Maud Tusseau and Alexandre Belot highlight work from He et al. showing that P2RY8 prevents the expansion of DNA-reactive B cells by restraining B cell mobility and activation within the germinal center.
Abstract
In this issue of JEM, He et al. (2021. J. Exp. Med. https://doi.org/10.1084/jem.20211004) associate novel P2RY8 genetic variants to lupus, expanding the field of monogenic autoimmunity. The authors demonstrate that P2RY8 prevents the expansion of DNA-reactive B cells by restraining B cell mobility and activation within the germinal center.
In this issue of JEM, the Carola Vinuesa and Jason Cyster laboratories (He et al., 2021) characterize the genetic contribution of germline P2RY8 variants to systemic lupus erythematosus (SLE) and further expand the role of P2RY8 in tolerance maintenance. P2RY8 serves as a G-protein receptor for S-geranyl-geranyl-L-glutathione (GGG) that drives B cell clustering inside germinal centers (GCs; Lu et al., 2019). GCs are transient structures within lymphoid organs enabling the antibody diversification and affinity maturation of activated B cells, thus representing a critical checkpoint for terminal B cell differentiation (Jacob et al., 1991; Berek et al., 1991). P2RY8 mutations have been previously described in GC-related lymphoma such as Burkitt lymphoma and diffuse large B cell lymphoma (DLBCL), but no germline variant was identified so far. The authors perform a detailed immune and genetic phenotyping to characterize new genetic variants of P2RY8 occurring in SLE patients, and they demonstrate the critical role of P2RY8 in preventing systemic autoimmunity.

Insights from Maud Tusseau and Alexandre Belot.
The work is exciting and shows at least three major findings.
First, the authors identify P2RY8 germline variants associated with SLE or antiphospholipid syndrome. Using next generation sequencing, He et al. (2021) depict two rare and one new genetic variants in P2RY8 in a total of eight patients with lupus or antiphospholipid syndrome. Among these genetic findings, the p.L257F variant is the most severe and best characterized variant. L257F is a de novo heterozygous P2RY8 variant identified in an early-onset lupus patient and is associated with a decreased protein expression in all lymphocyte subsets. Using various transfection experiments combining WT and L257F variants, the authors demonstrate a gene dosage effect with a reduced GGG-mediated inhibition of both cell migration and AKT/ERK activation to CXCL12 exposure when using the L257F variant. Interestingly, a reduction of P2RY8 B cell expression was also identified in lupus patients with greater lupus nephritis frequency, suggesting that P2RY8 defect can also occur under nongenetic influence and is associated to severity.
Second, the investigators nicely demonstrate that B cells carrying P2RY8 variants were not sensitive to GGG-related cell migration inhibition. P2RY8 has no orthologue in rodent, preventing the use of knockout mice models, but the authors elegantly circumvent this difficulty by transducing mouse B cells with fluorescent WT or L257F, N97K, E323G variants and further reinjected these cells in mice. Using this strategy, the authors show that the L257F variant was unable to promote B cell confinement to GCs, whereas a milder or no effect was seen in N97K or E323K variants. They also reveal that P2RY8 expression reduces plasma cell generation, a regulatory function that is lost in P2RY8L257F-expressing B cells.
Third, P2RY8 expression restricts B cell autoreactivity. To explore the role of P2RY8 in the B cell negative selection, He et al. (2021) took advantage of VH3H9 transgenic mice expressing a heavy (H) chain that was isolated from anti-dsDNA antibodies from MRL-lpr/lpr mice. Because the VH3H9 H chain can pair with endogenous lambda light (L) chains to generate anti-DNA reactive B cells, this model allows the analysis of B cell tolerance by measuring lambda (especially lambda-1)–expressing follicular B cells as a marker of DNA-specific autoreactivity. Investigators tested whether P2RY8 can modulate the autoreactive B cell generation in this context. Again, WT P2RY8 reduces the frequencies of lambda-1+–expressing follicular B cells, while P2RY8L257F-expressing B cells failed to promote this peripheral tolerance.
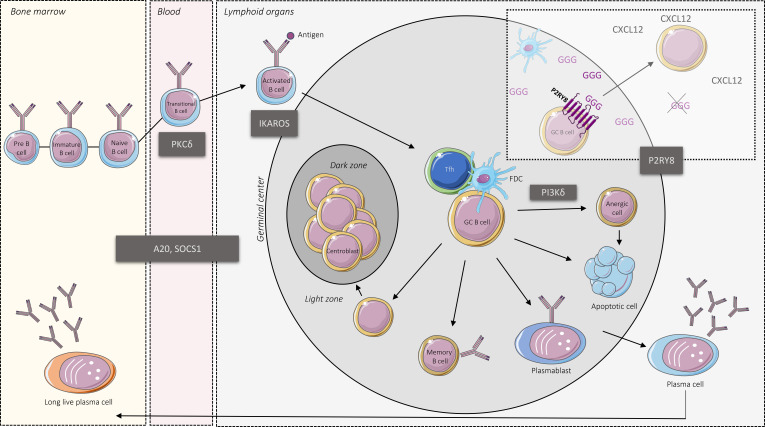
Interestingly, P2RY8 mutations have been previously described in GC-related lymphoma such as Burkitt lymphoma and DLBCL (Lohr et al., 2012). Other somatic genetic alterations identified in DLBCL (Lacy et al., 2020) are also reported as Mendelian cause of lupus, such as SOCS1 or TNFAIP3, responsible for SOCS1 or A20 haploinsufficiency, respectively (Li et al., 2020; Hadjadj et al., 2020). Variable dysregulation of AKT and ERK pathways were also reported in germline gain-of-function PIK3CD mutations defining the activated PI3K-δ syndrome and in protein kinase C-δ deficiency, while SOCS1 and A20 refer to JAK/STAT or NFKβ signaling inhibition (Li et al., 2020; Belot et al., 2013; Hadjadj et al., 2020); all these genetic conditions can drive lymphoproliferation and monogenic lupus (see figure). Actually, DLBCL shares more genetic anomalies with autoimmune disease than solid cancer (Din et al., 2019), and, as exemplified in the current work of He et al. (2021), DLBCL and lupus may represent two sides of the same coin. Thus, it is likely that new germline susceptibilities to lupus will be recognized in genes identified with recurrent somatic mutation in DLBCL, as illustrated here with germline mutations of P2RY8. While activation and cell migration are often linked, the specific role of spatial distribution versus cell activation in lymphomagenesis or tolerance break remains to be explored.
Monogenic causes of lupus and B cells tolerance break. GCs are specialized compartments within B cell follicles divided into two distinct zones: the dark zone, which is the site of proliferation and somatic hypermutation, and the light zone, where the cooperation between B cell, follicular dendritic cells (FDC), and T follicular helper (Tfh) allow for B cell selection. GCs represent critical checkpoints for terminal B cell differentiation. The main genetic causes of Mendelian lupus are represented together with P2RY8, which acts as a receptor for GGG enabling the B cell confinement into GC.
Lupus is a complex disease and heterogeneous with patients, encompassing a broad range of lesions that can damage virtually all tissues. While the cause of this autoimmune disease is still elusive, genetic factors have been underscored by two opposite genetic strategies. First, genome-wide association studies have identified in large cohorts of patients >100 susceptibility loci, often within the noncoding region (Bentham et al., 2015; Sun et al., 2016). Second, exploration of familial or early-onset or syndromic lupus have enabled the detection of rare and predicted pathogenic variants causal for the disease with a various level of penetrance (Omarjee et al., 2019). This study highlights the changes in the way we understand the complexity of lupus. Early-onset lupus is probably mainly driven by genetic factors. We have estimated that ≥7% of juvenile SLE is associated with the Mendelian genotype in a recent study (Belot et al., 2020), and the majority of genetic alteration detected in this population concerned innate immunity-related genes. In addition, accumulation of rare and predicted pathogenic variants was also seen in nonmonogenic SLE, suggesting an oligogenic nature of juvenile SLE. Disease-modifier genes are also called with stringent filtering in ∼20% of these cases, adding to the complexity of the disease. The assessment of the frequency of P2RY8 mutations in different population of lupus will be of great interest. To date, the main monogenic cause of lupus related to B cell tolerance breakdown is PKC-δ deficiency, in which the lupus phenotype is fully penetrant. In rare case of primary immunodeficiencies, SLE phenotype was variably reported in genes involved in recombination (RAG1, RAG2), BCR signaling (BTK, CD19, CD81, TNFRSF13B/TACI, PLCG2, PIK3CD), class switch recombination, and somatic hypermutation (AID, MHC-II genes, TNFRSF5/CD40, TNFSF5/CD40L), or in other molecules involved in hematopoietic differentiation (IKZF1/IKAROS), apoptosis regulation (CD95, CD95L, CASP10), or actin cytoskeleton biology (WAS), but in all cases the bona fide lupus phenotype is infrequent (Demirkaya et al., 2020). Following this work, P2RY8 will be added to the genetic screening for monogenic lupus diagnosis, and it will be interesting to explore whether other ethnicities can be also affected by deleterious variants in this gene, here identified in Australian and Asian populations.
This work also participates in changing the view of research and care for SLE. While a large number of patients with immune-mediated diseases have benefited from anti-cytokine therapies (Isenberg and Merrill, 2016), belimumab and anifrolumab are the unique biologicals that have been approved by the Food and Drug Administration over the last 30 yr in SLE. These disappointing results probably reflect the wide heterogeneity of the disease. Investigating early-onset lupus represents a powerful strategy in understanding the key factors driving the lupus pathogenesis (Omarjee et al., 2019). Here, the authors illustrate how genetic dissection of early-onset SLE can highlight new causal immunological pathways that can be also involved in adults. In P2RY8 deficiencies, inhibition of ERK activation or B cell–targeted therapies may be instrumental in treating the disease and offering tailored therapies. The growing description of Mendelian autoimmunity also raises interest in gene therapy strategies to ultimately cure these monogenic autoimmune diseases.
References
- Belot, A., et al. 2013. Arthritis Rheum. 10.1002/art.38008 [DOI] [Google Scholar]
- Belot, A., et al. 2020. Lancet Rheumatol. 10.1016/S2665-9913(19)30142-0 [DOI] [Google Scholar]
- Bentham, J., et al. 2015. Nat. Genet. 10.1038/ng.3434 [DOI] [Google Scholar]
- Berek, C., et al. 1991. Cell. 10.1016/0092-8674(91)90289-B [DOI] [Google Scholar]
- Demirkaya, E., et al. 2020. J. Clin. Med. 10.3390/jcm9030712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Din, L., et al. 2019. Genet. Epidemiol. 10.1002/gepi.22242 [DOI] [Google Scholar]
- Hadjadj, J., et al. 2020. Nat. Commun. 10.1038/s41467-020-18925-4 [DOI] [Google Scholar]
- He, Y., et al. 2021. J. Exp. Med. 10.1084/jem.20211004 [DOI] [Google Scholar]
- Isenberg, D.A., and Merrill J.T.. 2016. Expert Rev. Clin. Immunol. 10.1586/1744666X.2016.1112270 [DOI] [PubMed] [Google Scholar]
- Jacob, J., et al. 1991. Nature. 10.1038/354389a0 [DOI] [Google Scholar]
- Lacy, S.E., et al. 2020. Blood. 10.1182/blood.2019003535 [DOI] [Google Scholar]
- Li, G., et al. 2020. Medicine (Baltimore). 10.1097/MD.0000000000020232 [DOI] [Google Scholar]
- Lohr, J.G., et al. 2012. Proc. Natl. Acad. Sci. USA. 10.1073/pnas.1121343109 [DOI] [Google Scholar]
- Lu, E., et al. 2019. Nature. 10.1038/s41586-019-1003-z [DOI] [Google Scholar]
- Omarjee, O., et al. 2019. Autoimmun. Rev. 10.1016/j.autrev.2019.102361 [DOI] [PubMed] [Google Scholar]
- Sun, C., et al. 2016. Nat. Genet. 10.1038/ng.3496 [DOI] [Google Scholar]