Abstract
Pneumococcal conjugate vaccines will eventually be licensed after favorable results from phase III efficacy trials. After licensure of a conjugate vaccine for invasive pneumococcal disease in infants, new conjugate vaccines will likely be licensed primarily on the basis of immunogenicity data rather than clinical efficacy. Analytical methods must therefore be developed, evaluated, and validated to compare immunogenicity results accurately within and between laboratories for different vaccines. At present no analytical technique is uniformly accepted and used in vaccine evaluation studies to determine the acceptable level of agreement between a laboratory result and the assigned value for a given serum sample. This multicenter study describes the magnitude of agreement among 12 laboratories quantifying an identical series of 48 pneumococcal serum specimens from 24 individuals (quality-control sera) by a consensus immunoglobulin G (IgG) enzyme-linked immunosorbent assay (ELISA) developed for this study. After provisional or trial antibody concentrations were assigned to the quality-control serum samples for this study, four methods for comparison of a series of laboratory-determined values with the assigned concentrations were evaluated. The percent error between assigned values and laboratory-determined concentrations proved to be the most informative of the four methods. We present guidelines that a laboratory may follow to analyze a series of quality-control sera to determine if it can reproduce the assigned antibody concentrations within an acceptable level of tolerance. While this study focused on a pneumococcal IgG ELISA, the methods that we describe are easily generalizable to other immunological assays.
Pneumococcal conjugate vaccines will eventually be licensed after favorable results from phase III efficacy trials (S. Black, H. Shinefield, P. Ray, E. Lewis, B. Fireman, The Kaiser Permanente Vaccine Study Group, R. Austrian, G. Siber, J. Hackell, R. Kohberger, and I. Chang, Abstr. 38th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1398, p. 379, 1999). After licensure of a conjugate vaccine for invasive pneumococcal disease in infants, however, new conjugate vaccines will likely be licensed primarily on the basis of immunogenicity data (2, 13) rather than clinical efficacy. Serum antibody concentrations measured by an immunoglobulin G (IgG) enzyme-linked immunosorbent assay (ELISA) and functional antibody activity measured in a subset of serum samples by an opsonophagocytic assay will likely be used to evaluate and compare the immunogenicities of these vaccines. Analytical methods must be developed, evaluated, and validated in order to accurately compare immunogenicity results within and between laboratories for different vaccines. At present no analytical technique is uniformly accepted and used in vaccine evaluation studies to determine the acceptable level of agreement between a laboratory result and the assigned value for a given serum sample. One possible approach was presented by Concepcion and Frasch (2), who compared cross-standardized values for pneumococcal polysaccharide reference serum with those concentrations previously assigned by calculating the 20% ranges bracketing the cross-standardized and previously assigned concentrations and observing whether these ranges intersected.
A number of multicenter studies have been conducted in an effort to standardize ELISAs and the quantitation of serum antibody levels from a series of shared distributed specimens (1, 4, 7). Basic statistical techniques (e.g., means, standard deviations, and coefficients of variation) with bar and line graphs were used in those investigations to compare antibody levels within and among participating laboratories. While those trials provided insight into the variability of calculated antibody levels within and among laboratories, they did not focus on the development of methods which could be used to judge if the laboratory-determined values were sufficiently close to a set of assigned antibody concentrations.
This multicenter study describes the magnitude of agreement among 12 laboratories quantifying an identical series of 48 pneumococcal serum specimens from 24 individuals (quality-control sera) by a consensus IgG ELISA developed for this study. Each of these laboratories is highly experienced at performing ELISAs for bacterial pathogens, including Streptococcus pneumoniae, and quantifying antibody concentrations. In the absence of known antibody concentrations for the serum specimens analyzed in this study, provisional or trial values were assigned to facilitate the analysis of interlaboratory variability. After assigning trial antibody concentrations to the quality-control sera, a data set was created that was useful for designing a protocol to judge whether each laboratory was able to estimate these values within an acceptable degree of variability. The present investigation evaluates Concepcion and Frasch's intersecting range technique (2) along with three other methods for comparison of a series of laboratory-determined values with assigned antibody concentrations. This paper concludes with a series of guidelines that describe a protocol that a laboratory may follow to analyze a series of quality-control sera to determine if it can reproduce the assigned antibody concentrations within an acceptable level of tolerance. While this study focused on a pneumococcal IgG ELISA, the methods that we describe are easily generalizable to other immunological assays.
MATERIALS AND METHODS
Study design.
Total IgG anticapsular antibody concentrations were measured in 24 paired serum samples from adults before and after vaccination with a licensed 23-valent pneumococcal polysaccharide vaccine (Pneumovax II; Pasteur Mérieux, Lyon, France) for nine pneumococcal serotypes (1, 4, 5, 6B, 9V, 14, 18C, 19F, and 23F) by a consensus ELISA. The ELISA protocol was developed by consensus drafted during the World Health Organization-sponsored Pneumococcal ELISA Workshop, held 15 and 16 May 1996 at the Centers for Disease Control and Prevention (CDC) in Atlanta, Ga. Coded serum samples were prepared and sent to 12 participating laboratories by the National Institute for Biological Standards and Control in Hertfordshire, United Kingdom. Table 1 lists the name of each participating laboratory, in alphabetical order, and the number of assays for each serum sample submitted for final analysis. Unless otherwise specified, all serum samples were assayed manually. CDC, in addition, assayed serum samples robotically, using a Zymark System Robot (Zymark Corporation, Hopkington, Mass.). Whether assayed manually or robotically, the same consensus ELISA protocol was used.
TABLE 1.
Laboratories participating in the multicenter pneumococcal ELISA study and number of antibody determinations for each serum sample submitted for analysis
| Laboratory | No. of assays |
|---|---|
| CDC, Atlanta, Ga. (manual) | 1 |
| CDC, Atlanta, Ga. (robot) | 3 |
| Food and Drug Administration, Center for Biologics Evaluation and Research, Division of Bacterial Products, Bethesda, Md. | 1 |
| Institute of Child Health, Division of Cell and Molecular Biology, London, England | 1 |
| Merck Research Laboratories, Developmental Human Vaccine Serology, West Point, Pa. | 3 |
| National Public Health Institute, Helsinki, Finland | 3 |
| National University Hospital, Department of Immunology, Reykjavik, Iceland | 1 |
| Pasteur Mérieux Connaught, Clinical Sero-Immunology Laboratory, Val de Reuil, France | 1 |
| Pasteur Mérieux Connaught, Clinical Serology Laboratory, Swiftwater, Pa. | 1 |
| Statens Serum Institut, Division of Microbiology, Copenhagen, Denmark | 4 |
| University of Minnesota, Department of Pediatrics, Minneapolis | 3 |
| University of Rochester, School of Medicine, Rochester, N.Y. | 1 |
| Wyeth-Lederle Vaccines and Pediatrics, West Henrietta, N.Y. | 3 |
ELISA.
The ELISA used to quantitate IgG anti-capsular antibody concentrations was adapted from the methods described by Koskela (6) and Quataert et al. (12). This consensus ELISA was developed for the present study as an analytical model and is not presented as a standard ELISA to be used for all pneumococcal antibody concentration determinations. The antipneumococcal standard reference serum (89-SF) was provided by one of the authors (C.E.F.). Water of the highest purity available in each laboratory was used to prepare all reagents. Block titrations of each polysaccharide in 0.01 M sodium phosphate-buffered saline (PBS; pH 7.1 to 7.2) were done by each laboratory to determine optimal polysaccharide coating concentrations. In addition, each laboratory did block titrations of 89-SF and the enzyme conjugate. It was recommended that all serum samples be diluted to a final starting dilution of 1:50. All dilutions of 89-SF, serum samples, quality-control sera, cell-wall polysaccharides (C-Ps), and enzyme conjugate were made in 0.01 M PBS–0.05% Tween 20 with enzyme immunoassay-grade 1% bovine serum albumin. The standard reference serum (89-SF), each serum sample, and quality-control sera were neutralized for at least 30 min at room temperature with C-Ps (C-Ps; Statens Serum Institut, Copenhagen, Denmark) at a final concentration of 500 μg/ml in undiluted serum. Once neutralized, 89-SF and each serum sample were serially diluted. The standard recommended dilution scheme was six, two- or threefold serial dilutions.
The consensus ELISA consisted of coating each well of Nunc Immuno plates (Maxisorp; PGC Scientifics Corp., Gaithersburg, Md.) with 100 μl of pneumococcal polysaccharide antigen (American Type Culture Collection, Manassas, Va.) at 37°C. The coated plates were stored at 4°C and were used within 1 month. The coated plates were washed five times with 0.01 M PBS–0.1% Tween 20 (pH 7.4 to 7.6). Duplicate C-Ps neutralized antibody, serially diluted 89-SF, and serum samples (50 μl/well) were added. In-house quality-control wells as well as background wells (wells with all reagents except test serum) were included in all assays. Specimen dilutions were incubated for 2 h at room temperature. The plates were washed as described above, and 50 μl of the proper dilution of horseradish peroxidase-labeled enzyme conjugate (HP6043; Hybridoma Reagent Laboratory, Baltimore, Md.) was added to each well. The conjugate was incubated for at least 2 h at room temperature. Next, depending upon the manufacturer's instructions, 3,3′,5,5′-tetramethylbenzidine substrate was added to each well, the plate was incubated at room temperature, the enzyme reaction was stopped, and the optical density of each well was read at 450 nm. The average optical density of the blank wells was subtracted from the optical densities of all wells on a plate. Optical density data were analyzed either by the CDC data analysis ELISA program (9) or by each laboratory, using its comparable in-house data analysis program. The type- or group-specific total IgG antibody concentrations in each serum sample and the quality-control samples were determined relative to that in the standard reference serum, 89-SF. The standard reference serum had previously been assigned total type- or group-specific IgG antibody concentrations by one of the authors (12).
Serum antibody concentrations were calculated by using software selected by the laboratory. Several investigators have shown that calculated concentrations may vary, depending on the type of analysis used for antibody quantitation (5, 8, 11). Standard curves formed by using a four-parameter logistic-log model have been shown to deliver highly accurate and reproducible results (11). Consequently, laboratories were offered software distributed by CDC (9), which implements this model to estimate antibody concentrations. The consensus ELISA protocol stipulated that laboratories were free to use their own software if they had previously demonstrated that their results would be comparable to those obtained with CDC software. Laboratory-determined antibody concentrations were submitted to CDC for further analysis.
Data analysis.
Descriptive statistical methods were used to measure intra- and interlaboratory variability. Interassay coefficients of variation (CVs; calculated as [standard deviation/mean] × 100) were calculated from results submitted by laboratories that performed multiple assays. Median antibody concentrations were calculated for each serum sample from the 13 data sets for each serotype, and these were used as trial-assigned values for these sera.
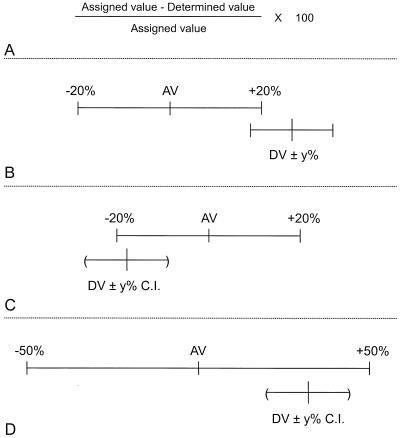
Twelve serum samples were selected to evaluate four methods used to determine the magnitude of agreement between antibody concentrations submitted by the participating laboratories and trial values assigned to the sera. These 12 serum samples were chosen on the basis of the low degree of variability of their laboratory-determined antibody concentrations across the 12 laboratories that evaluated them, as measured from their interassay CVs. They were also selected to span the full range of antibody concentrations measured in the study. The four methods of comparison are diagramed, schematically, in Fig. 1.
FIG. 1.
Schematic diagrams of four methods used to determine the magnitude of agreement between laboratory-determined values (DV) and values assigned to the sera (AV). (A) Percent error; (B) intersecting ranges; (C) intersecting range and confidence interval (C.I.); (D) overlapping range and confidence interval. See text for explanation.
Percent error.
Percent error measures the degree of error between a laboratory's determined value and the assigned value for the serum. This is expressed as a percentage of the serotype-specific median or assigned value and is defined as (Fig. 1A) [(assigned value − laboratory-determined value)/assigned value] × 100.
Intersecting ranges.
In an evaluation of previously assigned antibody concentrations in pneumococcal polysaccharide reference serum, Concepcion and Frasch (2) compared cross-standardized values with those concentrations previously assigned by calculating the 20% ranges bracketing both the cross-standardized and the previously assigned concentrations and observing whether these ranges intersected. This study records the presence or absence of an intersection between a 20% range bracketing the assigned value and an unspecified range (±y percent) bracketing the laboratory-determined value (Fig. 1B). The data in the present study will be used to optimize the range bracketing the laboratory determined value.
Intersecting range and confidence interval.
The intersecting range and confidence interval record the presence or absence of an intersection between a 20% range bracketing the assigned value and an unspecified confidence interval calculated from the laboratory-determined values (Fig. 1C). The data in the present study will be used to optimize the confidence bound for the laboratory-determined values.
Overlapping range and confidence interval.
The overlapping range and confidence interval record whether a 50% range bracketing the assigned value overlaps an unspecified confidence interval calculated from the laboratory-determined values (Fig. 1D). The data in the present study will be used to optimize the confidence bound for the laboratory-determined values.
The range bracketing the assigned value was held fixed, while the ranges and confidence intervals for the individual laboratory-determined values were varied and the percentages of intersections and overlaps were tabulated. This provided the necessary information to determine practical ranges and confidence intervals for the laboratory-determined values which led to maximum percentages of intersections and overlaps with the set ranges bracketing the assigned values.
RESULTS
Forty-eight quality-control serum samples were evaluated for nine serotypes, which led to 432 calculated serum antibody determinations. Twelve laboratories submitted results (one laboratory assayed the series twice), yielding 13 data sets. Six data sets contained values from multiple assays: five laboratories assayed their samples in triplicate, and one laboratory assayed its samples in quadruplicate. Mean assay values were used as point estimates for laboratories that submitted multiple results.
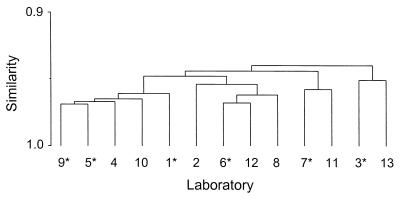
There were strong interlaboratory correlations, with the Spearman rank correlation coefficient being greater than or equal to 0.92 for all pairwise comparisons of laboratory results (Fig. 2). This indicated a broad sense of agreement among the laboratories.
FIG. 2.
Dendrogram displaying the level of agreement of ELISA-determined antibody concentrations among 12 laboratories. One laboratory evaluated the series twice. Laboratories denoted with an asterisk submitted three or more assay determinations for each serum sample. These values were averaged prior to generation of the dendrogram.
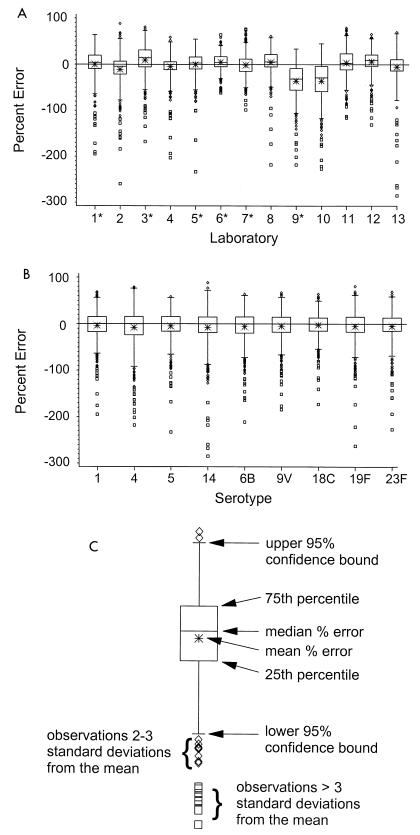
Serotype-specific medians of the laboratory-determined values from the 13 data sets were calculated for each serum sample and were used as trial-assigned values for subsequent calculations and tests. To gain a sense of laboratory variability, percent errors were calculated by using the assigned value as a reference point for all 432 specimens. The distribution of these percent errors for each laboratory are shown graphically by using box plots in Fig. 3A. The data are definitely skewed, with a large majority of the outliers greater in magnitude than the serotype-specific median or assigned value. Figure 3B displays similar box plots stratified by serotype, where the skewed nature of the data is also apparent. Figure 3C presents a schematic diagram that defines the different aspects of the box plots. These plots have been slightly altered from the traditional form to assist in interpretation. The one difference is that the whiskers define 95% confidence bounds. Inspection of Fig. 3A indicates that antibody concentrations submitted by laboratories 9 and 10 were consistently greater than those submitted by the other 11 laboratories. Table 2 details the frequencies of the outlying observations (values beyond the whiskers) stratified by the pre- and postvaccination status of the sera. The combined rate of outlying concentrations is 4.4%. Differences in the distributions of pre- and postvaccination percent errors were not statistically significant globally or when stratified by serotype. However, four laboratories did exhibit some differences between distributions of pre- and postvaccination percent errors (data not shown).
FIG. 3.
(A) Distribution of percent errors stratified by laboratory. One laboratory evaluated the series twice. The number of assay determinations from each laboratory is 432. Laboratories denoted with an asterisk submitted three or more assay determinations for each serum sample. These values were averaged prior to generation of the box plots. (B) Percent error distributions stratified by serotype. The number of assay determinations for each serotype is 624. (C) Diagram of the individual components of the box plots.
TABLE 2.
Percent error statistics as frequency of outlying assay determinations
| Assay | No. (%) of assay determinations with the following no. of SDs from the meana:
|
|
|---|---|---|
| 2–≤3 | >3 | |
| Prevaccination (nb = 2,808) | 89 (1.6) | 65 (1.2) |
| Postvaccination (n = 2,808) | 64 (1.1) | 28 (0.5) |
| Total (n = 5,616) | 153 (2.7) | 93 (1.7) |
Data were calculated over the 13 data sets for each type.
n, number of assay determinations.
To gauge within-laboratory variability, interassay CVs were calculated for the six laboratories that submitted multiple results. Table 3 outlines the results by tabulating the percentage of samples with CVs less than or equal to 30, 35, and 40% from each laboratory. As an example, Table 3 reveals that laboratory number 5 reported that 78% of its values had CVs of 30% or less. Additionally, all six laboratories reported that at least 89% of their serum antibody concentrations had CVs of 35% or less.
TABLE 3.
Frequencies of reported antibody concentrations at three levels of variability (CV) for laboratories submitting multiple antibody concentrations
| %CV | % of values for the following laboratory (na = 432):
|
||||||
|---|---|---|---|---|---|---|---|
| 1 | 3 | 5 | 6 | 7 | 9 | Total (n = 2,592) | |
| ≤30 | 97 | 97 | 78 | 86 | 89 | 93 | 90 |
| ≤35 | 98 | 98 | 89 | 89 | 92 | 97 | 94 |
| ≤40 | 99 | 99 | 94 | 93 | 94 | 98 | 96 |
n, number of specimens assayed from each laboratory.
Calculated antibody concentrations for several of the quality-control serum samples were highly variable. In an effort to reduce the number of serum samples to a manageable number that most laboratories could reasonably be expected to process, 12 of the most stable serum samples were chosen for final analyses. These sera were selected on the basis of their low variability, as measured from their interassay CVs. These sera were also selected because their antibody concentrations spanned the full range of antibody concentrations measured in this study. These were evaluated for the nine serotypes, which resulted in a subset of 108 determined values from each laboratory. Since the determined antibody concentrations from laboratories 9 and 10 were consistently greater than those from the remaining laboratories, we chose to remove these data and recalculate the trial-assigned values using the serotype-specific medians of the laboratory-determined concentrations from the remaining 11 data sets. The following analysis incorporates these revised trial-assigned values.
The distribution of percent errors across serotype as displayed in Fig. 3B was not significantly different when tested by a mixed-model analysis of variance that incorporated a repeated-measures design. However, when stratified by laboratory, the distribution of percent errors across serotypes did differ in 4 of the 11 data sets examined. Among these four data sets, no serotype(s) stood out as being uniformly atypical. Consequently, we chose to measure the magnitude of agreement between laboratory-determined concentrations and the assigned values without stratification by serotype.
Percent error.
Percent error calculations were referenced to the trial-assigned values as diagramed in Fig. 1A. These calculations were combined and summarized over all serotypes. With one exception, all laboratories reported that 85% or more of their specimens had percent errors of 40% or less (range, 19 to 40%; mean, 29%). Laboratory 11 reported that 84% of its concentrations had percent errors less than 41.5%. For this laboratory, 5 of 12 serum samples displayed percent errors greater than 40% for one serotype, serotype 4, suggesting that the 12 serum samples be reevaluated for this one serotype.
Intersecting ranges.
The range bracketing the assigned value was fixed at 20%. The range bracketing the individual laboratory-determined concentration was varied to discover the optimal value needed to describe at least 85% of the concentrations reported by each laboratory (Fig. 1B). With one exception, all laboratories achieved this goal when the range bracketing the reported value was 41%. Eighty-three percent of antibody concentrations reported by laboratory 13 intersected with the 20% range bracketing the assigned value when the range bracketing the reported value was set at 41%.
Intersecting range and confidence interval.
An alternative strategy to intersecting ranges is to continue bracketing the assigned value with a 20% range and calculate confidence intervals from the laboratory-determined concentrations and then look for intersections (Fig. 1C). This technique accounts for the actual variability from the reported values and requires multiple observations to calculate the confidence interval. Of the 11 data sets at hand, 5 offered multiple values for each serum sample and were eligible for inclusion in this method. At least 85% of the values from each laboratory intersected the assigned value ranges when the confidence level was set at 97%.
Overlapping range and confidence interval.
One final procedure entails calculation of a fixed range bracketing the assigned values (e.g., ±50%) and confidence intervals from the laboratory-determined values and then noting if the range overlaps the confidence intervals (Fig. 1D). Since the confidence interval from the laboratory-determined concentrations must be fully enclosed within the assigned value's range, the range was widened to 50% so that the method would not be too restrictive. Concentrations far removed from the assigned value must have a small degree of variability (short confidence interval) to be fully enclosed within the assigned value's range. This effectively penalizes concentrations distant from the assigned value by requiring them to have shorter confidence intervals to be judged in agreement with the assigned value. At least 85% of the values from each laboratory are overlapped by the 50% range bracketing the assigned value when the confidence level was set at 63%.
DISCUSSION
Our laboratories have participated in several multicenter immunogenicity studies over the past several years. These investigations have spanned a variety of bacterial pathogens including Haemophilus influenzae (7) and Neisseria meningitidis serogroups A (1) and C (4). With each study it has become increasingly clear that attention needs to be paid to ELISA protocols and subsequent antibody quantitation and data analysis to effectively compare results from several participating laboratories. These issues are amplified in the present study, in which the goal was to derive an analytical model that might be applied in future multicenter collaborative studies that may involve pathogens other than S. pneumoniae. The consensus ELISA developed and applied in the present study served as a mechanism for development of such an analytical model and is not meant to be used as a global standardized ELISA for all pneumococcal antibody determinations. In this study 12 laboratories evaluated a collection of sera for nine serotypes of S. pneumoniae, yielding 13 sets of data for analysis. These data provided an ample collection of results that may be used to develop such an analytical model. This model may, in turn, be used to evaluate the performance of succeeding laboratories, not involved in the original study, to assess whether they are able to estimate the same serum antibody concentrations within recommended limits.
This exploratory study was designed to quantify the degree of error associated with pneumococcal ELISAs. The laboratories in this study applied a consensus ELISA to a set of quality-control sera without assigned antibody concentrations. Trial-assigned values were estimated by using the determined values submitted by the participating laboratories. These trial-assigned values were then used to measure the performance of each laboratory. It is possible that some of the parameters measured in the course of this study will change as research continues. Eventually, concentrations will be assigned to these quality-control sera, which will give laboratories target values which may be used to optimize their individual ELISAs. It is our belief that this exercise will improve pneumococcal ELISAs and lessen the degree of variability measured in this study.
There was broad agreement in antibody concentrations among the laboratories, as evidenced by the dendrogram in Fig. 2. Even so, the box plots of percent errors in Fig. 3 indicated that two sets of results (those for laboratories 9 and 10) were sufficiently far removed from the remaining data to warrant exclusion from some aspects of the analysis. While the box plots revealed the data to be slightly skewed, the combined rate of outlying observations was less than 5%.
The percent error distributions across serotypes, as displayed in Fig. 3B, were not significantly different when tested by a mixed-model analysis of variance that incorporated a repeated-measures design. There were differences in 4 of 11 data sets when this same comparison was made within each individual laboratory. These differences were not systematic and did not implicate any particular serotype(s). Additionally, there were no significant differences in the distributions of pre- and postvaccination percent errors globally or when stratified by serotype. However, four laboratories did exhibit some differences between distributions of pre- and postvaccination percent errors. We did not wish to form separate analytical protocols with separate guidelines for each serotype or for each pre- or postvaccination status, so we pooled the data across serotype and vaccination status when describing interlaboratory variability and comparing laboratory-determined antibody concentrations to the assigned values. The resulting guidelines should be applicable to all serotypes and vaccination status, collectively.
Within-laboratory variability may be measured by inspecting within-assay coefficients of variation. Sera must be assayed multiple times for this, and in our study six laboratories generated results for which it was necessary to perform these calculations. Table 3 provides data necessary to select a CV that may be used to monitor assay variability in future studies. This CV must be both reasonable in magnitude and achievable in a regular laboratory setting. Each of these laboratories assayed the sera three or more times and reported CVs of 35% or less for 89% or more of their samples. This indicates one guideline for assessment of intralaboratory variability: the samples must be assayed at least in triplicate and at least 85% of the serum samples must be quantified to have CVs of 35% or less. Those samples with CVs greater than 35% should be distributed evenly across the nine serotypes.
Processing of 48 samples for nine serotypes to evaluate assay performance is a burden that few laboratories would or could undertake. In an effort to encourage investigators in other laboratories to use this protocol and reproduce these results, a subset of 12 serum samples was selected for subsequent analysis. These sera were chosen on the basis of their low variability, as measured from their interassay CVs, and because their antibody concentrations spanned the full range of antibody concentrations measured in this study. They were selected from the original collection of 48 serum samples to define a set of parameters necessary to establish pertinent bioassay and analysis protocols.
While quantifying interlaboratory variability and the four techniques for judging the level of agreement between a laboratory's determined value and the trial-assigned value, we selected the most stringent parameters possible that would successfully describe each of the data sets contributed by the participating laboratories. Given the level of experience with pneumococcal ELISA analysis and antibody quantitation that these laboratories possess, we believed that it was important to form guidelines that encompass all the laboratories in this study. Even so, we believe that the thresholds recommended in the guidelines provide reasonable targets which succeeding laboratories should be able to incorporate into their protocols.
We explored four techniques for judging the level of agreement between each laboratory's determined values and the trial-assigned values. An interval was fixed around the assigned values; the range and confidence interval about the laboratory-determined values were varied to find the setting that would capture or describe at least 85% of the data for each method. We believed that this percentage was large enough to describe a convincing majority of the data in the sample, while offering enough flexibility to tolerate a small number of outlying observations. Prior to the evaluation of these four models, data sets from two laboratories were excluded because their determined values were systematically greater than those submitted from the remaining laboratories. One goal of this study was to evaluate models that measured agreement between a laboratory's determined values and the respective trial-assigned values. A second goal was to develop a series of guidelines that would outline a protocol useful for future studies. Inclusion of the data sets from these two laboratories would have adversely influenced the model evaluations and the ensuing guidelines. Inclusion of these data sets would have resulted in a higher percentage of outlying observations overall and would have elevated the parameters in the model comparison discussed below to the extent that they would be unacceptably broad and unusable. These two laboratories whose data sets contained outlying values subsequently examined their ELISA conditions and, after optimizing a single assay parameter, were able to derive antibody concentrations in agreement with those derived by the remaining laboratories in the study (data not shown).
The following comments address the advantages and disadvantages of each comparison method listed in increasing order of applicability.
Overlapping range and confidence intervals.
A variant of the overlapping range and confidence interval procedure was developed by the National Institute of Occupational Safety and Health and is used to measure analytical accuracy in laboratory instruments (3). This method is perhaps the most attractive, statistically, as it considers the interassay variability of the serum antibody concentration as expressed by a confidence interval. This confidence interval, usually defined at the 95% level, must be totally enclosed by the 50% range about the assigned value to conclude that the two values are in agreement. One appealing attribute of this technique is that as the laboratory-determined values deviate from the assigned value, their 95% confidence intervals must be shorter to be fully enclosed within the range about the assigned value. Thus, a calculated concentration distant from the assigned value can be judged in agreement only if it is estimated with a high degree of precision (a low degree of variability that leads to a short 95% confidence interval). In this study the confidence level would need to be set at 63% to classify at least 85% of the antibody concentrations from each laboratory as being in agreement with the assigned values. This technique is lacking, primarily due to the questionable practice of calculating standard errors and confidence intervals from such a small number of observations. The 63% confidence level required to accept 85% of the concentrations is also excessively small, indicating that the data are too variable to apply this procedure with certainty. Given that the number of multiple assays needed for each serum sample to produce credible estimates of standard errors is beyond the capacity of most laboratories, this approach is too restrictive for these types of data and is better suited to monitoring of laboratory instrumentation.
Intersecting range and confidence intervals.
In this study at least 85% of the values from each laboratory intersected the 20% range bracketing the assigned value when the confidence level was set at 97%. This approach suffers from the same constraint of small sample sizes for multiple assays for each serum sample as the overlapping range and confidence interval technique. Credible estimates of standard errors cannot be made with such a small number of repeated observations. Also, confidence intervals calculated from highly variable serum antibody concentrations will more readily intersect with the assigned-value range. These confidence intervals will be excessively wide due to their inflated standard errors, which, in turn, will have a greater chance of intersecting the assigned-value range. These observations would be judged to be in agreement with the assigned values, while similar, more precise estimates, would not. This, as well as the weaknesses of calculating confidence intervals with such small numbers of multiple assay results, leads us to conclude that this technique was also ill-suited for the present application.
Intersecting ranges.
In the present study, the range bracketing the assigned value was set to 20%. The prescribed range bracketing the serum antibody concentration required to classify at least 85% of each of the laboratories' determined values as being in agreement with the assigned values was found to be 41%. Ten of the 11 data sets achieved this goal. It may be argued that this method disregards the inherent variability of the serum antibody concentrations. However, if a laboratory's interassay CVs are consistently less than or equal to 35%, the laboratory may feel reasonably assured that its concentration estimates are stable.
When the upper limit of the range bracketing the assigned value just intersects the lower limit of the range bracketing a laboratory-determined value, which is greater than the assigned value, the two values are judged to be in agreement. The maximum percent error between the two point estimates may then be calculated to gauge how similar (or different) they actually are and still be classified as in agreement. An analogous algebraic relationship holds for laboratory-determined values less than the assigned value. The present case, in which the range bracketing the assigned value is 20% and the range bracketing the laboratory-determined value is 41%, translates to maximum percent errors of 103% for antibody concentrations greater than the assigned value and 43% for antibody concentrations less than the assigned value. The range about the individual laboratory-determined value is defined in terms of the value itself, leading to these asymmetrical percent error calculations (e.g., 103 versus 43% above). With the intersecting ranges model, concentrations greater than the assigned value may be more distant from the assigned value than those less than the assigned value and still be judged in agreement, simply because their calculated ranges are broader. These considerations make this method less attractive than the percent error technique described below.
Percent error.
The percent error calculation is symmetrical in that laboratory-determined values which are equally spaced, both greater than and less than the assigned value, will express the same percent error and yield the same outcome with regard to agreement with the assigned value. With the exception of one laboratory, 85% of the concentrations were judged to be in agreement with the assigned values when the maximum percent error was set to 40% (range, 19 to 40%; mean, 29%). The one laboratory that failed this test displayed the greatest degree of error for serotype 4 results, suggesting that these samples should be reexamined. Given the symmetry of this calculation and the fact that this technique does not overextend the data by computing statistics for a small number of repeat observations, this procedure is our method of choice among the four examined.
If the data for the entire set of 48 serum samples were examined by the percent error technique, all 11 laboratories would have reported a percent error of 44% or less for at least 85% of the specimens (range, 28 to 44%; mean, 36%). Although the survey of a subset of 12 serum samples may give the impression that we are attempting to maximize the performance of this method, the variability associated with the percent error technique does not increase appreciably when the complete data set is analyzed.
These results and discussions suggest a series of analytical steps that may be followed in a multicenter collaborative study to assess the level of agreement between laboratory-determined serum antibody concentrations and assigned values for a set of quality-control specimens. Once a set of quality-control sera has been compiled and antibody concentration values assigned, this protocol may be used to judge whether a laboratory is able to determine these values within an acceptable level of tolerance. These steps may easily be adapted to any multicenter ELISA protocol. After a laboratory demonstrates its ability to reproduce the assigned values for a set of standardized sera within a suitable degree of variability, it can process regular unknown samples with a heightened degree of confidence.
We propose the following guidelines as an outline for an analytical protocol for pneumococcal ELISA studies. These guidelines are meant to help qualify a laboratory that wishes to routinely process serum specimens for antibodies to S. pneumoniae. They may be easily generalized to ELISAs that involve different organisms, once similar studies are conducted to optimize the protocol parameters and analytical thresholds.
Guidelines. (i) Guideline 1.
Use a set of quality-control specimens chosen for their low interassay and interlaboratory CVs and range of antibody concentrations. Evaluate the serum antibody concentrations for nine serotypes.
(ii) Guideline 2.
Use a standard reference serum sample to form standard or characteristic curves for quantitation of antibodies in the quality-control specimens.
(iii) Guideline 3.
Use assay protocol parameters that are performance based and optimized to eliminate systematic error within a laboratory. For example, if one laboratory's determined antibody concentrations are uniformly less than or greater than the assigned values, optimization of one or more segments of the assay protocol may bring these values into closer agreement.
(iv) Guideline 4.
Assay specimens at least in triplicate and use at least five dilutions.
(v) Guideline 5.
Calculate antibody concentrations by using a standardized software package or other software that will yield similar results. Jeffcoate and Das (5) and Pegg and Miner (8) have shown that differences in data processing techniques account for a significant portion of between-assay variability. Pegg and Miner (8) and Plikaytis et al. (10) have also demonstrated that different implementations of the same calibration formulas (the logit-log technique) give significantly different results.
(vi) Guideline 6.
Within a laboratory, ensure that at least 85% of the samples display an interassay CV of 35% or less. The results for the remaining 15% of the samples should be distributed evenly across the nine serotypes.
(vii) Guideline 7.
Within a laboratory, ensure that at least 85% of the specimens exhibit a percent error of 40% or less compared to the assigned values for the specimens. The results for the remaining 15% should be distributed evenly across the nine serotypes.
(viii) Guideline 8.
If more than 15% of specimens display errors greater than 40%, examine the results for each serotype. If one serotype displays a greater error rate than the others, repeat the assays for that serotype.
Once absolute antibody concentrations are assigned to these quality-control sera, a laboratory not involved with the original study may use this protocol as a guide to determine if it can conform to the performance standards set by the laboratories participating in this study. Since the assay parameters are performance based, a laboratory may also optimize one or more assay components to eliminate possible systematic error and bring its calculated antibody concentrations into closer agreement with the assigned values. The guidelines recommend an acceptable degree of variability that a laboratory must adhere to as it adjusts its assay characteristics. Once a laboratory demonstrates its ability to measure antibody concentrations for a series of quality-control specimens within an acceptable degree of tolerance, it may return to its regular method of processing specimens, including whatever quality-control procedures the laboratory routinely implements.
We recommend the use of the percent error calculation for comparison of a laboratory's determined values with an assigned antibody concentration. In the event that an assigned value does not exist and two or more laboratories are attempting to measure how well their values agree, the percent error calculation will lead to conflicting results. If there is no consensus as to which laboratory's determined values take precedence, this statistic will change, depending on the values and the order in which they are entered into the equation. In this case, more traditional descriptive statistics must be used, such as bivariate scatter plots, correlation coefficients, and linear regressions. A simple nonparametric test, the Wilcoxon signed rank sum test, may be used to test whether the distribution of values from the two laboratories are significantly different from each other.
In summary, this paper presents a series of guidelines that may be used to determine if a laboratory that is analyzing a series of quality-control sera with assigned antibody concentrations is able to estimate these values within an acceptable degree of tolerance. These criteria were developed by using data acquired from a multicenter pneumococcal ELISA study involving 12 laboratories and 13 data sets and trial antibody concentrations assigned to each serum sample. When final concentrations are assigned to these sera, these experiments should be repeated to confirm the CV and percent error parameters recommended in the guidelines. Once laboratories demonstrate that they can effectively quantify antibody concentrations in a set of quality-control sera within an acceptable degree of tolerance, they may initiate independent studies to evaluate vaccine formulations of their choice. If they retain the performance-based immunological assay components and analysis tools that they used in their quality-control trials, then we believe that their studies will be highly comparable and this will give regulatory agencies the means of evaluating and comparing the differences in immunogenicity levels elicited by new and developing vaccines. These guidelines form an analytical protocol for pneumococcal ELISA studies which is generalizable and which may be adapted to ELISA experiments that involve other pathogens. When the concentration is on different organisms, it may be necessary to conduct studies similar to the present one to optimize the protocol parameters and analytical thresholds incorporated in the guidelines.
REFERENCES
- 1.Carlone G M, Frasch C E, Siber G R, Quataert S, Gheesling L L, Turner S H, Plikaytis B D, Helsel L O, DeWitt W E, Bibb W F, Swaminathan B, Arakere G, Thompson C, Phipps D, Madore D, Broome C V. Multicenter comparison of levels of antibody to the Neisseria meningitidis group A capsular polysaccharide measured by using an enzyme-linked immunosorbent assay. J Clin Microbiol. 1992;30:154–159. doi: 10.1128/jcm.30.1.154-159.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Concepcion N, Frasch C E. Evaluation of previously assigned antibody concentrations in pneumococcal polysaccharide reference serum 89SF by the method of cross-standardization. Clin Diagn Lab Immunol. 1998;5:199–204. doi: 10.1128/cdli.5.2.199-204.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fischbach T, Song R, Shulman S. Some statistical procedures for analytical method accuracy tests and estimation. Am Ind Hyg Assoc J. 1996;57:440–451. doi: 10.1080/15428119691014783. [DOI] [PubMed] [Google Scholar]
- 4.Gheesling L L, Carlone G M, Pais L B, Holder P F, Maslanka S E, Plikaytis B D, Achtman M, Densen P, Frasch C E, Käyhty H, Mays J P, Nencioni L, Peeters C, Phipps D C, Poolman J T, Rosenqvist E, Siber G R, Thiesen B, Tai J, Thompson C M, Vella P P, Wenger J D. Multicenter comparison of Neisseria meningitidis serogroup C anticapsular polysaccharide antibody levels measured by a standardized enzyme-linked immunosorbent assay. J Clin Microbiol. 1994;32:1475–1482. doi: 10.1128/jcm.32.6.1475-1482.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jeffcoate S L, Das R E G. Interlaboratory comparison of radioimmunoassay results. Ann Clin Biochem. 1977;14:258–260. doi: 10.1177/000456327701400170. [DOI] [PubMed] [Google Scholar]
- 6.Koskela M. Serum antibodies to pneumococcal C polysaccharide in children: response to acute pneumococcal otitis media or to vaccination. J Immunol Methods. 1987;164:13–19. doi: 10.1097/00006454-198706000-00006. [DOI] [PubMed] [Google Scholar]
- 7.Madore D V, Anderson P, Baxter B D, Carlone G M, Edwards K M, Hamilton R G, Holder P, Käyhty K, Phipps D C, Peeters C C, Schneerson R, Siber G R, Ward J I, Frasch C E. Interlaboratory study evaluating quantitation of antibodies to Haemophilus influenzae type b polysaccharide by enzyme-linked immunosorbent assay. Clin Diagn Lab Immunol. 1996;3:84–88. doi: 10.1128/cdli.3.1.84-88.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pegg P J, Miner E M. The effect of data reduction technic on ligand assay proficiency survey results. Am J Clin Pathol. 1982;77:334–337. doi: 10.1093/ajcp/77.3.334. [DOI] [PubMed] [Google Scholar]
- 9.Plikaytis B D, Holder P F, Carlone G M. Program ELISA for Windows user's manual, version 1.00. Atlanta, Ga: Centers for Disease Control and Prevention; 1996. [Google Scholar]
- 10.Plikaytis B D, Holder P F, Pais L B, Maslanka S E, Gheesling L L, Carlone G M. Determination of parallelism and nonparallelism in bioassay dilution curves. J Clin Microbiol. 1994;32:2441–2447. doi: 10.1128/jcm.32.10.2441-2447.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Plikaytis B D, Turner S H, Gheesling L L, Carlone G M. Comparisons of standard curve-fitting methods to quantitate Neisseria meningitidis group A polysaccharide antibody levels by enzyme-linked immunosorbent assay. J Clin Microbiol. 1991;29:1439–1446. doi: 10.1128/jcm.29.7.1439-1446.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quataert S A, Kirch C S, Quackenbush L J, Phipps D C, Strohmeyer S, Cimino C O, Skuse J, Madore D V. Assignment of weight-based antibody units to a human antipneumococcal standard reference serum, lot 89-S. Clin Diagn Lab Immunol. 1995;2:590–597. doi: 10.1128/cdli.2.5.590-597.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robbins J B, Schneerson R, Szu S C, Bryla D A, Lin F Y. Measurement of human serum IgG antibodies or a surrogate is sufficient to standardize (predict efficacy) vaccines. Dev Biol Stand. 1998;95:221–222. [PubMed] [Google Scholar]