Abstract
Advances in sequencing technologies have tremendously reduced the time and costs associated with sequence generation, making genomic data an important asset for routine public health practices. Within this context, phylogenetic and phylogeographic inference has become a popular method to study disease transmission. In a Bayesian context, these approaches have the benefit of accommodating phylogenetic uncertainty and popular implementations provide the possibility to parameterize the transition rates between locations as a function of epidemiological and ecological data to reconstruct spatial spread while simultaneously identifying the main factors impacting the spatial spread dynamics. Recent developments enable researchers to make use of travel history data of infected individuals in the reconstruction of pathogen spread, offering increased inference accuracy and mitigating sampling bias. We here describe a detailed workflow on how to reconstruct the spatial spread of a pathogen through Bayesian phylogeographic analysis in discrete space using these novel approaches implemented in BEAST. The individual protocols focus on how to incorporate molecular data, covariates of spread and individual travel history data into the analysis.
INTRODUCTION
Bayesian Evolutionary Analysis Sampling Trees (BEAST) [Suchard et al., 2018] is a software package that provides a general framework for phylogenetic inference and evolutionary hypothesis testing using molecular sequence data [Drummond and Rambaut, 2007, Drummond et al., 2012, Suchard et al., 2018]. As such, BEAST employs a combination of different types of models (including but not limited to molecular clock models, coalescent models and substitution models) to infer time-calibrated phylogenetic trees from an alignment of time-stamped sequences. Phylogeographic analyses based on simple ancestral trait reconstruction models incorporate sampling locations as additional data. For discrete locations, the trait evolution process, modelled as a continuous-time Markov chain process, can be parameterized in terms of covariates to help uncover the key factors that facilitate or prevent the spread of a pathogen. Inference under these models is performed through Markov chain Monte Carlo (MCMC) sampling and the likelihood evaluations make use of BEAGLE, a high-performance computational library for statistical phylogenetics [Ayres et al., 2019]. BEAST is widely used in the field of phylodynamics and molecular epidemiology of infectious disease as it allows obtaining insights from molecular data through an expanding array of statistical models and estimation procedures.
Here, we focus on Bayesian phylogeographic inferences that aim to answer the question of “how did an epidemic spread through space and time?” through jointly reconstructing the evolutionary and geographical history of a pathogen population in the form of (geographically) annotated time-scaled phylogenies. Specifically, we focus on the discrete model in which transition rates are a function of potential predictors of spatial spread according to a simple generalized linear model (GLM). Such an approach has previously enabled researchers to, for example, assess the impact of air travel on the global spread of influenza [Lemey et al., 2014]. Importantly, the GLM formulation generally also offers a sparser parameterization of the spatial transition process as it avoids having to estimate all pairwise transitions rates, which scale quadratically with the number of locations and can be difficult to inform.
While phylogeographic analyses incorporate the location of sampling as a discrete trait associated with each pathogen genome, it is possible that sampled patients had recently travelled to different locations. In fact, during epidemics travelers may be specifically screened if they return from areas with high incidence. This has been the case for SARS-CoV-2 and it has motivated the development of model extensions that incorporate the specific times and locations of travel [Lemey et al., 2020]. This may be particularly useful for capturing diversity in the locations that remain under-sampled, and as such, it can mitigate bias associated with disparate sampling efforts. These are important considerations because the computationally convenient ancestral reconstructions are highly sensitive to sampling bias.
We here provide four related protocols to reproduce the travel history-aware phylogeographic reconstructions performed in [Lemey et al., 2020]. Protocol 1 introduces the GISAID database [Elbe and Buckland-Merrett, 2017] (https://www.gisaid.org/) and provides the required steps to construct a SARS-CoV-2 multiple sequence alignment from this database. In Protocol 2, we provide instructions on how to set up a discrete state phylogeographic inference under the GLM parametrization using BEAUti, a GUI tool shipped with BEAST. Protocol 3 introduces an automated script to modify a BEAUti-generated XML file to incorporate travel history data, which can subsequently be run using BEAST. Finally, in Protocol 4, we guide the user through the process of visualizing individual spatial dispersal histories from the posterior distribution of trees estimated by BEAST.
PROTOCOL 1: CREATING A SARS-CoV-2 MSA USING SEQUENCES FROM GISAID
The first step in any phylogenetic or phylogeographic analysis is to obtain a high-quality multiple sequence alignment (MSA). The largest publicly accessible repository of genomes is available through the GISAID database. The GISAID initiative provides a platform to openly share genomic data of influenza viruses as well as SARS-CoV-2 under specific rules [Elbe and Buckland-Merrett, 2017]. Access to the database is free (https://www.gisaid.org/registration/register/), but requires the user to register and agree to GISAID’s terms of use in order to obtain access.
This protocol describes how to construct a SARS-CoV-2 MSA from sequences downloaded from GISAID. Specifically, we will construct an alignment for the 282-taxa dataset analyzed in [Lemey et al., 2020].
Necessary Resources
Hardware
Standard computer running Linux, MacOS or Windows 10.
Software
A modern web browser (Google Chrome and Mozilla Firefox recommended).
The latest MAFFT [Katoh and Standley, 2013] version (v7.453 used in this protocol). The latest Aliview [Larsson, 2014] version (v1.26 used in this protocol).
A terminal emulator running a standard Unix shell.
Note: For Windows, a BASH shell can be installed through the Windows Subsystem for Linux. For more information, see https://docs.microsoft.com/en-us/windows/wsl/.
Files
A list of GISAID accession numbers/identifiers.
A FASTA file with the untranslated regions (UTR) from the SARS-CoV-2 reference genome sequences.
Example files can be found at
https://github.com/hongsamL/travHistProtocol/tree/main/files/Protocol1
Protocol steps and annotations
-
Search and download the desired sequences in the GISAID database.
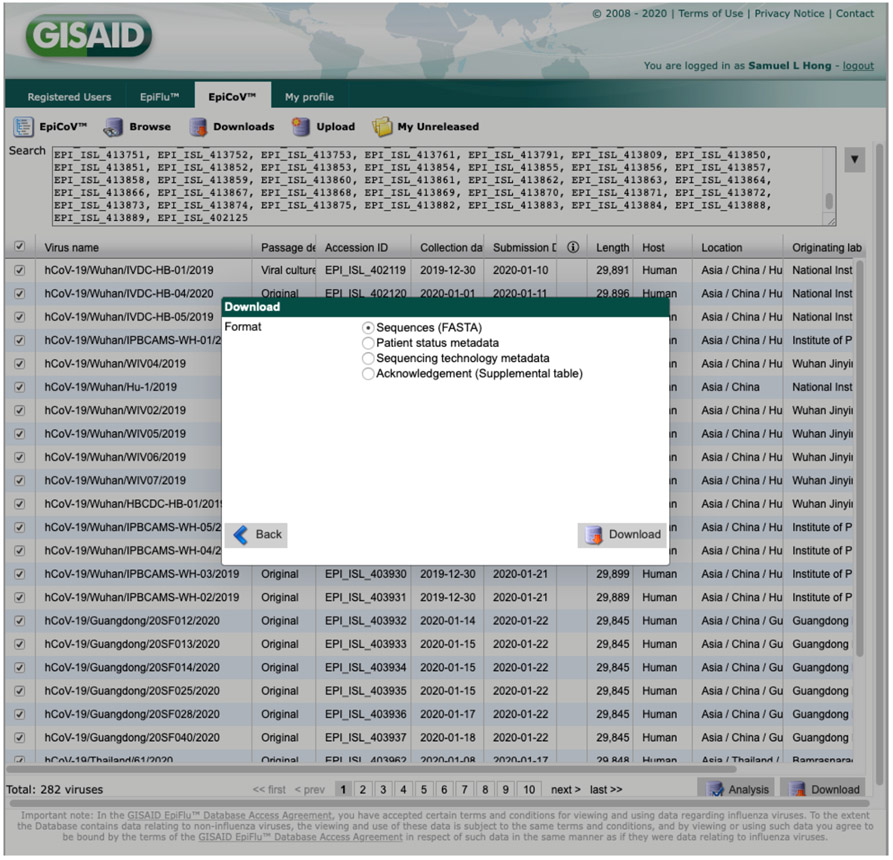
Log on to GISAID and click on EpiCoV™’s Browse tab to access a table with all available SARS-CoV-2 sequences in the database. To bulk download sequences by accession ID, click on the Fulltext button, paste your comma-separated list of accessions (see Files for an example) in the search box, and download the FASTA sequences using the download button (Figure 1). In this example, we will save the sequences in a file called gisaid_selection.fasta.
Note: We can also use the EpiCoV Browse portal to download a custom selection of genomes. On the header section of the table you will see multiple search fields and dropdown menus to filter sequences according to different criteria.
-
Remove whitespace from the FASTA file:
sed -i.bkp "s//_/g" gisaid_selection.fasta
Note: To avoid potential issues when parsing the FASTA headers with whitespaces (e.g. in country names), we replace all whitespace in the file with underscores using sed. We use the -i flag to find and replace the file in-place while keeping a backup of the original file.
-
Align the sequences using MAFFT
cat utr.fasta >> gisaid_selection.fasta
mafft --thread -1 --nomemsave gisaid_selection.fasta > gisaid_aln.fasta
Note: To remove potential sequencing errors in the error-prone 5’ and 3’ ends of the virus, we include the reference sequences for the 5’ and 3’ UTRs of the SARS-CoV-2 genome (see example files). We do this so that we can later trim these regions from the final alignment. We concatenate these sequences to the FASTA file containing our genomes of interest, and align all sequences using MAFFT.
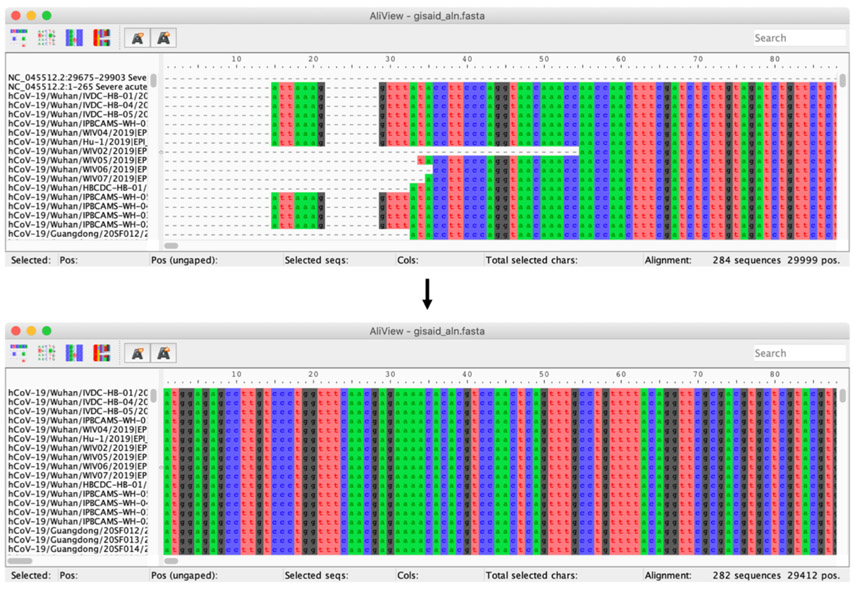
Manually trim UTRs in the MSA using Aliview. In Aliview, visually identify the UTR sequences and manually select the corresponding sites. Remove the selected sites using the Edit menu. Remove the now-empty reference sequences and save the trimmed MSA (Figure 2).
Figure 1:
EpiCoV GISAID portal. Here, we search for all 282 sequences analysed in [Lemey et al., 2020], and download them by selecting the checkbox on the left of the table and pressing the download button. We can also download metadata for the sequences and the corresponding GISAID acknowledgement table using the same approach.
Figure 2:
Alignment visualizations in Aliview before (top) and after (bottom) trimming the 5’ and 3’ UTR regions. Here, we remove a total of 286 sites on the 5’ side and 301 sites on the 3’ side, bringing the alignment length to 29,412 sites.
PROTOCOL 2: SETTING UP A DISCRETE TRAIT PHYLOGEOGRAPHIC RECONSTRUCTION IN BEAUTI
Performing Bayesian phylogeographic inference while accommodating individual travel history data constitutes an extension of the standard Bayesian ancestral reconstruction approach available in BEAST [Lemey et al., 2009]. We will first generate an XML file for the phylogeographic inference using the interactive BEAUti graphical application, which will serve as a basis for Protocol 3. BEAUti facilitates the design of the analysis you want to perform in BEAST and generates an XML file that will serve as the input file for BEAST or as the basis for manually specifying advanced models not available in BEAUti. The BEAUti-generated XML file contains all the required data, the models (and priors) that were selected in BEAUti, and the computational settings which will be used to run the MCMC algorithm that will collect samples of all relevant parameters (including the phylogenetic tree with annotated ancestral locations) from the joint density.
This protocol describes how to set up a phylogeographic analysis with a GLM extension to simultaneously reconstruct spatiotemporal history and test the contribution of potential predictors of spatial spread using BEAST [Lemey et al., 2014]. This extension parameterizes each rate of among-location movement in the phylogeographic model as a log linear function of the provided predictors. In this example, we will use the MSA generated in Protocol 1 to set up a phylogeographic GLM reconstruction using a flight connectivity matrix, a great-circle distance matrix and an asymmetry matrix as covariates (but see [Lemey et al., 2020] for more detailed information).
Necessary Resources
Hardware
Standard computer running Linux, MacOS, or Windows.
Software
Latest BEAUti version (v1.10.5)
Files
Multiple sequence alignment Tab-delimited metadata file
Covariate matrices in CSV format
Example metadata and covariate files available at https://github.com/hongsamL/travHistProtocol/tree/main/files/Protocol2
Protocol steps and annotations
-
Import the MSA from Basic Protocol 1 into BEAUti.
Load the MSA into BEAUti by selecting Import Data from the File menu (do not use Open…). You can also do this by dragging the FASTA file into the Partitions panel.
-
Specify the sampling dates for the tips.
Select the Tips tab and check the “Use tip dates” box. By default, all taxa will show as having a date of zero (i.e., all sequences were sampled at the same time in the present). To specify each sampling date, select “Import Dates” and load the metadata file. This metadata file contains a tab separated table mapping the FASTA header of each sequence in the alignment with the corresponding sampling date. When loading the file, select the “Parse calendar dates with variable precision” option. This allows for taxon dates to have different degrees of resolution (e.g., year-month-day vs. year-month). We estimate the sampling dates for those sequences without day-level resolution by selecting “Sampling uniformly from precision” in the “Tip date sampling” menu at the bottom of the table.
Note: It is also possible to specify the sampling dates without using a metadata file. This is done by parsing the FASTA headers of each sequence. To do this, click on “Parse Dates”, and specify the rules for delimiting the date from all taxon labels. For an example of what this looks like, see [Hill and Baele, 2019] Figure 2a.
-
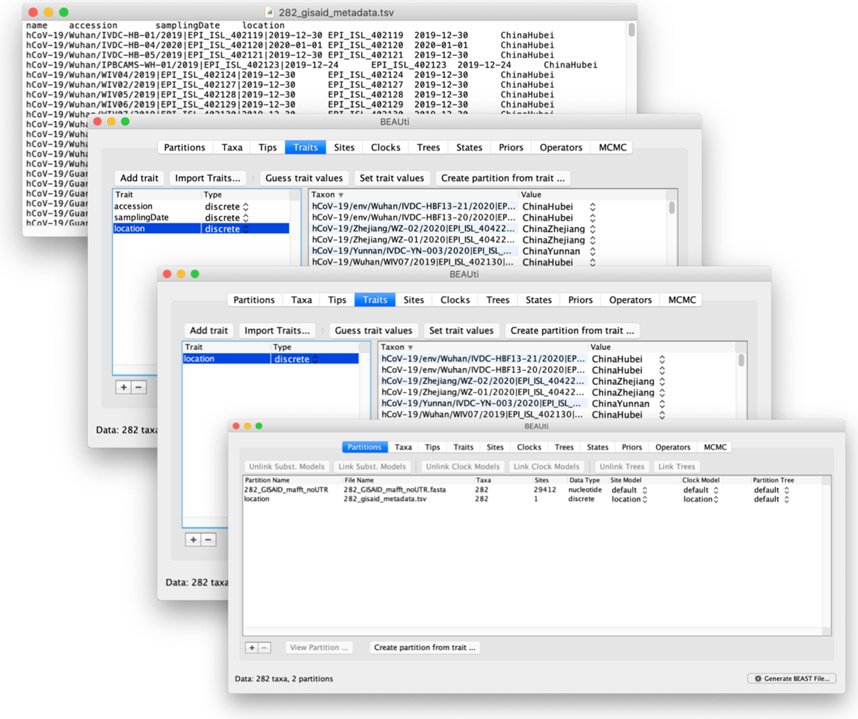
Specify the sampling location of each taxon as a discrete trait
Click on the Traits tab. To associate each taxon with a sampling location, click on “Import traits” and select the metadata file (Figure 3). This will create a new trait for each column on the metadata file. Delete all non-relevant traits by clicking on the “−” button at the bottom-left of the page (keep only the “location” trait for this example). Select the desired trait and click on “create partition from trait”. A new partition containing the trait data will be created under the Partitions tab.
Note: It is also possible to add a discrete trait without using a metadata file. This is done by parsing the FASTA headers of each sequence. To do this, click on “Add Trait” to create a new trait with a corresponding data partition. Select all taxa and click on “Guess trait” values to parse the trait values from the taxon labels.
-
Set up the nucleotide and trait substitution models.
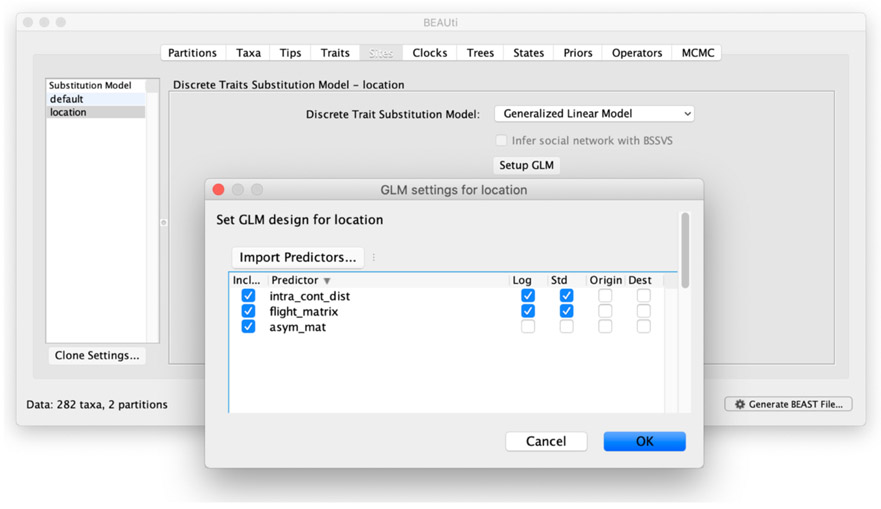
Click on the “Sites” tab. Following [Lemey et al., 2020], we will specify an HKY+Γ nucleotide substitution model, and a GLM-CTMC for the location trait. Load the covariates by clicking on “Setup GLM” and “Import Predictors” (Figure 4). In this example, we inform the rates of spread between locations using three predictors: a flight matrix containing air travel data between locations, a distance matrix containing intra-continental distances, and an asymmetry matrix where entries for transitions “from” and “to” Hubei are denoted with 1 and −1 respectively. Check the “Log” and “Std” boxes to log-transform and standardize the air travel and distance GLM predictors.
Note: In this context, the terms ‘predictor’ and ‘covariate’ are used interchangeably. By default, each predictor name will be the same as the name of the file it originates from. Non-pairwise covariates can also be setup as origin and destination predictors in this window. Predictor files are comma-separated value (CSV) files formatted in two different ways depending on the nature of the predictor. For pairwise predictors, the CSV file is in the form of a square matrix with the different locations as column and row names alphabetically ordered, with rows denoting the origin and columns the destination. For origin-destination predictors, the CSV file is in the form of a two-column table with location names alphabetically ordered and predictor values. You can also specify new predictor names by double clicking on each name, which allows you to enter a name of your choosing.
-
Specify a clock model.
Following the model specifications in [Lemey et al., 2020], click on the “Clocks” tab to specify a strict clock model for both the nucleotide and trait partitions.
Note: BEAST analyses operate under the assumption of a molecular clock to estimate time-stamped phylogenies from the molecular sequence data and associated sampling times. For this to hold valid, there needs to be sufficient temporal signal in the data (see the section Critical parameters and troubleshooting). The presence of temporal signal in a data set can be assessed using TempEst [Rambaut et al., 2016] and more formally tested using Bayesian Evaluation of Temporal Signal (BETS) [Duchene et al., 2020]. For more information on the different molecular clock models available in BEAST: https://beast.community/clocks
-
Specify a demographic model.
Click on the “Trees” tab to specify the Tree prior. Following [Lemey et al., 2020], we specify an exponential growth coalescent model parameterized with a growth rate parameter. We refer to http://beast.community/tree_priors for a more detailed explanation on a subset of the available coalescent models in BEAST.
Note: By default, BEAST will initialize the analysis with a randomly generated starting tree. It is also possible to start the analysis from a user-specified time-scaled phylogeny. This is usually done to reduce the burn-in time required for the tree topologies to converge. Specify a starting tree by clicking on “Import Data” under the “File” menu, to load a phylogenetic tree in Nexus format into BEAUti.
-
Set up the ancestral state reconstruction for the location trait.
Click on the “States” tab and select the location partition. Check the “Reconstruct state change counts” and “Reconstruct complete change history on tree” boxes to save complete realizations of the spatial spread process on the output trees. Be warned that this latter option might lead to large file sizes.
-
Specify the priors.
Click on the “Priors” tab. Following [Lemey et al., 2020], we use default priors and only specify a Lognormal prior with mu=1 and sigma=10 for the effective population size (where mu and sigma represent the log-mean and log-standard deviation), and a Laplace prior with mean=0 and scale=100 for the exponential growth rate parameter.
Note: BEAST aims to offer sensible default priors when informative prior information is unavailable, most of which can be considered largely uninformative. A wide range of prior distributions is also available to customize each analysis as needed.
-
Set up the transition kernels.
Click on the “Operators” tab. Identify the tip-date sampling transition kernels in the table. These have the description “Uniform sample from precision of age of this tip”. The parameters associated with these transition kernels tend to converge rapidly and have good mixing (i.e., their effective sample size – or ESS – accumulates rapidly). Decrease the weight of these operators to 0.25 but leave the weights of the other transition kernels at their default values, to sample these parameters less frequently so that the analysis gets to spend more time on estimating other parameters of interest.
Note: Transition kernels (called “operators” in BEAST) are used to propose new values for each parameter being estimated during the analysis. Different combinations of transition kernels can be used to customize the analysis as needed. For example, we can remove transition kernels to fix the value of certain parameters to their starting values (e.g. fixing the rate of the molecular clock or estimating spatial spread on a fixed user-provided tree).
-
Set up the MCMC options and generate the BEAST XML file.
Click on the “MCMC” tab. Set “Length of chain” to 200,000,000 states and “Log parameters every” to 100,000 states. This will thin the MCMC results so that only 2,000 samples are collected by the end of the run. Set your file name stem to generate the desired output file names (e.g., 282_GISAID_sarscov2), and click on “Generate BEAST File” to create the BEAST XML file for this analysis.
Note: Thinning consists of storing only every nth sample from an MCMC analysis. This subsampling is performed to decrease the autocorrelation in the posterior sample and reduce the file size of the output. This can be important since Bayesian phylogenetic analyses often require very long chains and storing every single state would be prohibitive for file storage.
Figure 3:
Adding the sampling locations as a trait. From back to front: i) tab-separated metadata file containing sequence names and sampling locations ii) columns in the metadata file other than “name” loaded as traits. iii) traits different from “location” removed from the analysis iv) partition corresponding to the “location” trait created.
Figure 4:
Setting up the predictors for the GLM-based spatial diffusion model. In this example, we load the air travel and intra-continental distance predictors, and log-transform and standardize them by checking the corresponding boxes.
PROTOCOL 3: PHYLOGEOGRAPHIC RECONSTRUCTION INCORPORATING TRAVEL HISTORY INFORMATION
Phylogeographic reconstruction using discrete locations has been shown to be sensitive to spatiotemporal sampling bias. The ancestral reconstruction of locations will depend on the availability of samples from each location. In practice, this means that over/under-sampling of sequences from a given location can greatly impact the estimated ancestral locations. One way to mitigate sampling bias is through the incorporation of available travel history information from infected individuals. Travel history data can be used to correct for gaps in sampling by allowing for ancestral nodes to be in a given location even when molecular sequence data for that location are not available.
This protocol explains how augment a phylogeographic analysis generated in BEAUti by incorporating individual travel history data. In this example, we will use the XML file generated in Protocol 2 and modify it to include the available travel history data (See [Lemey et al., 2020] for more detailed information). Importantly, BEAST requires the high-performance BEAGLE library [Ayres et al., 2019] to be installed in order to optimize computational performance on a variety of hardware resources.
Necessary Resources
Hardware
Standard computer running Linux, MacOS, or Windows.
A CUDA- or OpenCL-compatible GPU is optional, but recommended for speeding up the analyses
Software
Python v3.6+ with packages numpy and lxml BEAGLE v3+
Latest BEAST jar file (v1.10.5) (provided with this protocol)
add_travel_history.py Python script (provided with this protocol)
Files
XML file for a phylogeographic analysis (using a GLM parameterization) set up in BEAUti Travel history metadata CSV file Augmented covariate data files
Example files are provided in the repository https://github.com/hongsamL/travHistProtocol/tree/main/files/Protocol3.
Note: The example in this protocol assumes an XML file for a phylogeographic GLM reconstruction with new travel history locations not present in the original BEAST XML file generated by BEAUti. Covariate files augmented to include the new locations are thus required to accommodate for the increase in location state space.
Protocol steps and annotations
-
Update the BEAST XML file to incorporate travel history data
python add_travel_history.py --xml 282_GISAID_sarscov2.xml
--hist travel_metadata.csv
--covariate augmented_flight_matrix.csv
--covariate augmented_intra_cont_dist.csv
--out 282_GISAID_sarscov2_travelHist.xml
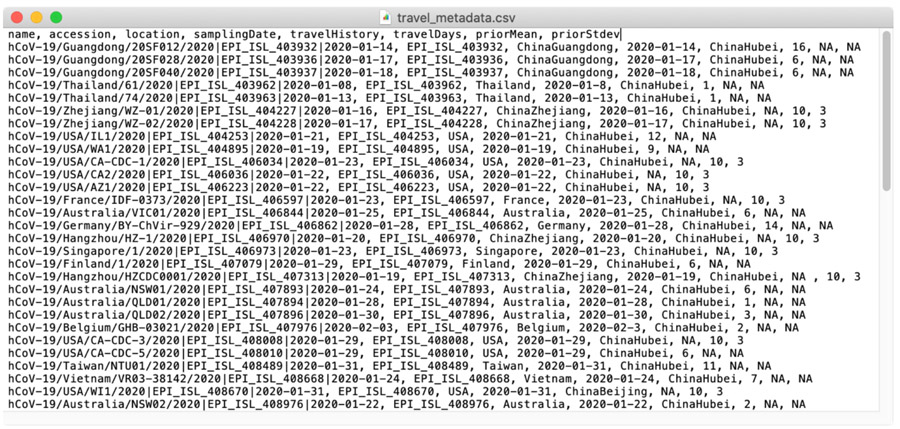
Note: Although this example uses a phylogeographic GLM analysis, the add_travel_history.py script also works for standard discrete phylogeographic models using symmetric and asymmetric CTMC parametrizations. For such cases, run the same command without the --covariate flags. This script also requires for the travel history metadata to follow a specific format (Figure 5). The metadata file is in CSV format and must contain the following columns: “name” (taxon name), “travelHistory” (travel location), “travelDays” (date of travel as days before to sampling date), “priorMean” and “priorStdev” (prior specifications for the mean and standard deviation of a normal prior on travel dates when exact data are unavailable).
-
Run the updated XML file using BEAST
java -cp beast.jar dr.app.beast.BeastMain -seed 2020
-beagle_double
-beagle_gpu
-save_every 1000000
-save_state travelHist.checkpoint
282_GISAID_sarscov2_travelHist.xml
Note: Here, we run BEAST on the command line using the latest build of BEAST. You can create your own beast.jar file by checking out and compiling the main branch of the beast-mcmc GitHub repository. We specify a starting seed with the -seed flag, and use the -beagle gpu flag to accelerate the likelihood computations using a graphics processing unit (GPU) (only applicable if you have a powerful GPU with sufficient double precision – or FP64 – compute performance available). This option is recommended when available, as using a GPU reduces runtime by accelerating likelihood computations when performing phylogeographical analyses on large datasets. Be sure to check the technical specifications of the GPU you want to use; ideally, at least 3 TFLOPS FP64 performance is recommend. We also take advantage of the BEAST checkpointing functionality [Gill et al., 2020] to save a snapshot of the MCMC run into travelHist. checkpoint every 1,000,000 states. This allows us to resume the analysis from the checkpoint in case the run becomes interrupted, or more iterations are required than initially anticipated.
Figure 5:
Travel history metadata. The metadata file must contain the columns “name”, “travelHistory”, “travelDays”, “priorMean” and “priorStdev”. Other columns can be included but will not be parsed to update the XML. For sequences where either exact travel dates are not available, we set the “travelDays” column to NA, such that the MCMC samples from a range of possible travel dates with a Gaussian prior distribution of mean “priorMean” (in units of days) and standard deviation “priorStdev”. For sequences where exact travel dates are available, we set the prior columns to NA.
PROTOCOL 4: VISUALIZING ANCESTRAL SPATIAL TRAJECTORIES FOR SPECIFIC TAXA
Necessary Resources
Hardware
Standard computer running Linux, MacOS, or Windows.
Software
Latest BEAST jar file (v1.10.5) (provided with this protocol)
R with package MarkovJumpR (https://github.com/beast-dev/MarkovJumpR)
Files
Trees output with Markov jump annotations from a BEAST phylogeographic analysis with travel history
Example .trees file provided in the repository https://github.com/hongsamL/travHistProtocol/tree/main/files/Protocol4
Protocol steps and annotations
-
Extract all Markov jump histories for an isolate of interest
java -cp beast.jar dr.app.tools.TaxaMarkovJumpHistoryAnalyzer
-taxaToProcess "hCoV-19/Brazil/SP-02/2020∣EPI_ISL_413016∣2020-02-28"
-StateAnnotation location
-burnin 100
-msrd 2020.1748633879781
282_GISAID_GLM.location.history.trees EPI_ISL_412975_MJhist.csv
Note: The BEAST jar file packages a number of standalone applications that can be accessed by using the -cp flag when calling Java from the command line. Here, we use the TaxaMarkovJumpHistoryAnalyzer application to extract all Markov jump histories for isolate EPIJSL 413016, into a CSV file. This application takes a trees file with complete Markov jump or state change history as an input (which is being generated by running the XML constructed in the protocol), and outputs the posterior set of spatial trajectories for a taxon or selection of taxa. We specify the desired taxon labels through the -taxaToProcess flag and specify the annotation name of the discrete trait that was reconstructed using -StateAnnotation. We can also remove a number of trees corresponding to the burn-in using the -burnin flag and scale the output results to reflect chronological time instead of node heights using the -msrd flag by specifying the most recent sampling date. An example output file can be found in https://github.com/hongsamL/travHistProtocol/tree/main/files/Protocol4
-
Load spatial trajectories into R.
library(MarkovJumpR)
spatial_paths <- loadPaths(fileName = "EPI_ISL_413016_MJhist.csv")
-
Inspect spatial trajectories reconstructed spatial_paths$minTime yields the earliest time along a spatial path across all trees 2019.892. To look at the frequency of locations visited across all spatial paths we type:
loc_freq <- table(spatial_paths$paths$location)
loc_freq[order(loc_freq,decreasing = T)]
which yields a frequency table of locations in descending order (Table 1). In this example, we see that Italy, Brazil, ChinaHubei and Switzerland appear most commonly across the spatial paths.
-
Set up plot colors.
We here specify four colors of choice corresponding to the four locations of interest that make up the spatial trajectory of isolate EPI_ISL_413016.
locations <- c("ChinaHubei", "Italy", "Brazil", "Switzerland")locationColors <-c("#E3272F", "#31B186", "#931ECF", "#C695BD")locationMap <- data.frame(location = locations,
position = c(1, 2, 3, 4))
locationMap$color <- sapply(locationColors,as.character)
-
Set up plot labels
dateLabels <- c("01-Dec-19", "15-Dec-19", "01-Jan-20", "15-Jan-20","01-Feb-20", "15-Feb-20", "01-Mar-20")
-
Plot path spatial trajectories
plotPaths(travelHistPaths$paths, locationMap = locationMap,
yJitterSd = 0.1, alpha = 0.1, minTime = spatial_paths$minTime,
addLocationLine = TRUE,
xAt = decimal_date(dmy(dateLabels)),
xLabels = dateLabels,
mustDisplayAllLocations = TRUE)
Table 1.
Frequency Table of Locations
| Italy | Brazil | ChinaHubei | Switzerland | Finland |
| 444 | 437 | 436 | 85 | 23 |
| ChinaBeijing | UK | Australia | Germany | USA |
| 9 | 8 | 7 | 6 | 6 |
| France | Singapore | Spain | ChinaHongKong | Japan |
| 5 | 5 | 5 | 4 | 3 |
| Netherlands | Sweden | Vietnam | ChinaGuangdong | ChinaShandong |
| 3 | 3 | 3 | 2 | 2 |
| NewZealand | Thailand | Belgium | Cambodia | Canada |
| 2 | 2 | 1 | 1 | 1 |
| ChinaChongqing | ChinaFujian | ChinaYunnan | India | Iran |
| 1 | 1 | 1 | 1 | 1 |
| Mexico | Nepal | Portugal | SouthKorea | Taiwan |
| 1 | 1 | 1 | 1 | 1 |
GUIDELINES FOR INTERPRETING RESULTS
The BEAST software package offers a flexible approach for combining demographic, molecular clock, nucleotide and trait evolution models to infer time-scaled trait-annotated phylogenies. BEAST employs Bayesian inference through MCMC to sample trees and all of the model parameters from the joint density (often simply called the posterior). Protocols 2 and 3 show how to set up the different models and run the corresponding BEAST analyses to collect samples from the posterior. The phylogenetic trees that are sampled from the posterior are stored in a .trees file and samples of the model parameters are stored in a .log file. Here we present some of the standard applications that are commonly used to interpret the output that BEAST generates.
Assessing convergence
The MCMC sampling strategy is to construct a Markov chain that (eventually) converges to a stationary distribution, which is the joint density in the case of Bayesian inference. For complex models and data sets, it may take considerable time for a chain to converge. We can visually assess the convergence of a BEAST run by inspecting the sampled parameter values across an MCMC analysis. To do so, we can load a .log file into Tracer (https://beast.community/tracer, [Rambaut et al., 2018]), and visually inspect the trace plot, which shows a time series of the parameter values sampled throughout the analysis. A detailed guide on how to use Tracer can be found at https://beast.community/tracer_convergence.html
Effective sample size and parameter estimates
A characteristic of inference through MCMC is that the samples collected tend to be correlated. This in turn poses a challenge, since having a large number of samples does not guarantee a considerable reduction of the uncertainty in our posterior estimates. A way to control for this is to look at the effective sample size (ESS) value associated with parameter estimation. The ESS of a parameter sampled from an MCMC method is the number of effectively independent draws from the posterior distribution that the Markov chain is equivalent to. ESS values are expected to increase as an increasing number of samples are collected. Higher ESS values will result in more precise posterior estimates but require higher computational resources. Loading a .log file in Tracer will also show the ESS values and estimates for each model parameter. Note that ESS values are only defined for continuous parameters that are being estimated. Tracer will automatically calculate the ESS for all parameters part of the log file, and flag values above 100 and 200 (preferable).
Summarizing trees and phylogeographic estimates
Individually inspecting every tree from the posterior obviously constitutes an impractical way to interpret the MCMC results. We are thus required to summarize the distribution of trees sampled as a point estimate with associated uncertainties. The TreeAnnotator application in BEAST enables creating a maximum clade credibility (MCC) tree to summarize the sampled trees for this purpose. For every tree in the distribution, a posterior clade probability for each node (i.e., the support for a node) is calculated by computing the frequency of the clustering that is defined by the relevant node. The MCC tree is then defined as the tree that maximizes the product of the posterior clade probabilities across the tree. Instructions on how to use TreeAnnotator can be found at http://beast.community/second_tutorial.
In some data sets, the posterior support for all nodes in a tree is such that many clusters in sampled topologies do not end up represented in the MCC tree. When that is the case, a point estimate of a tree is unable to capture the diverse phylogeographic histories compatible with the data. Protocol 4 allows us to inspect individual spatial trajectories by summarizing across all possible phylogenetic ancestries in the joint density. This was proposed as a visual summary for SARS-CoV-2 phylogeographic inferences that are poorly resolved. Each spatial trajectory in the joint density is represented by a stepwise curve, where vertical lines represent transitions between two locations, and horizontal lines time intervals during which the pathogen remains in the same location. The relative density of lines reflects the posterior uncertainty in spatiotemporal ancestry.
COMMENTARY
Background Information
Modern models for phylogeographic inference can be broadly categorized into two classes or types, depending on the assumptions used to model the spatial spread of a pathogen. The first type consists of structured coalescent approaches that model movement in terms of a migration matrix and relate migration rates with population sizes in a location and therefore involve population size dynamics estimates for each location [De Maio et al., 2015, Müller et al.,2018]. Under this type of approach, the shape and structure of the genealogy (i.e., the pattern of time intervals between coalescent, migration and sampling events) will inform the geographic reconstruction. The second type consists of diffusion approaches that consider sampling locations as observed traits independent from the tree-generative process, and model movement across space using a random walk process [Lemey et al., 2009, Lemey et al., 2010]. Structured coalescent models are more robust to sampling bias, but due to the difficulty to scale this approach to larger datasets diffusion methods remain popular. Currently, BEAST v1.10.5 focuses on providing phylogeographic inference using CTMC diffusion models for discrete location data.
Diffusion models for phylogeographic inference can be further categorized depending on the location data type used. Discrete locations are parameterized using the same CTMC models as used for the sequence substitution process [Lemey et al., 2009]. In contrast, continuous locations are modeled using Brownian diffusion-based random walk models [Lemey et al., 2010]. Much of the genomic data collected has a spatial resolution coarser than latitude and longitude, which restricts phylogeographic applications to the discrete approach.
The CTMC parameterization models movement between K discrete locations in terms of a K x K infinitesimal rate matrix Λ, where Λij is the instantaneous movement rate from location i to j. Parameterization is available for both symmetrical and asymmetrical transition rates, and extensions adopt Bayesian Stochastic Search Variable Selection (BSSVS) to limit the number of rates to only those that adequately explain the phylogeographic diffusion process. The GLM parameterization models the transition rates as a log linear combination of P of potential explanatory predictors (xij1, … , xijP), with corresponding coefficients (β1, …, βP) and indicator variables (δ1, …, δP) such that:
| Equation 1 |
This model specification allows us to use BSSVS to explore the space of 2P predictor combinations and obtain a posterior probability on the indicator variables in order to determine the support for inclusion of each predictor in the model.
Incorporating individual travel history data does not require the use of a GLM and can be used with standard CTMC models [Lemey et al., 2009]) by augmenting the available dataset to include ancestral nodes associated with a known state but not necessarily with a known sequence [Lemey et al., 2020]. This provides a richer source of information for phylogeographic reconstructions as compared to only using sampling location. Ambiguous ancestral locations can also be allowed by integrating over all possible locations with equal or user specified weights [Scotch et al., 2019]. An example would be the case where an individual traveled to multiple locations prior to being sampled. Given that the individual may have become infected in any of the visited countries, we can integrate this uncertainty by marginalizing over all possible locations for the unsampled ancestor when performing phylogeographic inference.
CRITICAL PARAMETERS AND TROUBLESHOOTING
A critical assumption for any BEAST analysis is that the data set under consideration constitutes a sample from a measurably evolving population (MEP). MEPs [Drummond et al., 2003, Biek et al., 2015] refer to time-stamped sequence data where a sufficient amount of molecular evolution has occurred throughout the sampling period to establish a statistical relationship between genetic divergence and time. A data set conforming to this criterion is said to contain sufficient temporal signal. A lack of temporal signal may result in poor behavior and unreliable divergence time estimates. Popular ways to assess temporal signal include explorations through root-to-tip regression of genetic divergence and sampling times based on maximum likelihood trees [Rambaut et al., 2016] and permuting tip date labels through date-randomization [Ramsden et al., 2009]. Recently, a formal way to assess temporal signal has been developed in a Bayesian framework using model comparison through Bayes factors [Duchene et al., 2020]. In cases where the temporal signal is deemed insufficiently strong, one can resort to adding more data to increase temporal coverage or using prior knowledge to inform the molecular clock rate or specific divergence times.
Another commonly encountered issue is that of low ESS values for parameters relevant to the analysis. At the end of a BEAST analysis, some parameters may have much higher ESS values associated to them compared to others. One way to increase the ESS values of a parameter is to increase the weight of the relevant operator in order to increase the sampling frequency of the (problematic) parameter (e.g., Protocol 2, step 9). Other ways to obtain more samples – and as a result increase the ESS value – include increasing the MCMC chain length and combining the output of multiple independent BEAST analyses, i.e., analyzing the same XML file using BEAST but with different starting seeds.
Figure 6:
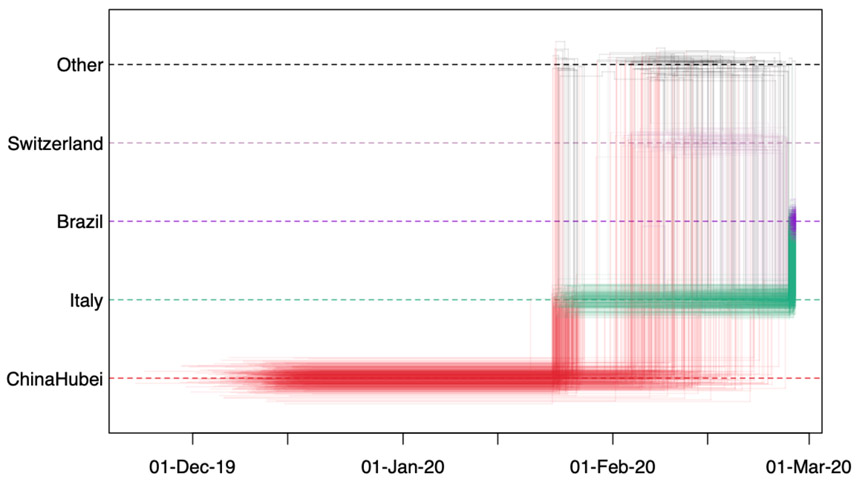
Spatial trajectory plot of isolate EPI_ISL_413016. The plot depicts the posterior spatiotemporal ancestral transition history for a single isolate. Each line represents a single Markov jump history in the posterior distribution. The time spent in each location is denoted in the horizontal dimension, and transitions between two locations are depicted with vertical lines. The relative density of lines reflects the posterior uncertainty in location state and transition time between states. Here we see that the most supported ancestral history for isolate EPI_ISL_413016 is that of an origin in Hubei, with a jump into Italy late January, and an introduction into Brazil close to March 1st.
ACKNOWLEDGEMENTS
The research leading to these results has received funding from the European Research Council under the European Union’s Horizon 2020 research and innovation programme (grant agreement no. 725422-ReservoirDOCS) and from the European Union's Horizon 2020 project MOOD (grant agreement no. 874850). The Artic Network receives funding from the Wellcome Trust through project 206298/Z/17/Z. PL acknowledges support by the Research Foundation - Flanders (‘Fonds voor Wetenschappelijk Onderzoek - Vlaanderen’, G066215N, G0D5117N and G0B9317N). GB acknowledges support from the Interne Fondsen KU Leuven / Internal Funds KU Leuven under grant agreement C14/18/094, and the Research Foundation – Flanders (‘Fonds voor Wetenschappelijk Onderzoek - Vlaanderen’, G0E1420N, G098321N). MAS acknowledges support from the National Institutes of Health grant U19 AI135995 and R01 AI153044.
References
- [Ayres et al. , 2019]Ayres DL, Cummings MP, Baele G, Darling AE, Lewis PO, Swofford DL, Huelsenbeck JP, Lemey P, Rambaut A, and Suchard MA (2019). BEAGLE 3: Improved performance, scaling, and usability for a high-performance computing library for statistical phylogenetics. Syst. Biol 68(6):1052–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [Biek et al. , 2015]Biek R, Pybus OG, Lloyd-Smith JO, and Didelot X (2015). Measurably evolving pathogens in the genomic era. Trends Ecol. Evol, 30(6):306–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [De Maio et al. , 2015] De Maio N, Wu C-H, O’Reilly KM, and Wilson D (2015). New routes to phylogeography: A Bayesian structured coalescent approximation. PLoS Genet., 11(8):e1005421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [Drummond et al. , 2003] Drummond AJ, Pybus OG, Rambaut A, Forsberg R, and Rodrigo AG (2003). Measurably evolving populations. Trends Ecol. Evol, 18(9):481–488. [Google Scholar]
- [Drummond and Rambaut, 2007] Drummond AJ and Rambaut A (2007). BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol, 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [Drummond et al. , 2012] Drummond AJ, Suchard MA, Xie D, and Rambaut A (2012). Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol, 29(8):1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [Duchene et al. , 2020] Duchene S, Lemey P, Stadler T, Ho SYW, Duchene DA, Dhanasekaran V, and Baele G (2020). Bayesian evaluation of temporal signal in measurably evolving populations. Mol. Biol. Evol [DOI] [PMC free article] [PubMed] [Google Scholar]
- [Elbe and Buckland-Merrett, 2017] Elbe S and Buckland-Merrett G (2017). Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob Chall, 1(1):33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [Gill et al. , 2020] Gill MS, Lemey P, Suchard MA, Rambaut A, and Baele G (2020). Online Bayesian phylodynamic inference in BEAST with application to epidemic reconstruction. Mol. Biol. Evol, 37(6):1832–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [Hill and Baele, 2019] Hill V and Baele G (2019). Bayesian estimation of past population dynamics in BEAST 1.10 using the skygrid coalescent model. Mol. Biol. Evol [DOI] [PMC free article] [PubMed] [Google Scholar]
- [Katoh and Standley, 2013] Katoh K and Standley DM (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol, 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [Larsson, 2014] Larsson A (2014). AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics, 30(22):3276–3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [Lemey et al. , 2020] Lemey P, Hong SL, Hill V, Baele G, Poletto C, Colizza V, O’Toole Á, McCrone JT, Andersen KG, Worobey M, Nelson MI, Rambaut A, Suchard MA (2020). Accommodating individual travel history and unsampled diversity in Bayesian phylogeographic inference of SARS-CoV-2. Nat. Commun, 11(1):5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [Lemey et al. , 2014] Lemey P, Rambaut A, Bedford T, Faria N, Bielejec F, Baele G, Russell CA, Smith DJ, Pybus OG, Brockmann D, and Suchard MA (2014). Unifying viral genetics and human transportation data to predict the global transmission dynamics of human influenza H3N2. PLoS Pathog., 10(2):e1003932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [Lemey et al. , 2009] Lemey P, Rambaut A, Drummond AJ, and Suchard MA (2009). Bayesian phylogeography finds its roots. PLoS Comput. Biol, 5(9):e1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [Lemey et al. , 2010] Lemey P, Rambaut A, Welch JJ, and Suchard MA (2010). Phylogeography takes a relaxed random walk in continuous space and time. Mol. Biol. Evol, 27(8):1877–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [Müller et al. , 2018] Müller NF, Rasmussen D, and Stadler T (2018). MASCOT: parameter and state inference under the marginal structured coalescent approximation. Bioinformatics, 34(22):3843–3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [Rambaut et al. , 2018] Rambaut A, Drummond AJ, Xie D, Baele G, and Suchard MA (2018). Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol, 67(5):901–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [Rambaut et al. , 2016] Rambaut A, Lam TT, Max Carvalho L, and Pybus OG (2016). Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol, 2(1):vew007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [Ramsden et al. , 2009] Ramsden C, Holmes EC, and Charleston MA (2009). Hantavirus evolution in relation to its rodent and insectivore hosts: no and evidence for codivergence. Mol. Biol. Evol, 26(1):143–153. [DOI] [PubMed] [Google Scholar]
- [Scotch et al. , 2019] Scotch M, Tahsin T, Weissenbacher D, O’Connor K, Magge A, Vaiente M, Suchard MA, and Gonzalez-Hernandez G (2019). Incorporating sampling uncertainty in the geospatial assignment of taxa for virus phylogeography. Virus Evol, 5(1):vey043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [Suchard et al. , 2018] Suchard MA, Lemey P, Baele G, Ayres DL, Drummond AJ, and Rambaut A (2018). Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol, 4(1):vey016. [DOI] [PMC free article] [PubMed] [Google Scholar]