bioline, Taunton, MA, USA) for sample preparation, following the manufacturer’s instructions. For each sample, 10 ng of cDNA was used. The results were analyzed using SDS 2.4 software (Applied Biosystems) to obtain the Ct values. Four replicate samples were prepared for each target, and the average and standard deviation of the Ct values obtained in the 4 replicate samples were calculated. The fold-change of expression levels in the TFR1 silenced samples (CtTFR1) with respect to the averaged Ct values of cells silenced with a scramble RNA as a negative control (CtNeg) was calculated as follows:
| 3 |
Statistical Analysis
In order to compare the uptake kinetics of the different corona-coated nanoparticles, a linear regression two-tailed Student’s t-test was applied to the uptake kinetic results up to 7 h of exposure, where uptake increases linearly with time (Figure 2).30 A nonparametric two-tailed Mann–Whitney test was used to determine statistical differences between two groups (Figure 5). A Kruskal–Wallis test was used to determine statistical differences between multiple groups (Figure 4), followed by a Mann–Whitney test with Bonferroni correction for multiple testing to compare the uptake level of single protein corona-coated nanoparticles to the uptake of bare nanoparticles (Bare NP) or nanoparticles coated with 15 μg mL–1 human plasma (human plasma). p ≤ 0.05 was considered significant. Mann–Whitney and Kruskal–Wallis tests were run on Statistics Kingdom (https://www.statskingdom.com/).
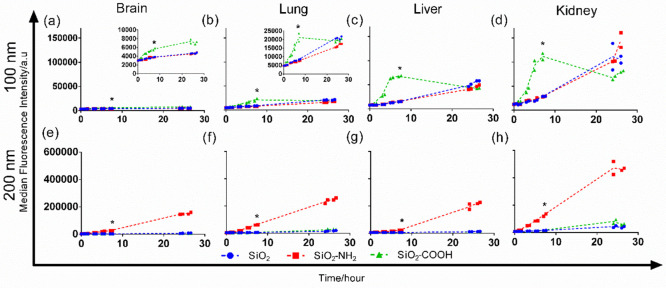
Figure 2.
Uptake kinetics of the corona-coated nanoparticles isolated from full human plasma. Brain (a and e), lung (b and f), liver (c and g), and kidney endothelium (d and h) were exposed to 50 μg mL–1 of 100 nm (a–d) or 30 μg mL–1 of 200 nm (e–h) corona-coated SiO2, SiO2–NH2, or SiO2–COOH in serum-free medium, isolated from full human plasma as described in the Materials and Methods section. The results show the median cell fluorescence intensity of two replicate samples, together with a line that passes through their average. A linear regression two-tailed Student’s t-test was applied to compare the uptake of the different corona-coated nanoparticles. Statistically significant differences up to a 7 h uptake (indicated by an asterisk) are observed for all cell types on 100 nm nanoparticles for SiO2–COOH as compared to the other functionalizations and on 200 nm nanoparticles for SiO2–NH2. p ≤ 0.05 was considered significant.
Results and Discussion
Protein Corona Characterization
Silica nanoparticles have been used as a well-characterized model for uptake studies in cells, including studies on protein corona.7,37,38 Here, we selected spherical silica nanoparticles of two different sizes (100 and 200 nm) and each with three different surface functionalizations (plain, amino modified (−NH2), and carboxylated (−COOH)) in order to obtain different coronas after dispersion in biological fluid. Nevertheless, any other nanomaterials with varying properties would be suitable to form a panel of different coronas for similar correlation analysis, including, for instance, gold, silver or biomaterials such as PLGA or liposomes.8,27,28,39 The silica nanoparticles are labeled with a chemically cross-linked fluorescent dye, thus excluding the possibility of dye leakage which could confuse cell uptake studies. SDS-PAGE confirmed that no or little free label was present for the 200 and 100 nm nanoparticles, while residual free dye could be easily removed with a washing step, as we performed here for isolation of corona-coated nanoparticles (Figure S1, Supporting Information). Instead of low concentrations of FBS, pooled human plasma was used as a more relevant source of proteins for corona formation when using human cells. In order to resemble better the physiological condition, full (100%) human plasma was used. For similar reasons, human plasma was chosen over human serum because it includes all blood components except the blood cells. Nevertheless, it is worth mentioning that the anticoagulant used for plasma preparation (which was citrate in the present study) is known to affect the final plasma composition, thus also corona formation.40,41 Similar effects are avoided when using serum, although serum lacks the coagulation factors. Additionally, other factors are known to affect corona formation in vivo, such as the presence of proteins secreted by cells and the presence of shear stress due to blood flow.10 Thus, ultimately, it would be important to perform similar studies on the in vivo cell uptake of nanoparticles in conjunction with protein corona analysis on nanoparticles recovered following injection in animal models.
As a first step, here, the corona-coated nanoparticles formed in full human plasma were isolated by repeated centrifugations and washes, following standard procedures to remove all unbound and loosely bound proteins. Control experiments confirmed that proteins did not pellet when full human plasma was centrifuged using the same procedure (Supporting Figure S2). Thus, prior to exposure to cells, the dispersions of the isolated corona-coated nanoparticles were characterized by dynamic light scattering (DLS) and zeta potential measurements (see Figure S3, Supporting Information, for details). It was previously shown that silica nanoparticle morphology is not affected upon corona formation,11 a part of the expected increase in average size, as we observed. DLS results showed that the isolated corona-coated nanoparticles were well redispersed and had a slight increase in average size in comparison to the bare nanoparticles in PBS (see Figure S3, Supporting Information), as expected upon protein adsorption and corona formation. For all nanoparticles, including the amino-modified nanoparticles, a negative zeta potential was measured for the dispersions in PBS. In fact, the zeta potential of both the 100 and 200 nm amino-modified nanoparticles is positive only at a pH lower than 7.4 (data from the manufacturer, not shown).
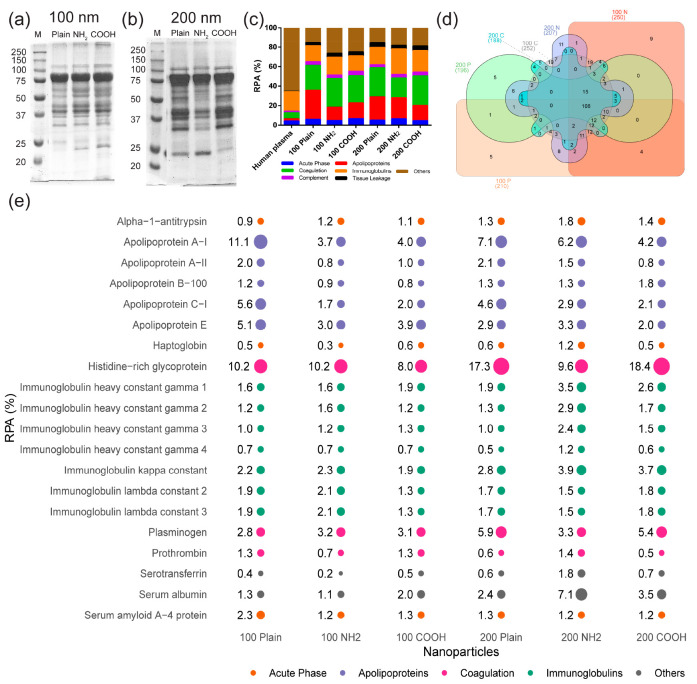
As a next step, the proteins in the different coronas were separated by SDS-PAGE in order to compare their composition. Different bands were observed for each nanoparticle, confirming that they adsorbed different types of proteins on their surface (Figure 1a and 1b, also Figure S4, Supporting Information, for protein corona on nanoparticles incubated in different concentrations of human plasma). Additionally, band intensities also differed, suggesting that for common corona proteins the absolute protein amounts also differed. Next, the hard corona proteins in each sample were identified by mass spectrometry (Figure 1c–e and the complete results in the Supporting Information). Approximately 300 proteins were detected for each sample, and around 100 of them were present in all coronas (Figure 1d). Figure 1c shows the distribution of protein classes in the different coronas and in human plasma. As also observed in other studies,5,6,29,42 corona formation led to the enrichment of different low abundant proteins on the nanoparticle surface, which was particularly high for apolipoproteins and coagulation factors and lower for complement factors and tissue leakage proteins. Figure 1e shows the list of the 20 most abundant proteins in each corona, which represented 40–60% of the total proteins recovered, together with their relative protein abundance, RPA (see the Materials and Methods section for details). As previously observed,5,6,29,42 several proteins were highly enriched in the different corona in comparison to their abundance in plasma. For instance, histidine-rich glycoprotein (HRG), which in plasma had an RPA of only 0.01% (see full results for plasma in the Supporting Information), was highly enriched in the corona formed on all different particles types, where alone it constituted roughly 8 to 18% of all corona proteins. This was further confirmed by Western blot of HRG, which showed a strong band at around 75 kDa (Figure S5, Supporting Information), which was also clearly visible in the SDS-PAGE gel image (Figure 1a and 1b). Additionally, as already visible by SDS-PAGE (Figure 1a,b), the relative amount of the different corona proteins differed for the nanoparticles with different surface functionalization, as well as for the different nanoparticle sizes.23,25,38 Some unique proteins present in only some of the coronas were also identified. Overall, these results confirmed that different coronas were formed.
Figure 1.
Characterization of the corona formed on 100 and 200 nm SiO2 (plain), SiO2–NH2 (NH2), or SiO2–COOH (COOH) in full human plasma. SDS-PAGE gel image of the proteins recovered on nanoparticle-corona complexes of 100 nm (a) or 200 nm (b) silica in full human plasma. The corona formed on all silica nanoparticles was prepared and isolated as described in the Materials and Methods section. The gel shows that different bands were present in the corona formed on the different silica nanoparticles. M: molecular weight size marker. Relative abundance (RPA%, see the Materials and Methods section for details) of the major protein groups identified by mass spectrometry in full human plasma and in the protein corona formed on the different silica nanoparticles (c). Venn diagram of the total amount of proteins identified by mass spectrometry in the nanoparticle-corona complexes formed in full human plasma (d). List of the top 20 most abundant corona proteins and their RPA (%) on the indicated silica nanoparticles, as measured by mass spectrometry (e). Proteins are ordered alphabetically. Different colors are used for the different protein families, and the spot size indicates their RPA (%).
Uptake of Corona-Coated Nanoparticles by Endothelial Cells from Different Organs
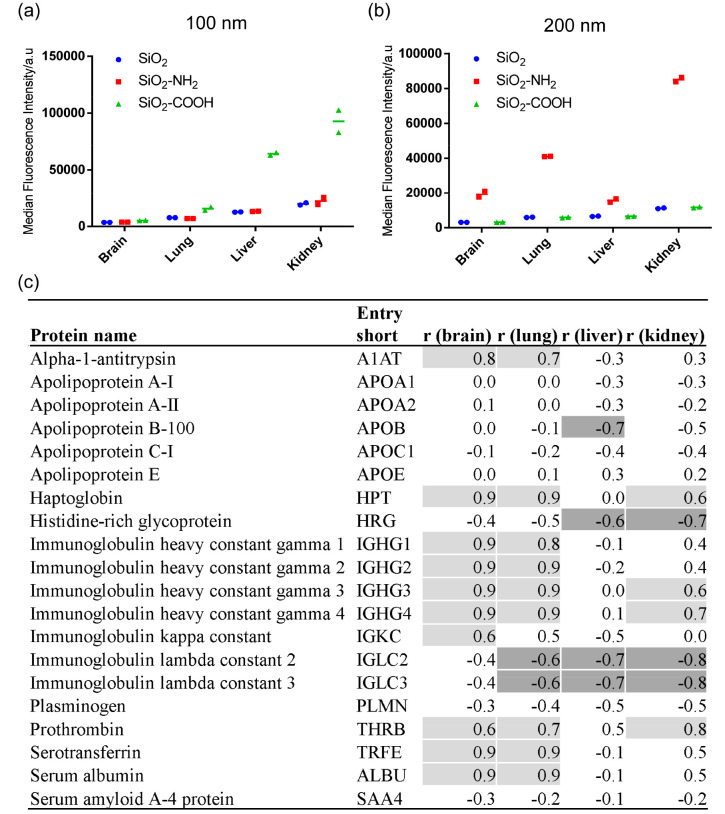
As a next step, we compared the uptake efficiency of the different corona-coated nanoparticles in different endothelial cell types. To this end, we used four endothelial cell lines derived from different organs (brain, lungs, liver, and kidneys). In order to be able to identify corona proteins that modulate uptake by cells and exclude additional effects due to the presence of free serum proteins in solutions, cells were exposed to the isolated corona-coated nanoparticles in serum-free medium.13,43 For this purpose, in order to limit preparation times for cell uptake studies, corona-coated nanoparticles were added to cells after only one centrifugation to remove the excess free proteins in solution, as opposed to the more extended washing procedure required for corona identification by mass spectrometry. A direct comparison of uptake levels after the two different procedures showed that the same trend was observed for the particles with different functionalization (see Figure S6, Supporting Information). This confirmed that the different washing procedures did not affect uptake levels by cells. Silica nanoparticles are known to be generally well tolerated by cells, especially when coated with a protein corona.37 In line with this, no evident toxicity and no differences in cell numbers were observed upon exposure to the different nanoparticles (see Figure S7, Supporting Information). Since all bare silica nanoparticles of the same size had comparable fluorescence intensity (Figure S8, Supporting Information), uptake levels for the nanoparticles with different surface functionalization could be directly compared (Figure 2). As shown in Figure 2, all endothelial cell types showed a preference for the same nanoparticle type. Specifically, when comparing the results for nanoparticles of the same size, SiO2–COOH (100 nm) and SiO2–NH2 (200 nm) showed higher uptake in all endothelial cell types. Nevertheless, each endothelium showed different uptake efficiency, with the brain endothelium showing the lowest nanoparticle uptake efficiency and liver and kidney endothelium showing the highest. These differences may reflect their different physiological functions, where the blood-brain barrier, in general, has a more selective uptake than liver sinusoids and kidney glomeruli. Similarly, after 5-h exposure (Figure 2 and Figure 3a,b) for the 100 nm carboxylated silica, cell fluorescence in the liver endothelium was higher than in the lung endothelium, while the opposite was observed for the 200 nm amino-modified silica, i.e., higher fluorescence in the lung endothelium. These differences in absolute uptake efficiency may relate to differences in the expression level and activity of the receptors and mechanisms that are responsible for the uptake of the corona-coated nanoparticles between different endothelial cell types. Exploiting such differences may enable novel targeting strategies.
Figure 3.
Correlation between nanoparticle uptake in brain, lung, liver, and kidney endothelium after 5h exposure and the relative protein abundance of adsorbed corona proteins for all six silica nanoparticles tested. The uptake level of the nanoparticle-corona complexes formed on 100 (a) and 200 nm (b) silica nanoparticles in full human serum after 5 h. The results show the median cell fluorescence intensity of two replicate samples, together with their average indicated with a line. (c) Corona proteins correlating with uptake. The table shows the results of the correlation analysis between the 5-h nanoparticle uptake in brain, lung, liver, and kidney endothelium and the relative protein abundance of adsorbed corona proteins, performed as described in the Materials and Methods section. Positive correlation coefficients (r) ≥ 0.6 are shaded in light gray, and negative correlation coefficients (r) ≤ 0.6 are shaded in dark gray.
Correlation between Protein Corona Composition and Nanoparticle Uptake
In order to identify corona proteins which affected nanoparticle uptake, we performed correlation analysis between cellular uptake levels after 5-h exposure (Figure 3a and 3b) and the corona composition of all 6 investigated silica nanoparticles using Pearson correlation (Figure 3c). A positive correlation (r ≥ 0.6), which indicates that a high amount of a certain protein in the corona correlated with higher uptake by cells, was observed in brain and lung endothelium for alpha-1-antitrypsin, haptoglobin, immunoglobulins, human serum albumin (HSA), prothrombin, and transferrin. Interestingly, in the liver endothelium, a strong negative correlation (r ≤ −0.6) was observed for some proteins including histidine-rich glycoprotein (HRG) and no positive correlation. This lack of discrimination between proteins for uptake may be explained by the clearance function of scavenger endothelial cells of the liver.
In addition, we also performed correlation analysis based on nanoparticle uptake levels at 24 h (Table S1, Supporting Information). Given that the uptake behavior, especially for the 100 nm nanoparticles, was different after 24 h, this expectedly led to different results in the correlation analysis. In this case, all endothelial cell types showed a similar profile of positively correlated proteins, and no negative correlation was observed for any of the proteins in all endothelial cell types, including for the liver. Many proteins in liver and kidney endothelium, including albumin and transferrin, which showed minimal correlation based on the 5-h uptake, now turned out to be positively correlated. These differences may reflect observed differences in nanoparticle distribution in vivo over time, when nanoparticles first may accumulate in certain organs and then are released again and accumulate in other organs at later times. Further studies are required to explain this observation and to determine, depending on the application, the most appropriate exposure time to use for similar correlation analysis. Similarly, it would be interesting to study effects on uptake due to corona composition evolution over time, e.g., during exposure to cells, where secreted cellular proteins may adsorb.38,44−47
Role of the Correlated Corona Proteins in Nanoparticle Uptake
In order to verify if the high abundant proteins in the corona that correlate with higher cellular uptake do have a role in nanoparticle uptake by the endothelial cells, i.e., to exclude coincidence, we performed validation experiments using artificial single protein-coronas (Figure 4) and competition studies (Figure S9, Supporting Information). For practical reasons, this validation study was performed using the 200 nm SiO2–NH2 since they showed higher uptake in all cell lines, thus making it easier to observe a possible inhibition in uptake. For similar reasons, validation was performed in brain and liver endothelium to compare the effects of the identified proteins in cells that showed the lowest and highest uptake, respectively. Thus, in order to verify whether individual corona proteins identified by the correlation analysis did affect nanoparticle uptake, we prepared artificial corona made of the single correlated proteins. These were made by simply exposing the nanoparticles to a solution of the protein of interest at an excess concentration, in order to ensure that enough protein to cover all available nanoparticle surface was present. Then, the uptake of nanoparticles with a corona made of the single correlated proteins was compared to the uptake levels of nanoparticles with a human plasma corona and bare nanoparticles. In this way, we could demonstrate if those single corona proteins alone could increase or decrease the uptake, as suggested by the correlation analysis. As shown in Figure 4, precoating 200 nm SiO2–NH2 with positively correlated proteins, namely HSA, transferrin, and alpha-1 antitrypsin, did not affect cellular uptake in either endothelium. Instead, coating the nanoparticles with HRG alone strongly reduced uptake in both endothelia to the same extent as with the natural full human plasma corona. This result is of particular interest because the correlation of the positively correlated proteins was stronger than the negative correlation of HRG, yet they showed no significant effect on the uptake. This example illustrates the importance of validating with other methods the role of the correlated proteins identified with this type of analysis.29
Next, to investigate the role of the cellular receptors for the correlated corona proteins (rather than the corona proteins themselves), we also performed a competition study in brain and liver endothelium using HSA and transferrin. In this approach, 200 nm SiO2–NH2 were again used as model nanoparticles, and the uptake of the corona-coated nanoparticles isolated from full human plasma was measured after adding free HSA or transferrin in the medium to test possible competition for the same receptors. As shown in Figure S9, Supporting Information, neither of the proteins were able to compete with nanoparticle uptake. Instead, contrary to our expectations, in liver endothelium (Figure S9a, Supporting Information), we observed a substantial increase in the uptake of 200 nm SiO2–NH2 in the presence of transferrin, i.e., the presence of free transferrin somehow stimulated nanoparticle internalization.
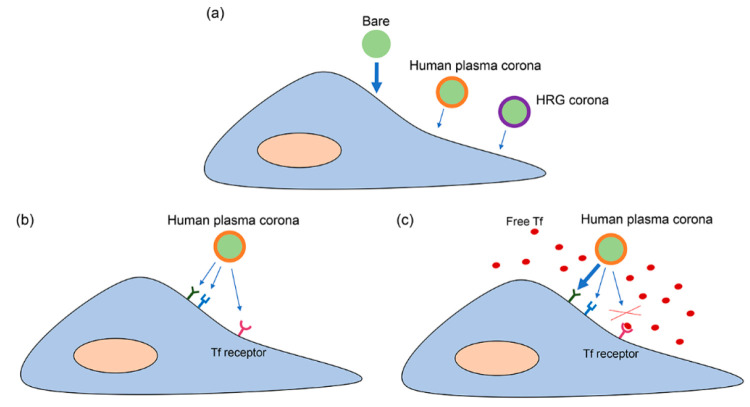
In order to understand this observation, further studies were performed to elucidate the effects of free transferrin on nanoparticle uptake in liver endothelium (Figure 5). Additional competition experiments where free transferrin was added at a range of increasing concentrations showed that nanoparticle uptake was increased by the presence of free transferrin in a concentration-dependent manner (Figure 5a). Interestingly, this increase was no longer observed when the free transferrin was removed again from the medium in which nanoparticles were dispersed (Figure S10, Supporting Information). This suggested that the effect was due to the presence of free transferrin in solution. SDS-PAGE of the human plasma corona-coated nanoparticles isolated after a second incubation with free transferrin did not show any evident increase in the intensity of the band corresponding to transferrin (also in Figure S10, Supporting Information). This suggested that no additional transferrin was adsorbed in the corona or that it was only loosely associated with the nanoparticles, thus washed away in the isolation procedure. Importantly, we also tried the reverse experiment to see if transferrin uptake could be completed by the addition of increasing amounts of corona-coated nanoparticles (Figure 5b). In this case, we observed a strong concentration-dependent competition, suggesting the involvement of the transferrin receptor in nanoparticle uptake. Instead, silencing the transferrin receptor TFR1 reduced transferrin uptake by ∼70%, confirming efficient reduction of its activity upon silencing (Figure S11, Supporting Information, also including RT-PCR results in silenced cells), but it did not have any effect on nanoparticle uptake (Figure 5c). A previous study suggested that the uptake of transferrin is predominantly mediated by TFR2 when extracellular transferrin concentration is high.48 However, this was not the case in our study, since silencing of TFR2 also did not have any effect on nanoparticle uptake (data not shown). A possible explanation for all these observations is that nanoparticles entered via multiple pathways,49 including via transferrin receptors. Blocking one pathway led to an increase in uptake through other pathways, which also had higher uptake efficiency.50−52 On the contrary, transferrin enters cells only via transferrin receptors. Therefore, the strong reduction in transferrin uptake in the presence of nanoparticles suggested that the nanoparticles occupy (and are internalized via) transferrin receptors. Because of this, we sought to investigate if clathrin-mediated endocytosis (CME) was involved in nanoparticle uptake, given that transferrin receptors are usually internalized by cells via this pathway. To this end, we blocked CME in liver endothelial cells using the pharmacological inhibitor chlorpromazine and observed its effect on nanoparticle uptake. As a first step, we tested this compound on cells to make sure it did not affect cell viability (Figure S12, Supporting Information).13,36,53,54 The results showed that addition of chlorpromazine blocked the uptake of transferrin and LDL, known to enter via this pathway, while it greatly increased nanoparticle uptake (Figure S12a–c, Supporting Information), similar to when adding free transferrin. The same result was obtained when we blocked CME through overexpression of the RFP-tagged AP180 C-terminus (Figure S12e–h, Supporting Information). The transfected cells showed higher nanoparticle uptake compared to nontransfected cells (Figure S12g,h, Supporting Information). Overall, as we illustrate in the scheme of Figure 6, these results suggested that the nanoparticles are taken up via the transferrin receptor via CME, possibly stimulated by the presence of transferrin in their corona. In addition, we speculate that when free transferrin is added, the nanoparticles are displaced (from transferrin receptors) to other receptors, possibly triggering uptake via clathrin-independent pathways, with higher uptake efficiency.
Figure 6.
Scheme illustrating the proposed mechanism of uptake for 200 nm SiO2–NH2 nanoparticles in liver endothelium. The thickness of the arrows represents uptake efficiency. (a) Uptake of particles with a human plasma corona is lower than for bare particles, but the same effect is obtained with a corona made by HRG alone. (b) Particles with a human plasma corona enter cells via multiple receptors including the transferrin (Tf) receptor. However, if free Tf is added (c), it competes with the particles for the Tf receptors, and the particles are displaced to a different receptor which has higher uptake efficiency, leading to a higher uptake.
Conclusions
In this study, we correlated protein corona composition and nanoparticle uptake in endothelial cells to identify corona proteins that are involved in the uptake and investigated the role of their receptors. For this, a panel of six nanoparticles and four different endothelial cells was used. For all silica nanoparticles tested, we observed a high enrichment of apolipoproteins and coagulation factors in the corona, especially HRG, which also argues for performing experiments in plasma rather than serum. Using correlation analysis between the protein corona composition and cellular uptake, we were able to identify proteins that might regulate uptake. Interestingly, correlation analysis showed that HRG presence in the corona negatively correlated with uptake, and indeed, covering the nanoparticles with HRG alone decreased nanoparticle uptake up to 80–90% in brain and liver endothelium (this is also illustrated in the scheme of Figure 6). As reported in recent studies, HRG has a high affinity for silica surfaces,13,55 and it had a similar effect on decreasing nanoparticle uptake also on macrophages due to its dysopsonin activity.55 This masking effect of HRG could be potentially exploited as an alternative strategy to obtain a “stealth” layer on nanomedicines.
Next to HRG, we also discovered that free transferrin had an effect on the uptake of 200 nm SiO2–NH2. In liver endothelium, transferrin uptake was reduced when these nanoparticles were added, suggesting a role for the transferrin receptor in nanoparticle binding and/or uptake. However, when free transferrin was added, nanoparticle uptake strongly increased. This was possibly due to redirection of nanoparticles to other receptors with higher uptake efficiency when the transferrin receptor was occupied by free transferrin. In other words, nanoparticles may have multiple ways to be taken up, possibly mediated via interaction of different proteins adsorbed on their surface with different receptors. Similar results were also reported in a recent study in which different pathways were involved in the uptake of hard corona-coated silica nanoparticles in HeLa cells.13 Further studies and novel methods are needed in order to identify all alternative receptors or pathways involved in nanoparticle uptake.
Overall, these results clearly highlight the complexity of the protein corona in the way it can affect the interactions of nanoparticles with cells. Correlation analysis between corona composition and cell uptake allows the discovery of specific proteins that are critically involved in nanoparticle uptake.8,16,26−29 However, it is important to stress that correlation alone does not demonstrate a role of the identified proteins in nanoparticle uptake, and validation via other methods is required to confirm whether the correlated proteins do affect uptake. Additionally, one needs to take into account the possibility that cell receptors may recognize more complex surfaces formed by multiple corona proteins all together in addition to single corona components.12 Therefore, future studies are needed to better understand protein corona-cell interactions, and at the same time, better methods need to be developed to identify which proteins have potential for the targeting of nanocarriers. Importantly, the presence of other biomolecules in solution also affects nanoparticle uptake, and their composition will vary between different cellular microenvironments. This adds another level of complexity to discerning possible ligand–receptor pairs for targeted delivery, which should be taken into account in the design of targeted nanoparticles.
While the role of individual proteins identified in this work, such as, for instance, eventual effects of HRG should be explicitly tested in vivo in order to determine their potential application, our results confirm that corona correlation analysis allows the discovery of proteins that have an impact on nanoparticle uptake by cells. The discovery of proteins promoting uptake can be used to develop novel targeting strategies directed at their receptors (thus, for instance, to make nanoparticles with antibodies or targeting ligands for the identified receptors). Similarly, the identification of corona proteins that block nanoparticle uptake in specific cell types could be used to design peptides for nanoparticle functionalization to evade internalization by those cell types. First studies have performed a similar analysis directly in vivo on nanoparticles injected and recovered from blood,56 where other factors such as the presence of blood flow and biomolecules released by cells are known to affect the corona forming on nanoparticles.10 These studies further confirm the suitability of this approach to discover new ways to improve nanoparticle targeting by a better understanding of the nanoparticle corona.
Acknowledgments
The hCMEC/D3 cells were kindly provided by Pierre-Olivier Couraud (Institute Cochin), and Ronald E. Unger (Johannes Gutenberg University of Mainz) is acknowledged for providing HPMEC-ST1.6R cells. Birke Bartosch and Romain Parent (INSERM) are acknowledged for providing TRP3 cells. Simon Satchell (University of Bristol) is acknowledged for providing CiGENC cells. Yvonne Vallis and Harvey T. McMahon are acknowledged for providing the pDNA encoding RFP-tagged C-terminal of AP180. Hjalmar Permentier and Margot Jeronimus-Stratingh (Interfaculty Mass Spectrometry Center, UMCG) are acknowledged for the helpful support with mass spectrometry.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsbiomaterials.1c00804.
Additional methods, physicochemical characterization of the silica nanoparticles tested (size, zeta potential, fluorescence), controls for free dye leakage and protein precipitation, SDS-PAGE gel image of proteins recovered on nanoparticle-corona complexes, Western blot of HRG in the corona, competition study, correlation analysis at 24 h, silencing efficiency of TFR1, and effect of inhibition of clathrin-mediated endocytosis on nanoparticle uptake in liver endothelium (PDF)
Complete mass spectrometry results (XLSX)
Funding from the H2020 European Research Council (ERC) grant NanoPaths (grant agreement no. 637614) to A.S. partially supported this work.
The authors declare no competing financial interest.
Supplementary Material
References
- Lee H.; Lytton-Jean A. K. R.; Chen Y.; Love K. T.; Park A. I.; Karagiannis E. D.; Sehgal A.; Querbes W.; Zurenko C. S.; Jayaraman M.; Peng C. G.; Charisse K.; Borodovsky A.; Manoharan M.; Donahoe J. S.; Truelove J.; Nahrendorf M.; Langer R.; Anderson D. G. Molecularly Self-Assembled Nucleic Acid Nanoparticles for Targeted in Vivo SiRNA Delivery. Nat. Nanotechnol. 2012, 7 (6), 389–393. 10.1038/nnano.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Mello S. R.; Cruz C. N.; Chen M. L.; Kapoor M.; Lee S. L.; Tyner K. M. The Evolving Landscape of Drug Products Containing Nanomaterials in the United States. Nat. Nanotechnol. 2017, 12 (6), 523–529. 10.1038/nnano.2017.67. [DOI] [PubMed] [Google Scholar]
- Cheng C. J.; Tietjen G. T.; Saucier-Sawyer J. K.; Saltzman W. M. A Holistic Approach to Targeting Disease with Polymeric Nanoparticles. Nat. Rev. Drug Discovery 2015, 14 (4), 239–247. 10.1038/nrd4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheon J.; Chan W.; Zuhorn I. The Future of Nanotechnology: Cross-Disciplined Progress to Improve Health and Medicine. Acc. Chem. Res. 2019, 17, 2405. 10.1021/acs.accounts.9b00423. [DOI] [PubMed] [Google Scholar]
- Cedervall T.; Lynch I.; Lindman S.; Berggard T.; Thulin E.; Nilsson H.; Dawson K. A.; Linse S.; Berggård T.; Thulin E.; Nilsson H.; Dawson K. A.; Linse S. Understanding the Nanoparticle-Protein Corona Using Methods to Quantify Exchange Rates and Affinities of Proteins for Nanoparticles. Proc. Natl. Acad. Sci. U. S. A. 2007, 104 (7), 2050–2055. 10.1073/pnas.0608582104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monopoli M. P.; Åberg C.; Salvati A.; Dawson K. A. Biomolecular Coronas Provide the Biological Identity of Nanosized Materials. Nat. Nanotechnol. 2012, 7 (12), 779–786. 10.1038/nnano.2012.207. [DOI] [PubMed] [Google Scholar]
- Monopoli M. P.; Walczyk D.; Campbell A.; Elia G.; Lynch I.; Baldelli Bombelli F.; Dawson K. A. Physical–Chemical Aspects of Protein Corona: Relevance to in Vitro and in Vivo Biological Impacts of Nanoparticles. J. Am. Chem. Soc. 2011, 133 (8), 2525–2534. 10.1021/ja107583h. [DOI] [PubMed] [Google Scholar]
- Walkey C. D.; Olsen J. B.; Song F.; Liu R.; Guo H.; Olsen D. W. H.; Cohen Y.; Emili A.; Chan W. C. W. Protein Corona Fingerprinting Predicts the Cellular Interaction of Gold and Silver Nanoparticles. ACS Nano 2014, 8 (3), 2439–2455. 10.1021/nn406018q. [DOI] [PubMed] [Google Scholar]
- Yan Y.; Gause K. T.; Kamphuis M. M. J.; Ang C. S.; O’Brien-Simpson N. M.; Lenzo J. C.; Reynolds E. C.; Nice E. C.; Caruso F. Differential Roles of the Protein Corona in the Cellular Uptake of Nanoporous Polymer Particles by Monocyte and Macrophage Cell Lines. ACS Nano 2013, 7 (12), 10960–10970. 10.1021/nn404481f. [DOI] [PubMed] [Google Scholar]
- Hadjidemetriou M.; Al-Ahmady Z.; Mazza M.; Collins R. F.; Dawson K.; Kostarelos K. In Vivo Biomolecule Corona around Blood-Circulating, Clinically Used and Antibody-Targeted Lipid Bilayer Nanoscale Vesicles. ACS Nano 2015, 9 (8), 8142–8156. 10.1021/acsnano.5b03300. [DOI] [PubMed] [Google Scholar]
- Lara S.; Alnasser F.; Polo E.; Garry D.; Lo Giudice M. C.; Hristov D. R.; Rocks L.; Salvati A.; Yan Y.; Dawson K. A. Identification of Receptor Binding to the Biomolecular Corona of Nanoparticles. ACS Nano 2017, 11 (2), 1884–1893. 10.1021/acsnano.6b07933. [DOI] [PubMed] [Google Scholar]
- Lara S.; Perez-Potti A.; Herda L. M.; Adumeau L.; Dawson K. A.; Yan Y. Differential Recognition of Nanoparticle Protein Corona and Modified Low-Density Lipoprotein by Macrophage Receptor with Collagenous Structure. ACS Nano 2018, 12 (5), 4930–4937. 10.1021/acsnano.8b02014. [DOI] [PubMed] [Google Scholar]
- Francia V.; Yang K.; Deville S.; Reker-Smit C.; Nelissen I.; Salvati A. Corona Composition Can Affect the Mechanisms Cells Use to Internalize Nanoparticles. ACS Nano 2019, 13 (10), 11107–11121. 10.1021/acsnano.9b03824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvati A.; Pitek A. S.; Monopoli M. P.; Prapainop K.; Bombelli F. B.; Hristov D. R.; Kelly P. M.; Åberg C.; Mahon E.; Dawson K. A. Transferrin-Functionalized Nanoparticles Lose Their Targeting Capabilities When a Biomolecule Corona Adsorbs on the Surface. Nat. Nanotechnol. 2013, 8 (2), 137–143. 10.1038/nnano.2012.237. [DOI] [PubMed] [Google Scholar]
- Zuhorn I. S.; Visser W. H.; Bakowsky U.; Engberts J. B. F. N.; Hoekstra D. Interference of Serum with Lipoplex-Cell Interaction: Modulation of Intracellular Processing. Biochim. Biophys. Acta, Biomembr. 2002, 1560 (1–2), 25–36. 10.1016/S0005-2736(01)00448-5. [DOI] [PubMed] [Google Scholar]
- Caracciolo G.; Cardarelli F.; Pozzi D.; Salomone F.; Maccari G.; Bardi G.; Capriotti A. L.; Cavaliere C.; Papi M.; Laganà A. Selective Targeting Capability Acquired with a Protein Corona Adsorbed on the Surface of 1,2-Dioleoyl-3-Trimethylammonium Propane/DNA Nanoparticles. ACS Appl. Mater. Interfaces 2013, 5 (24), 13171–13179. 10.1021/am404171h. [DOI] [PubMed] [Google Scholar]
- Kreuter J.; Shamenkov D.; Petrov V.; Ramge P.; Cychutek K.; Koch-Brandt C.; Alyautdin R. Apolipoprotein-Mediated Transport of Nanoparticle-Bound Drugs across the Blood-Brain Barrier. J. Drug Target. 2002, 10 (4), 317–325. 10.1080/10611860290031877. [DOI] [PubMed] [Google Scholar]
- Dobrovolskaia M. A.; Shurin M.; Shvedova A. A. Current Understanding of Interactions between Nanoparticles and the Immune System. Toxicol. Appl. Pharmacol. 2016, 299, 78–89. 10.1016/j.taap.2015.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. A.; Salvati A.; Åberg C.; Dawson K. A. Suppression of Nanoparticle Cytotoxicity Approaching in Vivo Serum Concentrations: Limitations of in Vitro Testing for Nanosafety. Nanoscale 2014, 6 (23), 14180–14184. 10.1039/C4NR04970E. [DOI] [PubMed] [Google Scholar]
- Schöttler S.; Becker G.; Winzen S.; Steinbach T.; Mohr K.; Landfester K.; Mailänder V.; Wurm F. R. Protein Adsorption Is Required for Stealth Effect of Poly(Ethylene Glycol)- and Poly(Phosphoester)-Coated Nanocarriers. Nat. Nanotechnol. 2016, 11 (4), 372–377. 10.1038/nnano.2015.330. [DOI] [PubMed] [Google Scholar]
- Akinc A.; Maier M. A.; Manoharan M.; Fitzgerald K.; Jayaraman M.; Barros S.; Ansell S.; Du X.; Hope M. J.; Madden T. D.; Mui B. L.; Semple S. C.; Tam Y. K.; Ciufolini M.; Witzigmann D.; Kulkarni J. A.; van der Meel R.; Cullis P. R. The Onpattro Story and the Clinical Translation of Nanomedicines Containing Nucleic Acid-Based Drugs. Nat. Nanotechnol. 2019, 14 (12), 1084–1087. 10.1038/s41565-019-0591-y. [DOI] [PubMed] [Google Scholar]
- Nel A. E.; Mädler L.; Velegol D.; Xia T.; Hoek E. M. V.; Somasundaran P.; Klaessig F.; Castranova V.; Thompson M. Understanding Biophysicochemical Interactions at the Nano-Bio Interface. Nat. Mater. 2009, 8, 543–557. 10.1038/nmat2442. [DOI] [PubMed] [Google Scholar]
- Lundqvist M.; Stigler J.; Elia G.; Lynch I.; Cedervall T.; Dawson K. A. Nanoparticle Size and Surface Properties Determine the Protein Corona with Possible Implications for Biological Impacts. Proc. Natl. Acad. Sci. U. S. A. 2008, 105 (38), 14265–14270. 10.1073/pnas.0805135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calatayud M. P.; Sanz B.; Raffa V.; Riggio C.; Ibarra M. R.; Goya G. F. The Effect of Surface Charge of Functionalized Fe3O4 Nanoparticles on Protein Adsorption and Cell Uptake. Biomaterials 2014, 35 (24), 6389–6399. 10.1016/j.biomaterials.2014.04.009. [DOI] [PubMed] [Google Scholar]
- Walkey C. D.; Olsen J. B.; Guo H.; Emili A.; Chan W. C. W. Nanoparticle Size and Surface Chemistry Determine Serum Protein Adsorption and Macrophage Uptake. J. Am. Chem. Soc. 2012, 134 (4), 2139–2147. 10.1021/ja2084338. [DOI] [PubMed] [Google Scholar]
- Liu R.; Jiang W.; Walkey C. D.; Chan W. C. W.; Cohen Y. Prediction of Nanoparticles-Cell Association Based on Corona Proteins and Physicochemical Properties. Nanoscale 2015, 7 (21), 9664–9675. 10.1039/C5NR01537E. [DOI] [PubMed] [Google Scholar]
- Bigdeli A.; Palchetti S.; Pozzi D.; Hormozi-Nezhad M. R.; Baldelli Bombelli F.; Caracciolo G.; Mahmoudi M. Exploring Cellular Interactions of Liposomes Using Protein Corona Fingerprints and Physicochemical Properties. ACS Nano 2016, 10 (3), 3723–3737. 10.1021/acsnano.6b00261. [DOI] [PubMed] [Google Scholar]
- Yang K.; Mesquita B.; Horvatovich P.; Salvati A. Tuning Liposome Composition to Modulate Corona Formation in Human Serum and Cellular Uptake. Acta Biomater. 2020, 106, 314–327. 10.1016/j.actbio.2020.02.018. [DOI] [PubMed] [Google Scholar]
- Ritz S.; Schöttler S.; Kotman N.; Baier G.; Musyanovych A.; Kuharev J.; Landfester K.; Schild H.; Jahn O.; Tenzer S.; Mailänder V. Protein Corona of Nanoparticles: Distinct Proteins Regulate the Cellular Uptake. Biomacromolecules 2015, 16 (4), 1311–1321. 10.1021/acs.biomac.5b00108. [DOI] [PubMed] [Google Scholar]
- Aliyandi A.; Satchell S.; Unger R. E.; Bartosch B.; Parent R.; Zuhorn I. S.; Salvati A. Effect of Endothelial Cell Heterogeneity on Nanoparticle Uptake. Int. J. Pharm. 2020, 587, 119699. 10.1016/j.ijpharm.2020.119699. [DOI] [PubMed] [Google Scholar]
- Weksler B. B.; Subileau E. a; Perrière N.; Charneau P.; Holloway K.; Leveque M.; Tricoire-Leignel H.; Nicotra A.; Bourdoulous S.; Turowski P.; Male D. K.; Roux F.; Greenwood J.; Romero I. a; Couraud P. O. Blood-brain Barrier-specific Properties of a Human Adult Brain Endothelial Cell Line. FASEB J. 2005, 19 (13), 1872–1874. 10.1096/fj.04-3458fje. [DOI] [PubMed] [Google Scholar]
- Krump-Konvalinkova V.; Bittinger F.; Unger R. E.; Peters K.; Lehr H.-A.; Kirkpatrick C. J. Generation of Human Pulmonary Microvascular Endothelial Cell Lines. Lab. Invest. 2001, 81 (12), 1717–1727. 10.1038/labinvest.3780385. [DOI] [PubMed] [Google Scholar]
- Parent R.; Durantel D.; Lahlali T.; Sallé A.; Plissonnier M.-L.; DaCosta D.; Lesca G.; Zoulim F.; Marion M.-J.; Bartosch B. An Immortalized Human Liver Endothelial Sinusoidal Cell Line for the Study of the Pathobiology of the Liver Endothelium. Biochem. Biophys. Res. Commun. 2014, 450 (1), 7–12. 10.1016/j.bbrc.2014.05.038. [DOI] [PubMed] [Google Scholar]
- Satchell S. C.; Tasman C. H.; Singh A.; Ni L.; Geelen J.; von Ruhland C. J.; O’Hare M. J.; Saleem M. A.; van den Heuvel L. P.; Mathieson P. W. Conditionally Immortalized Human Glomerular Endothelial Cells Expressing Fenestrations in Response to VEGF. Kidney Int. 2006, 69 (9), 1633–1640. 10.1038/sj.ki.5000277. [DOI] [PubMed] [Google Scholar]
- De Jong E.; Williams D. S.; Abdelmohsen L. K. E. A.; Van Hest J. C. M.; Zuhorn I. S. A Filter-Free Blood-Brain Barrier Model to Quantitatively Study Transendothelial Delivery of Nanoparticles by Fluorescence Spectroscopy. J. Controlled Release 2018, 289, 14–22. 10.1016/j.jconrel.2018.09.015. [DOI] [PubMed] [Google Scholar]
- Francia V.; Aliyandi A.; Salvati A. Effect of the Development of a Cell Barrier on Nanoparticle Uptake in Endothelial Cells. Nanoscale 2018, 10 (35), 16645–16656. 10.1039/C8NR03171A. [DOI] [PubMed] [Google Scholar]
- Lesniak A.; Fenaroli F.; Monopoli M. P.; Åberg C.; Dawson K. A.; Salvati A. Effects of the Presence or Absence of a Protein Corona on Silica Nanoparticle Uptake and Impact on Cells. ACS Nano 2012, 6 (7), 5845–5857. 10.1021/nn300223w. [DOI] [PubMed] [Google Scholar]
- Tenzer S.; Docter D.; Kuharev J.; Musyanovych A.; Fetz V.; Hecht R.; Schlenk F.; Fischer D.; Kiouptsi K.; Reinhardt C.; Landfester K.; Schild H.; Maskos M.; Knauer S. K.; Stauber R. H. Rapid Formation of Plasma Protein Corona Critically Affects Nanoparticle Pathophysiology. Nat. Nanotechnol. 2013, 8 (10), 772–781. 10.1038/nnano.2013.181. [DOI] [PubMed] [Google Scholar]
- Sobczynski D. J.; Charoenphol P.; Heslinga M. J.; Onyskiw P. J.; Namdee K.; Thompson A. J.; Eniola-Adefeso O. Plasma Protein Corona Modulates the Vascular Wall Interaction of Drug Carriers in a Material and Donor Specific Manner. PLoS One 2014, 9 (9), e107408. 10.1371/journal.pone.0107408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobczynski D. J.; Eniola-Adefeso O. Effect of Anticoagulants on the Protein Corona-Induced Reduced Drug Carrier Adhesion Efficiency in Human Blood Flow. Acta Biomater. 2017, 48, 186–194. 10.1016/j.actbio.2016.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöttler S.; Klein K.; Landfester K.; Mailänder V. Protein Source and Choice of Anticoagulant Decisively Affect Nanoparticle Protein Corona and Cellular Uptake. Nanoscale 2016, 8 (10), 5526–5536. 10.1039/C5NR08196C. [DOI] [PubMed] [Google Scholar]
- Yin B.; Chan C. K. W.; Liu S.; Hong H.; Wong S. H. D.; Lee L. K. C.; Ho L. W. C.; Zhang L.; Leung K. C.-F.; Choi P. C.-L.; Bian L.; Tian X. Y.; Chan M. N.; Choi C. H. J. Intrapulmonary Cellular-Level Distribution of Inhaled Nanoparticles with Defined Functional Groups and Its Correlations with Protein Corona and Inflammatory Response. ACS Nano 2019, 13 (12), 14048–14069. 10.1021/acsnano.9b06424. [DOI] [PubMed] [Google Scholar]
- Yang K.; Reker-Smit C.; Stuart M. C. A.; Salvati A. Effects of Protein Source on Liposome Uptake by Cells: Corona Composition and Impact of the Excess Free Proteins. Adv. Healthcare Mater. 2021, 10 (14), 2100370. 10.1002/adhm.202100370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albanese A.; Walkey C. D.; Olsen J. B.; Guo H.; Emili A.; Chan W. C. W. Secreted Biomolecules Alter the Biological Identity and Cellular Interactions of Nanoparticles. ACS Nano 2014, 8 (6), 5515–5526. 10.1021/nn4061012. [DOI] [PubMed] [Google Scholar]
- Casals E.; Pfaller T.; Duschl A.; Oostingh G. J.; Puntes V. Time Evolution of the Nanoparticle Protein Corona. ACS Nano 2010, 4 (7), 3623–3632. 10.1021/nn901372t. [DOI] [PubMed] [Google Scholar]
- Palchetti S.; Digiacomo L.; Pozzi D.; Peruzzi G.; Micarelli E.; Mahmoudi M.; Caracciolo G. Nanoparticles-Cell Association Predicted by Protein Corona Fingerprints. Nanoscale 2016, 8 (25), 12755–12763. 10.1039/C6NR03898K. [DOI] [PubMed] [Google Scholar]
- Barrán-Berdón A. L.; Pozzi D.; Caracciolo G.; Capriotti A. L.; Caruso G.; Cavaliere C.; Riccioli A.; Palchetti S.; Laganà A. Time Evolution of Nanoparticle–Protein Corona in Human Plasma: Relevance for Targeted Drug Delivery. Langmuir 2013, 29 (21), 6485–6494. 10.1021/la401192x. [DOI] [PubMed] [Google Scholar]
- Anderson G. J.; Powell L. W.; Halliday J. W. The Endocytosis of Transferrin by Rat Intestinal Epithelial Cells. Gastroenterology 1994, 106 (2), 414–422. 10.1016/0016-5085(94)90600-9. [DOI] [PubMed] [Google Scholar]
- Rehman Z. U.; Hoekstra D.; Zuhorn I. S. Protein Kinase A Inhibition Modulates the Intracellular Routing of Gene Delivery Vehicles in HeLa Cells, Leading to Productive Transfection. J. Controlled Release 2011, 156 (1), 76–84. 10.1016/j.jconrel.2011.07.015. [DOI] [PubMed] [Google Scholar]
- Harush-Frenkel O.; Debotton N.; Benita S.; Altschuler Y. Targeting of Nanoparticles to the Clathrin-Mediated Endocytic Pathway. Biochem. Biophys. Res. Commun. 2007, 353 (1), 26–32. 10.1016/j.bbrc.2006.11.135. [DOI] [PubMed] [Google Scholar]
- Damke H.; Baba T.; Van Der Bliek A. M.; Schmid S. L. Clathrin-Independent Pinocytosis Is Induced in Cells Overexpressing a Temperature-Sensitive Mutant of Dynamin. J. Cell Biol. 1995, 131 (1), 69–80. 10.1083/jcb.131.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman Z. U.; Zuhorn I. S.; Hoekstra D. How Cationic Lipids Transfer Nucleic Acids into Cells and across Cellular Membranes: Recent Advances. J. Controlled Release 2013, 166 (February), 46–56. 10.1016/j.jconrel.2012.12.014. [DOI] [PubMed] [Google Scholar]
- Zuhorn I. S.; Kalicharan R.; Hoekstra D. Lipoplex-Mediated Transfection of Mammalian Cells Occurs through the Cholesterol-Dependent Clathrin-Mediated Pathway of Endocytosis. J. Biol. Chem. 2002, 277 (20), 18021–18028. 10.1074/jbc.M111257200. [DOI] [PubMed] [Google Scholar]
- Georgieva J. V.; Kalicharan D.; Couraud P.-O. O.; Romero I. A.; Weksler B.; Hoekstra D.; Zuhorn I. S. Surface Characteristics of Nanoparticles Determine Their Intracellular Fate in and Processing by Human Blood–Brain Barrier Endothelial Cells In Vitro. Mol. Ther. 2011, 19 (2), 318–325. 10.1038/mt.2010.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedeli C.; Segat D.; Tavano R.; Bubacco L.; De Franceschi G.; De Laureto P. P.; Lubian E.; Selvestrel F.; Mancin F.; Papini E. The Functional Dissection of the Plasma Corona of SiO2-NPs Spots Histidine Rich Glycoprotein as a Major Player Able to Hamper Nanoparticle Capture by Macrophages. Nanoscale 2015, 7 (42), 17710–17728. 10.1039/C5NR05290D. [DOI] [PubMed] [Google Scholar]
- Lazarovits J.; Sindhwani S.; Tavares A. J.; Zhang Y.; Song F.; Audet J.; Krieger J. R.; Syed A. M.; Stordy B.; Chan W. C. W. Supervised Learning and Mass Spectrometry Predicts the in Vivo Fate of Nanomaterials. ACS Nano 2019, 13 (7), 8023–8034. 10.1021/acsnano.9b02774. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.