Using mice engineered to exhibit specific defects in distinct signaling pathways downstream of STING, Li et al. demonstrate that TBK1, but not IRF3, recruitment to STING mediates autoinflammatory arthritis caused by DNase II deficiency.
Abstract
Defective DNA clearance in DNase II−/− mice leads to lethal inflammatory diseases that can be rescued by deleting cGAS or STING, but the role of distinct signaling pathways downstream of STING in the disease manifestation is not known. We found that the STING S365A mutation, which abrogates IRF3 binding and type I interferon induction, rescued the embryonic lethality of DNase II−/− mice. However, the STING S365A mutant retains the ability to recruit TBK1 and activate NF-κB, and DNase II−/− STING-S365A mice exhibited severe polyarthritis, which was alleviated by neutralizing antibodies against TNF-α or IL-6 receptor. In contrast, the STING L373A mutation or C-terminal tail truncation, which disrupts TBK1 binding and therefore prevents activation of both IRF3 and NF-κB, completely rescued the phenotypes of DNase II−/− mice. These results demonstrate that TBK1 recruitment to STING mediates autoinflammatory arthritis independently of type I interferons. Inhibiting TBK1 binding to STING may be a therapeutic strategy for certain autoinflammatory diseases instigated by self-DNA.
Introduction
Autoimmune and inflammatory diseases, including systemic lupus erythematosus (SLE) and rheumatoid arthritis (RA), are characterized by the loss of immune self-tolerance and subsequent multiorgan inflammation and destruction. Although the causes of autoimmune diseases are often complex and involve multiple genetic and environmental factors, the accumulation of DNA from dying cells and defective DNA clearance are considered critical factors that contribute to the disease pathogenesis (Mahajan et al., 2016). Consistent with this idea, mutations in DNases were found to be associated with SLE and other autoimmune diseases (Keyel, 2017).
Among DNase-deficient mouse models, DNase II−/− mice show embryonic lethality accompanied by inflammation and anemia (Kawane et al., 2001). DNase II is an endonuclease in the lysosome, where it degrades exogenous DNA engulfed by macrophages. During development, DNase II–deficient macrophages fail to digest nuclear DNA expelled from erythroid precursor cells, and the subsequent DNA accumulation causes inflammation and embryonic lethality (Kawane et al., 2001). For SLE patients, DNase II polymorphisms were associated with a risk of renal disorder (Shin et al., 2005). In addition, biallelic loss-of-function mutations in the DNase II gene were found in patients displaying severe neonatal anemia, glomerulonephritis, liver fibrosis, and deforming arthropathy (Rodero et al., 2017).
Type I IFN production is responsible for the lethality of DNase II−/− mice; depleting the type I IFN receptor 1 (IFNAR1) allowed DNase II−/− mice to survive to adulthood (Kawane et al., 2006; Yoshida et al., 2005). However, DNase II−/− Ifnar1−/− mice developed chronic polyarthritis that resembles RA (Kawane et al., 2006). Inflammatory cytokines such as TNF-α, IL-1β, and IL-6 were major drivers of this polyarthritis (Kawane et al., 2006; Kawane et al., 2010). Strikingly, ablating members of the cytosolic DNA-sensing pathway such as cyclic GMP-AMP synthase (cGAS) or stimulator of IFN genes (STING) rescued DNase II−/− mice from both embryonic lethality and inflammatory arthritis (Ahn et al., 2012; Gao et al., 2015).
cGAS is a DNA sensor that detects double-stranded DNA invading the cytosol of a cell (Sun et al., 2013; Wu et al., 2013). cGAS binds DNA in a sequence-independent manner, allowing the detection of not only microbial DNA but also self-DNA that arises from cellular damage or defective DNA clearance (Ablasser et al., 2014; Ahn et al., 2012; Gall et al., 2012; Gao et al., 2015; Gray et al., 2015). Upon DNA binding, cGAS synthesizes a second messenger cyclic GMP-AMP (cGAMP) that binds to the downstream adaptor protein STING on the ER membrane (Ishikawa and Barber, 2008; Sun et al., 2009; Wu et al., 2013; Zhong et al., 2008). Activated STING proteins oligomerize and recruit TANK-binding kinase 1 (TBK1) to the [(D or E)xPxPLR(S or T)D] motif (in which “x” denotes any amino acid) at the C-terminal tail (CTT) of STING (Zhang et al., 2019). TBK1 phosphorylates the CTT of a neighboring STING, including the serine residue within the IFN regulatory factor 3 (IRF3) binding [pLxIS] motif (in which “p” represents a hydrophilic residue), thereby recruiting IRF3 to the phosphorylated STING (Liu et al., 2015; Tanaka and Chen, 2012; Zhang et al., 2019). Using STING as a scaffold, TBK1 then phosphorylates IRF3, which translocates to the nucleus to induce type I IFNs (Ishikawa and Barber, 2008; Sun et al., 2009; Zhong et al., 2008). TBK1 and its homologue IκB kinase ε (IKKε) also lead to activation of the IKK complex, which then activates the transcription factor NF-κB (Abe and Barber, 2014; Balka et al., 2020). NF-κB promotes IFN production by synergizing with IRF3 and also induces proinflammatory cytokines independently of IRF3. In addition, STING induces noncanonical autophagy to clear DNA or pathogens from the cytosol (Gui et al., 2019).
Inflammatory arthritis observed in DNase II−/− Ifnar1−/− mice (Kawane et al., 2006), but not in DNase II−/− Sting−/− mice (Ahn et al., 2012) or DNase II−/− cGas−/− mice (Gao et al., 2015), indicates that STING activation by self-DNA is responsible for the inflammatory cytokine production. Although NF-κB activation was suggested to induce inflammatory cytokines in vitro, the role of STING-induced cytokines by self-DNA in vivo remains elusive. We recently generated STING mutant mice with distinct signaling defects: S365A, L373A, and CTT truncation (Yum et al., 2021). The STING-S365A mutation disrupts the IRF3 binding site and thus selectively ablates IFN production while retaining TBK1 binding and NF-κB activation, as well as the ability to induce autophagy. The STING-ΔCTT and STING-L373A mutations disrupt the TBK1 binding site and are predicted to also disrupt IKKε binding based on sequence homology, although this remains to be tested. Cells harboring the STING-ΔCTT and STING-L373A mutations lacked phosphorylation of TBK1 and IKKε and the subsequent activation of IRF3 and NF-κB in response to stimulation (Balka et al., 2020; Yum et al., 2021).
To determine the molecular mechanism of inflammatory arthritis caused by defects in DNA clearance, we generated DNase II−/− STING mutant (S365A, L373A, or ΔCTT) mice. Here we show that all DNase II−/− STING mutant mice were rescued from lethality and survived into adulthood. However, DNase II−/− STINGS365A/S365A mice, but not STINGΔCTT/ΔCTT or STINGL373A/L373A mice, still displayed polyarthritis, footpad inflammation, and elevated levels of proinflammatory cytokines. Blocking TNF-α or IL-6 signaling using neutralizing antibodies alleviated arthritis manifestations in DNase II−/− STINGS365A/S365A mice. These results demonstrate that the recruitment of TBK1 but not IRF3 to STING is essential for the inflammatory polyarthritis caused by defective DNA clearance.
Results
DNase II−/− STINGS365A/S365A mice, but not DNase II−/− STINGΔCTT/ΔCTT or DNase II−/− STINGL373A/L373A mice, develop polyarthritis
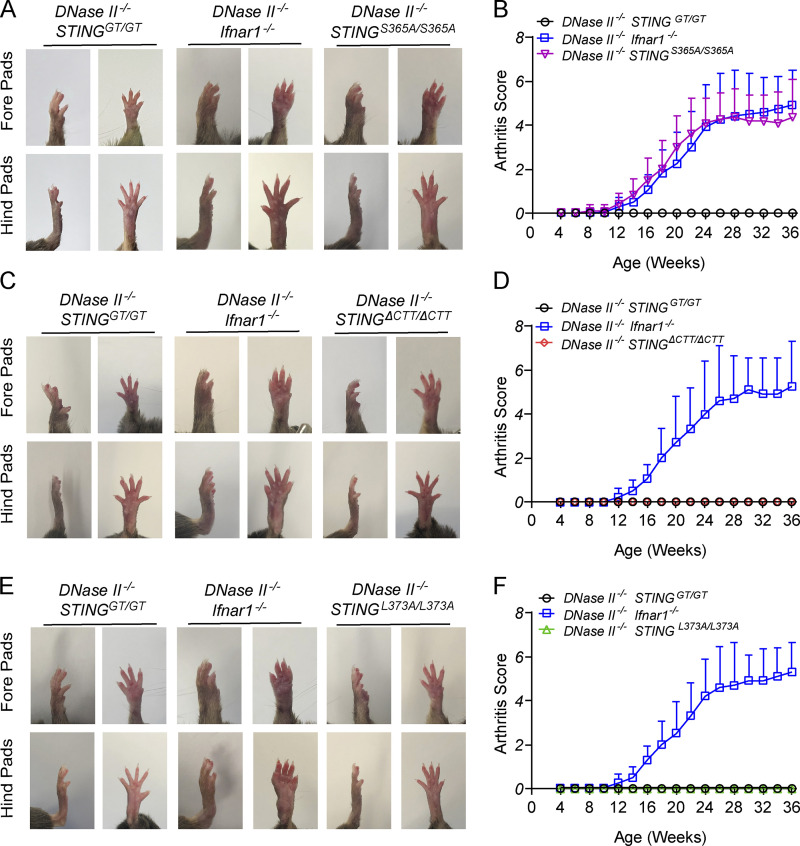
While DNase II−/− mice are embryonically lethal (Kawane et al., 2001), all DNase II−/− mice harboring STING mutations, including S365A, L373A, ΔCTT, or Goldenticket (gt; which lacks STING expression; Sauer et al., 2011), were born normally, similar to DNase II−/− Ifnar1−/− mice. However, DNase II−/− Ifnar1−/− and DNase II−/−STINGS365A/S365A mice still developed polyarthritis characterized by swelling of footpads starting from 10–12 wk of age (Fig. 1, A and B). In contrast, DNase II−/− STINGΔCTT/ΔCTT mice were completely rescued from the polyarthritis (Fig. 1, C and D), suggesting that the interaction of STING with downstream mediators through the CTT is required for the disease phenotypes. Similarly, DNase II−/− STINGL373A/L373A mice (Fig. 1, E and F) did not manifest any signs of arthritis, specifically indicating that TBK1 recruitment to STING mediates the arthritis development caused by defective DNA clearance. No apparent sex difference in phenotypes was observed for all mice used in this study.
Figure 1.
TBK1 recruitment to STING is required for polyarthritis in DNase II−/− mice. (A, C, and E) Representative images of 8-mo-old male mouse footpads. (B, D, and F) Arthritis scores of the forelimb and hindlimb (n = 10–12 in each genotype). Results are representative of two independent experiments.
TBK1 recruitment to STING causes inflammation in footpads of DNase II−/− mice
H&E staining of footpads of DNase II−/− Ifnar1−/− and DNase II−/− STINGS365A/S365A mice confirmed aggressive synovitis and bone erosion (Fig. 2, A–C). In addition, more osteoclasts were found in the joints of DNase II−/− STINGS365A/S365A mice, indicated by the staining of tartrate-resistant phosphatase (TRAP; Fig. 2, D and E). Infiltration of inflammatory cells, bone erosion, and osteoclast accumulation did not occur in DNase II−/− STINGGT/GT, STINGΔCTT/ΔCTT, or STINGL373A/L373A mice (Fig. 2, A–E). These data indicate that TBK1 recruitment to STING mediates inflammation of DNase II−/− mice.
Figure 2.
TBK1 recruitment to STING induces inflammation and bone erosion in DNase II−/− mouse footpads. (A–C) H&E-stained joint tissues from hind footpads of 8-mo-old male mice (n = 5–8 in each genotype). (A) Representative images. Scale bars, 200 µm. (B and C) Blinded analyses of histologic scores calculated as described in Materials and methods (mean ± SD, n = 5–8 per genotype). (D) Representative images of TRAP-stained joint sections from 8-mo-old male mice. Scale bars, 200 µm. (E) TRAP scores calculated as described in Materials and methods (mean ± SD, n = 5–8 per genotype). For B, C, and E, each dot represents an individual mouse. Statistical analysis was done using a two-tailed, unpaired Student’s test. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. The histologic scores were evaluated separately by two pathologists.
TBK1 recruitment to STING induces inflammatory cytokines and increases blood monocytes in DNase II−/− mice
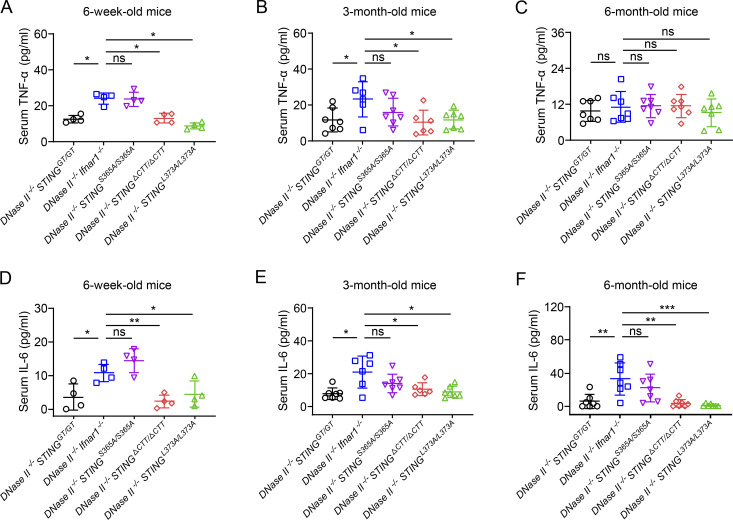
TNF-α and IL-6 are key cytokines that drive inflammation in arthritis; TNF-α levels were elevated in patients with loss-of-function mutations in the DNase II gene (Rodero et al., 2017), and both TNFα and IL-6 levels were high in DNase II−/− Ifnar1−/− mice (Kawane et al., 2006; Kawane et al., 2010). We detected systemic production of TNF-α and IL-6 in the sera of 6-wk-old DNase II−/− Ifnar1−/− and DNase II−/− STINGS365A/S365A mice before the development of arthritis (Fig. 3, A and D). This early production of STING-induced TNF-α and IL-6 was completely abrogated in DNase II−/− STINGΔCTT/ΔCTT and DNase II−/− STINGL373A/L373A mice (Fig. 3, A and D). The levels of serum TNF-α in DNase II−/− Ifnar1−/− and DNase II−/− STINGS365A/S365A mice gradually decreased in 3-mo-old (Fig. 3 B) and 6-mo-old (Fig. 3 C) mice, whereas the level of serum IL-6 gradually increased in older mice (Fig. 3, E and F). The kinetics of TNF-α and IL-6 production in these mice is consistent with previous reports suggesting that macrophages and synovial fibroblasts are the major sources of TNF-α and IL-6, respectively (Kinne et al., 2000; Yoshitomi, 2019).
Figure 3.
TBK1 recruitment to STING elevates serum TNF-α and IL-6 levels in DNase II−/− mice. (A–C) TNF-α levels in the serum of 6-wk-old male (A), 3-mo-old female (B), and 6-mo-old female (C) mice of the indicated genotypes (mean ± SD, n = 4–7 per genotype). (D–F) IL-6 levels in the serum of 6-wk-old male (D), 3-mo-old female (E), and 6-mo-old female (F) mice of the indicated genotypes (mean ± SD, n = 4–7 per genotype). Each dot represents an individual mouse. Statistical analysis was done using a two-tailed, unpaired Student’s test. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Results are representative of two independent experiments.
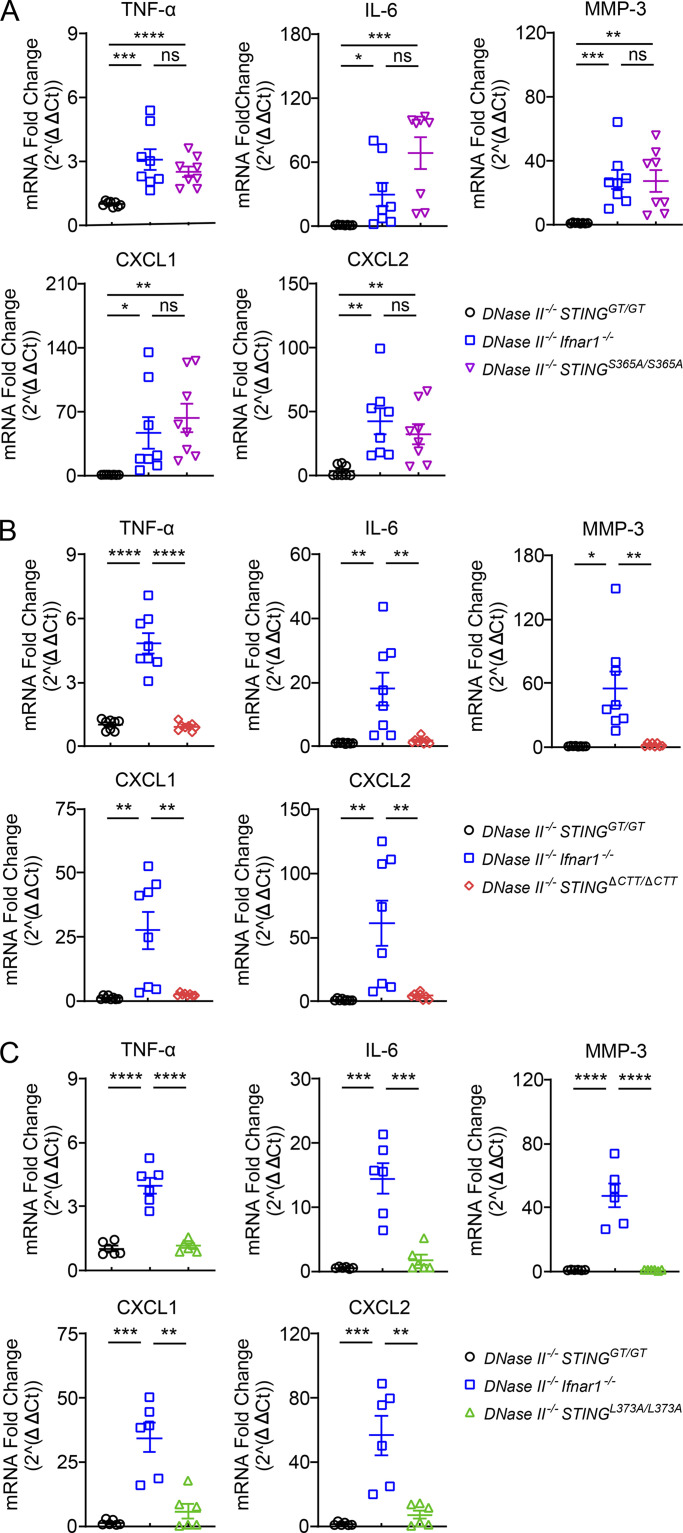
In addition to TNF-α and IL-6 that induce inflammation in the joints, matrix metallopeptidase 3 (MMP-3) degrades the extracellular matrix to drive bone erosion and is used as a marker of RA activity (Green et al., 2003; Tchetverikov et al., 2003). The expression of TNF-α, IL-6, and MMP-3 were all up-regulated in the paws of 8-mo-old DNase II−/− Ifnar1−/− and DNase II−/− STINGS365A/S365A mice but not in the paws of DNase II−/− STINGΔCTT/ΔCTT and DNase II−/− STINGL373A/L373A mice (Fig. 4, A–C). Our previous transcriptome analysis of bone marrow–derived macrophages showed that activation of STING-S365A induces high levels of C-X-C motif chemokine ligand 1 (CXCL1) and CXCL2 (Yum et al., 2021), which can recruit monocytes and neutrophils to the site of inflammation. Inflamed mouse paws from DNase II−/− Ifnar1−/− and DNase II−/− STINGS365A/S365A mice, but not from other STING mutant mice, expressed high levels of CXCL1 and CXCL2 (Fig. 4, A–C).
Figure 4.
TBK1 recruitment to STING induces inflammatory cytokines in DNase II−/− mouse paws. (A–C) Quantitative RT-PCR analysis of inflammatory cytokine levels in paws from 8-mo-old male mice of the indicated genotypes (mean ± SD, n = 6–8 per genotype). Each dot represents a mouse. Statistical analysis was done using a two-tailed, unpaired Student’s test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Results are representative of four independent experiments. Ct, cycle threshold.
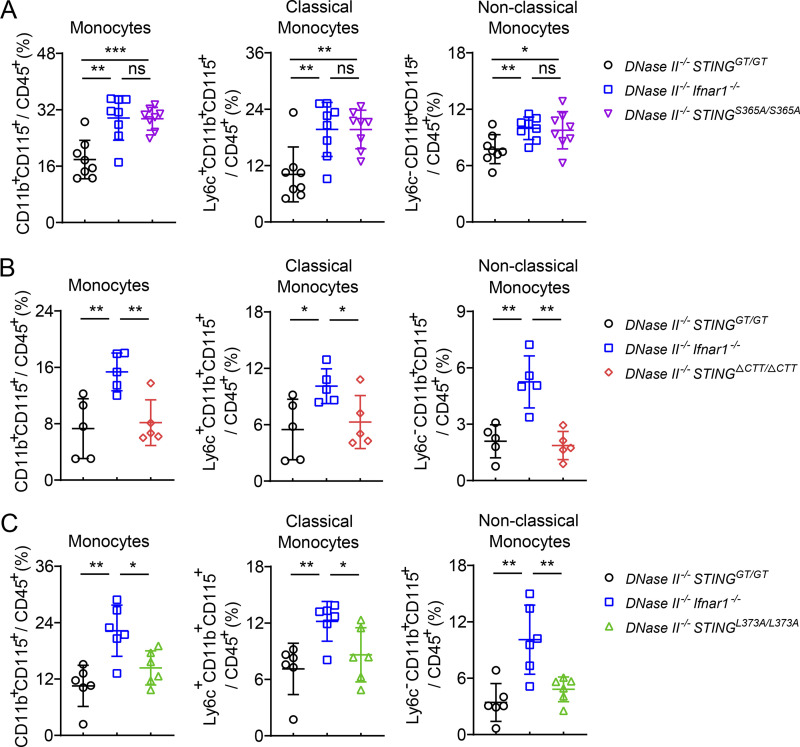
Blood circulating monocytes are recruited into the RA synovium and contribute to inflammatory and destructive processes of the synovial joints (Rana et al., 2018). Furthermore, nonclassic monocytes are the precursors of macrophages that drive inflammation and bone erosion in different inflammatory arthritis models (Misharin et al., 2014). We observed increased numbers of classic monocytes (Ly6c+) and nonclassic monocytes (Ly6c−) in DNase II−/− Ifnar1−/− and DNase II−/− STINGS365A/S365A mice compared to DNase II−/− STINGGT/GT mice (Fig. 5 A). These circulating monocyte levels were significantly decreased in DNase II−/− STINGΔCTT/ΔCTT and DNase II−/− STINGL373A/L373A mice (Fig. 5, B and C), suggesting that TBK1 recruitment to STING induces circulating monocytes that may lead to increased inflammatory cells and cytokines in the joints.
Figure 5.
TBK1 recruitment to STING increases blood monocyte populations in DNase II−/− mice. (A–C) FACS analysis of monocytes in the blood from 2–3-mo-old female mice of the indicated genotypes (mean ± SD, n = 5–8 in each genotype). Each dot represents a mouse. Statistical analysis was done using a two-tailed, unpaired Student’s test. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Results are representative of two independent experiments.
TNF-α and IL-6 mediate inflammatory arthritis in DNase II−/− STINGS365A/S365A mice
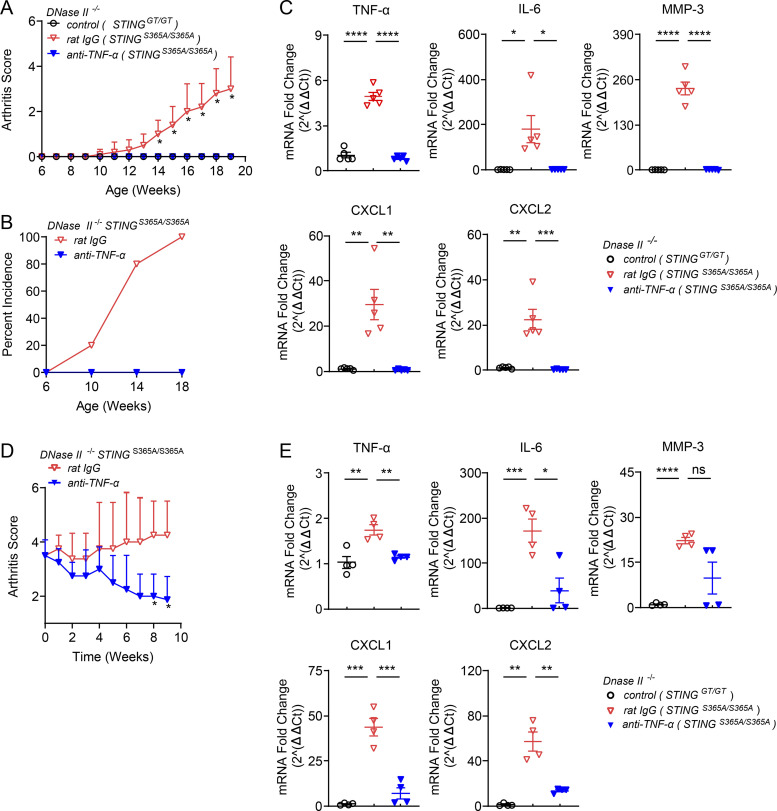
Various cytokines are abundant in RA tissues, and TNF-α was shown to drive the production of other cytokines to induce inflammation (Feldmann et al., 1996). Blocking TNF-α provided both protective and therapeutic effects against arthritis in DNase II−/− Ifnar1−/− mice (Kawane et al., 2006). To determine if TNF-α is responsible for developing inflammatory arthritis in DNase II−/− STINGS365A/S365A mice, we injected neutralizing antibodies against TNFα (anti–TNF-α) into mice once a week starting from 6 wk of age. The control IgG–treated group developed polyarthritis after 10 wk of age, but the anti–TNF-α–treated group was protected from arthritis development (Fig. 6, A and B). Blocking TNF-α also abrogated the induction of TNF-α, IL-6, MMP-3, CXCL1, and CXCL2 in mouse paws (Fig. 6 C), suggesting that TNF-α drives inflammatory gene expression in DNase II−/− STINGS365A/S365A mice.
Figure 6.
Blocking TNF-α prevents the development of inflammatory arthritis in DNase II−/− STINGS365A/S365A mice. (A–C) 6-wk-old DNase II−/− STINGS365A/S365A female mice received rat IgG or anti–TNF-α (40 µg/g body weight) i.p. once a week (n = 5 in each treatment group). (A) Arthritis scores were assessed every week (mean ± SD, n = 5 per group). Statistical analysis was done using a two-tailed, unpaired Student’s test. *, P < 0.05 between anti–TNF-α– and rat IgG–treated mice. (B) Incidence rates of arthritis (mean ± SD, n = 5 in each group). (C) qRT-PCR analysis of inflammatory cytokines in mouse paws (mean ± SD, n = 5 in each group). Each dot represents a mouse. (D and E) 5-mo-old male DNase II−/− STINGS365A/S365A mice displaying polyarthritis received rat IgG or anti–TNF-α (20 µg/g body weight) twice a week (n = 4 in each group). (D) Arthritis scores were assessed weekly (mean ± SD, n = 4 per group). *, P < 0.05 between rat IgG– and anti–TNF-α–treated mice. (E) qRT-PCR analysis of inflammatory cytokines in mouse paws (mean ± SD, n = 4 per group). Statistical analysis was done using a two-tailed, unpaired Student’s test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Results are representative of two independent experiments. Ct, cycle threshold.
To evaluate the therapeutic effect of TNF-α blockade, we treated 5-mo-old DNase II−/− STINGS365A/S365A mice that had already developed arthritis with anti–TNF-α (Fig. 6, D and E). Importantly, blocking TNF-α alleviated arthritis (Fig. 6 D) and greatly reduced the expression levels of inflammatory genes (Fig. 6 E).
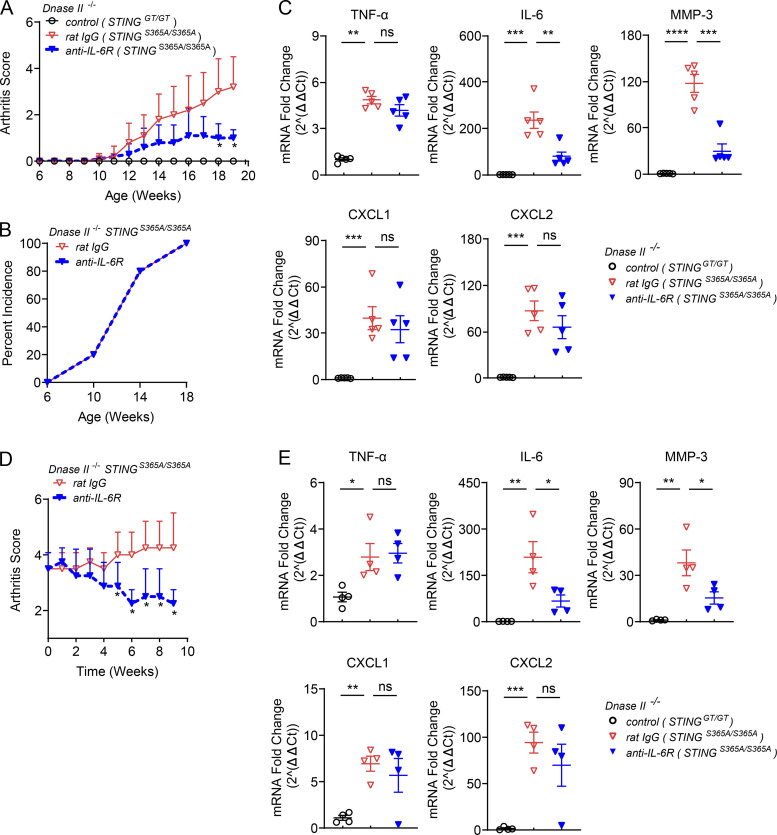
IL-6 is a mediator of multiple inflammatory diseases (Kishimoto, 2005), including arthritis in DNase II–deficient mice (Kawane et al., 2010). To determine the role of IL-6 in the development of arthritis, we treated 6-wk-old DNase II−/− STINGS365A/S365A mice with a neutralizing antibody against the IL-6 receptor (anti–IL-6R) once a week. Blocking IL-6 signaling reduced arthritis scores (Fig. 7 A) but did not block the incidence of arthritis (Fig. 7 B). The expression levels of IL-6 and MMP-3 in mouse paws were reduced by anti–IL-6R treatment; however, levels of TNF-α, CXCL1, and CXCL2 remained unchanged (Fig. 7 C). Treating the 5-mo-old DNase II−/− STINGS365A/S365A mice with anti–IL-6R reduced the arthritis score (Fig. 7 D) and the expression of IL-6 and MMP-3 but did not reduce the expression of TNF-α, CXCL1, and CXCL2 (Fig. 7 E).
Figure 7.
Blocking IL-6R alleviates polyarthritis and joint inflammation in DNase II−/− STINGS365A/S365A mice. (A–C) 6-wk-old DNase II−/− STINGS365A/S365A female mice received rat IgG or anti–IL-6R (100 µg/g body weight) once i.v., followed by i.p. treatment of antibodies (25 µg/g body weight) once a week (n = 5 in each treatment group). (A) Arthritis scores (mean ± SD, n = 5 per group). Statistical analysis was done using a two-tailed, unpaired Student’s test. *, P < 0.05 between rat IgG– and anti–IL-6R–treated mice. (B) Incidence rates of arthritis (n = 5 per group). (C) qRT-PCR analysis of inflammatory cytokines in mouse paws (mean ± SD, n = 5 in each group). Each dot represents a mouse. (D and E) 5-mo-old male DNase II−/− STINGS365A/S365A mice displaying polyarthritis received rat IgG or anti–IL-6R (100 µg/g body weight) once, followed by i.p. treatment of antibodies (25 µg/g body weight) once a week (n = 4 in each group). (D) Arthritis scores. *, P < 0.05 between rat IgG– and anti–IL-6R–treated mice. (E) qRT-PCR analysis of inflammatory cytokines in mouse paws (mean ± SD, n = 4 per group). Statistical analysis was done using a two-tailed, unpaired Student’s test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Results are representative of two independent experiments. Ct, cycle threshold.
Altogether, these results demonstrate a critical role for TNF-α and IL-6 in driving inflammatory arthritis in DNase II−/− STINGS365A/S365A mice downstream of TBK1 recruitment to STING. Blocking these cytokines was shown to improve the symptoms of human arthritis (Choy et al., 2020; Ma and Xu, 2013) but did not fully control the disease, probably due to other cytokines that can also cause inflammation (Brennan and McInnes, 2008). Treatments that can block the upstream STING-TBK1 interaction may provide benefits for self-DNA–induced inflammatory arthritis by inhibiting the production of most if not all inflammatory cytokines.
Discussion
Previous studies in mouse models with defective DNA clearance revealed a critical role for the cGAS–STING pathway in mediating autoimmunity (Ablasser et al., 2014; Ahn et al., 2012; Gall et al., 2012; Gao et al., 2015; Gray et al., 2015). Excessive type I IFN signaling mediated embryonic lethality in DNase II–deficient mice (Kawane et al., 2006; Yoshida et al., 2005); however, DNase II−/− Ifnar1−/− mice still developed severe polyarthritis (Kawane et al., 2006). In this study, we used three different DNase II−/− STING mutant mice that have distinct signaling defects (S365A, L373A, and ΔCTT) to determine the molecular mechanism downstream of STING activation that is necessary for inducing cytokines and inflammatory arthritis. We found that DNase II−/− STINGS365A/S365A mice, but not DNase II−/− STINGΔCTT/ΔCTT or DNase II−/− STINGL373A/L373A mice, developed polyarthritis characterized by inflammation and bone erosion due to the production of inflammatory cytokines such as TNF-α and IL-6. As the STING-L373A mutation in DNase II−/− mice completely abrogated disease phenotypes, these results show that TBK1 recruitment to STING not only mediates IFN-dependent lethality but also drives the IFN-independent inflammatory arthritis caused by defective DNA clearance. The L373A mutation in the TBK1-binding motif of STING may also disrupt IKKε recruitment, because the sequence of TBK1 involved in binding to STING is highly conserved in IKKε. IKKε is an inducible gene, and its expression levels are usually lower than those of TBK1 in most cells. Nevertheless, IKKε and TBK1 function redundantly in activating NF-κB downstream of STING in myeloid cells (Balka et al., 2020). Thus, the observed phenotypes of the STING-L373A mice are likely caused by impairment of both TBK1 and IKKε recruitment; however, this has yet to be demonstrated directly.
Because the STING-L373A and S365A mutants still induce autophagy, our data shows that STING-induced autophagy does not promote or prevent disease pathogenesis during DNase II deficiency. Although STING activation was shown to cause cell death in some cells such as T lymphocytes in an IFN-independent manner (Gaidt et al., 2017; Gulen et al., 2017; Larkin et al., 2017), the contribution of STING-induced cell death in this arthritis model may be minor, as blocking TNF-α alone prevented the development of arthritis in DNase II−/− STINGS365A/S365A mice (Fig. 6, A and B). Therefore, The NF-κB pathway induced by TBK1 recruitment to STING is the most likely candidate for inducing these inflammatory cytokines.
While DNase II degrades DNA in the lysosome, three-prime repair exonuclease 1 (TREX1 or DNase III) degrades DNA in the cytosol (Yang et al., 2007). Loss-of-function mutations of the TREX1 gene caused autoinflammatory diseases such as Aicardi-Goutières syndrome and familial chilblain lupus (Crow et al., 2006; Rice et al., 2015; Richards et al., 2007; Tüngler et al., 2012). TREX1-deficient mice survived until adulthood but developed severe autoinflammatory disease (Morita et al., 2004; Stetson et al., 2008). Similar to DNase II−/− mice, disease phenotypes of Trex1−/− mice such as mortality and inflammation were rescued by the loss of cGAS or STING (Gall et al., 2012; Gao et al., 2015; Gray et al., 2015). These results suggest that cGAS–STING pathway activation is a major cause of autoinflammatory diseases triggered by self-DNA.
Unlike DNase II−/− mice that develop inflammatory arthritis in an IFN-independent manner, Trex1−/− mice lacking IRF3 or IFNAR1 were rescued from lethality and inflammation (Stetson et al., 2008). What causes these discrepancies of IFN dependence in these DNase-deficient models? It is conceivable that more cytoplasmic DNA may accumulate in DNase II−/− cells than in Trex1−/− cells. When cytoplasmic DNA is limited, intracellular cGAS concentrations (i.e., one allele vs. two alleles of the cGAS gene expression) become critical for cGAS activation, because its activation requires DNA-induced liquid phase separation, which depends on the concentrations of both cGAS and DNA (Du and Chen, 2018). DNase II−/− mice required the deletion of both alleles of the cGAS gene to rescue mice from lethality, whereas deleting one allele of the cGAS gene was sufficient to largely eliminate disease phenotypes in Trex1−/− mice (Gao et al., 2015), suggesting abundant levels of cytoplasmic DNA and stronger activation of the cGAS–STING pathway in DNase II−/− mice. Therefore, robust activation of the STING-induced NF-κB pathway may contribute to the disease phenotypes more strongly in DNase II−/− mice than in Trex1−/− mice. In addition, the types of cells being activated may differ in the absence of DNase II versus TREX1. In DNase II–deficient mice, macrophages are believed to be the major source of inflammation, as they are responsible for engulfing and digesting nuclei from apoptotic cells (Nagata, 2005). The contribution of macrophage-induced inflammation to arthritis remains to be determined in vivo by depleting macrophages or specifically deleting the STING gene in macrophages during DNase II deficiency. In this regard, DNase II−/− STINGS365A/S365A mice, which survive into adulthood and develop arthritis, provide a good model to study the cell types driving the inflammation. The role of neutrophils or monocytes in arthritis also needs to be tested, as the chemoattractant CXCL1 and CXCL2 were up-regulated in DNase II−/− STINGS365A/S365A mouse paws (Fig. 4, A–C). The cell types responsible for the disease phenotypes of Trex1−/− mice require further investigation.
cGAS, activated by cytosolic DNA, produces cGAMP that binds to the cytosol-facing cGAMP-binding site of STING. Defects in TREX1 functions lead to the accumulation of cytosolic DNA that can directly bind to cGAS, whereas defects in DNase II result in undigested DNA in lysosomes. Abnormal macrophages carrying undigested DNA were found in the fetal liver and thymus of DNase II−/− mice (Kawane et al., 2001; Kawane et al., 2003). How lysosomal DNA reaches the cytosol to activate cGAS is still unclear, but studies have suggested that the lysosome is a vulnerable organelle that can undergo ruptures (Terman et al., 2006). As excessive amounts of DNA accumulate in lysosomes in the absence of DNase II, some lysosomes may undergo rupture and release DNA into the cytosol.
It is surprising that DNase II−/− mice do not display any overt phenotypes in the absence of cGAS or STING, suggesting that these mice can tolerate excess DNA that is not digested in the lysosome. One would have expected that the excess cellular DNA could activate other DNA sensors, such as TLR9 and absent in melanoma 2 (AIM2), to cause inflammatory diseases. The embryonic lethality of the DNase II−/− mice could not be rescued by deleting the AIM2 gene, although AIM2 deficiency reduced inflammatory arthritis in DNase II−/− Ifnar1−/− mice (Baum et al., 2015; Jakobs et al., 2015). Ablating TLR9 failed to rescue the lethality of DNase II−/− mice (Okabe et al., 2005) and failed to reduce the arthritis of DNase II−/− Ifnar1−/− mice (Kawane et al., 2006). However, 10-mo-old DNase II−/− Ifnar1−/− mice have been reported to develop splenomegaly and produce antinuclear antibodies in a STING- or AIM2-independent manner; instead, Unc93b, which regulates the localization of several TLRs (e.g., TLR3, TLR7, and TLR9) to the endosome, is required (Baum et al., 2015; Pawaria et al., 2015). Mice may elicit other signaling pathways including the TLR9 pathway after the long-term presence of lysosomal DNA during DNase II−/− deficiency. Further research is needed to determine why defective DNA clearance in the DNaseII−/− mice primarily activates cGAS but not AIM2 or TLR9.
In summary, using DNase II−/− STING mutant mouse models, our study provides the molecular mechanism by which STING activation causes autoinflammatory arthritis during DNase II deficiency. TBK1 recruitment to STING is required not only for IFN signaling that causes embryonic lethality but also for producing inflammatory cytokines that drive arthritis. These data suggest that an inhibitor interfering with TBK1 binding to STING may provide therapeutic benefits for self-DNA–induced inflammatory diseases such as arthritis. Interfering with TBK1 recruitment does not affect STING-induced autophagy but specifically abrogates cytokine production including, but not limited to, type I IFNs, TNF-α, and IL-6. This molecular mechanism may also apply to other STING-dependent autoimmunity. In this regard, a mouse model of STING-associated vasculopathy with onset in infancy that harbors gain-of-function mutations of STING developed inflammation even in the absence of IRF3 (Warner et al., 2017). Further studies on the role and molecular mechanism of STING-driven inflammation will provide new insights into drug development for autoimmune diseases.
Materials and methods
Mice and treatments
All mice used in this study were on the C57BL/6 background. DNase II+/− mice were from Dr. Shigekazu Nagata (Kyoto University, Kyoto, Japan) and crossed with IFNAR1−/− mice from Dr. David Farrar (University of Texas Southwestern Medical Center, Dallas, TX). STINGΔCTT/ΔCTT, STINGL373A/L373A, and STINGS365A/S365A mice were generated by the CRISPR/Cas9 technology in our laboratory as previously described (Yum et al., 2021). STINGGT/GT mice were purchased from The Jackson Laboratory. Genomic DNA from toes or tails was extracted using KAPA Express Extract Kits (Roche) for genotyping. All mice were bred and maintained under specific pathogen–free conditions in the animal facility at the University of Texas Southwestern Medical Center according to experimental protocols approved by the Institutional Animal Care and Use Committee. Mice were not littermate controlled because of the relatively low number of pups in each litter and the need for a large number of mice for the experiments. Wild-type C57BL/6 mice were purchased from The Jackson Laboratory. Mice were gender- and age-matched for all experiments.
InVivoMAb anti–TNF-α (XT3.11), rat IgG (HRPN), and anti–IL-6R (15A7) were from BioXcell. 6- or 5-mo-old STINGS365A/S365A mice were treated with the indicated amounts of each antibody by i.p. injection once or twice a week as shown in the figure legends. For anti–IL-6R treatment, the mice were first treated with the antibody i.v., followed by i.p. injection once a week.
Clinical assessment and pathology
The arthritis score of the forelimb and hindlimb joints was assessed manually: 0, no swelling; 1, mild swelling; and 2, severe swelling or deformation of the limb or finger (Kawane et al., 2006). The scores of four limbs were combined to give a total arthritis score for each mouse. For pathology, joint tissues were fixed in 4% (wt/vol) paraformaldehyde, decalcified in 11.4% EDTA, embedded in paraffin, cut into 5-µm sections, and stained with H&E or TRAP. Histologic inflammation and bone erosion scores were measured as described previously (Scales et al., 2016). For inflammatory cell infiltrates, the scoring system was as follows: 0, no infiltrate detected; 1, modest leukocyte infiltration in synovial tissue; 2, moderate leukocyte infiltration in synovial tissue; and 3, gross leukocyte infiltration in synovial membrane with significant loss of synovial and articular architecture. For erosion of cartilage and bone: 0, no abnormalities; 1, fibrillation of cartilage and/or mild erosive infiltration of periosteal and subchondral bone; 2, moderate fibrillation and loss of cartilage and/or moderate erosive infiltration of periosteal and subchondral bone; and 3, significant loss of cartilage and/or erosive infiltration of periosteal and subchondral bone.
TRAP staining was scored on a scale of 0–5 as follows: 0, no TRAP-positive cells; 1, rare TRAP-positive cells in BM or soft tissues; 2, few TRAP-positive cells seen in areas of resorption; 3, moderate TRAP-positive cells at least one site of resorption or attached to periarticular sites without bone erosion; 4, moderate TRAP-positive staining at multiple areas of resorption or within resorption pits; and 5, marked TRAP-positive cells at most or all areas of bone resorption (O’Brien et al., 2016). Images were acquired using an E400 microscope (Nikon) and NES software.
Quantitative RT-PCR (qRT-PCR)
RNA was isolated from homogenized mouse paws using TRIzol Reagent (Invitrogen). Reverse transcription and qRT-PCR were done using the cDNA reverse transcription kit and SYBR green master mix from Applied Biosystems following the manufacturer’s instructions. The primer sequences used for qRT-PCR are listed in Table 1.
Table 1. Mouse qRT-PCR primers.
| Gene | Forward (5′ to 3′) | Reverse (5′ to 3′) |
|---|---|---|
| GAPDH | TGGCAAAGTGGAGATTGTTGCC | AAGATGGTGATGGGCTTCCCG |
| TNF-α | TCCCAGGTTCTCTTCAAGGGA | GGTGAGGAGCACGTAGTCGG |
| IL-6 | TCCAGTTGCCTTCTTGGGAC | GTACTCCAGAAGACCAGAGG |
| MMP-3 | ACTCTACCACTCAGCCAAGG | TCCAGAGAGTTAGACTTGGTGG |
| CXCL1 | TCCAGAGCTTGAAGGTGTTG | GTCTGTCTTCTTTCTCCGTTACTT |
| CXCL2 | ATGCCTGAAGACCCTGCCAAG | GGTCAGTTAGCCTTGCCTTTG |
Flow cytometry and ELISA
Blood was collected into EDTA anticoagulant tubes and stained with various fluorescent dye–conjugated antibodies targeting the following antigens from BioLegend: Ly6G-FITC (1A8), CD115-PE (AFS98), CD11b-PerCP (M1/70), Ly6c-AF647 (HK1.4), CD11c-BV605 (N418), and CD45-AF700 (30-F11). For FACS staining, anti–mouse CD16/32 (BioLegend) was added into 100 µl of anticoagulated blood in a 1:200 ratio and incubated for 10 min. Fluorescent dye–conjugated antibodies were then added to the blood in a 1:200 ratio and incubated at room temperature for 30 min in the dark. 2 ml of 1× RBC lysis solution (349202; Thermo Fisher Scientific) was directly added into the blood/antibody mixture and incubated at room temperature for 10 min. The mixture was then centrifuged at 350 g for 5 min, and the supernatant was discarded. The pellet was washed three times with FACS buffer (3% FBS in PBS with 0.5 M EDTA). Stained cells were analyzed using an LSRII (BD Biosciences) and FlowJo. In detail, CD45+ cells were gated for CD11b+ Ly6G− cells, which were further gated for CD115+ cells (monocytes). The CD115+ monocyte population was gated for ly6c+ monocytes and ly6c− monocytes. We confirmed that the gated CD115+ monocyte population did not contain CD11c+ cells. The serum levels of TNF-α and IL-6 were determined with ELISA kits (R&D Systems) according to the manufacturer’s instructions.
Statistical analysis
All statistical analyses were performed with GraphPad Prism 8 software using a two-tailed, unpaired Student’s t test. All data are presented as mean ± SD.
Acknowledgments
We thank the Pathology core at the University of Texas Southwestern Medical Center for preparing stained sections of mouse tissues.
This work was supported by grants from the Cancer Prevention and Research Institute of Texas (RP180725) and the Welch Foundation (I-1389). Z.J. Chen is an investigator of the Howard Hughes Medical Institute.
Author contributions: T. Li, S. Yum, X. Zuo, and Z.J. Chen designed the experiments; T. Li performed experiments and analyzed data; T. Li and S. Yum developed mouse models; X. Chen provided reagents and mice; T. Li, S. Yum, M. Li, and Z.J. Chen wrote the paper.
References
- Abe, T., and Barber G.N.. 2014. Cytosolic-DNA-mediated, STING-dependent proinflammatory gene induction necessitates canonical NF-κB activation through TBK1. J. Virol. 88:5328–5341. 10.1128/JVI.00037-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ablasser, A., Hemmerling I., Schmid-Burgk J.L., Behrendt R., Roers A., and Hornung V.. 2014. TREX1 deficiency triggers cell-autonomous immunity in a cGAS-dependent manner. J. Immunol. 192:5993–5997. 10.4049/jimmunol.1400737 [DOI] [PubMed] [Google Scholar]
- Ahn, J., Gutman D., Saijo S., and Barber G.N.. 2012. STING manifests self DNA-dependent inflammatory disease. Proc. Natl. Acad. Sci. USA. 109:19386–19391. 10.1073/pnas.1215006109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balka, K.R., Louis C., Saunders T.L., Smith A.M., Calleja D.J., D’Silva D.B., Moghaddas F., Tailler M., Lawlor K.E., Zhan Y., et al. 2020. TBK1 and IKKε Act Redundantly to Mediate STING-Induced NF-κB Responses in Myeloid Cells. Cell Rep. 31:107492. 10.1016/j.celrep.2020.03.056 [DOI] [PubMed] [Google Scholar]
- Baum, R., Sharma S., Carpenter S., Li Q.Z., Busto P., Fitzgerald K.A., Marshak-Rothstein A., and Gravallese E.M.. 2015. Cutting edge: AIM2 and endosomal TLRs differentially regulate arthritis and autoantibody production in DNase II-deficient mice. J. Immunol. 194:873–877. 10.4049/jimmunol.1402573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan, F.M., and McInnes I.B.. 2008. Evidence that cytokines play a role in rheumatoid arthritis. J. Clin. Invest. 118:3537–3545. 10.1172/JCI36389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy, E.H., De Benedetti F., Takeuchi T., Hashizume M., John M.R., and Kishimoto T.. 2020. Translating IL-6 biology into effective treatments. Nat. Rev. Rheumatol. 16:335–345. 10.1038/s41584-020-0419-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow, Y.J., Hayward B.E., Parmar R., Robins P., Leitch A., Ali M., Black D.N., van Bokhoven H., Brunner H.G., Hamel B.C., et al. 2006. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 cause Aicardi-Goutières syndrome at the AGS1 locus. Nat. Genet. 38:917–920. 10.1038/ng1845 [DOI] [PubMed] [Google Scholar]
- Du, M., and Chen Z.J.. 2018. DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science. 361:704–709. 10.1126/science.aat1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann, M., Brennan F.M., and Maini R.N.. 1996. Role of cytokines in rheumatoid arthritis. Annu. Rev. Immunol. 14:397–440. 10.1146/annurev.immunol.14.1.397 [DOI] [PubMed] [Google Scholar]
- Gaidt, M.M., Ebert T.S., Chauhan D., Ramshorn K., Pinci F., Zuber S., O’Duill F., Schmid-Burgk J.L., Hoss F., Buhmann R., et al. 2017. The DNA Inflammasome in Human Myeloid Cells Is Initiated by a STING-Cell Death Program Upstream of NLRP3. Cell. 171:1110–1124.e18. 10.1016/j.cell.2017.09.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall, A., Treuting P., Elkon K.B., Loo Y.M., Gale M. Jr., Barber G.N., and Stetson D.B.. 2012. Autoimmunity initiates in nonhematopoietic cells and progresses via lymphocytes in an interferon-dependent autoimmune disease. Immunity. 36:120–131. 10.1016/j.immuni.2011.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, D., Li T., Li X.D., Chen X., Li Q.Z., Wight-Carter M., and Chen Z.J.. 2015. Activation of cyclic GMP-AMP synthase by self-DNA causes autoimmune diseases. Proc. Natl. Acad. Sci. USA. 112:E5699–E5705. 10.1073/pnas.1516465112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, E.E., Treuting P.M., Woodward J.J., and Stetson D.B.. 2015. Cutting Edge: cGAS Is Required for Lethal Autoimmune Disease in the Trex1-Deficient Mouse Model of Aicardi-Goutières Syndrome. J. Immunol. 195:1939–1943. 10.4049/jimmunol.1500969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, M.J., Gough A.K., Devlin J., Smith J., Astin P., Taylor D., and Emery P.. 2003. Serum MMP-3 and MMP-1 and progression of joint damage in early rheumatoid arthritis. Rheumatology (Oxford). 42:83–88. 10.1093/rheumatology/keg037 [DOI] [PubMed] [Google Scholar]
- Gui, X., Yang H., Li T., Tan X., Shi P., Li M., Du F., and Chen Z.J.. 2019. Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature. 567:262–266. 10.1038/s41586-019-1006-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulen, M.F., Koch U., Haag S.M., Schuler F., Apetoh L., Villunger A., Radtke F., and Ablasser A.. 2017. Signalling strength determines proapoptotic functions of STING. Nat. Commun. 8:427. 10.1038/s41467-017-00573-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa, H., and Barber G.N.. 2008. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 455:674–678. 10.1038/nature07317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobs, C., Perner S., and Hornung V.. 2015. AIM2 Drives Joint Inflammation in a Self-DNA Triggered Model of Chronic Polyarthritis. PLoS One. 10:e0131702. 10.1371/journal.pone.0131702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawane, K., Fukuyama H., Kondoh G., Takeda J., Ohsawa Y., Uchiyama Y., and Nagata S.. 2001. Requirement of DNase II for definitive erythropoiesis in the mouse fetal liver. Science. 292:1546–1549. 10.1126/science.292.5521.1546 [DOI] [PubMed] [Google Scholar]
- Kawane, K., Fukuyama H., Yoshida H., Nagase H., Ohsawa Y., Uchiyama Y., Okada K., Iida T., and Nagata S.. 2003. Impaired thymic development in mouse embryos deficient in apoptotic DNA degradation. Nat. Immunol. 4:138–144. 10.1038/ni881 [DOI] [PubMed] [Google Scholar]
- Kawane, K., Ohtani M., Miwa K., Kizawa T., Kanbara Y., Yoshioka Y., Yoshikawa H., and Nagata S.. 2006. Chronic polyarthritis caused by mammalian DNA that escapes from degradation in macrophages. Nature. 443:998–1002. 10.1038/nature05245 [DOI] [PubMed] [Google Scholar]
- Kawane, K., Tanaka H., Kitahara Y., Shimaoka S., and Nagata S.. 2010. Cytokine-dependent but acquired immunity-independent arthritis caused by DNA escaped from degradation. Proc. Natl. Acad. Sci. USA. 107:19432–19437. 10.1073/pnas.1010603107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyel, P.A. 2017. Dnases in health and disease. Dev. Biol. 429:1–11. 10.1016/j.ydbio.2017.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinne, R.W., Bräuer R., Stuhlmüller B., Palombo-Kinne E., and Burmester G.R.. 2000. Macrophages in rheumatoid arthritis. Arthritis Res. 2:189–202. 10.1186/ar86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto, T. 2005. Interleukin-6: from basic science to medicine--40 years in immunology. Annu. Rev. Immunol. 23:1–21. 10.1146/annurev.immunol.23.021704.115806 [DOI] [PubMed] [Google Scholar]
- Larkin, B., Ilyukha V., Sorokin M., Buzdin A., Vannier E., and Poltorak A.. 2017. Cutting Edge: Activation of STING in T Cells Induces Type I IFN Responses and Cell Death. J. Immunol. 199:397–402. 10.4049/jimmunol.1601999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S., Cai X., Wu J., Cong Q., Chen X., Li T., Du F., Ren J., Wu Y.T., Grishin N.V., and Chen Z.J.. 2015. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 347:aaa2630. 10.1126/science.aaa2630 [DOI] [PubMed] [Google Scholar]
- Ma, X., and Xu S.. 2013. TNF inhibitor therapy for rheumatoid arthritis. Biomed. Rep. 1:177–184. 10.3892/br.2012.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan, A., Herrmann M., and Muñoz L.E.. 2016. Clearance Deficiency and Cell Death Pathways: A Model for the Pathogenesis of SLE. Front. Immunol. 7:35. 10.3389/fimmu.2016.00035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misharin, A.V., Cuda C.M., Saber R., Turner J.D., Gierut A.K., Haines G.K. III, Berdnikovs S., Filer A., Clark A.R., Buckley C.D., et al. 2014. Nonclassical Ly6C(-) monocytes drive the development of inflammatory arthritis in mice. Cell Rep. 9:591–604. 10.1016/j.celrep.2014.09.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita, M., Stamp G., Robins P., Dulic A., Rosewell I., Hrivnak G., Daly G., Lindahl T., and Barnes D.E.. 2004. Gene-targeted mice lacking the Trex1 (DNase III) 3′-->5′ DNA exonuclease develop inflammatory myocarditis. Mol. Cell. Biol. 24:6719–6727. 10.1128/MCB.24.15.6719-6727.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata, S. 2005. DNA degradation in development and programmed cell death. Annu. Rev. Immunol. 23:853–875. 10.1146/annurev.immunol.23.021704.115811 [DOI] [PubMed] [Google Scholar]
- O’Brien, W., Fissel B.M., Maeda Y., Yan J., Ge X., Gravallese E.M., Aliprantis A.O., and Charles J.F.. 2016. RANK-Independent Osteoclast Formation and Bone Erosion in Inflammatory Arthritis. Arthritis Rheumatol. 68:2889–2900. 10.1002/art.39837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe, Y., Kawane K., Akira S., Taniguchi T., and Nagata S.. 2005. Toll-like receptor-independent gene induction program activated by mammalian DNA escaped from apoptotic DNA degradation. J. Exp. Med. 202:1333–1339. 10.1084/jem.20051654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawaria, S., Moody K., Busto P., Nündel K., Choi C.H., Ghayur T., and Marshak-Rothstein A.. 2015. Cutting Edge: DNase II deficiency prevents activation of autoreactive B cells by double-stranded DNA endogenous ligands. J. Immunol. 194:1403–1407. 10.4049/jimmunol.1402893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana, A.K., Li Y., Dang Q., and Yang F.. 2018. Monocytes in rheumatoid arthritis: Circulating precursors of macrophages and osteoclasts and, their heterogeneity and plasticity role in RA pathogenesis. Int. Immunopharmacol. 65:348–359. 10.1016/j.intimp.2018.10.016 [DOI] [PubMed] [Google Scholar]
- Rice, G.I., Rodero M.P., and Crow Y.J.. 2015. Human disease phenotypes associated with mutations in TREX1. J. Clin. Immunol. 35:235–243. 10.1007/s10875-015-0147-3 [DOI] [PubMed] [Google Scholar]
- Richards, A., van den Maagdenberg A.M., Jen J.C., Kavanagh D., Bertram P., Spitzer D., Liszewski M.K., Barilla-Labarca M.L., Terwindt G.M., Kasai Y., et al. 2007. C-terminal truncations in human 3′-5′ DNA exonuclease TREX1 cause autosomal dominant retinal vasculopathy with cerebral leukodystrophy. Nat. Genet. 39:1068–1070. 10.1038/ng2082 [DOI] [PubMed] [Google Scholar]
- Rodero, M.P., Tesser A., Bartok E., Rice G.I., Della Mina E., Depp M., Beitz B., Bondet V., Cagnard N., Duffy D., et al. 2017. Type I interferon-mediated autoinflammation due to DNase II deficiency. Nat. Commun. 8:2176. 10.1038/s41467-017-01932-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer, J.D., Sotelo-Troha K., von Moltke J., Monroe K.M., Rae C.S., Brubaker S.W., Hyodo M., Hayakawa Y., Woodward J.J., Portnoy D.A., and Vance R.E.. 2011. The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect. Immun. 79:688–694. 10.1128/IAI.00999-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scales, H.E., Ierna M., Smith K.M., Ross K., Meiklejohn G.R., Patterson-Kane J.C., McInnes I.B., Brewer J.M., Garside P., and Maffia P.. 2016. Assessment of murine collagen-induced arthritis by longitudinal non-invasive duplexed molecular optical imaging. Rheumatology (Oxford). 55:564–572. [DOI] [PubMed] [Google Scholar]
- Shin, H.D., Park B.L., Cheong H.S., Lee H.S., Jun J.B., and Bae S.C.. 2005. DNase II polymorphisms associated with risk of renal disorder among systemic lupus erythematosus patients. J. Hum. Genet. 50:107–111. 10.1007/s10038-004-0227-3 [DOI] [PubMed] [Google Scholar]
- Stetson, D.B., Ko J.S., Heidmann T., and Medzhitov R.. 2008. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 134:587–598. 10.1016/j.cell.2008.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, W., Li Y., Chen L., Chen H., You F., Zhou X., Zhou Y., Zhai Z., Chen D., and Jiang Z.. 2009. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc. Natl. Acad. Sci. USA. 106:8653–8658. 10.1073/pnas.0900850106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, L., Wu J., Du F., Chen X., and Chen Z.J.. 2013. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 339:786–791. 10.1126/science.1232458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, Y., and Chen Z.J.. 2012. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci. Signal. 5:ra20. 10.1126/scisignal.2002521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchetverikov, I., Lard L.R., DeGroot J., Verzijl N., TeKoppele J.M., Breedveld F.C., Huizinga T.W., and Hanemaaijer R.. 2003. Matrix metalloproteinases-3, -8, -9 as markers of disease activity and joint damage progression in early rheumatoid arthritis. Ann. Rheum. Dis. 62:1094–1099. 10.1136/ard.62.11.1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terman, A., Kurz T., Gustafsson B., and Brunk U.T.. 2006. Lysosomal labilization. IUBMB Life. 58:531–539. 10.1080/15216540600904885 [DOI] [PubMed] [Google Scholar]
- Tüngler, V., Silver R.M., Walkenhorst H., Günther C., and Lee-Kirsch M.A.. 2012. Inherited or de novo mutation affecting aspartate 18 of TREX1 results in either familial chilblain lupus or Aicardi-Goutières syndrome. Br. J. Dermatol. 167:212–214. 10.1111/j.1365-2133.2012.10813.x [DOI] [PubMed] [Google Scholar]
- Warner, J.D., Irizarry-Caro R.A., Bennion B.G., Ai T.L., Smith A.M., Miner C.A., Sakai T., Gonugunta V.K., Wu J., Platt D.J., et al. 2017. STING-associated vasculopathy develops independently of IRF3 in mice. J. Exp. Med. 214:3279–3292. 10.1084/jem.20171351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J., Sun L., Chen X., Du F., Shi H., Chen C., and Chen Z.J.. 2013. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 339:826–830. 10.1126/science.1229963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y.G., Lindahl T., and Barnes D.E.. 2007. Trex1 exonuclease degrades ssDNA to prevent chronic checkpoint activation and autoimmune disease. Cell. 131:873–886. 10.1016/j.cell.2007.10.017 [DOI] [PubMed] [Google Scholar]
- Yoshida, H., Okabe Y., Kawane K., Fukuyama H., and Nagata S.. 2005. Lethal anemia caused by interferon-beta produced in mouse embryos carrying undigested DNA. Nat. Immunol. 6:49–56. 10.1038/ni1146 [DOI] [PubMed] [Google Scholar]
- Yoshitomi, H. 2019. Regulation of Immune Responses and Chronic Inflammation by Fibroblast-Like Synoviocytes. Front. Immunol. 10:1395. 10.3389/fimmu.2019.01395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yum, S., Li M., Fang Y., and Chen Z.J.. 2021. TBK1 recruitment to STING activates both IRF3 and NF-κB that mediate immune defense against tumors and viral infections. Proc. Natl. Acad. Sci. USA. 118:e2100225118. 10.1073/pnas.2100225118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C., Shang G., Gui X., Zhang X., Bai X.C., and Chen Z.J.. 2019. Structural basis of STING binding with and phosphorylation by TBK1. Nature. 567:394–398. 10.1038/s41586-019-1000-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, B., Yang Y., Li S., Wang Y.Y., Li Y., Diao F., Lei C., He X., Zhang L., Tien P., and Shu H.B.. 2008. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 29:538–550. 10.1016/j.immuni.2008.09.003 [DOI] [PubMed] [Google Scholar]