Abstract
Viperin is a member of the radical S-adenosylmethionine superfamily and has been shown to restrict the replication of a wide range of RNA and DNA viruses. We recently demonstrated that human viperin (HsVip) catalyzes the conversion of CTP to 3′-deoxy-3′,4′-didehydro-CTP (ddhCTP or ddh-synthase), which acts as a chain terminator for virally encoded RNA-dependent RNA polymerases from several flaviviruses. Viperin homologues also exist in non-chordate eukaryotes (e.g., Cnidaria and Mollusca), numerous fungi, and members of the archaeal and eubacterial domains. Recently, it was reported that non-chordate and non-eukaryotic viperin-like homologues are also ddh-synthases and generate a diverse range of ddhNTPs, including the newly discovered ddhUTP and ddhGTP. Herein, we expand on the catalytic mechanism of mammalian, fungal, bacterial, and archaeal viperin-like enzymes with a combination of X-ray crystallography and enzymology. We demonstrate that, like mammalian viperins, these recently discovered viperin-like enzymes operate through the same mechanism and can be classified as ddh-synthases. Furthermore, we define the unique chemical and physical determinants supporting ddh-synthase activity and nucleotide selectivity, including the crystallographic characterization of a fungal viperin-like enzyme that utilizes UTP as a substrate and a cnidaria viperin-like enzyme that utilizes CTP as a substrate. Together, these results support the evolutionary conservation of the ddh-synthase activity and its broad phylogenetic role in innate antiviral immunity.
Graphical Abstract
Mammalian viperins [virus-inhibitory protein, endoplasmic reticulum-associated, interferon (IFN)-inducible] are IFN-inducible innate immune proteins,1,3,4 which inhibit and/or are involved in the replication of a remarkable range of viruses, including chikungunya virus,5 Bunyamwera virus,6 Dengue virus,7,8 tick-borne encephalitis virus,9 influenza A virus,10 West Nile virus,7,11 human cytomegalovirus,12 hepatitis C virus,13 Sindbis virus,14 Japanese encephalitis virus,14 HIV-1,15 and other DNA and RNA viruses.4,16 In addition, viperin has been implicated in the restriction of the bacterial pathogens Shigella f lexneri and Listeria monocytogenes in cells through a currently unknown mechanism.17 The viperin-associated restriction mechanisms employed against different viruses have been proposed to involve interactions between viperin and a wide range of viral and host proteins involved in diverse cellular functions, including metabolism,18,19 signaling, 20 iron–sulfur cluster formation,9 lipid raft perturbation,10 targeted protein degradation,21 and isoprenoid biosynthesis. 10,18,22 Two recent reviews have discussed these topics in depth.16,23 Independent of its antiviral functions, viperin has also been demonstrated to play a physiological role in regulating thermogenesis and lipogenesis.24,25 The breadth of these functions is not immediately suggestive of a common or shared mechanism for antiviral function. Furthermore, mechanistic details governing these putative interactions remain incomplete and are predominately based on indirect methods (e.g., yeast two-hybrid, colocalization, and immunoprecipitation), without direct validation by quantitative biochemical approaches, though several recent studies have begun to dissect these interactions in a quantitative manner.26,27
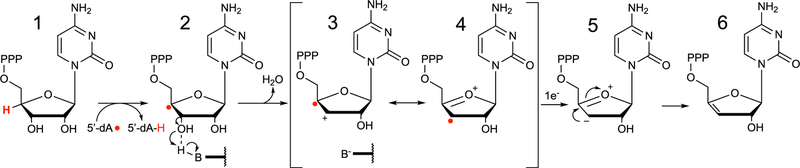
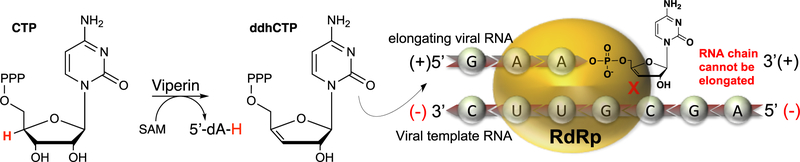
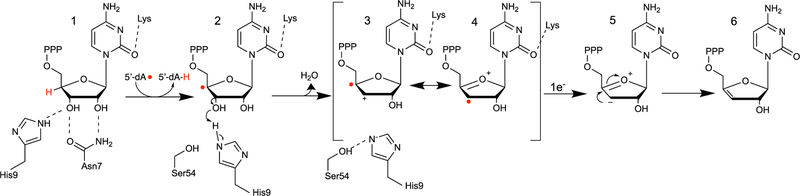
As a member of the radical S-adenosyl-l-methionine (SAM) superfamily of metalloenzymes,28 viperin employs a [4Fe-4S] (FeS) cluster cofactor that binds and supports homolytic cleavage of SAM, yielding methionine and a 5′-deoxyadenosyl radical (5′-dA•).29,30 The only member of the radical SAM (RS) superfamily that does not catalyze this reaction is the cobalamin-dependent RS methyltransferase TsrM involved in the biosynthesis of thiostrepton.31,32 In most reactions catalyzed by RS enzymes, SAM is consumed during turnover, making it a co-substrate;33,34 however, a few examples in which SAM is a true cofactor and regenerated at the end of each catalytic cycle exist.35–37 Almost all RS enzymes utilize 5′-dA• to abstract a target hydrogen atom from the substrate, initiating radical-based rearrangements that lead to product formation.38,34 We recently demonstrated that viperin uses SAM as a co-substrate and that the 5′-dA• acts directly on the substrate, cytidine triphosphate (CTP).1 In our proposed mechanism, the 5′-dA• abstracts the hydrogen atom from the 4′ position of the CTP ribose, resulting in a 4′ radical (Figure 1, intermediate 1 → 2). The presence of the radical at the 4′ position facilitates the loss of the 3′-OH group through acid–base catalysis to generate a radical–carbocation intermediate (Figure 1, intermediate 2 → 3). This mechanism has precedent in previous studies of ribonucleotide reductase (RNR) and model studies of the radiolytic cleavage of single-stranded DNA.1,39,40 A subsequent one-electron reduction of intermediate 4, presumably supplied by the FeS cluster, and tautomerization would result in the formation of 3′-deoxy-3′,4′-didehydro-CTP (ddhCTP), a previously undescribed biologically active molecule.1 We also established that ddhCTP acts as a chain terminator for the RNA-dependent RNA polymerases (RdRp) from multiple members of the Flavivirus genus and demonstrated that ddhCTP directly inhibits replication of Zika virus in vivo1 (Scheme 1). In addition, a recent preprint shows that ddhCTP can efficiently terminate the template-directed RNA synthesis catalyzed by SARS-CoV-2 RdRp.41 However, ddhCTP does not inhibit the RdRp from all viruses (e.g., picorna viruses);1 thus, we underscore the likelihood that other mechanisms for viperin-mediated viral restriction likely exist, including targeted protein degradation, and modulation of host membrane properties.5,10,12,13,20,21,26,42,43
Figure 1.
Proposed mechanism of viperin-catalyzed formation of ddhCTP. Hydrogen atom abstraction at the 4′ position of CTP by 5′-dA radical leads to general acid-assisted loss of the 3′-OH group. Loss of the 3′-OH group produces a carbocation/radical intermediate reminiscent of an intermediate formed during ribonucleotide reduction by RNR.2 Intermediate 4 is reduced by one electron to yield the ddhCTP product.
Scheme 1. Viral Restriction Mechanism of Mammalian Viperina.
aConversion of CTP to ddhCTP by mammalian viperins. ddhCTP then acts as a small molecule chain terminator against the RNA-dependent RNA-polymerases (RdRp) from multiple flaviviruses, inhibiting the replication of the viral genomes.1
Viperin is highly conserved, with homologues found throughout the phylum Chordata, which includes mammals, fish, birds, reptiles, and amphibians. Viperin-like genes, which we define as homologues that occur in species not containing an obvious interferon-inducible innate immune system and/or endoplasmic reticulum, are also present in several species of non-chordate eukaryotes, such as the sea anemone Nematostella vectensis and the beneficial plant fungus Trichoderma virens. The viperin-like enzymes from these two organisms share 73% and 55% sequence identity, respectively, with human viperin and 53% identity between each other. Similarly, several bacteria harbor viperin-like genes, including Lacinutrix marinif lava (38% identity with human viperin) and Shewanella baltica (29% identity), as well as multiple archaea, such as Methanofollis liminatans (35% identity). A recent report suggested that eukaryotic viperin likely originated from a clade of bacterial and archaeal proteins that protect against phage infection.44 That report and others also demonstrated that viperin-like enzymes, in addition to ddhCTP, are able to produce ddhUTP44,45 and ddhGTP.44 The mechanisms responsible for the observed range of substrate preferences are not understood.
Herein, we provide enzymatic and crystallographic evidence that expands upon previous studies to describe the mechanisms underlying these substrate preferences. We demonstrate that viperin-like enzymes from fungi, cnidaria, bacteria, and archaea all utilize the 3′-deoxy-3′,4′-didehydronucleoside triphosphate synthase (ddh-synthase) mechanism described for mammalian viperin. We further delineate the residues within the catalytic site that are required for viperin catalysis. Interestingly, we also find that mammalian viperin can catalyze the conversion of 2′-deoxy-CTP to its cognate ddh product, demonstrating that the 2′-OH is not essential for catalysis and functions as an element to facilitate proper substrate positioning. Taken together, these data support the view that distant viperin-like enzymes are capable of performing the ddh-synthase reaction as an evolutionarily conserved component of host defense against viral infection.
RESULTS
Cloning, Expression, and Purification of Viperin-like Enzymes.
Viperin-like genes were cloned from the fungus T. virens, the lower eukaryote N. vectensis (Cnidaria), the bacteria L. marinif lava and S. baltica, and the archaeon M. liminatans by creating synthetic DNA constructs that were codon-optimized for expression in Escherichia coli. The viperin from Mus musculus was cloned by creating synthetic DNA constructs from the wild-type sequence (see the Supporting Information for details and gene sequences). The constructs were cloned into a pSGC-His vector, which allows for production of proteins with an N-terminal hexahistidine tag. The resulting constructs were termed TvVip, NvVip, LmVip, SbVip, MlVip, and MmVip and were expressed and purified as previously described for Rattus norvegicus viperin (RnVip).1 Variants of MmVip, HsVip, and RnVip were generated by synthetic gene constructs. All variants were produced as described for the wild-type constructs.1 All of these expression constructs yielded highly soluble and stable proteins (see Figures S1 and S2).
Substrate Specificity of Nucleoside Triphosphates.
We previously reported that the mammalian viperins from human and rat convert CTP to ddhCTP.1 It has also been previously reported that mouse viperin can act on both CTP and UTP when provided in separate reactions.46 A recent study by Bernheim et al. provided the first evidence that prokaryotic viperins can catalyze ddh formation, from CTP, UTP, or GTP, through analysis of cell extracts from bacteria expressing an array of viperin and viperin-like enzymes, as well as in vitro end-point assays to confirm ddhNTP production.44 Furthermore, Ebrahimi et al. demonstrated that the viperin-like enzyme from Thielavia terrestris can act on CTP, UTP, and 5-bromo-UTP to convert them to their ddhNTP products.45
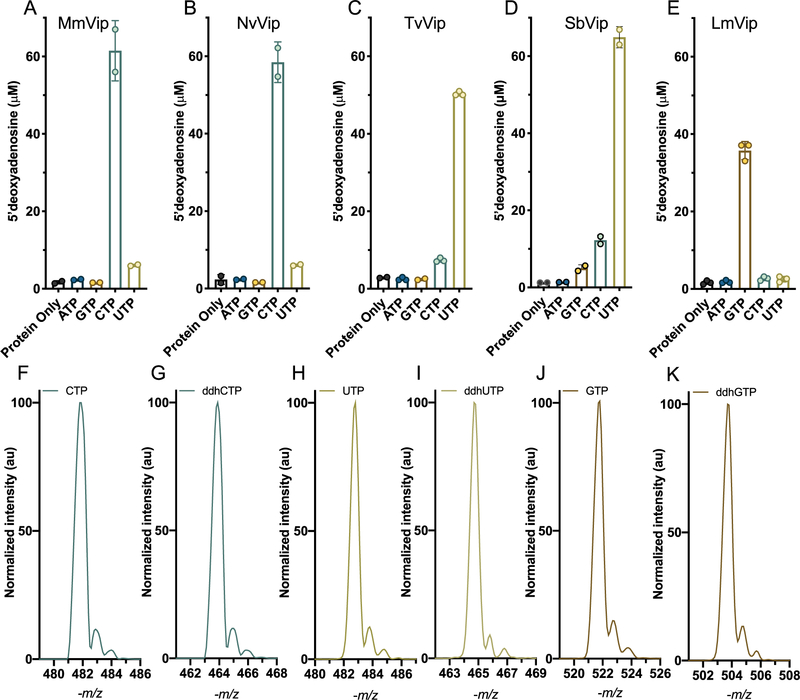
To begin evaluating the ddh-synthase reaction mechanism from different viperin-like enzymes, we first determined the catalytic rates of ddhNTP formation by fungal, bacterial, and archaeal viperin-like enzymes. To that end, we examined the ability of MmVip, NvVip, TvVip, LmVip, and SbVip to utilize ATP, GTP, CTP, and UTP as potential substrates, looking for enhanced 5′-dA formation as an indicator of substrate utilization and/or preference. In a typical assay, 50 μM enzyme was mixed with 2 mM SAM, 1 mM substrate (ATP, GTP, CTP, or UTP), and 5 mM dithionite (exogenous reductant), and the reaction allowed to proceed for 5 min. As shown in panels A and B of Figure 2, in the presence of CTP, MmVip generated ~60 μM 5′-dA, with a rate constant (Vmax/Etotal) of 0.23 ± 0.02 min−1, comparable to that reported by Fenwick et al.46 In addition, the formation of ddhCTP by NvVip, a eukaryotic viperin-like enzyme, was similar to that of MmVip (0.25 ± 0.03 min−1). On the basis of the production of 5′-dA by NvVip and MmVip, the rate of utilization for other NTPs as substrates was 10–100-fold lower. In contrast, the fungal TvVip and the bacterial SbVip produced 5′-dA only in the presence of UTP, though with similar rate constants (Vmax/Etotal values of 0.20 ± 0.01 and 0.24 ± 0.01 min−1, respectively), demonstrating that these viperin-like enzymes preferentially utilize UTP as a substrate (Figure 2C,D). Interestingly, when a similar reaction was performed with LmVip, only GTP directed production of 5′-dA (Figure 2E, Vmax/Etotal of 0.14 ± 0.01 min−1). These kinetic constants are similar to those previously published for RnVip (turnover rate of 0.185 ± 0.007 min−1).1 It was recently reported that the fungal viperin-like enzyme from T. terrestris catalyzed formation of ddhUTP at a rate of 0.0015 μmol min−1 (mg of protein)−1 (or Vmax/Etotal of 0.055 ± 0.008 min−1).45
Figure 2.
Determination of nucleotide preference and mass spectra of NTP and ddhNTP products. Each viperin homologue at 50 μM (MmVip, A; NvVip, B; TvVip, C; SbVip, D; LmVip, E) was separately mixed with 1 mM NTP, 5 mM dithionite, and 2 mM SAM and reacted for 5 min at room temperature. 5′-dA production is plotted for each nucleotide. ESI– mass spectra of (F) CTP and (G) the corresponding product of NvVip, and MmVip, ddhCTP. ESI– mass spectra of (H) UTP and (I) the corresponding product of TvVip and SbVip, ddhUTP. ESI– mass spectra of (J) GTP and (K) the corresponding product of LmVip, ddhGTP. N = 2 for MmVip, NvVip, and SbVip, and N = 3 for TvVip and LmVip.
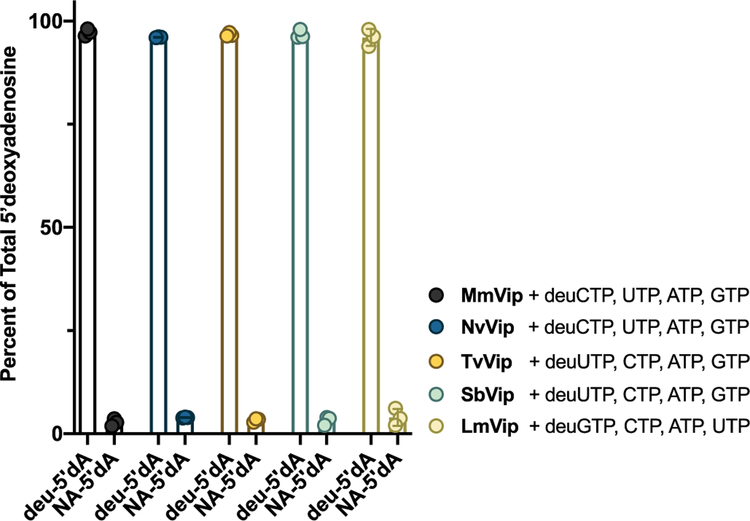
Taken together, these data suggest that viperin and viperin-like enzymes possess distinct substrate preferences, though it is possible that the increased level of 5′-dA production is due to nucleotide-dependent enhancement of background abortive cleavage, in which the generation of 5′-dA is uncoupled from subsequent hydrogen atom abstraction.47 To directly validate substrate preferences and utilization, we co-incubated each protein in the presence of their presumed deuterated NTP at 1 mM [5,6-2H2-2′,3′,4′,5′,5″−2H5-CTP (deuCTP), 5,6-2H2-2′,3′,4′,5′,5″−2H5-UTP (deuUTP), or 3′,4′,5′−2H3-GTP (deuGTP)] and each of the remaining NTPs at 1 mM (e.g., deuCTP with GTP, ATP, and UTP). In the proposed mechanism (Figure 1), viperin utilizes 5′-dA• to abstract a hydrogen atom at the 4′ position of the ribose. In previous work, when RnVip was provided with CTP site-specifically labeled with deuterium at the 4′ position, 5′-dA produced from the reaction was >99% enriched with a single deuterium atom, indicating that (1) the target hydrogen of 5′-dA• is the 4′ position of the CTP ribose and (2) 5′-dA production is tightly coupled to product formation (i.e., for every molecule of 5′-dA produced, one molecule of ddhCTP is produced).1 Product coupling by RnVip was further validated with assays in which ddhCTP production and 5′-dA production were directly monitored. Due to the difficulty of producing all four ribonucleotides labeled at the 4′ position, we opted for a commercially available deuNTP labeled at the positions identified above, with the expectation that all viperin-like enzymes will abstract the 4′-ribose hydrogen atom as demonstrated in our earlier report.1 The appearance of 5′-dA with a positive mass-to-charge ratio (+m/z) of 253.1 (natural abundance 5′-dA has a +m/z of 252.1), in the presence of competing unlabeled nucleotides, would demonstrate that each protein selectively abstracts deuterium from their presumed substrates, and on the basis of previous data, formation of products would be tightly coupled. As illustrated in Figure 3, the ratio of deuterated 5′-dA (deu5′-dA) to 5′-dA for the reaction catalyzed by MmVip and NvVip in the presence of deuCTP in competition assays is >99%, consistent with high selectivity for CTP as the substrate. TvVip, SbVip, and LmVip all produced deu5′-dA in >95% excess relative to natural abundance 5′-dA, indicating that these enzymes are selective for UTP, UTP, and GTP, respectively. Furthermore, LC-MS analysis of the NTP-derived products produced by the enzymes mentioned above displays an −m/z that is 18 Da less than that of their respective substrate and exhibits greater retention times on a C-18 reverse phase column, consistent with the formation of 3′-deoxy-3′,4′-didehydro-NTPs (Figure 2F–K).
Figure 3.
Nucleotide selectivity. The indicated enzyme (50 μM) was mixed with 2 mM SAM and 1 mM deuterium-labeled CTP (deuCTP) for MmVip or NvVip, deuterium-labeled UTP (deuUTP) for TvVip and SbVip, or deuterium-labeled GTP (deuGTP) for LmVip, in addition to 1 mM of each of three remaining unlabeled nucleotides. The resulting 5′-dA is determined, and the ratio of heavy to light 5′-dA is plotted. The transfer of a deuteron to 5′-dA (deu5′-dA) will occur if the enzyme is selective for the deuterium-containing substrate. All enzymes are selective for their respective substrates and do not abortively produce 5′-dA under these conditions. N = 3.
Interestingly, when the viperin-like enzyme from M. liminatans was incubated with SAM, dithionite, and any of the individual NTPs, all reactions showed an increase in the level of 5′-dA production. When the same reaction was performed with each individual deuNTP, only deuCTP resulted in enhanced deu5′-dA formation, albeit only ~65% of the total 5′-dA contained deuterium, suggesting a sizable fraction of uncoupled cleavage of SAM that does not result in product formation (Figure S3A). When these reactions were monitored for ddhNTP products by LC-MS, only deuCTP was converted to the respective ddhNTP product by MlVip (Figure S3B,C). On the basis of these data, it is likely that MlVip prefers CTP as a substrate, though we cannot exclude the interesting possibility that the in vivo substrate is actually a modified form of CTP or another related nucleotide-like molecule.
To confirm that the products produced by TvVip and NvVip are indeed ddhUTP and ddhCTP, respectively, anion exchange chromatography was used to purify the nucleotide products from a 3 mL reaction mixture of NvVip with SAM and 13C9,15N3-CTP, or TvVip with SAM and 13C9,15N2-UTP (Figure S4; see the Supporting Information for details). Comparison of the 1H–13C two-dimensional (2D) heteronuclear single-quantum coherence (HSQC) chemical shift data of CTP with those of the product from the NvVip reaction clearly demonstrates the loss of the C4′-hydrogen correlation peak at 85.4 and 4.1 ppm (Figure S5, top and bottom left). Comparison of the HSQC spectra of the NvVip product to that of authentic ddhCTP produced by RnVip reveals proton–carbon single-bond correlations that are essentially identical (Tables S7 and S8, chemical shifts). Similarly, when the 1H–13C 2D HSQC spectra of the TvVip reaction product are compared with those of UTP, the absence of the C4′-hydrogen correlation peak at 86.1 and 4.2 ppm is readily apparent (Figure S5, top and bottom right). The 1H–13C 2D HSQC spectra of ddhCTP and ddhUTP are similar to the notable exception of the 1H–13C correlation peak arising from the C5 position of the base, which is shifted upfield in the ddhUTP spectra relative to ddhCTP (Figure S5, bottom left and bottom right). The nuclear magnetic resonance spectra, along with the MS data presented above, are consistent with the products of NvVip and TvVip being ddhCTP and ddhUTP, respectively. Co-injection on a reverse phase C-18 HPLC column of the heavy labeled ddhCTP produced by MmVip or NvVip and an authentic ddhCTP standard produced by RnVip revealed identical retention times (Figure S6).
Potential Non-nucleoside Triphosphate Substrates.
It was previously reported that viperin-like enzymes from the fungal species T. terrestris and T. virens utilize UDP-glucose and isopentenyl pyrophosphate (IPP) as substrates, catalyzing covalent coupling with 5′-dA to yield UDP-glucose-5′-dA and IPP-5′-dA, respectively.22,48 Although mammalian viperins do not use UDP-glucose or IPP as substrates,1 we carefully examined whether these nonmammalian viperin-like enzymes were promiscuous in terms of the substrates they recognized and the types of reactions they catalyzed. We performed competition assays with TvVip, MmVip, and NvVip, wherein 1 mM nucleotide substrate (UTP for TvVip and CTP for MmVip and NvVip) was mixed with either 10 mM UDP-glucose or 10 mM IPP. If UDP-glucose or IPP were capable of effectively competing for the catalytic site of these enzymes, then the amount of 5′-dA produced would be predicted to decrease due to formation of the coupled products UDP-glucose and 5′-dA or IPP and 5′-dA. As illustrated in Figure S7, the formation of 5′-dA is unaffected by the presence of UDP-glucose or IPP, compared to UTP or CTP alone. This behavior indicates that these viperin-like enzymes are highly selective for their respective nucleotide substrates (i.e., ddhNTP synthase activity) and, like the mammalian viperins, are unlikely to catalyze the coupling of 5′-dA to UDP-glucose or IPP.1 In addition, on the time scale of our assays, no uncharacterized peaks were observed in the HPLC traces of reaction products, which is also consistent with the inability of these viperin-like enzymes to catalyze 5′-dA-associated coupling reactions.
In Vivo Selectivity for Nucleoside Triphosphates.
While it has been reported that MmVip can produce ddhUTP as well as ddhCTP,46 our ability to prepare the 13C9,15N2-ddhUTP standard from TvVip reactions allowed for the evaluation of whether mammalian viperin expressed in human cells exhibits selectively for CTP relative to UTP in vivo. We therefore expressed the full length HsVip in HEK293 cells, extracted the metabolites in the presence of both 13C9,5N3-ddhCTP and 13C9,15N2-ddhUTP as internal standards, and quantified the amount of natural abundance ddhCTP and ddhUTP, using our previously described methods.1 Under these conditions, HsVip produces 194 μM ddhCTP and only 0.2 μM ddhUTP, a 970-fold selectivity for CTP in vivo (Figure S8).
Structural Characterization of Nucleoside Triphosphate Substrate Recognition in NvVip and TvVip.
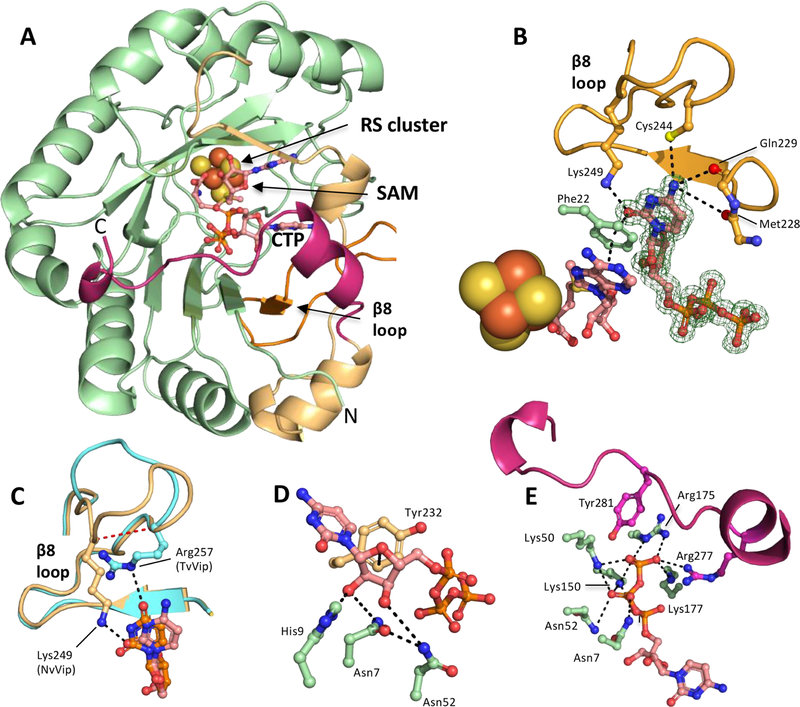
Fenwick and co-workers recently reported structures of MmVip bound to only SAM or SAH [Protein Data Bank (PDB) entry 5VSL or 5VSM, respectively]49 and more fully liganded structures containing SAH as well as CTP or UTP (PDB entry 6Q2P or 6Q2Q, respectively).46 As anticipated, in all cases, MmVip adopts a partial (β/α)6-TIM barrel commonly found in RS enzymes, with the RS [4Fe-4S] cluster located at the base of the catalytic site.46,49 Interestingly, this work demonstrated that MmVip’s catalytic site can accommodate both CTP and UTP. The major difference between these structures was the disposition of Lys319 (mouse numbering), which adopts a well ordered pose in the CTP-bound structure, as a consequence of a hydrogen bond with the C2 exocyclic oxygen of the cytidine base, but which is disordered in the UTP-bound structure. Similar to RnVip, and consistent with our data, MmVip strongly prefers CTP as a substrate.46 To determine how selectivity arises in different viperin-like enzymes and MmVip, we determined the crystal structures of NvVip and TvVip (Figure 4A and Figure S9A). To generate crystals of NvVip, the purified protein was treated with TEV protease to remove the N-terminal hexahistidine tag, followed by sparse matrix crystallization trials using sitting drop vapor diffusion in the presence of SAM and CTP under anaerobic conditions (see the Supporting Information for details). These screening efforts yielded crystals exhibiting diffraction consistent with orthorhombic space group P212121 that extended to a resolution of 1.42 Å. The structure was determined by molecular replacement using the MmVip structure (PDB entry 5VSL, ligands removed) as the search model. The final model of NvVip has one molecule in the asymmetric unit (Figure 4A), and omit maps show strong density for CTP in the catalytic site (Figure 4B). NvVip and MmVip share 60% sequence identity and exhibit highly similar overall structures, as superposition of the liganded NvVip and unliganded MmVip structures results in a root-mean-square deviation (RMSD) of 0.44 Å over 214 Cα atoms (Figure S9B), while comparison with the liganded MmVip (6Q2P; CTP, SAM) structure results in an RMSD of 0.25 Å over 246 Cα atom.
Figure 4.
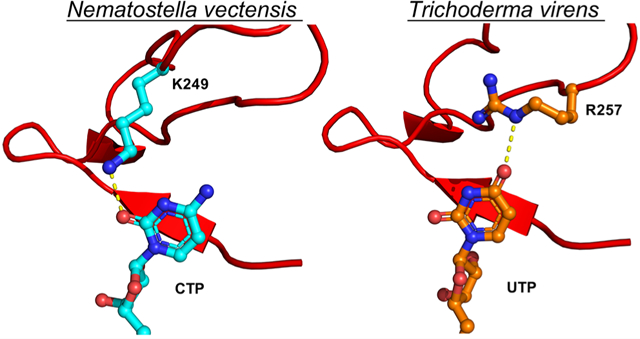
Structural basis for catalysis of the CTP analogue ddhCTP. (A) The asymmetric unit of NvVip contains one protein chain, a 4Fe-4S cluster, CTP, and SAM. Radical SAM domain (green), C-terminal domain (gold), the β−8 loop (orange), and the C-terminal tail (maroon). (B) SAM, Phe22, and CTP participate in a π-stacking network within the catalytic site. Lys249 coordinates the C2 exocyclic oxygen of the cytidine base, which is analogous to Lys318 in HsVip. The C4 exocyclic amine interacts with the side chain of Cys244 and the carbonyl backbones of Met228 and Gln229. The Fo – Fc omit map is contoured at 2σ (green mesh). (C) Overlay of the β−8 loop from NvVip (orange) and TvVip (teal). Lys249 in NvVip coordinates the CTP base, and Arg257 from TvVip coordinates the UTP base. The red line denotes the five-residue shift between NvVip and TvVip in the coordination of their respective nucleobase, CTP or UTP. (D) The 2′- and 3′-OH of the ribose of CTP hydrogen bond with His9, Asn7, and Asn52. An additional van der Waals interaction is provided by Tyr232. (E) Hydrogen-bonding network responsible for recognizing the triphosphate moiety of NTP by NvVip. The C-terminal tail of viperin caps the active site and provides the final γ-phosphate with residues Tyr281 and Arg 277.
The pyrimidine base of CTP is anchored to the NvVip catalytic site through a π-stacking interaction network involving the cytosine ring, Phe22 (supplied by the RS domain), and the adenine base of SAM (Figure 4B). In the NvVip and MmVip structures, the exocyclic amine of the pyrimidine base of CTP is recognized via contacts involving carbonyl backbone functionalities (Met228 and Gln229 for NvVip and Met298 and Lys299 for MmVip) and the side chain of Cys244 (NvVip) located in the β−8 loop (Figure 4B). Additionally, Lys249 in the NvVip β−8 loop forms an additional specific side-chain interaction with the C2-keto functionality of CTP, analogous to Lys319 in MmVip. A multiple-sequence alignment shows that among eukaryotes, lysine is only partially conserved at this position. We propose that members of the viperin family harboring this β−8 loop lysine can effectively utilize CTP as a substrate and that a lack of lysine at this position might suggest different specificity. For example, TvVip does not have a lysine residue in its β−8 loop but instead uses an arginine to coordinate the uridine base [discussed in more detail below (Figure 4C and Figure S10)]. To determine the functional significance of this lysine residue, we generated a lysine to alanine variant in RnVip (K317A, residue number equivalent to NvVip Lys249, MmVip Lys319, and HsVip Lys318). In a reaction mixture containing CTP, SAM and dithionite, the level of product formation is reduced by 4.3-fold compared to that of the wild type (Figure S11A). In addition, when HEK293T cells were transfected with human viperin constructs harboring this lysine to alanine variant (K318A, human numbering), a nearly 20-fold difference in the level of ddhCTP formation is observed when compared to cells expressing wild-type human viperin [p < 0.001 (Figure S11B)]. This reduction in activity is likely due to impaired CTP binding.
In NvVip, the ribose of CTP is coordinated by three invariant residues supplied by the RS domain: Asn7, His9, and Asn52 (Figure 4D). The Asn7 (MmVip Asn77) side chain oxygen atom forms hydrogen bonds with the adjacent Asn52 (MmVip Asn122) and 3′-OH of the ribose, while the Asn 7 side chain nitrogen atom forms a hydrogen bond with an oxygen atom from the β-phosphate of CTP. On the opposite side of the ribose, His9 (MmVip His79 and RnVip His77) forms a hydrogen bond with the 2′-OH of the ribose (Figure 4D). The observed orientation of Asn7 (as well as Asn77 in MmVip) in the crystal structure does not appear to be compatible with participation in acid–base catalysis; we are also not aware of literature precedent for Asn side-chain functionalities acting as general acids. Thus, His9 is an attractive candidate for supplying the proton during catalysis.1 To determine if this histidine residue plays a role in catalysis, we relied on the mammalian viperins from rat and mouse, RnVip and MmVip, respectively, for kinetic assays as the human version is recalcitrant to expression in bacterial cells. We generated the RnVip variant H77A and the MmVip variant H79A, which both exhibit an ~6-fold reduction in the level of ddhCTP formation relative to that of the wild-type protein (Figures S11A and S12A). Notably, this mutation is associated with stronger effects in vivo. When we transfected HEK293T cells with the analogous variant of human viperin (HsVip H78A), the level of ddhCTP production was reduced by ~40-fold (p < 0.001) compared to that of cells expressing wild-type human viperin (Figure S11B), suggesting that this catalytic site histidine (NvVip His9 and HsVip His78) plays a role in ddh-synthase chemistry. Ser54 (NvVip) is also close to the nucleotide substrate in the catalytic site (although it is not conserved), so we tested the MmVip S124A variant, which yielded an ~1.7-fold reduction in the level of ddhCTP formation. Notably, the MmVip variant N77A (NvVip Asn7) did not produce detectable amounts of ddhCTP after 5 or 60 min (Figure S12), suggesting the unusual possibility that Asn could be acting as an acid in this reaction, though it would require the need to invoke a minor conformational reorganization to a state not observed in the crystal structure. Alternatively, the lack of activity may also arise from a decreased or nonproductive level of binding of the substrate, as the side chain of Asn7 (MmVip Asn77) may aid in orienting the substrate and provide a hydrogen bond to the 3′-OH of the ribose, which could serve to accommodate the charge redistribution associated with dehydration. The considerable catalytic activity exhibited by MmVip H79A (NvVip His7) might be accounted for if Ser124 (NvVip Ser54) acted as a substitute proton donor, or if Asn7 (NvVip) served to position a water molecule to act as an acid. Further experimentation is needed to determine the precise functional contributions of these residues.
To determine if the catalytic site histidine plays a role in discriminating between ribose and deoxyribose substrates (i.e., CTP vs 2′-deoxyCTP), reactions were performed with 25 μM wild-type or H79A MmVip, 1 mM SAM, 10 mM 2′-deoxyCTP, and dithionite. The appearance of a species with a molecular mass corresponding to 2′,3′-deoxy-3′,4′-didehydroCTP (m/z 448) was monitored over time. Interestingly, both MmVip and the H79A variant were able to catalyze the formation of 2′,3′-deoxy-3′,4′-didehydroCTP, with the H79A variant exhibiting an only ~2-fold reduction in relative rate compared with that of the wild-type enzyme (Figure S13A). Absolute quantification of 2′,3′-deoxy-3′,4′-didehydroCTP was not possible because (1) we do not have a synthetic standard for quantitation and (2) the starting material and product separate poorly under our separation conditions (Figure S13B). Due to the low intracellular concentration of 2′-deoxyCTP in cells (29 μM),50 the 2′,3′-deoxy-3′,4′-didehydroCTP product is unlikely to be produced at significant levels under physiological conditions. On the basis of the crystal structures, this His residue clearly provides binding contacts to the ribose; however, substitution of this residue did not have a large impact on catalysis as would be expected for a catalytic acid.
The endo-face of the ribose ring of CTP also makes face-to-face van der Waals contacts with the side chain of Tyr232 contributed by the C-terminal domain (Figure 4D). The presence of an aromatic amino acid at this position is invariant among mammalian viperins (~20% have Phe in place of Tyr), which we postulate is involved in a π-donating interaction that stabilizes the radical cation intermediate formed during catalysis (Figure 1, intermediate 3). A recent report suggested that a tyrosyl radical, with features that were similar to those of other tyrosyl radicals, formed when the viperin-like enzyme from T. terrestris was provided with UTP and 5′-bromo-UTP;45 and that a double variant (S251A and Y252F) of this enzyme was not active.45 To determine whether this conserved tyrosine (analogous to Y302 in MmVip) acts as an electron donor (generating a tyrosyl radical) or participates in a π–cation interaction with intermediate 3/4 (Figure 5), we created Y302A and Y302F variants in MmVip. One would predict the Y302F variant to be inactive if a tyrosyl radical is required, whereas only reduced activity would be observed if a π–cation interaction contributes to catalysis. Indeed, the Y302F variant is capable of making ddhCTP, albeit with ~20-fold less activity than the wild type [V/ET of 0.14 vs 0.008 min−1 (Figure S12B)]. Furthermore, the Y302A variant did not produce detectable levels of ddhCTP even at extended reaction times (Figure S12B). These data suggest that this tyrosine plays a major role in catalysis. On the basis of our data, we favor the π–cation stabilization during catalysis; however, additional studies are needed to confirm the role of the conserved tyrosine in catalysis. It is important to note that these variants do not affect FeS cluster formation or the overall stability of purified RnVip or MmVip (Figures S1B and S2)
Figure 5.
Mechanistic proposal for ddhCTP formation by viperin and viperin-like RS enzymes. This mechanism is likely operative in all viperin and viperin-like reactions.
In the absence of a substrate, the C-terminal tail of MmVip is disordered (residues 336–362), which, upon the binding of CTP, forms a cap over the top of the catalytic site and provides hydrogen bonds to the polyphosphate tail of CTP.46,49 Similarly, a C-terminal tail is present in the structure of NvVip (residues 266–288), the sequence of which aligns well with the C-terminal tail of MmVip (Figure 4E), and also participates in binding of the triphosphate moiety of CTP. Asn7 and Asn52 play dual roles in CTP binding as they participate in hydrogen bonding interactions with both the β-phosphate and the ribose (Figure 4E). A total of five lysine and arginine side chains (NvVip numbering; Lys50, Lys150, Lys177, Arg175, and Arg277) participate in polar interactions with the α-, β-, and γ-phosphate groups, in addition to Asn7 and Asn52 mentioned above, with equivalent residues present in MmVip (Lys120, Lys220, Lys247, Arg245, Arg347, Asn77, and Asn122). Sequence comparisons show that a second C-terminal tyrosine residue (NvVip Tyr281, MmVip Tyr351, or HsVip Tyr 350), which forms the terminal hydrogen bond with the γ-phosphate, is present in >93% of the available sequences, suggesting a conserved mechanism for discrimination between NDPs and NTPs.
Crystals of TvVip bound to SAM and UTP exhibited diffraction consistent with trigonal space group P31, which extended to a resolution of 3.2 Å, with three molecules in the asymmetric unit (Figure S9A). TvVip shares sequence identity of 45% and 54% with NvVip and MmVip, respectively, as well as overall structural similarity; superposition of TvVip with NvVip (over 240 Cα atoms) and MmVip (PDB entry 6Q2P, 219 Cα atoms) results in RMSDs of 0.63 and 0.73 Å, respectively (Figure S9B). In the TvVip structure, UTP is bound in a pose nearly identical to that of CTP in the NvVip structure, with the ribose and phosphate sharing similar side-chain interactions (Figures S9C–F and S10). The greatest difference between the liganded NvVip and liganded TvVip structures is within the β−8 loop, TvVip 245–259 and NvVip 232–251, found near the C-terminus of all viperin and viperin-like enzymes, which contributes the residues responsible for discrimination between pyrimidine bases, CTP and UTP, respectively. These differences do not arise from crystal packing interactions, as these regions are devoid of interactions with symmetry mates in the NvVip and TvVip structures. In the TvVip structure, the β−8 loop contains a unique Arg257 (not found in eukaryotic viperins), which forms a hydrogen bond to the C4 exocyclic oxygen of UTP (Figure 4C and Figures S9C and S10). Interestingly, TvVip utilizes a distinct part of the β−8 loop relative to NvVip (or MmVip), where Arg257 occupies a site that is five amino acids closer to the beginning of the loop (i.e., Lys249 in TvVip) (Figure 4C, red dotted line) and occupies the position of Cys244 from NvVip. Unlike MmVip and NvVip, TvVip contains no additional side-chain specific H-bonding interactions with the pyrimidine base in UTP. Thus, these highly similar proteins achieve substrate specificity by exploiting distinct determinants for recognition of unique substrate functionalities. Together, these structures begin to reveal how these viperin-like enzymes discriminate between CTP and UTP.
A sequence alignment reveals that an analogue of Arg257 in the β−8 loop found in TvVip is not present in the bacterial viperin-like homologue SbVip, even though both of these proteins utilize UTP as a substrate (Figure S14A). These observations highlight the variability found in this loop region and underscore the difficulty in predicting nucleotide substrate selectivities for the viperin-like enzymes on the basis of sequence analysis alone. Because of these challenges, the β−8 loop determinants responsible for the selective recognition of GTP by LmVip and UTP by SbVip are not readily predictable.
Structural Insight into the Catalytic Mechanisms of NvVip and TvVip.
The NvVip, TvVip, and MmVip structures provide insight into the catalytic mechanisms utilized by the nonmammalian viperin-like enzymes for the transformation of NTPs to ddhNTPs. We previously described a provisional mechanism in which mammalian viperins utilize the 5′-dA• to abstract the hydrogen atom at the C4′ position of the ribose of CTP (Figure 1).1 The structures reported herein are consistent with the C4′ position of the ribose being the site of hydrogen abstraction; as in the NvVip and TvVip structures, the 5′ carbon of SAM (i.e., the site of the 5′-dA•) is positioned ~3.5 Å from the 4′-H of the NTP-ribose (Figure S9F). This distance is consistent with that found in the liganded MmVip structures and in the structures of other RS enzymes bound to both SAM and their respective substrates.37,51,52 The second step in the reaction, dehydration of the ribose ring, involves the elimination of the 3′-OH, which requires the action of a general acid to accommodate the leaving group. The closest functionality to the 3′-OH is the carboxamide side chain of an invariant Asn residue (NvVip Asn7, MmVip Asn77, and TvVip Asn24) (~2.5 Å in the substrate-bound structures), implicating this residue as an important determinant for binding and stabilizing the ribose moiety of the substrate. Asparagine residues are not typically invoked as proton donors in acid-base catalysis; however, a conserved His residue (NvVip His9, TvVip His26, or HsVip H78) and a Ser residue (not conserved) are in close proximity of the 3′-OH group (3.3 and 3.2 Å, respectively). Interestingly, the His residue forms a hydrogen bond with the 2′-OH at a distance of ~2.8 Å. While the His and Ser variants decreased activity by only ~6- and ~1.7-fold, respectively, these results do not rule out the possibility that they may still act as acids. Additionally, these results offer the possibility of an Asn acting as an acid in this reaction. However, based on the placement of this residue in crystal structures, it is unlikely that the Asn is acting as a catalytic acid. Therefore, we propose that after hydrogen atom abstraction, a modest side-chain rearrangement places the His residue proximal to the 3′-OH and, with the assistance of the Asn, facilitates its release. In our proposed mechanism for catalysis (Figure 5), a radical carbocation intermediate is generated after the 3′-OH group is eliminated as water (intermediate 3). One can envision that the delocalized π-electrons of the aromatic ring of Tyr232 (NvVip) could participate in a π-cation interaction that stabilizes this radical intermediate. Our data support this mechanism as the Y302F variant, with a decrease in π–cation donation,53 still catalyzes formation of ddhCTP, while the Y302A variant is characterized by a complete loss of catalytic activity. However, as previously suggested, these data do not completely rule out the possibility of a Y302 acting as an electron donor and generating a tyrosyl radical45 to yield the ddhCTP product. In our preferred mechanism, where Y302 acts only as a π–cation interaction partner, the source of the electron that reduces intermediate 3 or 4 in the viperin-catalyzed reaction is currently unclear; however, ketyl radicals (e.g., intermediate 4 in the proposed viperin reaction) are potent oxidants with potentials in the range of 2 V, making this step thermodynamically favorable.54 We propose that, similar to other RS enzyme reactions,55–57 the electron ultimately derives from a reduced FeS cluster, suggesting that viperin requires two electrons to complete each turnover (two successive one-electron reductions): one to generate the 5′-dA• and another to reduce intermediate 3/4.
Sequence Similarity Network of Viperin and Viperin-like Enzymes.
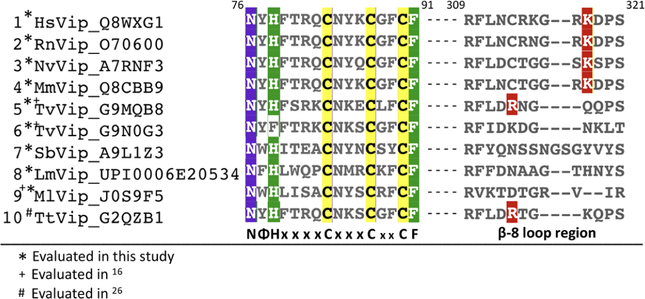
Although the substrate selectivities of the viperin-like enzymes are difficult to determine on the basis of primary sequence, their ability to catalyze the ddh-synthase reaction can be predicted with considerable confidence due to the presence of the highly conserved sequence motif NΦHX4CX3CX2CF (Figure 6). In this motif, Φ represents either Trp, Tyr, or Phe, X represents any amino acid, N and H are proposed to function as hydrogen bond acceptor/proton donors or determinants of substrate pose (e.g., Asn7 and His9 in NvVip), and F engages in a π-stacking network involving SAM and the nucleotide base. The hallmark cysteines from the RS cluster motif, CX3CX2C, ligate the three iron ions of the [4Fe-4S] cluster, positioning the open iron site proximal to the sulfonium of SAM. Together, these residues allow viperin and viperin-like enzymes to catalyze the ddh-synthase reaction, and perhaps related reactions. The NΦHX4CX3CX2CF sequence motif is invariant in all of the viperin and viperin-like enzymes for which we demonstrated ddh-synthase activity (Figure 6) and is conserved in all three domains of life. Thus, the NΦHX4CX3CX2CF motif serves as a powerful signature for identifying candidate viperin family members within the RS superfamily of enzymes.
Figure 6.
Sequence determinants of ddh-synthase activity. Key residues within the highly conserved NΦHX4CX3CX2CF motif responsible for ddh-synthase chemistry are highlighted. In this motif, NΦHX4CX3CX2CF, Φ represents either Trp, Tyr, or Phe, X represents any amino acid, N and H are the required hydrogen bond acceptor/proton donor (e.g., Asn7 and His9 in NvVip), and F participates in a π-stacking network with SAM and the CTP base. The hallmark cysteines, CX3CX2C, ligate three iron ions of a [4Fe-4S] cluster, positioning the open iron site proximal to the sulfonium of SAM. Together, these residues allow viperin and viperin-like enzymes to catalyze the 3′-deoxy-3′,4′-didehydronucleoside triphosphate synthase reaction. The β−8 loop is a variable region in viperin and viperin-like enzymes that participates in sequestration and selectivity of a nucleotide triphosphate. A conserved lysine residue (highlighted in red) (HsVip K318 or NvVip K249) coordinates the exocyclic amine of the pyrimidine base of CTP. A lysine residue in this position is only partially conserved among eukaryotes. TvVip and TtVip do not have a lysine residue in their β−8 loop and instead use an arginine (highlighted in red) to coordinate the uridine base. Numbering (above) according to HsVip.
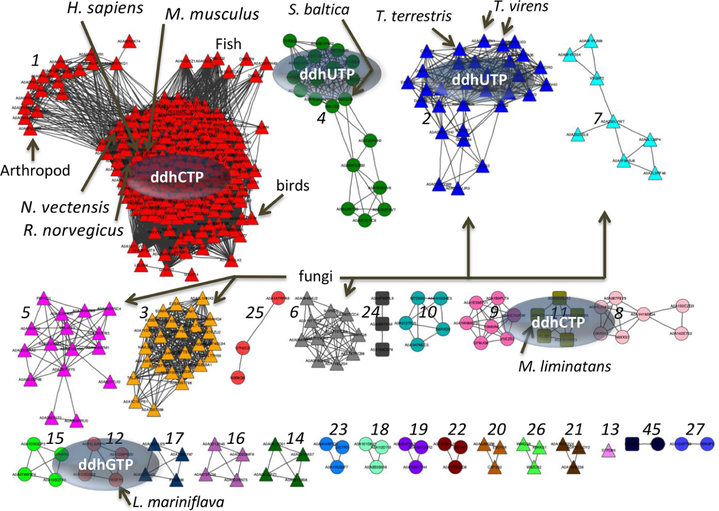
Genes that share sequence similarity to human viperin can be found throughout all three domains of life. To fully evaluate the phylogenetic distribution of these viperin-like genes in fungal, bacterial, and archaeal species, we generated a sequence similarity network (SSN; publicly available service from the Enzyme Function Initiative, https://efi.igb.illinois.edu/efiest)58 using the amino acid sequence from HsVip as the seed sequence. The resulting SSN (Figure 7) shows well-ordered clusters, with the largest cluster (cluster 1) representing multicellular eukaryotes, including mammals, birds, fish, and arthropods, as well as the cnidarian NvVip sequence. On the basis of the tight clustering, suggesting a high degree of sequence similarity, especially within the lysine-containing β−8 loop region (i.e., HsVip Lys318), and the biochemical data described herein, we infer that all proteins in cluster 1 likely catalyze the conversion of CTP to ddhCTP. In addition, we can assign functions to other clusters, including cluster 2 (fungal species) and cluster 4 (bacterial species) that catalyze the formation of ddhUTP, and cluster 11 (archaeal), which produces ddhCTP, and cluster 12 (bacterial), which produces ddhGTP (Figure 7).
Figure 7.
Sequence similarity network generated by using human viperin. Human viperin, Uniprot entry Q8WXG1, was used as the seed sequence (e value of 5, edge alignment score of 120) to generate a representative SSN. Eukaryotes (triangle nodes), bacteria (circle nodes), and archaea (square nodes) cluster largely with respect to evolutionary lineage, and each distinct cluster is represented by a unique color. Viperin-like enzymes characterized in this paper are labeled, and their corresponding ddhNTP is shown in the oval.
It is interesting to note that T. virens has two proteins (Uniprot entries G9N0G3 and G9MQB8) that are similar to the mammalian viperins (Figure 6 and Figure S14B). Above, we demonstrated that TvVip (G9MQB8) is indeed a ddh-synthase (viperin-like); however, as shown in Figure 6 and Figure S14B, the second viperin-like sequence possesses a Phe instead of a His in the NΦHX4CX3CX2CF motif [i.e., NΦFX4CX3CX2CF (see the Supporting Information)]. When this protein was purified and assayed, 5′-dA was formed at comparable levels in the presence and absence of NTPs, and no ddhNTP products were observed. Furthermore, in the presence of deuNTPs, no deuterium was transferred to 5′-dA. It is likely that dehydration of the ribose is not observed due to nonproductive poses in the catalytic site or steric occlusion of substrate binding. To date, the activity and function of this protein remain unknown. The His → Phe viperin-like enzyme is present in only a small number of fungi (mainly within the Trichoderma genus); however, within organisms containing a homologue with the His → Phe substitution, there is always a viperin-like enzyme with the canonical viperin motif (NΦHX4CX3CX2CF), suggesting that additional enzyme-mediated transformations that utilize a viperin-like enzyme scaffold may exist.
DISCUSSION
We previously reported that RnVip catalyzes the transformation of CTP to ddhCTP through a radical-based mechanism.1 Viperin is a highly conserved gene, with homologues present in all chordates, in non-chordate eukaryotes, and in a modest number of bacteria and archaea.44 Studies have reported that mammalian viperin can catalyze the formation of both ddhCTP and ddhUTP,46 whereas viperin-like enzymes can catalyze the formation of ddhCTP, ddhUTP, or ddhGTP.44,45 Bernheim et al. made the interesting observation that while ddhNTPs were capable of restricting phage replication, they had no effect on the viability of the bacteria used in their experiments, demonstrating that these nucleotide analogues are selective for phage-replication machinery.44 Others have reported that viperin-like enzymes can utilize non-nucleotide substrates such as UDP-glucose59 and isopentyl pyrophosphate.22 Herein, we sought to provide X-ray crystallographic and enzymatic evidence to support and expand upon these previous studies. We demonstrate that viperin-like enzymes from the sea anemone N. vectensis and the archaeon M. liminatans are indeed capable of performing the same reaction as the human enzyme (i.e., ddhCTP production). We find that viperin-like enzymes from the plant fungus T. virens as well as from the soil bacterium S. baltica prefer UTP as a substrate, and as previously reported, the viperin-like enzyme from L. marinif lava, another soil bacterium, operates on GTP44 while the methane-producing archaeon M. liminatans preferentially operates on CTP, though it was activated (increased uncoupled production of 5′-dA) by all NTPs. The activity of MlVip suggests that viperin-like enzymes may be capable of promiscuous ddh-synthase activity and that other substrates, in addition to the canonical NTPs, may exist. However, we find that this substrate promiscuity does not extend to the viperin-like enzymes from T. virens or N. vectensis. Although they were previously proposed to utilize UDP-glucose and isopentyl pyrophosphate as substrates in condensation reactions, we provide evidence that, as in the case with mammalian viperins,1 these putative substrates are unable to outcompete nucleotides for catalysis by these viperin-like enzymes.
Human viperin exhibits an ~1000-fold preference for CTP over UTP as the substrate in vivo. While CTP and UTP concentrations are generally similar in mammalian cells (e.g., in HEK-293 cells CTP and UTP are present at concentrations of ~0.8–1.0 and ~1.0–1.5 mM, respectively),1 CTP is likely to be significantly preferred, with little or no UTP utilization; thus, the apparent selectivity in vivo arises from the endogenous properties of viperin and is not the consequence of substrate levels. It is important to note that the high level of substrate selectivity observed in human viperin in vivo may not be a universal feature of bacterial, archaeal, and fungal viperin-like enzymes. Future work will be required to define the in vivo selectivities of viperin-like enzymes within the cellular environments of their host organisms. Nevertheless, on the basis of our studies herein, we provide evidence that the viperin-like enzymes from N. vectensis, T. virens, S. baltica, and L. marinif lava possess strong preference for their respective NTP substrates in vitro, even in the presence of high concentrations of competing nucleotides.
Direct structural analyses defined the determinants responsible for the selective binding and turnover of CTP and UTP within the catalytic sites of the nonmammalian viperin-like enzymes from NvVip and TvVip, respectively. These structures identified an invariant asparagine that plays a central role in coordinating the ribose moiety of NTPs, and in particular, the 3′-OH group that is eliminated as water. Furthermore, these structures and complementary enzymatic studies suggest that the proton needed for the liberation of the 3′-OH group may derive from the histidine found in the highly conserved motif, NΦHX4CX3CX2CF, where Φ represents Trp, Tyr, or Phe and X represents any amino acid. However, we cannot ignore the unusual possibility of the asparagine acting as the proton source responsible for 3′-OH liberation. This proposal is consistent with our mutagenesis studies demonstrating that His-Ala and Asn-Ala substitutions significantly reduced ddhNTP synthase activity in mammalian viperin. Because viperin catalyzes a reaction highly analogous to RNR, important analogies can be drawn from previous studies of this enzyme. For instance, a conserved asparagine (Asn437) is required for ribonucleotide reduction and serves as a stabilizing factor in the catalytic site that orients the substrate and catalytic residues, Cys439 and Glu441.60 Mutation of this residue abolished activity.60 The asparagine in viperin’s catalytic site is oriented in a similar fashion, which makes proton donation unlikely. The asparagine (NvVip Asn7) is oriented with the nitrogen hydrogen bonding to the 2′-OH and the β-phosphate of the CTP substrate, while the carbonyl is hydrogen bonding to the 3′-OH of CTP. We believe this organization leaves only the histidine (NvVip His9) to act as an acid in this reaction (the adjacent serine is not conserved in viperin or viperin-like enzymes). Enzymatic studies have also confirmed that the conserved tyrosine is important for catalysis, as the Y302A variant of MmVip had no detectable activity. However, the MmVip variant Y302F could still catalyze ddhCTP formation, albeit at reduced rates. This tyrosine (TvVip Y245, MmVip Y302, or NvVip Y232) has previously been proposed to act as the source of the second electron required to reduce intermediate 3 or 4, via formation of a tyrosyl radical.45 However, this may not be an on-pathway process, as there does not appear to be an electron transport chain for shuttling electron equivalents, which is required to complete the catalytic cycle. On the basis of our activity data, we propose that this tyrosine acts primarily as a π-donor to stabilize the carbocation intermediate (Figure 5, intermediate 3/4). In particular, the activity of the Tyr → Phe mutation strongly argues against the proposed role of the tyrosyl radical; however, further work is needed to fully delineate the role of this residue and to identify the catalytic acid that protonates the leaving group hydroxyl. Enzymatic studies of 2′-deoxycytidine 5′-triphosphate also demonstrate that the 2′-OH group is not necessary for formation of the 3′,4′-deoxydidehydro bond, though because quantification of the 2′,3′-deoxy-3′,4′-didehydroCTP product was not possible; therefore, we could not determine rate constants.
The NΦHX4CX3CX2CF motif is present in all viperin and viperin-like enzymes that have been demonstrated to catalyze ddhNTP formation and provides a diagnostic signature for predicting distant homologues likely to carry out similar chemical transformations (e.g., abstraction of 4′-hydrogen, followed by dehydration and loss of the 3′-OH). While the presence of this NΦHX4CX3CX2CF consensus sequence is predictive of the chemistry responsible for converting nucleoside triphosphate ddh-containing analogues, the considerable variation in the β−8 loop responsible for nucleobase selectivity precludes the prediction of substrate specificity for the distantly related viperin-related molecules in unicellular eukaryotes, bacteria, and archaea. This variability leaves open the possibility that proteins containing the related NΦFX4CX3CX2CF motif may operate on a wider range of substrates, as we propose for the His → Phe variant in T. virens.
A recent report suggested that eukaryotic viperin originated from a clade of bacterial and archaeal proteins and demonstrated that several of these viperin-like enzymes from non-eukaryotic species were protective against phage infection, consistent with the roles of viperin in our own innate immune system as a restriction factor against bacteria and viruses.44 These viperin-like enzymes function, at least in part, as antiviral/antiphage restriction factors by selectively targeting pathogen-encoded nucleic acid polymerases via their ddhNTP products.1 This behavior is analogous to that described in our previous report demonstrating that mammalian viperin-generated ddhCTP effectively restricts flaviviral replication by acting as an RNA-dependent RNA-polymerase chain terminator, with no discernible effect on the host polymerases.1 These proposals are fully consistent with the recent report that viperin expression in mammalian cells selectively inhibits bacteriophage T7 RNA polymerase, while not affecting the host polymerases.61 On the basis of the conservation of this mechanism, we predict that all organisms with viperin-related sequences are insensitive to the action of their associated ddhNTP products (i.e., the host nucleic acid polymerases are not sensitive to ddhNTPs, or other protective mechanisms exist). Notably, not all mammalian viruses that elicit an IFN-dependent stimulation of viperin expression are inhibited by the action of ddhCTP on their replicative polymerases (e.g., picornaviruses), and alternative viperin-associated protective mechanisms have been proposed, including targeted ubiquitination-driven proteasomal degradation of innate immune molecules,21 disruption of plasma membrane properties (i.e., cholesterol production) required for efficient viral budding,10 and the direct modulation of innate immune machinery.13,20,26 Notably, cell lysis, not budding, is the exit mechanism used almost exclusively by bacterial viruses.62,63 Furthermore, most components of the mammalian innate immune response that have been reported to interact with mammalian viperin (e.g., TRAF6 and IRAK126) are not present in eubacteria or archaea. These observations are consistent with the hypothesis that within the archaeal and eubacterial domains, the viperin-like enzymes function as antiviral/antiphage restriction factors by generating ddhNTPs to target the pathogen nucleic acid polymerases.
CONCLUSION
Future efforts will need to examine the selective pressures that guided the evolution of viperin-like enzymes with distinct substrate preferences, such as the inherent (and likely orthogonal) sensitivities of the nucleic acid polymerases encoded by the hosts and their associated pathogens. For example, it will be important to determine the toxicity of ddhUTP upon its application to mammalian cells and whether higher-order eukaryotes developed the ability to selectively utilize CTP versus UTP to avoid toxicities associated with the uridine analogue. Furthermore, the selectivities exhibited by the fungal and bacterial viperin-like enzymes might point to potential antifungal and antibacterial leads based on ddhNTPs, for which the pathogen-encoded polymerases are sensitive. Finally, viperin and the associated ddhCTP are important components of the mammalian antiviral innate immune system; the studies presented herein support the idea that this role in antiviral host defense extends to all three domains of life, similar to the broad protective functions associated with other systems such as the Toll-like receptor proteins.64
Supplementary Material
ACKNOWLEDGMENTS
The authors acknowledge the Albert Einstein Anaerobic Structural and Functional Genomics Resource (http://www.nysgxrc.org/psi3/anaerobic.html) and the Einstein Macromolecular Therapeutics Development Facility. This research used beamline FMX of the National Synchrotron Light Source II, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Brookhaven National Laboratory under Contract No. DE-SC0012704. The Center for BioMolecular Structure (CBMS) is primarily supported by the National Institutes of Health, National Institute of General Medical Sciences (NIGMS) through a Center Core P30 Grant (P30GM133893), and by the DOE Office of Biological and Environmental Research (KP1605010).
Funding
This work was supported by National Institutes of Health Grants R21-AI133329 (T.L.G. and S.C.A.), P01-GM118303-01 (J. A. Gerlt and S.C.A.), U54-GM093342 (J. A. Gerlt and S.C.A.), U54-GM094662 (S.C.A.), T32 GM007491 (J.C.L.), and the Price Family Foundation (S.C.A.).
Footnotes
Notes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.biochem.0c00958.
Detailed procedures for cloning, expression, and purification of viperin proteins, assays, cell culture assays, and crystallization and structure determination (PDF)
Accession Codes
TvVip, Uniprot entry G9MQB8; NvVip, Uniprot entry A7RNF3; SbVip, Uniprot entry A9L1Z3; MlVip, Uniprot entry J0S9F5; LmVip, UniParc entry UPI0006E20534; MmVip, Uniprot entry Q8CBB9; RnVip, Uniprot entry O70600; HsVip, Uniprot entry Q8WXG1. The coordinates of NvVip and TvVip with substrates have been deposited in the Protein Data Bank as entries 7N7H and 7N7I, respectively.
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.biochem.0c00958
Contributor Information
Jake C. Lachowicz, Department of Biochemistry, Albert Einstein College of Medicine, Bronx, New York 10461, United States
Anthony S. Gizzi, Department of Biochemistry, Albert Einstein College of Medicine, Bronx, New York 10461, United States.
Steven C. Almo, Department of Biochemistry and Department of Biophysics, Albert Einstein College of Medicine, Bronx, New York 10461, United States
Tyler L. Grove, Department of Biochemistry, Albert Einstein College of Medicine, Bronx, New York 10461, United States.
REFERENCES
- (1).Gizzi AS, Grove TL, Arnold JJ, Jose J, Jangra RK, Garforth SJ, Du Q, Cahill SM, Dulyaninova NG, Love JD, Chandran K, Bresnick AR, Cameron CE, and Almo SC (2018) Publisher Correction: A naturally occurring antiviral ribonucleotide encoded by the human genome. Nature 562 (7725), E3. [DOI] [PubMed] [Google Scholar]
- (2).Eklund H, Uhlin U, Farnegardh M, Logan DT, and Nordlund P (2001) Structure and function of the radical enzyme ribonucleotide reductase. Prog. Biophys. Mol. Biol 77 (3), 177–268. [DOI] [PubMed] [Google Scholar]
- (3).Chin KC, and Cresswell P (2001) Viperin (cig5), an IFN-inducible antiviral protein directly induced by human cytomegalovirus. Proc. Natl. Acad. Sci. U. S. A 98 (26), 15125–15130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Seo JY, Yaneva R, and Cresswell P (2011) Viperin: A Multifunctional, Interferon-Inducible Protein that Regulates Virus Replication. Cell Host Microbe 10 (6), 534–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Teng TS, Foo SS, Simamarta D, Lum FM, Teo TH, Lulla A, Yeo NKW, Koh EGL, Chow A, Leo YS, Merits A, Chin KC, and Ng LFP (2012) Viperin restricts chikungunya virus replication and pathology. J. Clin. Invest 122 (12), 4447–4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Carlton-Smith C, and Elliott RM (2012) Viperin, MTAP44, and protein kinase R contribute to the interferon-induced inhibition of Bunyamwera Orthobunyavirus replication. J. Virol 86 (21), 11548–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Jiang D, Weidner JM, Qing M, Pan XB, Guo H, Xu C, Zhang X, Birk A, Chang J, Shi PY, Block TM, and Guo JT (2010) Identification of five interferon-induced cellular proteins that inhibit west nile virus and dengue virus infections. J. Virol 84 (16), 8332–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Helbig KJ, Carr JM, Calvert JK, Wati S, Clarke JN, Eyre NS, Narayana SK, Fiches GN, McCartney EM, and Beard MR (2013) Viperin Is Induced following Dengue Virus Type-2 (DENV-2) Infection and Has Anti-viral Actions Requiring the C-terminal End of Viperin. PLoS Neglected Trop. Dis 7 (4), No. e2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Upadhyay AS, Vonderstein K, Pichlmair A, Stehling O, Bennett KL, Dobler G, Guo JT, Superti-Furga G, Lill R, Overby AK, and Weber F (2014) Viperin is an iron-sulfur protein that inhibits genome synthesis of tick-borne encephalitis virus via radical SAM domain activity. Cell. Microbiol 16 (6), 834–848. [DOI] [PubMed] [Google Scholar]
- (10).Wang XY, Hinson ER, and Cresswell P (2007) The interferon-inducible protein viperin inhibits influenza virus release by perturbing lipid rafts. Cell Host Microbe 2 (2), 96–105. [DOI] [PubMed] [Google Scholar]
- (11).Szretter KJ, Brien JD, Thackray LB, Virgin HW, Cresswell P, and Diamond MS (2011) The Interferon-Inducible Gene viperin Restricts West Nile Virus Pathogenesis. Journal of Virology 85 (22), 11557–11566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Seo JY, Yaneva R, Hinson ER, and Cresswell P (2011) Human Cytomegalovirus Directly Induces the Antiviral Protein Viperin to Enhance Infectivity. Science 332 (6033), 1093–1097. [DOI] [PubMed] [Google Scholar]
- (13).Wang SS, Wu XF, Pan TT, Song WH, Wang YH, Zhang F, and Yuan ZH (2012) Viperin inhibits hepatitis C virus replication by interfering with binding of NS5A to host protein hVAP-33. J. Gen. Virol 93, 83–92. [DOI] [PubMed] [Google Scholar]
- (14).Chan YL, Chang TH, Liao CL, and Lin YL (2008) The Cellular Antiviral Protein Viperin Is Attenuated by Proteasome-Mediated Protein Degradation in Japanese Encephalitis Virus-Infected Cells. J. Virol 82 (21), 10455–10464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Rivieccio MA, Suh HS, Zhao YM, Zhao ML, Chin KC, Lee SC, and Brosnan CF (2006) TLR3 ligation activates an antiviral response in human fetal astrocytes: A role for viperin/cig5. J. Immunol 177 (7), 4735–4741. [DOI] [PubMed] [Google Scholar]
- (16).Rivera-Serrano EE, Gizzi AS, Arnold JJ, Grove TL, Almo SC, and Cameron CE (2020) Viperin Reveals Its True Function. Annu. Rev. Virol 7 (1), 421–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Helbig KJ, Teh MY, Crosse KM, Monson EA, Smith M, Tran EN, Standish AJ, Morona R, and Beard MR (2019) The interferon stimulated gene viperin, restricts Shigella. f lexneri in vitro. Sci. Rep 9 (1), 15598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Makins C, Ghosh S, Roman-Melendez GD, Malec PA, Kennedy RT, and Marsh ENG (2016) Does Viperin Function as a Radical S-Adenosyl-L-methionine-dependent Enzyme in Regulating Farnesylpyrophosphate Synthase Expression and Activity? J. Biol. Chem 291 (52), 26806–26815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Dumbrepatil AB, Zegalia KA, Sajja K, Kennedy RT, and Marsh ENG (2020) Targeting viperin to the mitochondrion inhibits the thiolase activity of the trifunctional enzyme complex. J. Biol. Chem 295 (9), 2839–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Hee JS, and Cresswell P (2017) Viperin interaction with mitochondrial antiviral signaling protein (MAVS) limits viperin-mediated inhibition of the interferon response in macrophages. PLoS One 12 (2), No. e0172236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Panayiotou C, Lindqvist R, Kurhade C, Vonderstein K, Pasto J, Edlund K, Upadhyay AS, and Overby AK (2018) Viperin Restricts Zika Virus and Tick-Borne Encephalitis Virus Replication by Targeting NS3 for Proteasomal Degradation. J. Virol 92 (7), e02054–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Chakravarti A, Selvadurai K, Shahoei R, Lee H, Fatma S, Tajkhorshid E, and Huang RH (2018) Reconstitution and substrate specificity for isopentenyl pyrophosphate of the antiviral radical SAM enzyme viperin. J. Biol. Chem 293 (36), 14122–14133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Ghosh S, and Marsh ENG (2020) Viperin: an ancient radical-SAM enzyme finds its place in modern cellular metabolism and innate immunity. J. Biol. Chem 295, 11513–11528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Eom J, Kim JJ, Yoon SG, Jeong H, Son S, Lee JB, Yoo J, Seo HJ, Cho Y, Kim KS, Choi KM, Kim IY, Lee HY, Nam KT, Cresswell P, Seong JK, and Seo JY (2019) Intrinsic expression of viperin regulates thermogenesis in adipose tissues. Proc. Natl. Acad. Sci. U. S. A 116 (35), 17419–17428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Qi Z, Xia J, Xue X, Liu J, Liu W, and Ding S (2017) Targeting viperin improves diet-induced glucose intolerance but not adipose tissue inflammation. Oncotarget 8 (60), 101418–101436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Dumbrepatil AB, Ghosh S, Zegalia KA, Malec PA, Hoff JD, Kennedy RT, and Marsh ENG (2019) Viperin interacts with the kinase IRAK1 and the E3 ubiquitin ligase TRAF6, coupling innate immune signaling to antiviral ribonucleotide synthesis. J. Biol. Chem 294 (17), 6888–6898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Grunkemeyer TJ, Ghosh S, Patel AM, Sajja K, Windak J, Basrur V, Kim Y, Nesvizhskii AI, Kennedy RT, and Marsh ENG (2021) Viperin inhibits cholesterol biosynthesis and interacts with enzymes in the cholesterol biosynthetic pathway. bioRxiv, DOI: 10.1101/2021.02.19.431989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Duschene KS, and Broderick JB (2010) The antiviral protein viperin is a radical SAM enzyme. FEBS Lett. 584 (6), 1263–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Broderick WE, and Broderick JB (2019) Radical SAM enzymes: surprises along the path to understanding mechanism. JBIC, J. Biol. Inorg. Chem 24 (6), 769–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Yang H, Impano S, Shepard EM, James CD, Broderick WE, Broderick JB, and Hoffman BM (2019) Photoinduced Electron Transfer in a Radical SAM Enzyme Generates an S-Adenosylmethionine Derived Methyl Radical. J. Am. Chem. Soc 141 (40), 16117–16124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Blaszczyk AJ, Knox HL, and Booker SJ (2019) Understanding the role of electron donors in the reaction catalyzed by Tsrm, a cobalamin-dependent radical S-adenosylmethionine methylase. JBIC, J. Biol. Inorg. Chem 24 (6), 831–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Knox HL, Chen PY-T, Blaszczyk AJ, Mukherjee A, Grove TL, Schwalm EL, Wang B, Drennan CL, and Booker SJ (2021) Structural basis for non-radical catalysis by TsrM, a radical SAM methylase. Nat. Chem. Biol 17, 485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Booker SJ (2012) Radical SAM enzymes and radical enzymology. Biochim. Biophys. Acta, Proteins Proteomics 1824 (11), 1151–3. [DOI] [PubMed] [Google Scholar]
- (34).Holliday GL, Akiva E, Meng EC, Brown SD, Calhoun S, Pieper U, Sali A, Booker SJ, and Babbitt PC (2018) Atlas of the Radical SAM Superfamily: Divergent Evolution of Function Using a “Plug and Play” Domain. Methods Enzymol. 606, 1–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Li L (2012) Mechanistic studies of the radical SAM enzyme spore photoproduct lyase (SPL). Biochim. Biophys. Acta, Proteins Proteomics 1824 (11), 1264–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Frey PA, Chang CH, Ballinger MD, and Reed GH (2002) Kinetic characterization of transient free radical intermediates in reaction of lysine 2,3-aminomutase by EPR lineshape analysis. Methods Enzymol. 354, 426–35. [DOI] [PubMed] [Google Scholar]
- (37).Dowling DP, Bruender NA, Young AP, McCarty RM, Bandarian V, and Drennan CL (2014) Radical SAM enzyme QueE defines a new minimal core fold and metal-dependent mechanism. Nat. Chem. Biol 10 (2), 106–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Broderick JB, Duffus BR, Duschene KS, and Shepard EM (2014) Radical S-Adenosylmethionine Enzymes. Chem. Rev 114 (8), 4229–4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Minnihan EC, Nocera DG, and Stubbe J (2013) Reversible, Long-Range Radical Transfer in E-coli Class la Ribonucleotide Reductase. Acc. Chem. Res 46 (11), 2524–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Giese B, Beyrich-Graf X, Erdmann P, Petretta M, and Schwitter U (1995) The chemistry of single-stranded 4′-DNA radicals: influence of the radical precursor on anaerobic and aerobic strand cleavage. Chem. Biol 2 (6), 367–75. [DOI] [PubMed] [Google Scholar]
- (41).Seifert M, Bera SC, van Nies P, Kirchdoerfer RN, Shannon A, Le T-T-N, Grove TL, Papini FS, Arnold JJ, Almo SC, Canard B, Depken M, Cameron CE, and Dulin D (2020) Signatures and mechanisms of efficacious therapeutic ribonucleotides against SARS-CoV-2 revealed by analysis of its replicase using magnetic tweezers. bioRxiv, DOI: 10.1101/2020.08.06.240325. [DOI] [Google Scholar]
- (42).Yuan Y, Miao Y, Qian L, Zhang Y, Liu C, Liu J, Zuo Y, Feng Q, Guo T, Zhang L, Chen X, Jin L, Huang F, Zhang H, Zhang W, Li W, Xu G, and Zheng H (2020) Targeting UBE4A Revives Viperin Protein in Epithelium to Enhance Host Antiviral Defense. Mol. Cell 77, 734–747.e7. [DOI] [PubMed] [Google Scholar]
- (43).Ghosh S, Patel AM, Grunkemeyer TJ, Dumbrepatil AB, Zegalia K, Kennedy RT, and Marsh ENG (2020) Interactions between Viperin, Vesicle-Associated Membrane Protein A, and Hepatitis C Virus Protein NS5A Modulate Viperin Activity and NS5A Degradation. Biochemistry 59 (6), 780–789. [DOI] [PubMed] [Google Scholar]
- (44).Bernheim A, Millman A, Ofir G, Meitav G, Avraham C, Shomar H, Rosenberg MM, Tal N, Melamed S, Amitai G, and Sorek R (2021) Prokaryotic viperins produce diverse antiviral molecules. Nature 589 (7840), 120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Honarmand Ebrahimi K, Rowbotham JS, Mccullagh J, and James WS (2020) Mechanism of Diol Dehydration by a Promiscuous Radical-SAM Enzyme Homologue of the Antiviral Enzyme Viperin (RSAD2). ChemBioChem 21 (11), 1605–1612. [DOI] [PubMed] [Google Scholar]
- (46).Fenwick MK, Su D, Dong M, Lin H, and Ealick SE (2020) Structural Basis of the Substrate Selectivity of Viperin. Biochemistry 59 (5), 652–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Lotierzo M, Raux E, Tse Sum Bui B, Goasdoue N, Libot F, Florentin D, Warren MJ, and Marquet A (2006) Biotin Synthase Mechanism: Mutagenesis of the YNHNLD Conserved Motif. Biochemistry 45 (40), 12274–12281. [DOI] [PubMed] [Google Scholar]
- (48).Honarmand Ebrahimi K, Carr SB, McCullagh J, Wickens J, Rees NH, Cantley J, and Armstrong FA (2017) The radical-SAM enzyme Viperin catalyzes reductive addition of a 5′-deoxyadenosyl radical to UDP-glucose in vitro. FEBS Lett. 591 (16), 2394–2405. [DOI] [PubMed] [Google Scholar]
- (49).Fenwick MK, Li Y, Cresswell P, Modis Y, and Ealick SE (2017) Structural studies of viperin, an antiviral radical SAM enzyme. Proc. Natl. Acad. Sci. U. S. A 114 (26), 6806–6811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Traut TW (1994) Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem 140 (1), 1–22. [DOI] [PubMed] [Google Scholar]
- (51).Liu WQ, Amara P, Mouesca JM, Ji X, Renoux O, Martin L, Zhang C, Zhang Q, and Nicolet Y (2018) 1,2-Diol Dehydration by the Radical SAM Enzyme AprD4: A Matter of Proton Circulation and Substrate Flexibility. J. Am. Chem. Soc 140 (4), 1365–1371. [DOI] [PubMed] [Google Scholar]
- (52).Vey JL, and Drennan CL (2011) Structural insights into radical generation by the radical SAM superfamily. Chem. Rev 111 (4), 2487–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Gallivan JP, and Dougherty DA (1999) Cation-pi interactions in structural biology. Proc. Natl. Acad. Sci. U. S. A 96 (17), 9459–9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Schwarz HA, and Dodson RW (1989) Reduction Potentials of Co2- and the Alcohol Radicals. J. Phys. Chem 93 (1), 409–414. [Google Scholar]
- (55).Silakov A, Grove TL, Radle MI, Bauerle MR, Green MT, Rosenzweig AC, Boal AK, and Booker SJ (2014) Characterization of a Cross-Linked Protein Nucleic Acid Substrate Radical in the Reaction Catalyzed by RlmN. J. Am. Chem. Soc 136 (23), 8221–8228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Grove TL, Radle MI, Krebs C, and Booker SJ (2011) Cfr and RlmN Contain a Single [4Fe-4S] Cluster, which Directs Two Distinct Reactivities for S-Adenosylmethionine: Methyl Transfer by S(N)2 Displacement and Radical Generation. J. Am. Chem. Soc 133 (49), 19586–19589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Grove TL, Livada J, Schwalm EL, Green MT, Booker SJ, and Silakov A (2013) A substrate radical intermediate in catalysis by the antibiotic resistance protein Cfr. Nat. Chem. Biol 9 (7), 422–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Gerlt JA, Bouvier JT, Davidson DB, Imker HJ, Sadkhin B, Slater DR, and Whalen KL (2015) Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST): A web tool for generating protein sequence similarity networks. Biochim. Biophys. Acta, Proteins Proteomics 1854 (8), 1019–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Honarmand Ebrahimi K, Carr SB, Mccullagh J, Wickens J, Rees NH, Cantley J, and Armstrong FA (2017) The radical-SAM enzyme Viperin catalyzes reductive addition of a 5′-deoxyadenosyl radical to UDP-glucose in vitro. FEBS Lett. 591 (16), 2394–2405. [DOI] [PubMed] [Google Scholar]
- (60).Kasrayan A, Persson AL, Sahlin M, and Sjöberg B-M (2002) The Conserved Active Site Asparagine in Class I Ribonucleotide Reductase Is Essential for Catalysis. J. Biol. Chem 277 (8), 5749–5755. [DOI] [PubMed] [Google Scholar]
- (61).Dukhovny A, Shlomai A, and Sklan EH (2018) The antiviral protein Viperin suppresses T7 promoter dependent RNA synthesis-possible implications for its antiviral activity. Sci. Rep 8, 8100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Rakonjac J, Bennett NJ, Spagnuolo J, Gagic D, and Russel M (2011) Filamentous bacteriophage: biology, phage display and nanotechnology applications. Curr. Issues Mol. Biol 13 (2), 51–76. [PubMed] [Google Scholar]
- (63).Buchmann JP, and Holmes EC (2015) Cell Walls and the Convergent Evolution of the Viral Envelope. Microbiol. Mol. Biol. Rev 79 (4), 403–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Doron S, Melamed S, Ofir G, Leavitt A, Lopatina A, Keren M, Amitai G, and Sorek R (2018) Systematic discovery of antiphage defense systems in the microbial pangenome. Science 359 (6379), No. eaar4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.