Abstract
Ligand-dependent exchange of coactivators and corepressors is the fundamental regulator of nuclear hormone receptor (NHR) function. The interaction surfaces of coactivators and corepressors are similar but distinct enough to allow the ligand to function as a switch. Multiple NHRs share features that allow corepressor binding, and each of two distinct corepressors (N-CoR and SMRT) contains two similar CoRNR motifs that interact with NHRs. Here we report that the specificity of corepressor-NHR interaction is determined by the individual NHR interacting with specific CoRNR boxes within a preferred corepressor. First, receptors have distinct preferences for CoRNR1 versus CoRNR2. For example, the retinoic acid receptor binds CoRNR1, while RXR interacts almost exclusively with CoRNR2. Second, the NHR preference for N-CoR or SMRT is due to differences in CoRNR1 but not CoRNR2. Moreover, within a single corepressor, affinity for different NHRs is determined by distinct regions flanking CoRNR1. The highly specific determinants of NHR-corepressor interaction and preference suggest that repression is regulated by the permissibility of selected receptor-CoRNR-corepressor combinations. Interestingly, different NHR surfaces contribute to binding of CoRNR1 and CoRNR2, suggesting a model to explain corepressor binding to NHR heterodimers.
Nuclear hormone receptors (NHRs) regulate transcription in response to hormones. Binding of hormones to NHRs recruits coactivator protein complexes that contain histone-modifying activity and/or components that interact with initiation complexes (reviewed in reference 12). In the absence of their cognate hormones, NHRs such as the thyroid hormone receptor (TR) and retinoic acid receptor (RAR) actively repress transcription by binding to corepressor protein N-CoR or SMRT (5, 18). NHRs are a small group of receptors within a larger superfamily. Members of this family without an identified ligand are called orphan receptors (reviewed in reference 27). Some of the orphan receptors, such as COUP-TF (38) and RevErb (41), also repress transcription.
N-CoR and SMRT are large modular proteins that contain separable N-terminal repression domains and C-terminal interaction domains. The N-terminal repression domains interact with histone deacetylase complexes and a transducin β-like protein that interacts with histones (15, 17, 21, 23, 24, 29). The C-terminal domain contains two interaction domains, ID1 and ID2 (7, 36, 41). We and others have found motifs called CoRNR (corepressor-nuclear receptor) boxes within ID1 and -2 that are responsible for interaction with NHRs (19, 30, 32). Based on the stoichiometry of corepressor binding, it is likely that each of these CoRNR boxes interacts with a single NHR in a DNA-bound dimer (7, 42). Each CoRNR box contains the motif 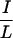 XX
XX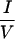 I, which is very similar to the LXXLL motifs (NR box) found in coactivator proteins (16). The crystal structures of several NHR ligand binding domains (LBDs) and associated coactivator LXXLL motif peptides have been solved (2, 8, 10, 31). The overall LBD contains 12 α-helices (H1 to H12). The binding surface for the coactivator peptide is formed by H3, H4, part of H5, and H12. The position of H12 is regulated by a ligand. In the liganded receptor, H12 folds back to form part of the coactivator-binding surface. By contrast, H12 inhibits corepressor binding to RXR and other NHRs (35, 43). The corepressor interaction surface does require H3, H4, and H5, thereby overlapping the coactivator interaction surface (19, 30, 32).
I, which is very similar to the LXXLL motifs (NR box) found in coactivator proteins (16). The crystal structures of several NHR ligand binding domains (LBDs) and associated coactivator LXXLL motif peptides have been solved (2, 8, 10, 31). The overall LBD contains 12 α-helices (H1 to H12). The binding surface for the coactivator peptide is formed by H3, H4, part of H5, and H12. The position of H12 is regulated by a ligand. In the liganded receptor, H12 folds back to form part of the coactivator-binding surface. By contrast, H12 inhibits corepressor binding to RXR and other NHRs (35, 43). The corepressor interaction surface does require H3, H4, and H5, thereby overlapping the coactivator interaction surface (19, 30, 32).
Repression by nuclear receptors plays very important roles in many biological processes and also plays a role in the mechanism of several diseases, including acute promyelocytic leukemia (APL) (13, 14, 25, 26). The most common type of APL is caused by fusion of the promyelocytic leukemia protein to RARα. Treatment of APL patients with all-trans retinoic acid, an RAR ligand, leads to complete remission (22). However, patients often relapse with RA-resistant disease due to mutations in the RAR moiety that prevent corepressor dissociation (reviewed in reference 28). The discovery that small CoRNR peptides block the interaction between NHRs and corepressors suggests an alternative way to treat this disease. However, N-CoR and SMRT are widely expressed and interact with many NHRs. To specifically block interaction between corepressors and RARs, we must understand in detail how corepressors interact with specific NHRs.
Here we have investigated the receptor, corepressor, and CoRNR specificity of a repression complex assembled on nuclear receptors. We have found that NHRs preferentially interact with one or the other CoRNR box and sometimes within a favored corepressor. CoRNR1 and CoRNR2 interactions bind in different positions to the coregulator-binding surface, mediated by distinct regions of different NHRs. In the case of CoRNR1, particular residues are required to interact with specific NHRs. These results suggest remarkable specificity in the permissibility of functional NHR-CoRNR-corepressor combinations. We propose a model to explain the recruitment of the two CoRNR boxes to the NHR heterodimer.
MATERIALS AND METHODS
Plasmids.
Nuclear receptor and corepressor constructs were created by PCR, endonuclease restriction digestion, Quick-change mutagenesis (Stratagene), or a combination of these techniques followed by ligation. All constructs were verified by sequencing. All receptor and corepressor expression constructs are in the CMX vector. The receptor constructs contain only the ligand binding domain. The luciferase reporter has five copies of Gal4 binding sites in front of the simian virus 40 (SV40) promoter. A detailed description of each construct is included in figure legends.
Interaction assay.
A mammalian two-hybrid (transfection and luciferase reporter) assay and glutathione S-transferase (GST) pull-down assay were performed as described previously (21). All mammalian two-hybrid experiments were done with 293T cells. Peptides were dissolved in dimethyl sulfoxide.
Gel shift assay.
Gel mobility shift assays were performed with proteins derived from an in vitro translation reaction. RevErb and a 5× concentration of Gal-N-CoR and Gal-SMRT fusion proteins were used. A RevDR2 containing a restriction fragment was labeled by 32P and used as a probe. Incubation was carried out at room temperature for 1.5 h with a binding buffer (20 mM Hepes [pH 7.5], 100 mM KCl, 7.5% glycerol, 2 mM dithiothreitol), and poly(dI-dC) (2 μg), with or without antibody (anti-Gal4–DNA-binding domain or anti-RevErb). The resulting complexes were resolved on a 4% nondenaturing gel, followed by autoradiography.
ChIP assay.
A chromatin immunoprecipitation (ChIP) assay was performed according to the protocols from Upstate Biotechnology with minor modifications. Corepressor, receptor, and reporter plasmids were transfected into 293T cells. Twenty-four hours after transfection, cells were cross-linked and harvested. Immunoprecipitations were conducted and DNA was purified. PCR was performed using two primers flanking the RevErb response element.
RESULTS
CoRNR and corepressor specificity of NHRs.
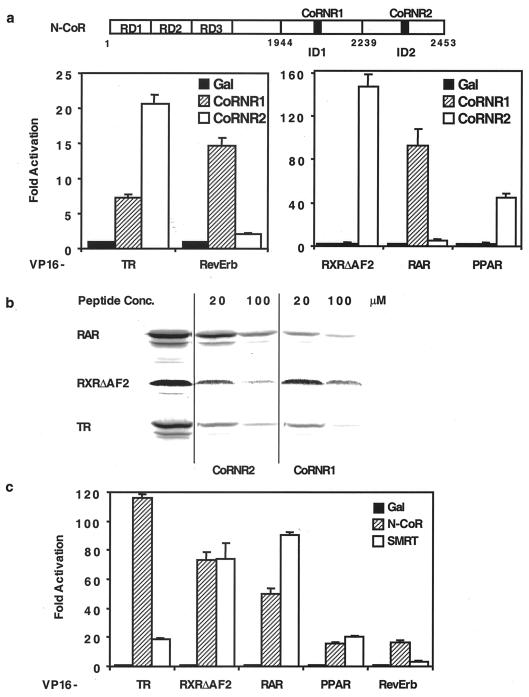
We previously identified the CoRNR boxes and showed that TR can bind both CoRNR1 and CoRNR2, while RXR interacts exclusively with CoRNR2 (19). To extend our knowledge to other receptors, we tested RAR, PPARγ, and RevErb in a standard mammalian two-hybrid protein-protein interaction assay. As shown in Fig. 1a, RAR interacts strongly with CoRNR1 and minimally with CoRNR2. PPAR interacts primarily with CoRNR2. By contrast, RevErb interacts exclusively with CoRNR1. To confirm these observations of CoRNR specificity, we tested the ability of CoRNR peptides to block interaction between NHRs and a GST fusion of the N-CoR interaction domain. As expected, CoRNR1 blocked most of the interaction between RAR and N-CoR at 20 and 100 μM, whereas CoRNR2 had less effect (Fig. 1b). By contrast, CoRNR2 peptides but not CoRNR1 peptides blocked RXR interaction with N-CoR (Fig. 1b). Also consistent with the in vivo interaction studies, both CoRNR1 and CoRNR2 peptides efficiently blocked TR interaction with corepressors.
FIG. 1.
CoRNR and corepressor specificity of NHRs. (a) Top: schematic diagram of N-CoR domains. RD, repression domains; ID, interaction domains. Bottom: in vivo assay of CoRNR specificity of NHRs. Gal4 fusion of the CoRNR peptide was tested for interaction with VP16 fusion of NHRs in a mammalian two-hybrid assay. (b) In vitro assay of CoRNR specificity of NHRs. Peptides were used in a GST pull-down assay to block interaction between NHR and GST–N-CoR (amino acids 1944 to 2453). The CoRNR1 peptide has 30 residues (mN-CoR, 2057 to 2086), and CoRNR2 has 14 (mN-CoR, 2274 to 2287), as shown in Fig. 3a and 2, respectively. (c) Corepressor specificity of NHRs. The interaction domains of N-CoR or SMRT were fused to Gal4 DBD and tested for interaction with NHRs.
We next assessed whether different NHRs preferentially bind to corepressors using Gal4 fusion to either the N-CoR or SMRT ID in the mammalian two-hybrid interaction assay. As shown in Fig. 1c, RXR and PPARγ showed no preference between N-CoR and SMRT. By contrast, TR and RevErb have much stronger binding to N-CoR while RAR prefers SMRT. Consistent with these results, Cohen et al. recently showed that RAR binds more strongly to SMRT in a gel shift assay (6).
CoRNR2 peptides of N-CoR and SMRT interact indistinguishably with NHRs.
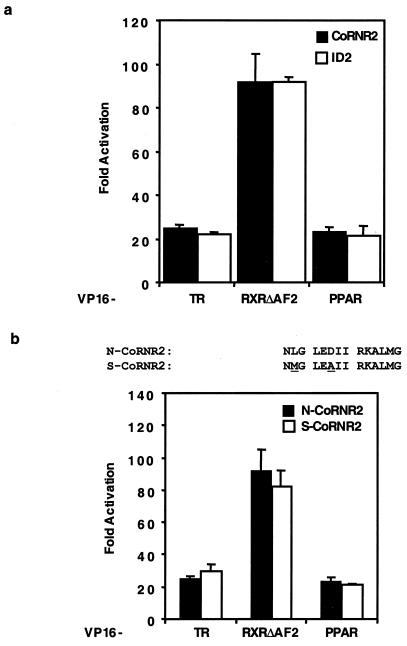
To discover what feature within the corepressor determines the specificity of NHR interaction, we first looked at the extremely C-terminal ID2. We have previously shown that only 14 amino acids of N-CoR CoRNR2 are sufficient to interact with TR and RXR. As shown in Fig. 2a, these 14 amino acids recapitulate the interactions of the entire ID2 polypeptide (around 200 amino acids) in a mammalian two-hybrid assay. These 14 amino acids are highly conserved between N-CoR and SMRT, with only 2 nonidentical residues, suggesting that CoRNR2 does not mediate corepressor binding specificity. Indeed, 14-residue CoRNR2 peptides from N-CoR and SMRT interact equally well with NHRs (Fig. 2b).
FIG. 2.
Fourteen-amino-acid CoRNR2 from either N-CoR or SMRT interacts efficiently with NHRs. (a) The 14-amino-acid CoRNR2 polypeptide interacts efficiently with NHRs in a mammalian two-hybrid assay. Gal4 fusion of CoRNR2 (N-CoR, amino acids 2274 to 2287) or ID2 (N-CoR, amino acids 2239 to 2453) was tested for interaction with VP16 fusion of NHRs. (b) Top: sequences of N-CoR CoRNR2 and SMRT CoRNR2. Bottom: N-CoRNR2 and S-CoRNR2 interact equally well with NHRs. S-CoRNR2 contains amino acids 2339 to 2352 of hSMRT.
Different residues in ID1 are required for interaction with different NHRs.
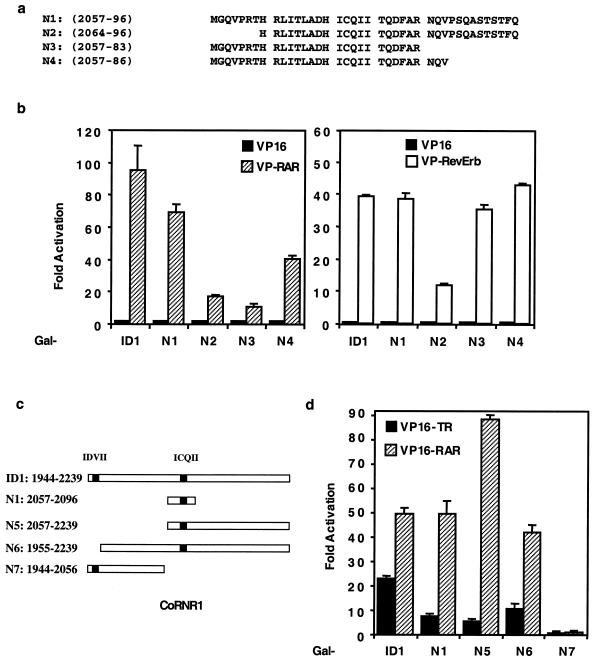
The above result suggests that ID1 or CoRNR1 contains determinants for corepressor specificity. As shown in Fig. 1a, the 40-residue CoRNR1 polypeptide is sufficient to interact with TR, RAR, and RevErb. To determine which residues in this CoRNR1 peptide are important for interaction with these different NHRs, we made further deletions in this region (Fig. 3a). RAR and RevErb interact equally well with either the entire ID1 or the 40-residue CoRNR1 (N1) in a mammalian two-hybrid assay (Fig. 3b), suggesting that determinants for RAR and RevErb interaction lie within these 40 residues. Deletion of the N-terminal residues (N2) decreased interaction with RAR and RevErb, whereas deletion of the C-terminal residues (N3 and N4) specifically reduced interaction with RAR but not RevErb (Fig. 3b). These deletion constructs were expressed at similar levels (data not shown). The above results suggest that both N- and C-terminal residues of CoRNR1 are required for efficient interaction with RAR, whereas only N-terminal residues are required for RevErb interaction.
FIG. 3.
Different residues in N-CoR ID1 are involved in interaction with different NHRs. (a) CoRNR deletion constructs. (b) Interaction between deletion constructs and VP16-RAR (left) or VP16-RevErb (right) in a mammalian two-hybrid assay. ID1, N-CoR (amino acids 1944 to 2239). (c) Schematic diagram of the ID1 deletions. The two 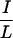 XX
XX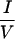 I motifs are shown as black. (d) TR and RAR interaction with these deletions in a mammalian two-hybrid assay.
I motifs are shown as black. (d) TR and RAR interaction with these deletions in a mammalian two-hybrid assay.
The 40-residue CoRNR1 interacts with TR very weakly (Fig. 1a). By contrast, the entire ID1 has much stronger interaction with TR, suggesting that residues outside the CoRNR1 region contribute to TR interaction. We therefore made a series of deletions in ID1 (Fig. 3c). Deleting the N terminus of ID1 (N5 in Fig. 3c) resulted in a decrease in TR interaction in a mammalian two-hybrid assay (Fig. 3d). Inspection of the deleted region revealed an 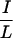 XX
XX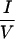 I motif (IDVII) six amino acids downstream of the extreme N terminus (Fig. 3c). Deletion of 11 residues containing this motif reduced TR interaction without affecting RAR interaction (Fig. 3d; compare ID1 with N6). Although this 11-residue
I motif (IDVII) six amino acids downstream of the extreme N terminus (Fig. 3c). Deletion of 11 residues containing this motif reduced TR interaction without affecting RAR interaction (Fig. 3d; compare ID1 with N6). Although this 11-residue 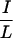 XX
XX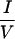 I-containing fragment was required for optimal TR interaction, a fragment containing this motif (N7) was not sufficient to interact with TR or RAR. Interestingly, this IDVII motif is not conserved in SMRT. Together, these results suggest that CoRNR1 is the major interaction surface for TR, with the N-terminal motif facilitating optimal binding to TR but not to other receptors.
I-containing fragment was required for optimal TR interaction, a fragment containing this motif (N7) was not sufficient to interact with TR or RAR. Interestingly, this IDVII motif is not conserved in SMRT. Together, these results suggest that CoRNR1 is the major interaction surface for TR, with the N-terminal motif facilitating optimal binding to TR but not to other receptors.
CoRNR1 contains the determinants for corepressor specificity.
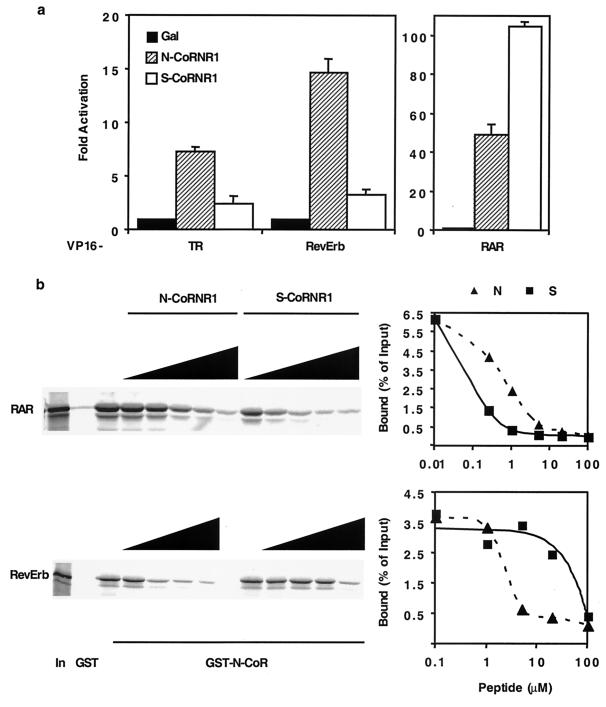
Thus far we have shown that corepressor specificity is not determined by CoRNR2 (Fig. 2) and that different regions flanking CoRNR1 specify NHR binding (Fig. 3). We hypothesized that CoRNR1 also determines NHR preferences for N-CoR versus SMRT; i.e., RAR prefers SMRT while TR and RevErb prefer N-CoR (Fig. 1). To test this, we studied the 40-residue SMRT CoRNR1 (S-CoRNR1) according to the homology alignment. As shown in Fig. 4a, S-CoRNR1 has weaker interaction with TR and RevErb than with RAR, thus recapitulating the corepressor specificity. Again, to confirm the in vivo mammalian two-hybrid result, we used peptides to block the interaction between receptor and N-CoR in the GST pull-down assay. S-CoRNR1 peptide blocks RAR and N-CoR interaction more efficiently than N-CoRNR1 (Fig. 4b). In contrast, N-CoRNR1 is much more effective than S-CoRNR1 in competing for corepressor binding to RevErb (Fig. 4b).
FIG. 4.
Determinants for corepressor specificity are in CoRNR1. (a) Preference for N-CoR or SMRT CoRNR1 by NHRs in 293T cells. Gal4 fusion of N-CoRNR1 (N-CoR, amino acids 2057 to 2096) or S-CoRNR1 (SMRT, amino acids 2123 to 2162) was tested for interaction with VP16 fusion of NHRs. (b) Affinities of N-CoR CoRNR1 and SMRT CoRNR1 peptides to RAR and RevErb. Peptides were used to block interaction between NHR and GST–N-CoR (amino acids 1944 to 2453). The results were quantified by PhosphorImager. In, 10% input. The N-CoRNR1 peptide contains amino acids 2057 to 2086 of N-CoR (N4 in Fig. 3a). The S-CoRNR peptide contains amino acids 2123 to 2152 of SMRT.
CoRNR1 residues determine corepressor specificity for different NHRs.
The results thus far show that the determinants for corepressor specificity are in the 40-residue CoRNR1. Within CoRNR1, the central 19 residues are highly conserved between N-CoR and SMRT (Fig. 5a). We first asked whether these 19 residues determine specificity by converting the 19 residues in S-CoRNR1 into the corresponding N-CoR sequences (SNS in Fig. 5a). For RAR, this swap did decrease interaction to the level of N-CoR CoRNR1 in the mammalian two-hybrid experiment (Fig. 5b). However, for RevErb this swap has no effect, suggesting that residues outside this 19-residue core region determine specificity. Within the core region, the most striking difference is around the 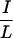 XX
XX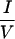 I motif. In N-CoR, the sequence is DHICOII, whereas it is OHISEVI in SMRT (the differing residues are underlined). To ask if these amino acid residues are determinants of specificity, we converted those in SMRT CoRNR1 to those of N-CoR sequences (D2 in Fig. 5a). Indeed, this change reduced RAR interaction and had no effect on RevErb interaction (Fig. 5b). Swapping the other differing residues in the core region had no effect (data not shown).
I motif. In N-CoR, the sequence is DHICOII, whereas it is OHISEVI in SMRT (the differing residues are underlined). To ask if these amino acid residues are determinants of specificity, we converted those in SMRT CoRNR1 to those of N-CoR sequences (D2 in Fig. 5a). Indeed, this change reduced RAR interaction and had no effect on RevErb interaction (Fig. 5b). Swapping the other differing residues in the core region had no effect (data not shown).
FIG. 5.
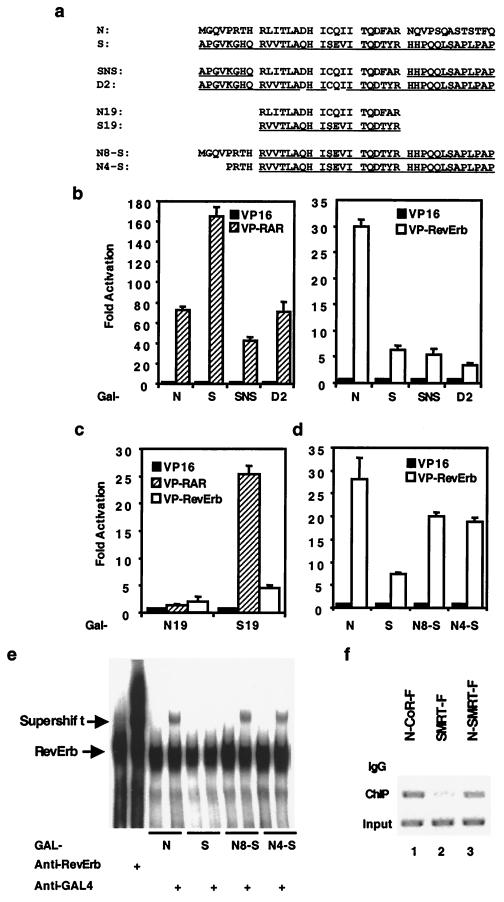
Residues in CoRNR1 that determine corepressor specificity. (a) Sequences of the constructs used in this figure. SMRT sequences are underlined. (b) Differences in the center of CoRNR1 determine corepressor specificity for RAR but not RevErb. (c) The 19-residue core regions of N-CoR and SMRT interact differently with RAR and RevErb. (d) The N terminus of N-CoR CoRNR1 enhances interaction with RevErb. Gal4 fusion of each deletion or mutation construct was tested for interaction with VP16 fusion of NHRs in a mammalian two-hybrid assay. (e) The N terminus of N-CoR CoRNR1 promotes binding of corepressor to DNA-bound RevErb in a gel shift assay. The positions of the RevErb band and the supershift band are indicated by arrows. The free probe ran off the gel. (f) The N terminus of N-CoR CoRNR1 increases binding of full-length SMRT to DNA-bound RevErb in vivo. N-CoR-F, SMRT-F, and N-SMRT-F (N4-S in full-length SMRT) were each transfected into 293T cells along with the RevDR2-containing reporter plasmid. IgG, precipitated by nonspecific IgG; ChIP, precipitated by Flag antibody; Input, aliquots of sample before immunoprecipitation. Bands are ethidium bromide staining of PCRs using primers spanning the RevErb binding site.
It has been shown that this 19-residue core region of SMRT is sufficient to interact with RAR (30). To our surprise, when we tried to compare the interactions of the 19-residue N-CoR (N19) and SMRT (S19) with RAR and RevErb (Fig. 5a), N19 lost the ability to interact with either receptor in the mammalian two-hybrid assay (Fig. 5c) despite comparable levels of expression (data not shown). In contrast, S19 can still interact with RAR or RevErb (Fig. 5c), although at a much reduced level with RAR (compare the fold activation with that in Fig. 4a). GST pull-down assays with the 19-residue peptides also confirmed this result in vitro (data not shown). These results suggest that N-CoR and SMRT CoRNR1 interact with receptors by very different means: N-CoR CoRNR1 requires the core and surrounding residues for interaction, whereas SMRT CoRNR1 mainly uses the core region.
Figure 5b also suggests that corepressor determinants for interaction with RevErb lie outside the core region. We also learned from Fig. 3a that the N terminus but not C terminus of N-CoR CoRNR1 is required for optimal interaction with RevErb. Taken together, we postulated that the N-terminal sequences of N-CoR CoRNR1 determine the corepressor specificity for interaction with RevErb. To test our hypothesis, we changed the N-terminal eight or four residues of S-CoRNR1 into the corresponding N-CoR sequences (N8-S and N4-S in Fig. 5a). Indeed, as shown in Fig. 5d, swapping either eight or four residues increased interaction with RevErb to the level of N-CoR CoRNR1 in the mammalian two-hybrid assay. This result suggests that the four residues just N terminal to the core region determine affinity for interaction with RevErb.
RevErb interacts with N-CoR but not SMRT on DNA (42). To address whether this is due to the four residues implicated above, we studied RevErb interactions with CoRNR1 peptides on DNA using a gel shift assay (Fig. 5e). As expected, RevErb bound to the RevDR2 element and the addition of antibody against RevErb supershifted the binding (lanes 1 and 2). The 40-residue N-CoR CoRNR1 was expected to bind to RevErb on DNA, but no apparent supershift was observed when this polypeptide was added as a Gal4 DNA-binding domain (DBD) fusion (Gal-N). However, the addition of antibody to Gal4 DBD (anti-Gal4) did supershift the complex, indicating that N-CoR CoRNR1 did indeed bind to RevErb on DNA, presumably because the gel mobility of the RevErb-DNA and the RevErb–Gal-N–DNA were indistinguishable or because antibody stabilizes the RevErb–Gal-N–DNA complex. However, the anti-Gal4 antibody did not supershift the RevErb DNA binding complex in the presence of SMRT CoRNR1 (Gal-S), indicating that S-CoRNR1 did not interact with RevErb on DNA. These results confirmed our earlier observations when using the entire interaction domain. We then tested the chimera N8-S and N4-S under the same conditions. As shown in Fig. 5e, the anti-Gal4 did supershift the RevErb complexes in the presence of Gal4–N8-S and Gal4–N4-S. These results show that the 4 residues just N terminal to the 19-amino-acid CoRNR1 core determine the selective binding of N-CoR to RevErb on DNA.
We then used the ChIP assay to determine whether the selectivity of RevErb for N-CoR over SMRT on DNA pertains to full-length corepressors in vivo. The four-amino-acid change that was shown earlier to allow SMRT CoRNR1 to interact with RevErb was made in full-length SMRT that was also tagged with the Flag peptide (N-SMRT-F). This construct (or N-CoR-F or SMRT-F) was transfected into 293T cells, along with full-length RevErb and a reporter gene containing the RevErb response element (Rev-DR2) that we have previously shown to be dramatically repressed by RevErb (41). Flag antibody was used to precipitate the corepressor complex after cross-linking, and in this ChIP assay bound DNA was amplified using specific primers surrounding the Rev-DR2 (Fig. 5f). As expected, nonspecific antibody immunoglobulin G (IgG) did not precipitate a significant amount of the Rev-DR2-containing DNA (Fig. 5f, top lanes). ChIP of N-CoR-F showed considerably more association with the RevDR2 site than with SMRT-F (compare middle lanes 1 and 2). However, the ChIP assay of N-SMRT-F indicated that the amount of Rev-DR2 associated with this chimeric corepressor containing only four amino acids from N-CoR CoRNR1 was much higher than that associated with wild-type SMRT and is comparable to that associated with N-CoR. Thus, the 4 residues just N terminal to the 19-residue core of CoRNR1 determine the specificity of corepressor binding to RevErb on its response element.
Specific residues in RAR H3-4-5 are required for interaction with CoRNR1.
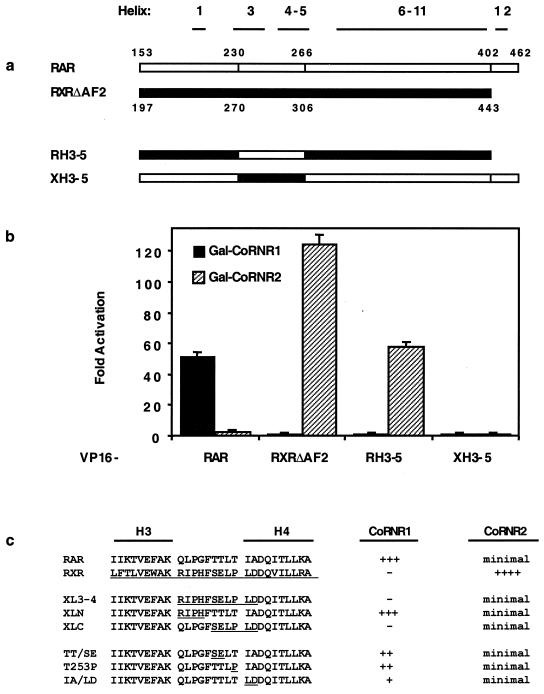
We next undertook an analysis of the molecular explanation for why some NHRs prefer CoRNR1 whereas other NHRs prefer CoRNR2. As RAR strongly prefers CoRNR1 and RXR almost exclusively interacts with CoRNR2 (Fig. 1a), we made chimeric RAR-RXR proteins to determine which features of the receptor LBD determines CoRNR preference. We and others have previously shown that mutations in helices 3, 4, and 5 of RXR, RAR, or TR abolish corepressor interaction (19, 30, 32), indicating that corepressors and coactivators utilize overlapping binding surfaces on the receptor LBD. To address whether specific residues in this binding surface determine CoRNR specificity, we made corepressor binding surface (H3-4-5) swaps between RAR and RXR (Fig. 6a). When the H3-4-5 binding surface of RXR was changed to that of RAR, the resulting chimera resembled RXR in its ability to interact with CoRNR2 and not CoRNR1 in a mammalian two-hybrid assay (RH3-5, Fig. 6b). This result suggested that the binding surface residues of RXR do not determine the preference for CoRNR2. The opposite swap, changing the H3-4-5 region of RAR to that of RXR, resulted in a chimera that cannot interact with either CoRNR box (XH3-5, Fig. 6b). However, this chimera also lost interaction with RXR, suggesting a major conformation defect (data not shown).
FIG. 6.
Effect of H3-4-5 swaps on CoRNR1 and CoRNR2 interaction. (a) Schematic diagram of the chimerical constructs. The corresponding amino acid numbers in hRARα and hRXRα are indicated. (b) Interactions with CoRNR1 and CoRNR2 in a mammalian two-hybrid assay. (c) CoRNR interactions of an RAR and RXR H3-4 region swap and mutant constructs. CoRNR peptides were used as a Gal4 fusion and RAR/RXR constructs were used as a VP16 fusion. Summaries of two-hybrid interaction with CoRNR1 and CoRNR2—the strength of interaction (fold activation in a mammalian two-hybrid assay)—are indicated. For CoRNR1 interaction: +++, >50-fold activation; ++, 25- to 40-fold; + 5- to 15-fold; −, no interaction. For CoRNR2 interaction: minimal, 2- to 3-fold interaction; ++++, >100-fold interaction.
To evaluate the role of RAR H3-4-5, we made subtle changes in this region of RAR. Only mutants that retained interaction with RXR (shown in Fig. 6c) were studied. Of note, when the amino acid residues between H3 and H4 plus two residues at the beginning of H4 were swapped, the chimera lost interaction with CoRNR1 in a mammalian two-hybrid assay (XL3-4, Fig. 6c). The residues required for interaction with CoRNR1 lie at the C-terminal half of this loop, as interaction with CoRNR1 was not abolished by an N-terminal swap (XLN) but was completely abrogated by a C-terminal swap (XLC) (Fig. 6c). Further point mutations in this C-terminal loop region showed that the most critical residues are the two residues at the beginning of H4. Changing these two residues from IA to LD greatly reduced interaction with CoRNR1 (IA/LD, Fig. 6c). Changing the other three residues in this region (TT/SE, T253P) also decreased interaction with CoRNR1. These results indicated that the specific sequences between H3 and H4 are required for CoRNR1 interaction. Note that these swaps led to a loss of function for CoRNR1 binding but not an increase in CoRNR2 interaction, indicating that key CoRNR2 binding determinants of RXR lie outside this region.
RAR H12 partially blocks corepressor interactions.
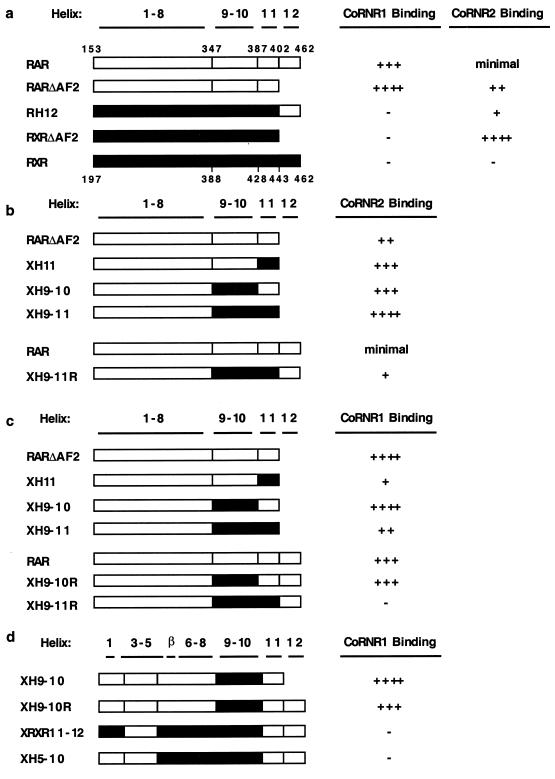
Since the H3-4-5 swaps did not change CoRNR specificity, we turned our attention to the extreme C-terminal region of the receptor LBD because structural studies indicate that this region (H11 and H12) is also in the vicinity of the H3-4-5 binding surface (1, 34, 39). Helix 12 of NHRs is required for NR box interaction and also regulates corepressor interaction (reviewed in reference 20). It has been shown previously that RXR H12 inhibits corepressor binding (43). A recent report that RXR H12 binds to the H3-4-5 region of an adjacent molecule in the RXR tetramer explains why RXR cannot interact with a corepressor (11). Although not as dramatic as for RXR, deletion of H12 from RAR (RARΔAF2) increased interaction with CoRNR1 in a mammalian two-hybrid assay (Fig. 7a). Moreover, this mutant RAR also showed increased interaction with CoRNR2 (Fig. 7a), although the preference for CoRNR1 was retained (250-fold versus 20-fold interaction). These results confirm earlier observations by Schulman et al. (35) that RAR H12 hinders interaction with corepressors. Interestingly, replacement of RXR H12 with RAR H12 allowed RXR to interact with CoRNR2, although this interaction was much less than that observed with RXRΔAF2. This was surprising, since it was shown previously that alanine residues could substitute for the ability of the naturally occurring RXR H12 to prevent corepressor binding (43). We speculate that this is due to the presence of proline residues in the loop between H11 and H12, as mutating residues in this loop of RXR to proline also partially release the masking of repression (J. Zhang and M. A. Lazar, unpublished result).
FIG. 7.
CoRNR specificity determinants are different for RAR and RXR. (a) RAR H12 partially blocks corepressor interaction. (b) RXR H9-11 determines CoRNR2 binding. (c) RXRH11 inhibits CoRNR1 binding. (d) H5-8 is required for CoRNR1 interaction. CoRNR peptides were used as a Gal4 fusion and RAR/RXR constructs were used as a VP16 fusion. The strength of interaction (fold activation in a mammalian two-hybrid assay) is indicated. For CoRNR1: ++++, >200-fold interaction; +++, 60- to 150-fold interaction; ++, 25- to 40-fold interaction; +, 5- to 15-fold interaction; −, no interaction. For CoRNR2: ++++, >100-fold interaction; +++, 35- to 70-fold interaction; ++, 15- to 30-fold interaction; +, 5- to 10-fold interaction; minimal, 2- to 3-fold interaction; −, no interaction.
RXR H9-11 enhances CoRNR2 interaction and RXR H11 inhibits CoRNR1 interaction.
We next tested the role of H11 in determining CoRNR specificity. We first changed H11 of RAR into that of RXR in the context of RARΔAF2. This swap increased interaction with CoRNR2 in a mammalian two-hybrid assay (XH11, Fig. 7b), suggesting that RXR H11 contains primary sequence information that favors CoRNR2 interaction. As this interaction was considerably less than that of RXRΔAF2, we further replaced H9-10 of RAR with that of RXR (XH9-11). This substitution further increased the interaction to the level of RXRΔAF2. Moreover, substitution of only H9-10 of RXR into RAR (XH9-10) also increased interaction. These constructs are all in the context of RARΔAF2. When RAR H12 is present, swapping H9-11 also resulted in an increase in interaction with CoRNR2 (compare with RAR). Together, these results suggested that H9-11 of RXR determines the specificity for CoRNR2 and that the extent of the interaction is governed by H12.
Since RXR H11 favors CoRNR2 but RXR does not interact with CoRNR1, we hypothesized that this helix also inhibits interaction with CoRNR1. Indeed, swapping RXR H11 into RAR (XH11) led to a sharp decrease in interaction compared to RARΔAF2 in a mammalian two-hybrid assay (Fig. 7c). In contrast, swapping RXRH9-10 into RAR had no effect on CoRNR1 interaction either in the presence or absence of RAR H12 (in Fig. 7c, compare XH9-10 with RARΔAF2 and XH9-10R with RAR). When H11 was further substituted either in the presence or absence of RAR H12, interaction with CoRNR1 was greatly decreased (in Fig. 7c, compare XH9-10 with XH9-11 and XH9-10R with XH9-11R). Taking these results together, we conclude that RXR H11 specifically inhibits CoRNR1 interaction.
RAR β-turn and H6-8 regions are required for CoRNR1 interaction.
We have shown that specific sequences between RAR H3 and H4 are required but not sufficient for CoRNR1 binding (Fig. 6b). Since RXR H11 inhibited CoRNR1 binding, we considered whether RAR H11 determines CoRNR1 specificity. To test this, we made a chimeric construct in which H3-4-5 and H11-12 are from RAR and the rest of the LBD derives from RXR. This construct (XRXR11-12) is unable to interact with CoRNR1 in a mammalian two-hybrid assay (Fig. 7d), suggesting that the determinants for CoRNR1 were not in H11. H9-10 is also not required for CoRNR1 binding, as XH9-10 can still interact with CoRNR1 (Fig. 7d). We then made further substitution into the region, including part of H5, the β-turn, and H6-8. The resulting chimera (XH5-10) lost interaction with CoRNR1 (Fig. 7d), suggesting that this region, in addition to H3-4-5, is required for CoRNR1 interaction by RAR. Based on these results together with those shown in Fig. 7d, we conclude that H3 to H8 of RAR is required for CoRNR1 interaction.
DISCUSSION
NHR-corepressor interactions are highly specific.
CoRNR2, which binds RXR, is relatively simple and does not determine specificity for N-CoR versus SMRT (Fig. 2). CoRNR1, on the other hand, is more complex and contains most of the sequence features that determine specificity for N-CoR or SMRT. TR and RevErb bind strongly to N-CoR and weakly to SMRT, while RAR does the opposite. Distinct regions of the corepressors enable these preferences. Although the 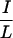 XX
XX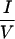 I motifs are required for interaction, corepressors utilize different residues within or outside the CoRNR box for efficient interaction with different NHRs (Fig. 3). RAR requires both N- and C-terminal sequences within the 40-residue CoRNR1 for optimal interaction. RevErb utilizes extremely proximal N-terminal sequences. RevErb has previously been shown to preferentially bind N-CoR on DNA even though it is capable of interacting with SMRT in solution (42). The present studies suggest that this is due to the affinity difference for N-CoR and SMRT. Four residues in the N terminal of CoRNR1 are responsible for this difference. Replacing these four residues in SMRT with corresponding N-CoR sequences resulted in a corepressor that binds RevErb on DNA both in vitro and in vivo (Fig. 5). TR, in sharp contrast, requires sequences about 100 residues N terminal to the core of CoRNR1 for efficient interaction. Within this region, there are
I motifs are required for interaction, corepressors utilize different residues within or outside the CoRNR box for efficient interaction with different NHRs (Fig. 3). RAR requires both N- and C-terminal sequences within the 40-residue CoRNR1 for optimal interaction. RevErb utilizes extremely proximal N-terminal sequences. RevErb has previously been shown to preferentially bind N-CoR on DNA even though it is capable of interacting with SMRT in solution (42). The present studies suggest that this is due to the affinity difference for N-CoR and SMRT. Four residues in the N terminal of CoRNR1 are responsible for this difference. Replacing these four residues in SMRT with corresponding N-CoR sequences resulted in a corepressor that binds RevErb on DNA both in vitro and in vivo (Fig. 5). TR, in sharp contrast, requires sequences about 100 residues N terminal to the core of CoRNR1 for efficient interaction. Within this region, there are 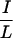 XX
XX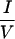 I-like sequences in N-CoR but not in SMRT that are likely to contribute to the TR's preference for N-CoR. Thus, sequences distant from the CoRNR box are required for high-affinity interaction with some but not all NHRs.
I-like sequences in N-CoR but not in SMRT that are likely to contribute to the TR's preference for N-CoR. Thus, sequences distant from the CoRNR box are required for high-affinity interaction with some but not all NHRs.
The fact that CoRNR peptides can block interactions between NHR and corepressors suggests novel ways to treat diseases such as promyelocytic leukemia. Since corepressor binding is a general feature of NHRs, such therapies could have unwanted toxicities. The present finding that interactions between CoRNR boxes and NHRs are highly specific suggests that it could be possible to develop peptidomimetic drugs that selectively block interaction between corepressors and a specific NHR, thereby targeting specific receptor-specific diseases without affecting the normal biological functions of other NHRs.
H3-4-5 is highly conserved among NHRs and is necessary but not sufficient for high-affinity interaction with corepressors (4, 19, 30, 32). Additional regions of RAR (H6-8) that are not in proximity to H3-4-5 in the LBD crystal structures (1, 34, 39) contribute to its affinity for CoRNR1 (Fig. 7). We postulate that H6-8 “frames” the corepressor interaction helices in RAR to bind selectively to CoRNR1. For RXR, the region distant from H3-4-5 that adds to the affinity for CoRNR2 is H9-10. This frame for CoRNR2 is likely to be different from the RAR H6-8-dependent frame for CoRNR1.
Model of corepressor binding to NHR heterodimers.
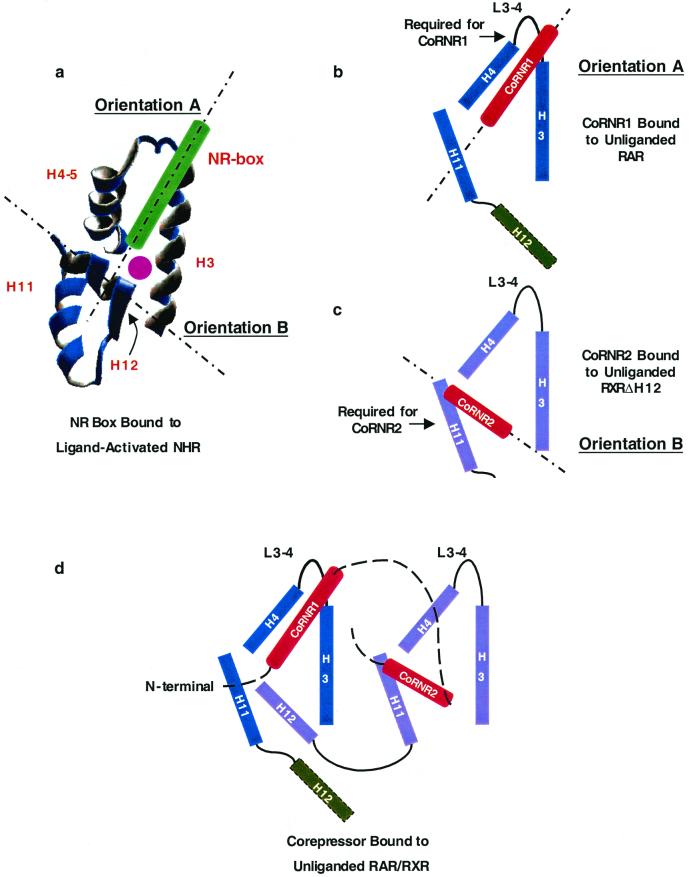
The structure of the CoRNR peptide bound to unliganded NHR has not yet been solved. The signature CoRNR box feature is an amphipathic α-helix that is not only similar to coactivator NR boxes but also to H12 of NHRs, which contains ΦXXΦΦ (Φ is a hydrophobic residue). In the crystal structures of NR box peptides bound to agonist-liganded NHRs, the NR box binds in a hydrophobic groove between H3 and H4-5, and H12 folds back to form part of the binding surface for the NR box (8, 31) (Fig. 8a). Thus a single NHR LBD has at least two distinct and differently oriented docking sites for amphipathic α-helices. In the following discussion, we will refer to the orientation of the NR box and of H12 bound to the liganded receptor as orientation A and orientation B, respectively (Fig. 8a). There is flexibility in the sequences that each docking site can accommodate, since in the antagonist-bound NHR crystal structure, H12 binds in orientation A to prevent interaction with coactivators (3, 37).
FIG. 8.
Model of corepressor-nuclear receptor interaction. (a) Crystal structure of the relative orientations of helices forming the coactivator-binding surface on the liganded NHR. The coactivator NR peptide is in green. The ligand is shown as a purple dot. The two orientations that an α-helix can bind are indicated as dotted lines. (b and c) The two possible orientations of CoRNR box peptides on NHR. The residues required for each CoRNR box are indicated by arrows. (b) The CoRNR1 peptide in the NR peptide orientation on RAR (orientation A). (c) The CoRNR2 peptide in the H12 orientation on RXRΔH12 (orientation B). Please note that these two orientations may not be the same as those in panel a. (d) Models of the positions of the two CoRNR boxes in the heterodimer of RAR-RXR. CoRNR box peptides are in red. The position of RAR H12 (shown as a dashed rectangle) is unknown.
CoRNR1 binding to RAR requires specific sequences in the loop between H3 and H4-5 (Fig. 6) but not H11 (data not shown), suggesting that it binds to the NHR in orientation A (Fig. 8b). On the other hand, CoRNR2 peptide binding to RXR requires specific sequences in H11 but is independent of the sequence of the loop between H3 and H4-5, similar to orientation B (Fig. 8c). We should point out that the two CoRNR binding orientations may not be exactly the same as those of the liganded H12 and NR box helices. They are probably also different on different NHRs and with different corepressors.
These considerations suggest a model to explain how a single molecule of N-CoR or SMRT might bind to RAR-RXR heterodimers. The NHR preferences dictate binding of CoRNR1 to RAR and CoRNR2 to RXR. We and others have previously shown that RXR H12 binds to the H3-4-5 region of its partner and that this unmasks the CoRNR binding site on RXR (40, 43). These findings created an apparent paradox, since both RXR H12 and the CoRNR box have to bind to the H3-4-5 region of RAR in the RAR-RXR heterodimer. This is explained by the model shown in Fig. 8d. In the RAR-RXR heterodimer, CoRNR1 and RXR H12 bind simultaneously to RAR in orientation A and orientation B, respectively. CoRNR2 binds to RXR in orientation B. It is possible that RAR H12 binds to RXR in orientation A (the “antagonist” position in the estrogen receptor structure). This working model of corepressor interaction with the RAR-RXR heterodimer will need to be tested by cocrystallization of RAR, RXR, and a peptide containing CoRNR1 and CoRNR2.
ACKNOWLEDGMENTS
We thank Helen Bayes, Yongfeng Shang, and Myles Brown for instructions on the ChIP assay. We also thank Matt Guenther, Eric Huang, Helen Bayes, and Orr Barak for valuable discussions.
This work was supported by NIH grants DK43806 and DK45586 to M.A.L. Peptides were synthesized by the Protein Chemistry Laboratory of the University of Pennsylvania Medical Center.
REFERENCES
- 1.Bourguet W, Ruff M, Chambon P, Gronemeyer H, Moras D. Crystal structure of the ligand-binding domain of the human nuclear receptor RXR-alpha. Nature. 1995;375:377–382. doi: 10.1038/375377a0. [DOI] [PubMed] [Google Scholar]
- 2.Bourguet W, Vivat V, Wurtz J M, Chambon P, Gronemeyer H, Moras D. Crystal structure of a heterodimeric complex of RAR and RXR ligand-binding domains. Mol Cell. 2000;5:289–298. doi: 10.1016/s1097-2765(00)80424-4. [DOI] [PubMed] [Google Scholar]
- 3.Brzozowski A M, Pike A C, Dauter Z, Hubbard R E, Bonn T, Engstrom O, Ohman L, Greene G L, Gustafsson J A, Carlquist M. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 4.Burke L J, Downes M, Laudet V, Muscat G E. Identification and characterization of a novel corepressor interaction region in RVR and Rev-erbA alpha. Mol Endocrinol. 1998;12:248–262. doi: 10.1210/mend.12.2.0061. [DOI] [PubMed] [Google Scholar]
- 5.Chen J D, Evans R M. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 6.Cohen R N, Putney A, Wondisford F E, Hollenberg A N. The nuclear corepressors recognize distinct nuclear receptor complexes. Mol Endocrinol. 2000;14:900–914. doi: 10.1210/mend.14.6.0474. [DOI] [PubMed] [Google Scholar]
- 7.Cohen R N, Wondisford F E, Hollenberg A N. Two separate NCoR (nuclear receptor corepressor) interaction domains mediate corepressor action on thyroid hormone response elements. Mol Endocrinol. 1998;12:1567–1581. doi: 10.1210/mend.12.10.0188. [DOI] [PubMed] [Google Scholar]
- 8.Darimont B D, Wagner R L, Apriletti J W, Stallcup M R, Kushner P J, Baxter J D, Fletterick R J, Yamamoto K R. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng W, Ribeiro R C, Wagner R L, Nguyen H, Apriletti J W, Fletterick R J, Baxter J D, Kushner P J, West B L. Hormone-dependent coactivator binding to a hydrophobic cleft on nuclear receptors. Science. 1998;280:1747–1749. doi: 10.1126/science.280.5370.1747. [DOI] [PubMed] [Google Scholar]
- 10.Gampe R T, Jr, Montana V G, Lambert M H, Miller A B, Bledsoe R K, Milburn M V, Kliewer S A, Willson T M, Xu H E. Asymmetry in the PPARgamma/RXRalpha crystal structure reveals the molecular basis of heterodimerization among nuclear receptors. Mol Cell. 2000;5:545–555. doi: 10.1016/s1097-2765(00)80448-7. [DOI] [PubMed] [Google Scholar]
- 11.Gampe R T, Jr, Montana V G, Lambert M H, Wisely G B, Milburn M V, Xu H E. Structural basis for autorepression of retimoid X receptor by tetramer formation and the AF-2 helix. Genes Dev. 2000;14:2229–2241. doi: 10.1101/gad.802300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glass C K, Rosenfeld M G. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- 13.Grignani F, De Matteis S, Nervi C, Tomassoni L, Gelmetti V, Cioce M, Fanelli M, Ruthardt M, Ferrara F F, Zamir I, Seiser C, Lazar M A, Minucci S, Pelicci P G. Fusion proteins of the retinoic acid receptor-alpha recruit histone deacetylase in promyelocytic leukaemia. Nature. 1998;391:815–818. doi: 10.1038/35901. [DOI] [PubMed] [Google Scholar]
- 14.Grignani F, Valtieri M, Gabbianelli M, Gelmetti V, Botta R, Luchetti L, Masella B, Morsilli O, Pelosi E, Samoggia P, Pelicci P G, Peschle C. PML/RARalpha fusion protein expression in normal human hematopoietic progenitors dictates myeloid commitment and the promyelocytic phenotype. Blood. 2000;96:1531–1537. [PubMed] [Google Scholar]
- 15.Guenther M G, Lane W S, Fischle W, Verdin E, Lazar M A, Shiekhattar R. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev. 2000;14:1048–1057. [PMC free article] [PubMed] [Google Scholar]
- 16.Heery D M, Kalkhoven E, Hoare S, Parker M G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 17.Heinzel T, Lavinsky R M, Mullen T M, Soderstrom M, Laherty C D, Torchia J, Yang W M, Brard G, Ngo S D, Davie J R, Seto E, Eisenman R N, Rose D W, Glass C K, Rosenfeld M G. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 18.Horlein A J, Naar A M, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass C K, et al. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 19.Hu X, Lazar M A. The CoRNR motif controls the recruitment of corepressors by nuclear hormone receptors. Nature. 1999;402:93–96. doi: 10.1038/47069. [DOI] [PubMed] [Google Scholar]
- 20.Hu X, Lazar M A. Transcriptional repression by nuclear hormone receptors. Trends Endocrinol Metab. 2000;11:6–10. doi: 10.1016/s1043-2760(99)00215-5. [DOI] [PubMed] [Google Scholar]
- 21.Huang E Y, Zhang J, Miska E A, Guenther M G, Kouzarides T, Lazar M A. Nuclear receptor corepressors partner with class II histone deacetylases in a Sin3-independent repression pathway. Genes Dev. 2000;14:45–54. [PMC free article] [PubMed] [Google Scholar]
- 22.Huang M E, Ye Y C, Chen S R, Chai J R, Lu J X, Zhoa L, Gu L J, Wang Z Y. Use of all-trans retinoic acid in the treatment of acute promyelocytic leukemia. Blood. 1988;72:567–572. [PubMed] [Google Scholar]
- 23.Kao H Y, Downes M, Ordentlich P, Evans R M. Isolation of a novel histone deacetylase reveals that class I and class II deacetylases promote SMRT-mediated repression. Genes Dev. 2000;14:55–66. [PMC free article] [PubMed] [Google Scholar]
- 24.Li J, Wang J, Nawaz Z, Liu J M, Qin J, Wong J. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 2000;19:4342–4350. doi: 10.1093/emboj/19.16.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin R J, Evans R M. Acquisition of oncogenic potential by RAR chimeras in acute promyelocytic leukemia through formation of homodimers. Mol Cell. 2000;5:821–830. doi: 10.1016/s1097-2765(00)80322-6. [DOI] [PubMed] [Google Scholar]
- 26.Lin R J, Nagy L, Inoue S, Shao W, Miller W H, Jr, Evans R M. Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature. 1998;391:811–814. doi: 10.1038/35895. [DOI] [PubMed] [Google Scholar]
- 27.Mangelsdorf D J, Evans R M. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 28.Melnick A, Licht J D. Deconstructing a disease: RARalpha, its fusion partners, and their roles in the pathogenesis of acute promyelocytic leukemia. Blood. 1999;93:3167–3215. [PubMed] [Google Scholar]
- 29.Nagy L, Kao H Y, Chakravarti D, Lin R J, Hassig C A, Ayer D E, Schreiber S L, Evans R M. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 30.Nagy L, Kao H Y, Love J D, Li C, Banayo E, Gooch J T, Krishna V, Chatterjee K, Evans R M, Schwabe J W. Mechanism of corepressor binding and release from nuclear hormone receptors. Genes Dev. 1999;13:3209–3216. doi: 10.1101/gad.13.24.3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nolte R T, Wisely G B, Westin S, Cobb J E, Lambert M H, Kurokawa R, Rosenfeld M G, Willson T M, Glass C K, Milburn M V. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature. 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 32.Perissi V, Staszewski L M, McInerney E M, Kurokawa R, Krones A, Rose D W, Lambert M H, Milburn M V, Glass C K, Rosenfeld M G. Molecular determinants of nuclear receptor-corepressor interaction. Genes Dev. 1999;13:3198–3208. doi: 10.1101/gad.13.24.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Renaud J P, Harris J M, Downes M, Burke L J, Muscat G E. Structure-function analysis of the Rev-erbA and RVR ligand-binding domains reveals a large hydrophobic surface that mediates corepressor binding and a ligand cavity occupied by side chains. Mol Endocrinol. 2000;14:700–717. doi: 10.1210/mend.14.5.0444. [DOI] [PubMed] [Google Scholar]
- 34.Renaud J P, Rochel N, Ruff M, Vivat V, Chambon P, Gronemeyer H, Moras D. Crystal structure of the RAR-gamma ligand-binding domain bound to all-trans retinoic acid. Nature. 1995;378:681–689. doi: 10.1038/378681a0. [DOI] [PubMed] [Google Scholar]
- 35.Schulman I G, Juguilon H, Evans R M. Activation and repression by nuclear hormone receptors: hormone modulates an equilibrium between active and repressive states. Mol Cell Biol. 1996;16:3807–3813. doi: 10.1128/mcb.16.7.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seol W, Mahon M J, Lee Y K, Moore D D. Two receptor interacting domains in the nuclear hormone receptor corepressor RIP13/N-CoR. Mol Endocrinol. 1996;10:1646–1655. doi: 10.1210/mend.10.12.8961273. [DOI] [PubMed] [Google Scholar]
- 37.Shiau A K, Barstad D, Loria P M, Cheng L, Kushner P J, Agard D A, Greene G L. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 38.Shibata H, Nawaz Z, Tsai S Y, O'Malley B W, Tsai M J. Gene silencing by chicken ovalbumin upstream promoter-transcription factor I (COUP-TFI) is mediated by transcriptional corepressors, nuclear receptor-corepressor (N-CoR) and silencing mediator for retinoic acid receptor and thyroid hormone receptor (SMRT) Mol Endocrinol. 1997;11:714–724. doi: 10.1210/mend.11.6.0002. [DOI] [PubMed] [Google Scholar]
- 39.Wagner R L, Apriletti J W, McGrath M E, West B L, Baxter J D, Fletterick R J. A structural role for hormone in the thyroid hormone receptor. Nature. 1995;378:690–697. doi: 10.1038/378690a0. [DOI] [PubMed] [Google Scholar]
- 40.Westin S, Kurokawa R, Nolte R T, Wisely G B, McInerney E M, Rose D W, Milburn M V, Rosenfeld M G, Glass C K. Interactions controlling the assembly of nuclear-receptor heterodimers and co-activators. Nature. 1998;395:199–202. doi: 10.1038/26040. [DOI] [PubMed] [Google Scholar]
- 41.Zamir I, Harding H P, Atkins G B, Horlein A, Glass C K, Rosenfeld M G, Lazar M A. A nuclear hormone receptor corepressor mediates transcriptional silencing by receptors with distinct repression domains. Mol Cell Biol. 1996;16:5458–5465. doi: 10.1128/mcb.16.10.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zamir I, Zhang J, Lazar M A. Stoichiometric and steric principles governing repression by nuclear hormone receptors. Genes Dev. 1997;11:835–846. doi: 10.1101/gad.11.7.835. [DOI] [PubMed] [Google Scholar]
- 43.Zhang J, Hu X, Lazar M A. A novel role for helix 12 of retinoid X receptor in regulating repression. Mol Cell Biol. 1999;19:6448–6457. doi: 10.1128/mcb.19.9.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]