Abstract
The cuticle, the outer covering of the nematode C. elegans, is synthesized five times during the worm’s life by the underlying hypodermis. Cuticle collagens, the major cuticle component, are encoded by a large family of col genes and, interestingly, many of these genes express predominantly at a single developmental stage. This temporal preference motivated us to investigate the mechanisms underlying col gene expression and here we focus on a subset of col genes expressed in the L4 stage. We identified minimal promoter regions of <300 bp for col-38, col-49, and col-63. In these regions, we predicted cis-regulatory sequences and evaluated their function in vivo via mutagenesis of a col-38p::yfp reporter. We used RNAi to study the requirement for candidate transcription regulators ELT-1 and ELT-3, LIN-29, and the LIN-29 co-factor MAB-10, and found LIN-29 to be necessary for the expression of four L4-specific genes (col-38, col-49, col-63, and col-138). Temporal misexpression of LIN-29 was also sufficient to activate these genes at a different developmental stage. The LIN-29 DNA-binding domain bound the col-38, col-49, and col-63 minimal promoters in vitro. For col-38 we showed that the LIN-29 sites necessary for reporter expression in vivo are also bound in vitro: this is the first identification of specific binding sites for LIN-29 necessary for in vivo target gene expression.
Keywords: development, nematode, temporal expression
1 |. INTRODUCTION
The temporal regulation of gene expression is an essential aspect of metazoan development. After embryogenesis, the ecdysozoan nematode C. elegans goes through four larval stages (L1–L4), molting its outer cuticle between each transition before becoming an adult. The major components of the cuticle are nematode-specific cuticle collagen proteins, which are expressed from a large gene family (Cox, 1992; Page & Johnstone, 2007). Previously, it was shown that several cuticle collagen (col) genes show a peak of expression in each larval stage (Johnstone, 2000; Johnstone & Barry, 1996), but examination of modENCODE project RNA-Seq temporal development data showed that many col genes (116/187) display a strong peak of expression in only one developmental stage (Gerstein et al., 2010; Jackson, Abete-Luzi, Krause, & Eisenmann, 2014). This set of temporally coregulated related genes provides a powerful system to study temporal regulation of gene expression. In C. elegans, the heterochronic pathway controls the timing of several developmental events (Moss, 2007). LIN-29, the most downstream effector of this pathway, is a zinc finger transcription factor that accumulates in hypodermal cells in the L4 stage and is required for a subset of developmental events at the larva-to-adult transition, including the expression of adult specific collagens col-7 and col-19 (Liu, Kirch, & Ambros, 1995). Our work is focused on a subset of col genes that peak during the L4 larval stage for which we identify regulatory sequences, transcription factor requirements and in vitro DNA-protein interactions with the heterochronic transcription factor LIN-29.
2 |. RESULTS AND DISCUSSION
To validate the modENCODE data analysis showing stage-specific col expression, we built several transcriptional YFP reporters using entire col upstream promoter regions and integrated them in single copy into the genome. Reporters for col genes with peak expression in the embryo (col-121, dpy-17), L2 stage (col-54, col-41), and L4 stage (col-49, col-63, and col-38) all began to show strong YFP expression at the expected time (Figure 1), indicating that members of this large gene family display specific temporal expression at defined points in the life cycle, and this temporal control is mediated by upstream genomic regions.
FIGURE 1.
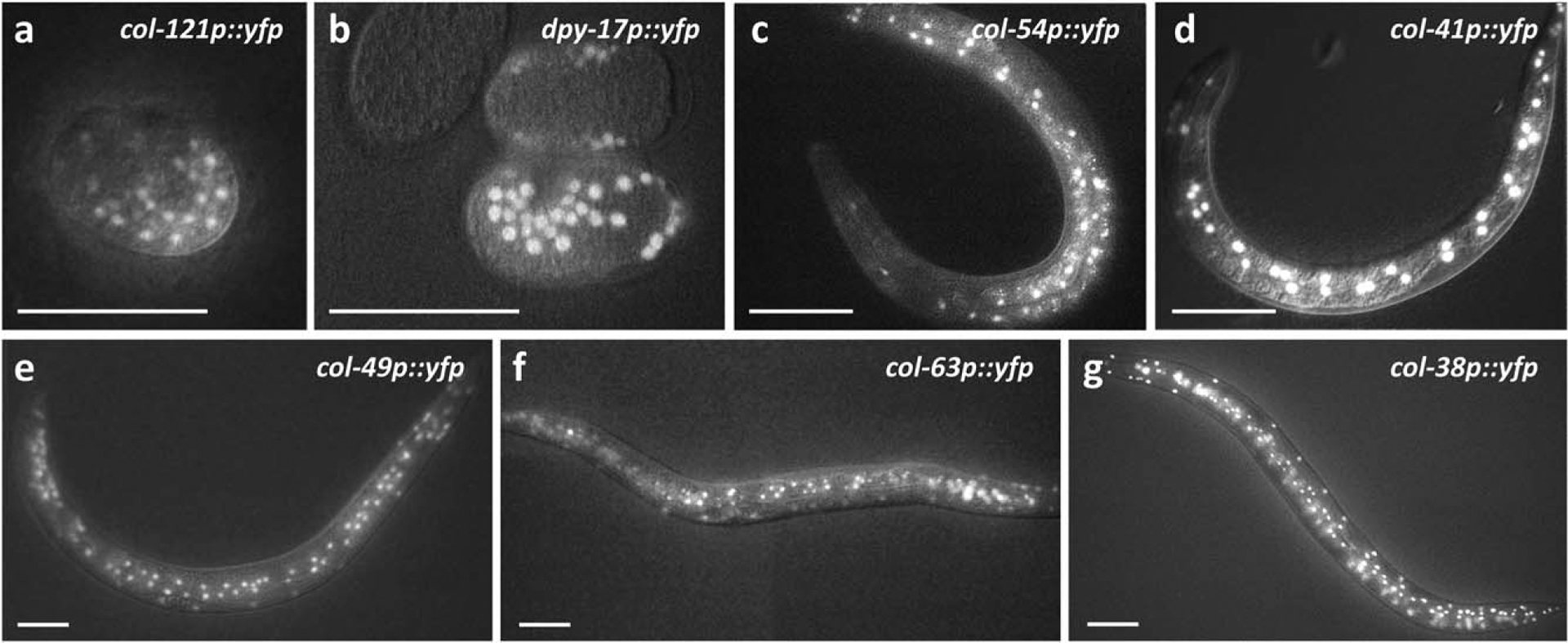
Stage-specific expression of cuticle collagen genes. Animals carrying the indicated YFP transcriptional reporters were imaged by epifluorescence and Nomarski microscopy at different stages: (a, b) embryo; (c, d) L2 stage; and (e–g) L4 stage. For each reporter, the figure shows the earliest timepoint when YFP was visible during development. YFP expression often perdured past this time. In the case of L2 and L4 stage col reporters, YFP was observed in hypodermal cells of the tail and head, hyp7, and seam cells. For details of col-38p::yfp developmental expression, see Methods. Scale bars indicate 50 μm
To understand this temporal control of col gene expression, we investigated the regulation of three col genes with peak expression in the L4 stage. Promoter deletion analysis on these reporters narrowed the elements necessary for correct temporal expression to regions 262, 282, and 222 bp upstream of the start codons of col-38, col-49, and col-63, respectively (Figure 2), remarkably small regions for C. elegans genes.
FIGURE 2.
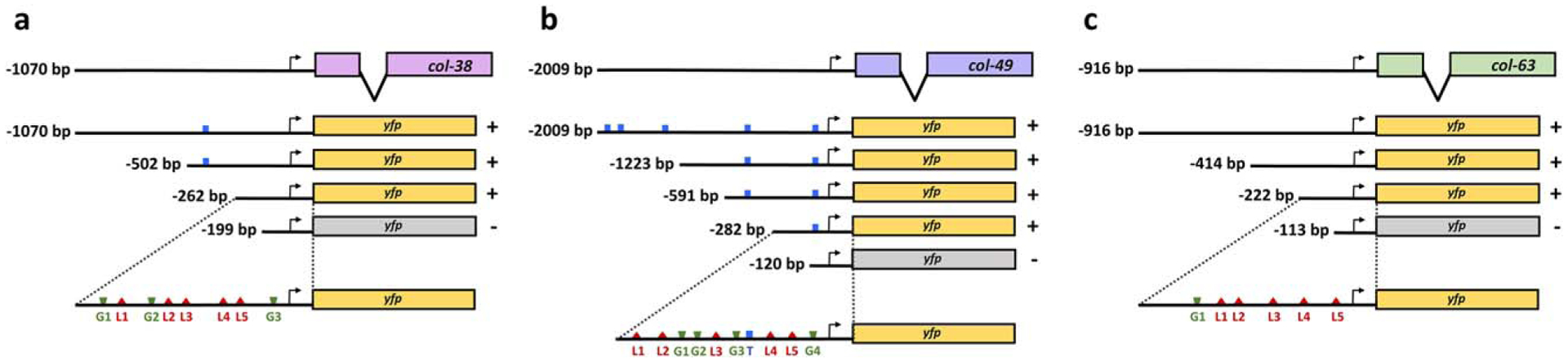
Identification of regulatory regions required for L4 expression in three L4 col YFP reporters. Promoter deletion analyses allowed the identification of minimal promoter regions of 262, 282, and 222 bp in (a) col-38, (b) col-49, and (c) col-63, respectively. For each construct ≥22 animals were assessed in at least two independent lines. In all cases, YFP expression in the L4 stage was either present in ≥80% of the animals (+), or undetectable (−), in which case p<0.001 (Fisher’s exact test) when compared to full length promoter. Locations of predicted binding motifs are shown for TCF/POP-1 (T, blue), GATA factors (G, green), and LIN-29 (L, red)
Previous work has identified four transcription factors regulating col gene expression. First, we showed that the L4 col genes col-38, col-49, and col-71 are regulated by the Wnt pathway transcription factor BAR-1 (beta-catenin) (Gorrepati et al., 2015; Jackson et al., 2014; Van Der Bent et al., 2015), which binds to its target genes via interaction with the TCF transcription factor POP-1 (Jackson & Eisenmann, 2012). Second GATA factors ELT-1 and ELT-3 are required for proper expression of the col genes dpy-7 (embryo peak), col-41 (L2 peak), and col-144 (no peak) (Budovskaya et al., 2008; Gilleard, Barry, & Johnstone, 1997; Gilleard & McGhee, 2001; Yin, Madaan, Park, Aftab, & Savage-Dunn, 2015). Finally, LIN-29, the terminal transcription factor in the heterochronic pathway, regulates expression of the adult col genes col-7 and col-19 and can bind large DNA fragments from the col-19 promoter region (Liu et al., 1995; Rougvie & Ambros, 1995). Therefore, we looked for POP-1(TCF), GATA and LIN-29 binding sites in the minimal promoters of col-38, col-49, and col-63.
Motif searching for a POP-1 binding motif ([T/C]TTTG[T/A][T/A]) (Jackson & Eisenmann, 2012) in the col gene minimal promoter regions showed a single site in col - 49p(−282)(Figure 2), which could mediate the BAR-1 responsiveness of this gene, however there were no POP-1 sites in the minimal fragments col - 38p(−262) and col - 63p(−222). This suggests that while two of these col genes may be responsive to BAR-1, their temporal pattern of expression is not likely to depend on BAR-1/POP-1 binding.
Unlike POP-1, there are putative LIN-29 and GATA sites in all three col minimal promoters. Narasimhan et al. showed that the LIN-29 DNA binding domain prefers sequences of 5As or 6As in vitro (Narasimhan et al., 2015). For each promoter, we identified five putative LIN-29 binding sites and called them L1 – L5 (Figure 2; Supporting Information File 1). In a like manner, we identified GATA factor binding sites (GATA[A/G]) and named them G1, G2, G3, and so forth (Figure 2). We evaluated the requirement of these putative LIN-29 and GATA binding sites for proper temporal expression of col-38 by mutating or deleting them in our col-38p(−262)::yfp minimal promoter reporter and assessing YFP expression in vivo.
Removal of predicted LIN-29 binding site L1 or simultaneous mutation of sites L2 and L3 had no effect; however, mutation of L1, L2 and L3 together caused a slight reduction in the number of animals showing YFP expression (L1L2L3m, Table 1). Interestingly, while neither the individual mutation of sites L4 or L5 showed an effect on col-38p(−262)::yfp expression, when L4 and L5 were simultaneously mutated YFP expression was completely abolished in vivo (L4L5m, Table 1). In addition, when either L4 or L5 was the only intact site, YFP expression was also absent (L1L2L3L4m; L1L2L3L5m, Table 1). Evolutionarily conserved, predicted LIN-29 binding sites in the position of L4 and L5 are found in the col-38 gene promoters from four other Caenorhabditis species (Supporting Information Figure 1). These results indicate that: (1) putative LIN-29 binding sites are necessary for col-38 expression in vivo, (2) these sites act redundantly; (3) sites L1, L2 and L3 enhance col-38 expression but are not sufficient; (4) loss of both L4 and L5 prevents col-38 expression; and (5) neither site L4 nor L5 alone is sufficient for activation of col-38.
TABLE 1.
In vivo expression of col-38 transcriptional reporters
| col-38p(−262) reporter | % YFP expression (n) |
|---|---|
| WT | 100 (34) |
| ΔL1G1 (−231) | 97 (26) |
| L2L3m | 87 (39) |
| L1L2L3m | 82* (46) |
| L4m | 100 (38) |
| L5m | 100 (48) |
| L4L5m | 0** (29) |
| L1L2L3L4m | 0** (39) |
| L1L2L3L5m | 0** (25) |
| G1G2m | 100 (28) |
| G1G2G3m | 0** (10) |
| G3m | 0** (28) |
Transgenic animals carrying the indicated constructs were scored for YFP expression in the late L4 larval stage. Except for the −231 bp deletion (ΔL1G1), all reporter variants were mutagenized versions of the col-38p −262 bp reporter (col-38p(262)::yfp), and their names indicate which LIN-29 or GATA sites were mutated. Animals were scored as either positive or negative for YFP expression, since animals at the L4 stage all showed similar YFP intensity.
p≤0.01 and
p<0.0001 (Fisher’s exact test) compared to WT.
We also tested the requirement of GATA sites for col-38p(−262):: yfp expression. While removal of G1 had no effect on YFP expression (ΔL1G1(−231), Table 1), deletion of the region containing G1 and G2 caused a loss of expression (−199; Figure 2a), suggesting these sites may be necessary. However, simultaneous mutation of G1 and G2 had no effect on YFP expression (G1G2m, Table 1), arguing that it is not the sites, but some other sequence in the region or the altered spacing in the −199 mutant that is important. Mutation of site G3 alone did completely abolish YFP reporter expression (G3m, Table 1) indicating this site is necessary for col-38 expression.
Because our analyses of cis-regulatory elements implicated GATA factors and the zinc finger transcription factor LIN-29 in L4-specific expression of col-38, we used RNAi to reduce function of these transcription factors. We examined col-38 expression via in vivo observation of our col-38p::yfp reporter strain and also performed qPCR to assay endogenous expression of L4-specific col genes col-38, col-49, col-63, and col-138 in an RNAi-hypersensitive background (rrf-3(pk1426)).
ELT-1 and ELT-3 are hypodermal-specific GATA factors that are essential for hypodermal cell fate specification during embryogenesis (Chisholm & Hsiao, 2012) and that positively regulate expression of col-41, col-144, and dpy-7 (Budovskaya et al., 2008; Gilleard & McGhee, 2001; Gilleard et al., 1997; Yin et al., 2015). Neither elt-1 (RNAi) nor elt-3(RNAi) caused a change in expression of col-38p(–262):: YFP or endogenous col-38 (data not shown). However, since elt-1 and elt-3 may act redundantly (Gilleard & McGhee, 2001), we also tested combined elt-1 + elt-3 RNAi. While we know this treatment was effective based on observation of expected phenotypes (see Methods), the col-38p(−262)::yfp reporter showed no observable change in expression when elt-1/elt-3 combined RNAi was performed on L1 stage animals, and qPCR analysis of endogenous gene expression showed no effect except for a slightly higher expression of col-63 in elt-1/elt-3(RNAi) animals (Table 2; Figure 3a). We repeated the double RNAi treatment on a strain containing the col-38(−262G1G2m)::YFP reporter in which only GATA site G3 is intact, and in an RNAi-hypersensitive background (col-38(−262)::yfp; eri-1(ok2683)), but still observed no change in YFP expression in vivo (Table 2). Finally, performing elt-1/elt-3 combined RNAi on RNAi-hypersensitive mothers and observing col-38p(−262)::yfp reporter expression in the surviving progeny also caused no obvious change in penetrance or expressivity of YFP expression in the hypodermis (data not shown). These data suggest that either our RNAi treatment was strong enough to cause embryonic and larval somatic phenotypes but was not strong enough to compromise col gene expression, or that ELT-1 and ELT-3 do not play a major role in expression of this gene and some other protein may bind and function at or near the G3 site.
TABLE 2.
LIN-29 is required for L4 expression of col-38
| % YFP expression (n) | ||||
|---|---|---|---|---|
| Strain | Treatment | Early L4 | Mid L4 | Late L4 |
| col-38p(−262)::yfp | control RNAi | 0 (4) | 84 (30) | 100 (15) |
| col-38p(−262)::yfp | lin-29 RNAi | 0 (5) | 0* (13) | 0* (24) |
| col-38p(−262)::yfp | mab-10 RNAi | 0 (5) | 92 (26) | 100 (31) |
| col-38p(−262)::yfp | elt-1/elt-3 RNAi | n.d. | 78 (32) | 100 (25) |
| col-38p(−262)::yfp; eri-1(ok2683) | control RNAi | 0 (4) | 77 (13) | 100 (10) |
| col-38p(−262)::yfp; eri-1(ok2683) | elt-1/elt-3 RNAi | 0 (10) | 81 (31) | 100 (25) |
| col-38p(−262G1G2m)::yfp | control RNAi | 0 (13) | 83 (23) | 97 (29) |
| col-38p(−262G1G2m)::yfp | elt-1/elt-3 RNAi | 0 (9) | 81 (42) | 96 (24) |
Transcription factor requirements for the regulation of col-38 expression were evaluated by in vivo imaging of YFP transcriptional reporters under different RNAi treatments. In all cases, RNAi was by the ‘L1 feeding’ method, and the efficiency of each RNAi treatment was corroborated by observation of known phenotypes or by the effect on the expression of known downstream target genes (see Methods). Hypodermal GATA requirements were tested by feeding animals with a mix of bacteria expressing elt-1 RNAi combined with bacteria expressing elt-3 RNAi constructs. This experiment was also conducted in an RNAi-sensitive background (eri-1(ok2683); Kennedy et al., 2004), and in animals carrying the col-38p reporter with G1 and G2 sites mutated, so G3 is the only functional GATA site (col-38p(−262G1G2m)). Animals were scored as either positive or negative for YFP expression, since animals at the same L4 stage all showed similar YFP intensity.
p<0.001 (Fisher’s exact test) compared to the corresponding control.
FIGURE 3.
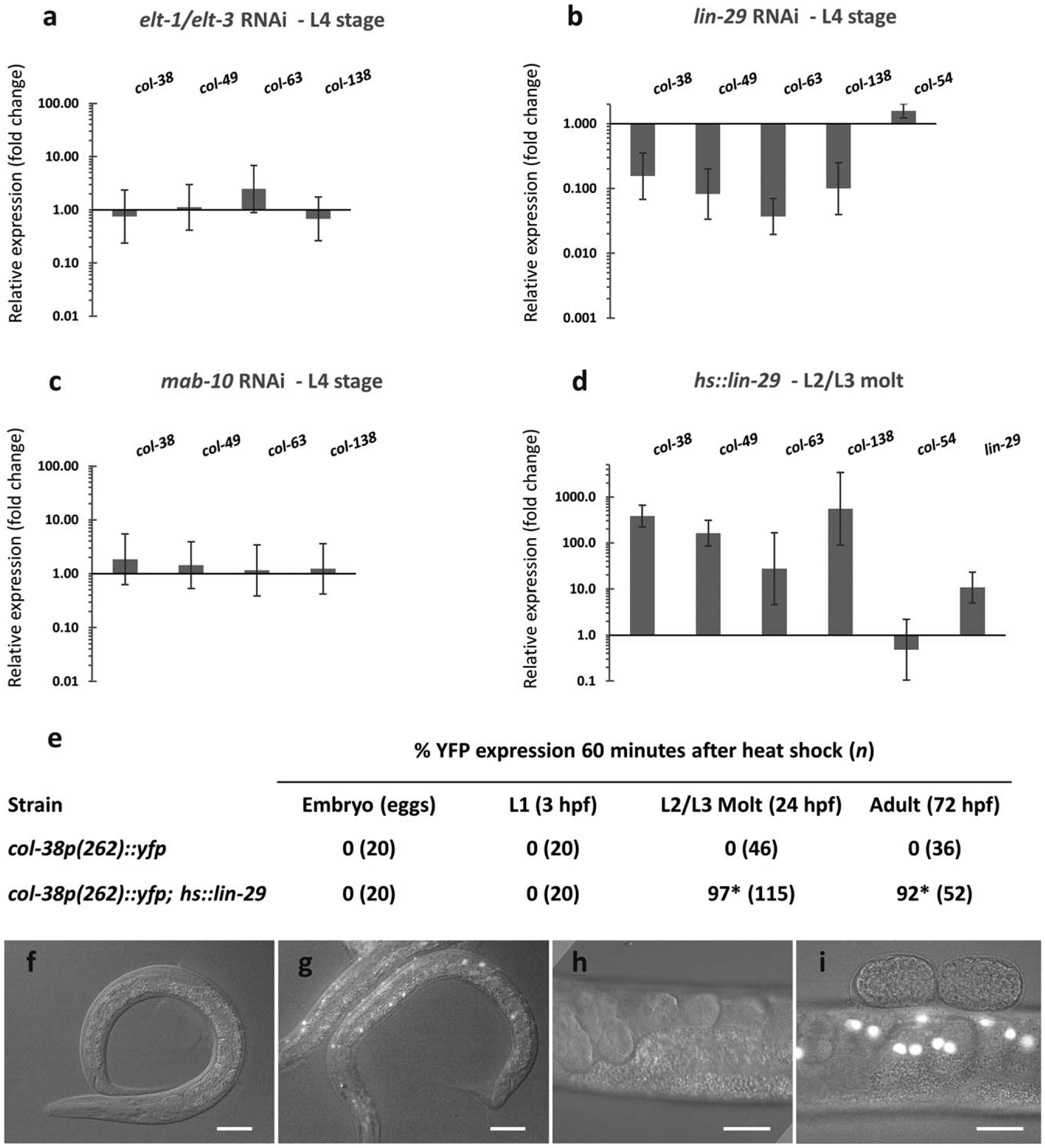
LIN-29 is necessary and sufficient for L4 col expression. Endogenous col gene expression in the L4 stage was assessed by RT-qPCR after different RNAi treatments: (a) combined GATA factors elt-1/elt-3 RNAi, (b) lin-29 RNAi and (c) mab-10 RNAi. Quantification was relative to expression in animals treated with empty vector RNAi control. (d) L4 col gene expression was evaluated 1 hr after inducing LIN-29 at the L2/L3 molt in a hs::lin-29 background. Quantification was relative to expression in hs::control animals. col-54 peaks in the L2 stage when lin-29 is not normally expressed, and served as a control. Error bars represent standard errors of the mean. (e) Expression of col-38p(−262)::yfp reporter was assessed after ectopic induction of LIN-29 in the embryo, in the L1, at the L2/L3 molt, and in the adult. (f–i) Epifluorescence and Nomarski microscopy of worms carrying either col-38p(−262)::yfp alone (f,h) or in a hs::lin-29 background (g, i) examined at the L2/L3 molt (f, g) and in the adult (h, i). Scale bars are 25 μm. *p < 0.001 (Fisher’s exact test) when compared to strains carrying col-38p(262)::yfp alone
On the other hand, YFP expression from our single copy col-38p (−262)::yfp strain was completely abolished when treated with lin-29 RNAi (Table 2). Similarly, transgenic strains carrying multicopy full length-promoter YFP reporters of the L4 col genes bli-1 and col-38 (Jackson et al., 2014) also showed strong YFP reduction in the L4 stage under the same conditions (Suppl. Table 1). In addition to these in vivo YFP reporter observations, endogenous levels of the L4 col genes col-38, col-49, col-63, and col-138, but not the L2 col gene col-54, were significantly reduced in the L4 when treated with lin-29 RNAi (Figure 3b). Together these results strongly implicate LIN-29 in the regulation of five cuticle collagen genes showing a peak of expression in the L4 stage.
Harris & Horvitz showed that the transcription co-factor MAB-10 physically interacts with LIN-29 and together they promote seam cell differentiation and prevent extra molting events in males during the larva-to-adult switch (Harris & Horvitz, 2011). Therefore, we investigated whether MAB-10 is also required for LIN-29 regulation of L4-expressed col genes in hermaphrodites. We examined col-38p(−262)::yfp expression in mab-10 RNAi treated animals and observed no effect (Table 2). Similarly, when we analyzed endogenous transcript levels of L4 col genes at the L4 stage under mab-10 RNAi, we did not observe major changes in expression (Figure 3c). These results suggest that while MAB-10 may interact with LIN-29 to regulate several processes in males, it is not required for the regulation of col gene expression by LIN-29 in the L4 stage hermaphrodite.
Because LIN-29 alone was strongly required for the regulation of L4 col expression, we investigated whether ectopically induced LIN-29 is sufficient for the misexpression of the L4 col genes. We generated transgenic lines carrying a single copy of either hs::lin-29 or hs::control (see Methods) and assessed endogenous L4 col gene expression after ectopic induction at the L2/L3 molt and in the adult. Unlike the L2 gene col-54, the L4 cols examined showed a strong increase when lin-29 was induced at the L2/L3 molt (Figure 3d), and a modest increase when induction was done in the adult (data not shown). Consistent with these results, we heat-shocked col-38p::yfp; hs::lin-29 animals and found that LIN-29 was sufficient to induce YFP expression at the L2/L3 molt and in the adult, but not in earlier stages (Figure 3e–i). This result shows that the provision of LIN-29 at a time it is not normally present is sufficient to induce expression of L4 col genes, suggesting it may be the major regulator of their expression at the normal L4 stage. Curiously, LIN-29 was not able to induce L4 col expression when provided in the embryo and L1 stage (Figure 3e), suggesting that at the earlier times either (1) some additional positive acting factor may be missing, (2) a repressor may be present, or (3) a nonpermissive chromatin state exists at these genes.
To determine whether LIN-29 may regulate L4 col gene directly, we tested LIN-29 binding to col promoter sequences in vitro. Notably, gel shift experiments showed that the LIN-29 DNA binding domain successfully binds the col-38, col-49, and col-63 minimal promoter pieces in vitro (Figure 4a), producing multiple shifted species, consistent with multiple predicted LIN-29 binding sites in these sequences. In all cases, the LIN-29-promoter interactions were competed by a 34-bp oligo containing the single LIN-29 site L5 from col-38p and its flanking sequence (Figure 4a). A smaller 155 bp fragment of col-38p containing only L4 and L5, the two sites necessary for expression in vivo, also showed direct interaction with LIN-29 in vitro. This interaction was reduced when the L5 site was mutated, and almost abolished when both L4 and L5 sites were mutated (Figure 4b). Lastly, an even smaller 139 bp col-38p fragment containing only the L5 site also bound LIN-29 protein. However, when we mutated only the L5 site in the probe, the interaction was abolished (Figure 4b).
FIGURE 4.
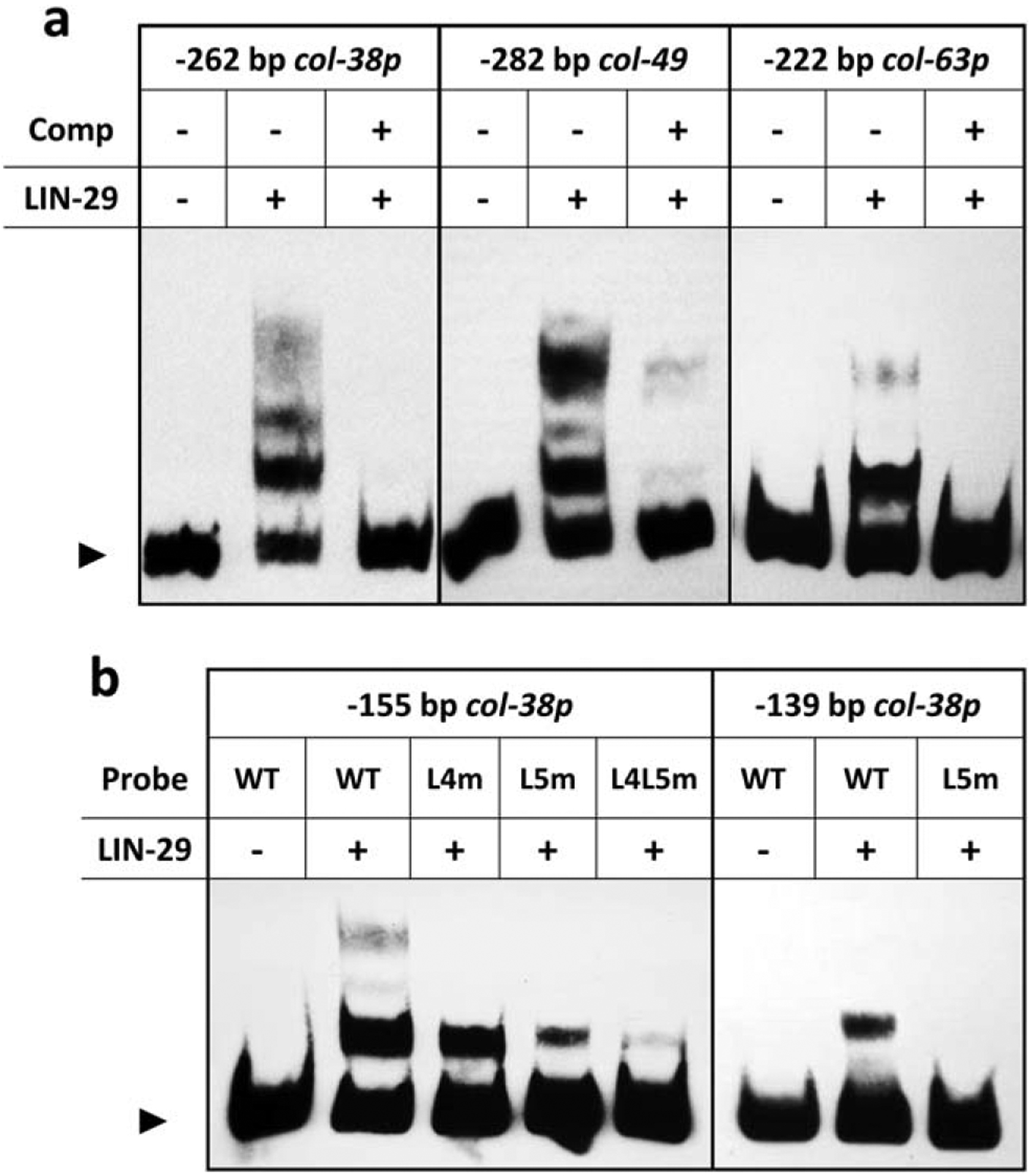
LIN-29 binds predicted DNA motifs in L4 col promoters in vitro. (a) Electrophoretic mobility shift assays done with LIN-29 DNA binding domain-GST fusion protein and the minimal promoters of col-38, col-49, and col-63 as probes. In each case, the binding was competed away by a 34 bp oligo consisting of the single col-38 LIN-29 site L5 and its flanking sequence (Comp). (b) Electrophoretic mobility shift assays with LIN-29 DNA binding domain-GST fusion protein and either a −155 bp region of col-38p containing predicted LIN-29 sites L4 and L5 (left) or a −139 bp fragment of col-38p containing only site L5 (right). Binding of LIN-29 to these fragments was reduced or abolished when sites L4 and L5 were mutated individually (L4m, L5m) or together (L4L5m). Arrowhead indicates free probe
In summary, in this work, we have (1) identified the heterochronic protein LIN-29 as a major regulator of the temporal expression of several cuticle collagen genes expressed during the larva-to-adult transition (col-38, col-49, col-63, and likely col-138 and bli-1), (2) validated the predicted LIN-29 binding motif derived by Narasimhan et al. (2015), and (3) in the case of col-38, have shown direct binding of LIN-29 to two sites in vitro, and the requirement for those sites for in vivo expression in the L4 stage. Although the demonstration of direct regulation of these genes will require proof of LIN-29 in vivo binding site occupancy in L4 animals, we believe this is the first identification of specific binding sites for LIN-29 necessary for in vivo target gene expression. These results should aid with future efforts to understand temporal regulation of gene expression by this heterochronic protein at the larval-to-adult transition.
3 |. METHODS
3.1 |. C. elegans growth and strains used
C. elegans animals were cultured using standard methods (Brenner, 1974). Worms were grown on NGM plates and fed with E. coli OP50, or HT115 in the case of RNAi experiments. Experiments were performed at 20°C unless indicated otherwise. The following strains were used in this work:
NL2099: rrf-3(pk1426) II
EG669: ttTi5605 II; unc-119(ed3) III
EG8078: oxTi185 I; unc-119(ed3) III
RB2025: eri-1(ok2683) IV.
3.2 |. Molecular cloning and mutagenesis
All col reporters and subsequent col promoter deletions were cloned upstream of 2XNLS::yfp in pDE350. pDE350 was created by removing the region containing [multicloningsite::2XNLS::yfp::unc-54–3’UTR] from pBJ101 (Jackson et al., 2014) and inserting it between the SbfI and SpeI sites of the MosSCI (ttTi5605) targeting vector pCFJ350 (Frøkjær-Jensen, Davis, Ailion, & Jorgensen, 2012; Addgene plasmid #34866). Different col promoter fragments were specified by PCR amplification using the primers listed in Supporting Information Table 2 and inserted into pDE350 digested with AvrII and SbfI, via Gibson Assembly® by New England Biolabs (NEB). All mutations of col-38p were generated with Q5® Site-Directed Mutagenesis Kit (NEB) except for col-38p (−262G1G2G3m), col-38p(−262G3m) and col-38p(−262L1L2L3L4m) which were built using gBlocks® from Integrated DNA Technologies (IDT) containing the desired sequence. Mutations of predicted LIN-29 binding sites were all 5’-AAAAA-3’ → 5’-AGGGA-3’ and mutations of predicted GATA binding sites were 5’-TTATC-3’ → 5’-GCAGC-3’. To express heat-shock inducible LIN-29 (hs::lin-29), we first used PCR to remove a fragment from heat shock vector pPD48.79 (a gift from Andrew Fire, Addgene plasmid # 1447) containing [hsp-16.2p:multicloning site:unc-54–3’UTR] and cloned it into MosSCI (ttTi5605) targeting vector pCFJ350 to create plasmid pPA4. We then introduced a lin-29a cDNA (gBlocks®, IDT) into pPA4 via Gibson Assembly® (NEB) to generate pPA5 (hsp-16.2p::lin-29::unc-54-3’UTR). Worms were injected with either these plasmids to create the single-copy integrated strains hs::control (pPA4) and hs::lin-29 (pPA5). Sequences of plasmids are available upon request.
3.3 |. Generation of transgenic strains
All single copy insertion strains carrying YFP transcriptional reporters, as well as inducible LIN-29 (hsp-16.2p::lin-29::unc-54-3’UTR) and control (hsp-16.2p::unc-54-3’UTR) were generated by microinjection of MosSCI targeting vectors specific for the ttTi5605 site, into insertion strain EG6699 (Mos site in LGII) following standard protocols for injection (Mello & Fire, 1995) and selection (Frøkjær-Jensen, 2015). The hsp-16.2p::lin-29::unc-54-3’UTR vector was also microinjected into EG8078 (Mos site on LGI) and integrated to facilitate crossing with the strain carrying col-38p(−262) reporter integrated on LGII.
3.4 |. Ectopic induction of LIN-29
Strains carrying either hsp-16.2p::lin-29::unc-54-3’UTR or hsp-16.2p:: unc-54-3’UTR (control) were grown at 20°C and induced by heat shock exposure of 30 minutes at 37°C followed by 60 min recovery at 20°C before sample collection or imaging. Inductions in the embryo were done in a mixed population of eggs. Inductions in the L1 stage, in the L2/L3 molt and in the adult, were done at 3, 26, and 66 hr-post-feeding, respectively.
3.5 |. Imaging and YFP expression recording
All transgenic animals carrying yfp reporters were imaged on a Zeiss Axioplan 2 and recorded with a Lumenera Infinity 3 camera and Infinity Analyze software. Every construct was assessed in at least two independent lines. Specific developmental stages in which animals were assessed were determined by the extent of gonad migration or vulval morphology. YFP expression results were evaluated in terms of penetrance and recorded as either positive or negative. In general, the intensity (expressivity) of the L4-stage YFP reporters used in this work showed minor variation in the mid L4 with a tendency to increase in brightness, and reached a stable maximum in the late L4. The expression of col-38p::yfp across all stages was as follows: late embryogenesis 0%; L1 stage 0%; L2 stage 0%; L3 stage 0%; early L4 stage 0%; mid L4 stage (Christmas tree) 81%, late L4 stage 100%; newly-gravid adult 0% (n≥15 in all cases). The developmental expression of col-49p::yfp and col-63p::yfp reporters was substantially similar.
3.6 |. RNAi
Synchronized L1-staged worms were incubated at 20°C (or at 25°C when indicated) and RNAi treated by feeding as described (Kamath, Martinez-Campos, Zipperlen, Fraser, & Ahringer, 2000). The lin-29 RNAi clone used in this work was obtained from the Ahringer RNAi library (Kamath & Ahringer, 2003). RNAi clones for elt-1, elt-3 and mab-10 were obtained from the Vidal library (Rual et al., 2004). The RNAi control was empty “feeding” vector L4440, a gift from Andrew Fire (Addgene plasmid # 1654). Effectiveness of lin-29 RNAi was monitored by lin-29(lf) related adult phenotypes of abnormal vulva and egg-laying defective (Kamath & Ahringer, 2003; Rual et al., 2004; Trent, Tsuing, & Horvitz, 1983) which were greater than 80% penetrant. Effectiveness of elt-1 + elt-3 RNAi was assessed by the somatic phenotype of herniation through the vulva at the L4 molt (Smith, McGarr, & Gilleard, 2005; penetrance 50%) and embryonic lethality of progeny (Baugh et al., 2005; Page, Zhang, Steward, Blumenthal, & Priess, 1997; penetrance >80%). Lastly, mab-10 RNAi causes no visible phenotypes in hermaphrodites, however it was shown that mab-10(lf) leads to a threefold increase of nhr-25 expression in the adult (Harris & Horvitz, 2011). We corroborated the effectiveness of mab-10(RNAi) experiments by including qPCR analysis of nhr-25 (three replicates) and observed a twofold increase of the latter in the mid L4, indicating that the treatment was effective to some degree. In all cases, vector control animals did not display these phenotypes. Maternal feeding of col-38p(−262)::yfp; eri-1 (RNAi hypersensitive) animals was also performed: P0 hermaphrodites were grown from the L1 stage on elt-1/3 RNAi plates and then surviving newly-hatched L1s were moved onto fresh elt-1/3 RNAi plates and scored for YFP expression in the late L4.
3.7 |. RT-qPCR
Synchronized and RNAi-treated animals were collected at the mid L4 stage; synchronized and heat-shocked animals were collected either at the L2/L3 molt stage or at the first day of adulthood, 1 hr after the heat shock. Each condition was assessed by two-step RT-qPCR in three independent biological replicates. In all cases, samples consisted of pellets of 50–100 μL of worms which were washed multiple times and resuspended (~600 μL) in DEPC water. Worms were homogenized with gentleMAC dissociator and used for RNA preparations with commercial kit Quick-RNA™ MiniPrep (Zymo Research). Total RNA was reverse transcribed with a blend of oligo(dT) and random primers provided by iScript cDNA synthesis kit (BioRad). Real-time PCRs were performed with exon-exon spanning primers (Supporting Information Table 3) and the iQ™ SYBR® Green Supermix system (BioRad). All Ct values were normalized to housekeeping gene gpd-2 and data was analyzed by the 2(delta-delta-Ct method) (Livak & Schmittgen, 2001).
3.8 |. Electrophoretic mobility shift assays
LIN-29 DNA-binding domain protein was made with the in vitro Protein Synthesis kit (PURExpress) using plasmid pTH9033 as a template, which was a gift from K. Narasimhan and T. Hughes (Narasimhan et al., 2015). Probes were made by PCR amplification using 5’ biotinylated primers (Eurofins Operon). Probe and competitor oligonucleotide sequences are listed in Supporting Information File 1. Electrophoretic mobility shift assays were done using the LightShift Chemiluminescent EMSA Kit (Thermo Scientific). Per 20 μL of [0.2 mM EDTA/3 mM MgCl2/50 μM Zinc Acetate/1 mg mL−1 BSA] binding reaction we used 5 fmol of probe and 2 μL of PURExpress in vitro protein synthesis reaction. Binding reactions were incubated at room temperature for 20 min, then run on a 4% acrylamide gel for 70 min at 100 V. Cold competitor oligonucleotide was included in the binding reaction at 1000-fold molar excess. Samples were then transferred to a nylon membrane (100 V for 40 min) and DNA was crosslinked by UV exposure. Detection was done by chemoluminescence and exposure on X-ray film following manufacturer’s protocol.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank K. Narasimhan and T. Hughes, C. Frøkjær-Jensen and E. Jorgensen, A. Fire, A. Golden, and K. O’Connell for providing plasmids, and A. Rougvie for providing plasmids and sharing unpublished results. They thank M. Negesse and D. Stonko for creation and sharing of reporter constructs. They thank members of the Eisenmann lab for advice and support. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). Funding for this work was provided by the University of Maryland Baltimore County.
Funding information
NIH Office of Research Infrastructure Programs, Grant Number: P40 OD010440; University of Maryland Baltimore County
Footnotes
SUPPORTING INFORMATION
Additional Supporting Information may be found online in the supporting information tab for this article.
REFERENCES
- Baugh LR, Wen JC, Hill AA, Slonim DK, Brown EL, & Hunter CP (2005). Synthetic lethal analysis of Caenorhabditis elegans posterior embryonic patterning genes identifies conserved genetic interactions. Genome Biology, 6, R45. 10.1186/gb-2005-6-5-r45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S (1974). The genetics of Caenorhabditis elegans. Genetics, 77, 71–94. Available at: http://www.ncbi.nlm.nih.gov/pubmed/4366476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budovskaya YV, Wu K, Southworth LK, Jiang M, Tedesco P, Johnson TE, & Kim SK (2008). An elt-3/elt-5/elt-6 GATA transcription circuit guides aging in C. elegans. Cell, 134, 291–303. 10.1016/j.cell.2008.05.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm AD, & Hsiao TI (2012). The C. elegans epidermis as a model skin. I: Development, patterning, and growth. Wiley Interdisciplinary Reviews: Developmental Biology, 1, 861–878. 10.1002/wdev.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox GN (1992). Molecular and biochemical aspects of nematode collagens. Journal of Parasitology, 78, 1–15. Retrieved from http://www.jstor.org/stable/3283678 [PubMed] [Google Scholar]
- Frøkjær-Jensen C (2015). Transposon-assisted genetic engineering with Mos1-mediated single-copy insertion (MosSCI) (pp. 49–58). Totowa, NJ: Humana Press. 10.1007/978-1-4939-2842-2_5 [DOI] [PubMed] [Google Scholar]
- Frøkjær-Jensen C, Davis MW, Ailion M, & Jorgensen EM (2012). Improved Mos1-mediated transgenesis in C. elegans. Nature Methods, 9, 117–118. 10.1038/nmeth.1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, … Trifonov T (2010). Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science, 330, 1775 LP–1787. 10.2436/20.7010.01.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilleard JS, Barry JD, & Johnstone IL (1997). cis regulatory requirements for hypodermal cell-specific expression of the Caenorhabditis elegans cuticle collagen gene dpy-7. Molecular and Cellular Biology, 17, 2301–2311. Retrieved from https://www.ncbi.nlm.nih.gov/pmc/articles/PMC232079/pdf/172301.pdf [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilleard JS, & McGhee JD (2001). Activation of hypodermal differentiation in the Caenorhabditis elegans embryo by GATA transcription factors ELT-1 and ELT-3. Molecular and Cellular Biology, 21, 2533–2544. 10.1128/MCB.21.7.2533-2544.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorrepati L, Krause MW, Chen W, Brodigan TM, Correa-Mendez M, & Eisenmann DM (2015). Identification of Wnt pathway target genes regulating the division and differentiation of larval seam cells and vulval precursor cells in Caenorhabditis elegans. Genes,Genomes, Genetics, 5(8), 1551–1566. 10.1534/g3.115.017715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris DT, & Horvitz HR (2011). MAB-10/NAB acts with LIN-29/EGR to regulate terminal differentiation and the transition from larva to adult in C. elegans. Development, 138, 4051–4062. 10.1242/dev.065417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson BM, Abete-Luzi P, Krause MW, & Eisenmann DM (2014). Use of an activated beta-catenin to identify Wnt pathway target genes in Caenorhabditis elegans, including a subset of collagen genes expressed in late larval development. Genes, Genomes, Genetics, 4, 733–747. 10.1534/g3.113.009522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson BM, & Eisenmann DM (2012). b-catenin-dependent Wnt signaling in C. elegans: Teaching an old dog a new trick. Cold Spring Harbor Perspectives in Biology, 4, a007948–a007948. 10.1101/cshperspect.a007948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone IL (2000). Cuticle collagen genes. Expression in Caenorhabditis elegans. Trends in Genetics, 16, 21–27. Available at: http://www.ncbi.nlm.nih.gov/pubmed/10637627 [DOI] [PubMed] [Google Scholar]
- Johnstone IL, & Barry JD (1996). Temporal reiteration of a precise gene expression pattern during nematode development. The EMBO Journal, 15, 3633–3639. Available at: http://www.ncbi.nlm.nih.gov/pubmed/8670866 [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, & Ahringer J (2003). Genome-wide RNAi screening in Caenorhabditis elegans. Methods (San Diego, Calif.).), 30, 313–321. Available at: http://www.ncbi.nlm.nih.gov/pubmed/12828945 [DOI] [PubMed] [Google Scholar]
- Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, & Ahringer J (2000). Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biology, 2, research0002.1. 10.1186/gb-2000-2-1-research0002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy S, Wang D, & Ruvkun G (2004). A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature, 427, 645–649. 10.1038/nature02302 [DOI] [PubMed] [Google Scholar]
- Liu Z, Kirch S, & Ambros V (1995). The Caenorhabditis elegans heterochronic gene pathway controls stage-specific transcription of collagen genes. Development (Cambridge, England), 121, 2471–2478. [DOI] [PubMed] [Google Scholar]
- Livak KJ, & Schmittgen TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods, 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Mello C, & Fire A (1995). DNA transformation. Methods in Cell Biology, 48, 451–82. Available at: http://www.ncbi.nlm.nih.gov/pubmed/8531738 [PubMed] [Google Scholar]
- Moss EG (2007). Heterochronic genes and the nature of developmental time. Current Biology, 17, R425–R434. 10.1016/j.cub.2007.03.043 [DOI] [PubMed] [Google Scholar]
- Narasimhan K, Lambert SA, Yang AWH, Riddell J, Mnaimneh S, Zheng H, … Hughes TR (2015). Mapping and analysis of Caenorhabditis elegans transcription factor sequence specificities. eLife, 4, 1–53. 10.7554/eLife.06967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page AP, & Johnstone IL (2007). The cuticle, WormBook, ed. The C. elegans Research Community, WormBook, 10.1895/wormbook.1.138.1, http://www.wormbook.org [DOI] [Google Scholar]
- Page BD, Zhang W, Steward K, Blumenthal T, & Priess JR (1997). ELT-1, a GATA-like transcription factor, is required for epidermal cell fates in Caenorhabditis elegans embryos. Genes & Development, 11, 1651–1661. 10.1101/gad.11.13.1651 [DOI] [PubMed] [Google Scholar]
- Rougvie AE, & Ambros V (1995). The heterochronic gene Lin-29 encodes a zinc-finger protein that controls a terminal differentiation event in Caenorhabditis elegans. Development, 121, 2491–2500. [DOI] [PubMed] [Google Scholar]
- Rual J-F, Ceron J, Koreth J, Hao T, Nicot A-S, Hirozane-Kishikawa T, … Vidal M (2004). Toward improving Caenorhabditis elegans phenome mapping with an orfeome-based RNAi library. Genome Research, 14, 2162–2168. 10.1101/gr.2505604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JA, McGarr P, & Gilleard JS (2005). The Caenorhabditis elegans GATA factor elt-1 is essential for differentiation and maintenance of hypodermal seam cells and for normal locomotion. Journal of Cell Science, 118, 5709–5719. 10.1242/jcs.02678 [DOI] [PubMed] [Google Scholar]
- Trent C, Tsuing N, & Horvitz HR (1983). Egg-laying defective mutants of the nematode Caenorhabditis elegans. Genetics, 104, 619–647. Available at: http://www.ncbi.nlm.nih.gov/pubmed/11813735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Bent ML, Sterken MG, Volkers RJM, Riksen JAG, Schmid T, Hajnal A, … Snoek LB (2015). Loss-of-function of bcatenin bar-1 slows development and activates the Wnt pathway in Caenorhabditis elegans. Scientific Reports, 4, 1–6. 10.1038/srep04926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, Madaan U, Park A, Aftab N, & Savage-Dunn C (2015). Multiple cis elements and GATA factors regulate a cuticle collagen gene in Caenorhabditis elegans. Genesis, 53, 278–284. 10.1002/dvg.22847 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


