Abstract
Background
Understanding the genetic interplay between human hosts and infectious pathogens is crucial for how we interpret virulence factors. Here, we tested for associations between HIV and host genetics, and interactive genetic effects on viral load (VL) in HIV-positive antiretroviral treatment-naive clinical trial participants.
Methods
HIV genomes were sequenced and the encoded amino acid (AA) variants were associated with VL, human single nucleotide polymorphisms (SNPs), and imputed HLA alleles using generalized linear models with Bonferroni correction.
Results
Human (388 501 SNPs) and HIV (3010 variants) genetic data were available for 2122 persons. Four HIV variants were associated with VL (P < 1.66 × 10–5). Twelve HIV variants were associated with a range of 1–512 human SNPs (P < 4.28 × 10–11). We found 46 associations between HLA alleles and HIV variants (P < 1.29 × 10–7). HIV variants and immunotypes when analyzed separately were associated with lower VL, whereas the opposite was true when analyzed in concert. Epitope binding predictions supported our observations.
Conclusions
Our results show the importance of immunotype specificity on viral antigenic determinants, and the identified genetic interplay emphasizes that viral and human genetics should be studied in the context of each other.
Clinical Trials Registration: NCT00867048.
Keywords: HIV, GWAS, genome-wide association study, viral genomics, host genomics, genome-to-genome analysis, viral load
We analyzed HIV and human host genetics in concert to resolve the interplay of viral and human genetics during infection. We showed that HIV genetic variants associated with different HLA alleles, and that genetic interactions affected viral load.
Human leukocyte antigens (HLA) present peptides from invading pathogens to help the generation of an immune response [1]. Genetic variants of HLA genes, that is different alleles, show differences in peptide presentation which translates into nonidentical immune response towards a given pathogen, and some HLA alleles will be selectively beneficial for the host to fight infection. Vice versa, pathogens are genetically diverse, and genetic variants are selected to evade the human immune response [2], but also to optimize other phenotypes such as antiretroviral therapy resistance [3–5]. Accordingly, it has been found that the genetic variation of human immunodeficiency virus 1 (HIV-1) is partly shaped by which HLA is encountered within the host [6]. Similarly, HLAs expressed by the host affect the replicative rate of the virus: some HLAs or even their functional groups are known to be protective while others are associated with worse outcomes [7, 8]. The proportion of variation in the HIV genome attributable directly to the interaction with the host genetics remains unclear. Furthermore, the molecular specificity of the interplay between viral genetic escape and host genomic control remains poorly defined [9].
In principle, the ability of HIV to replicate in the body can be quantified by the plasma viral load (VL), the key determinant to identify the risk of disease progression and death [3]. Large variations in VL are observed in HIV-positive persons and a sizable fraction hereof in early HIV infection is assumed to be due to variation in viral replicative ability, which is also influenced by the host immune response [10]. A fraction of variance in HIV replication and disease progression is attributable to viral [10–14] or host genetics [15–18]. However, viral and host genetic interactions are poorly understood [9]. Prior HIV and host genetic association studies encompassed data from persons with similar genetic background or individual subtypes of HIV [19–21]. Expanding to a cohort with multiple viral subtypes and diverse host demographics could elucidate specific factors that influence HIV pathogenesis or generalize previous findings from more homogenous cohorts.
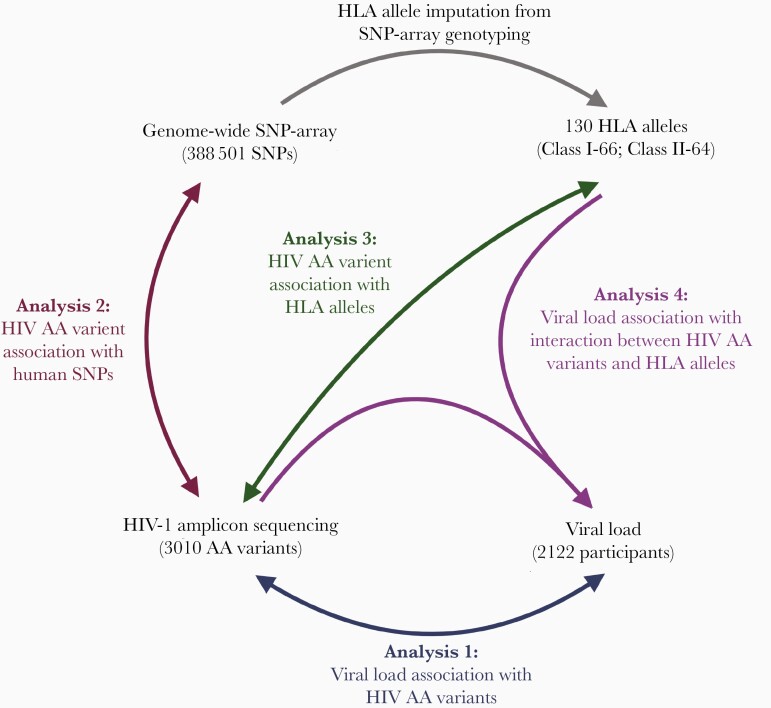
In this study we performed 4 analyses (Figure 1): (1) association of HIV AA variants with VL to estimate the effect of viral genetic variation on VL in HIV infection, (2) association of HIV AA variants with human single nucleotide polymorphisms (SNPs) to identify genome-to-genome associations, (3) association of HIV AA variants with HLA alleles to identify HLA alleles with the strongest association with HIV genetic variants, and (4) bivariate association and interaction of HIV AA variants and HLA alleles on VL to resolve the interplay of viral and human genetics during HIV infection. These associations were performed in a demographically diverse cohort of treatment-naive HIV-positive persons with different HIV subtypes from the Strategic Timing of Antiretroviral Treatment (START) cohort [22].
Figure 1.
Schematic visualization of the 4 association analyses performed in this study. Abbreviations: AA, amino acid; HIV, human immunodeficiency virus; SNP, single nucleotide polymorphism.
METHODS
Study Population
Samples analyzed in this study were derived from participants in the START trial (NCT00867048), conducted by the International Network for Strategic Initiatives in Global HIV Trials, which included 4685 antiretroviral therapy-naive and asymptomatic participants [22] who had a CD4+ cell count >500 cells/µL at baseline, no history of AIDS or of antiretroviral treatment, and were older than 18 years at study entry. For this study, we included participants that (1) had baseline plasma samples with a VL measurement ≥1000 copies/mL and viral genomes sequenced by next-generation sequencing (n = 3785), and (2) consented to human genotyping (n = 2546). All samples were derived from participants who provided written informed consent and all informed consents were reviewed and approved by participant site ethics review committees. The study was approved by the institutional review board or ethics committee at each contributing center.
HIV Sequencing, Alignment, and Variant Calling
The detailed laboratory procedure is described in the Supplementary Materials and Methods. In short, viral RNA was extracted from plasma and amplified using 2 amplicons spanning 7125 nucleotides of the 9719-nucleotide (74%) HIV genome and sequenced by next-generation sequencing. Reads were first aligned against the GRCh37.p13 human reference genome with Bowtie2 version 2.2.8 to retain read pairs with neither read aligning to the human genome. Virvarseq [23] was used for alignment of cleaned reads and AA variant calling with the HXB2 HIV-1 reference genome (GenBank accession number K03455.1) with default parameters. Virvarseq alignment, realignment, and AA variant calling were performed per gene basis (rev and tat genes were split into separate analyses for each reading frame). The median, minimum, maximum, first and third quartile read alignment coverage, and depth for HIV genes are visualized in Supplementary Figure 1. The 2 amplicons did not completely cover the genes gag, pol, and tat; therefore, variant calls for the gag gene were considered from HXB2 reference genome AA position 240, for the pol gene until AA position 983, and for the tat gene from position 47. No sequencing was available for the genes vpr and vif. Minimum Phred quality scores for all called codons were >20. Variant calling was performed only for samples that had at least 10% of the gene covered by aligned reads with minimum 10-fold coverage. Variants were called if there was a minimum of 10-fold coverage and a minimum of 50% aligned reads supported the variant, that is we used the consensus call for the viral quasispecies.
Genotyping and Quality Control
The 2546 study participants were genotyped by Affymetrix Axiom SNP-array enriched with markers related to immune dysfunction, consisting of 388 501 probes after excluding the following: (1) nonmonoallelic variants, (2) variants with >20% missingness, (3) <0.05 minor allele frequency, and (4) variants that deviated from the Hardy-Weinberg equilibrium (P < 1 × 10–6). The detailed procedure for host genotyping and HLA allele imputation is described elsewhere [15]. Only HLA alleles with ≥1% frequency in the population were used in the association analyses: 66 class I HLA alleles (20 HLA-A, 29 HLA-B, and 17 HLA-C alleles) and 64 class II HLA alleles (14 HLA-DPB1, 13 HLA-DQA1, 14 HLA-DQB1, and 23 HLA-DRB1 alleles). Individuals with (1) >20% missingness, (2) outlying heterozygosity rates (more than ±3 SD), (3) cryptic relatedness (PI Hat > 0.2) [24], and (4) missing baseline VL measurements were excluded.
Statistical Analysis
The association analysis was performed with PLINK2 [25]. A dominant linear regression model was fitted to assess associations of VL with HIV AA variants, and of VL with the bivariate association and interaction between HIV AA variants and HLA alleles. A generalized linear model with logit link function was applied for genome-to-genome and HIV AA variants with HLA allele association analysis. VL measurements were log10 transformed prior to analysis. Bonferroni correction for multiple testing with an adjusted threshold of 0.05 was used for all associations except the VL association with the interaction between viral and host genetics. Age, sex assigned at birth, and the first 4 principal components of the genetic data from both human and viral populations were used as covariates to account for population structures. The principal component analysis was performed using EIGENSOFT version 6.1.4 [26, 27]. R [28] was used for explained variance analysis and visualizations. The explained variance was analyzed by comparing the variable’s sum of squares with the total sum of squares from analysis of variance (ANOVA).
Epitope Binding Prediction
Epitope binding was predicted with NetMHCpan version 4.0 [29] considering all the 9-mer peptides with a 9-nucleotide window around the position of interest using the default binding affinity thresholds.
RESULTS
Study Participants
A total of 2172 participants consented for genetic testing and had matching HIV genomes sequenced. Three participants did not have baseline VL measured, 46 had outlying heterozygosity, and 1 had cryptic relatedness, leaving 2122 participants in the study, spanning 5 continents and 23 countries (Table 1).
Table 1.
Characteristics of 2122 Participants From the START Trial Included in the Analysis
| Characteristic | START Participants After Quality Control (n = 2122) |
|---|---|
| Age, y, median (IQR) | 36 (29–45) |
| Sex (%) | |
| Female | 365 (17) |
| Male | 1757 (83) |
| Self-reported race (%) | |
| Asian | 21 (1) |
| Black | 432 (20) |
| Hispanic | 382 (18) |
| White | 1250 (59) |
| Other | 37 (2) |
| Geographic region (%) | |
| Africa | 244 (11) |
| Australia | 85 (4) |
| Europe and Israel | 1030 (49) |
| Latin America | 392 (18) |
| United States | 371 (17) |
| Mode of HIV infection (%) | |
| Injection drug use | 34 (2) |
| Sex with same sex | 1445 (68) |
| Sex with opposite sex | 625 (29) |
| Other | 23 (1) |
| HIV subtype (%) | |
| A | 24 (1) |
| B | 1364 (64) |
| C | 101 (5) |
| AB | 35 (2) |
| AE | 17 (1) |
| AG | 38 (2) |
| BC | 85 (4) |
| BF | 113 (5) |
| BG | 11 (1) |
| Other | 67 (3) |
| Unknown | 268 (13) |
| Time since HIV diagnosis, y, median (IQR) | 1.08 (0.44–2.84) |
| Recent infection, within 6 mo (%) | 163 (7.7) |
| CD4+ cell count, cells/μL, median (IQR) | 643 (583–744) |
| HIV load, copies/mL, median (IQR) | 19 450 (6393–51 732) |
Data are No. (%) except where indicated.
Abbreviations: HIV, human immunodeficiency virus; IQR, interquartile range; START, Strategic Timing of Antiretroviral Treatment.
HIV Amino Acid Variants
A total of 16 040 AA variants were called with Virvarseq. The AA variants were filtered based on allele frequency: ≥5% and ≤95% of the study population. If more than 1 AA variant was observed in a position, the most abundant variant was designated as a reference and excluded from further analysis to reduce redundancy. This reduced the number of HIV AA variants to 3010. The AA variant distribution across the HIV genome was 215 (7%) variants in gag, 1041 (35%) in pol, 80 (2%) in tat, 144 (5%) in rev, 110 (4%) in vpu, 1184 (39%) in env, and 236 (8%) in nef genes.
Association of HIV Amino Acid Variants with Viral Load
Of the 3010 analyzed HIV AA variants, 4 were associated with VL (Bonferroni P value < 1.66 × 10–5). All HIV AA variants tested for association with VL are visualized in a Manhattan plot (Supplementary Figure 2). The 4 variants were Pol980E (integrase), Tat53R, Rev7D, and Env571W (gp41); details of these AA variants and their sequencing coverage are listed in Supplementary Tables 1 and 2. The effect size of the 4 HIV AA variants ranged from −0.084 to +0.097 log10 copies/mL. ANOVA of the 4 HIV AA variants shows that they account for 8.2% of the total variation in log10 VL. Pearson correlation coefficient of VL with the number of aligned reads was 0.21.
HIV Amino Acid Variant Association With Host SNPs
Association tests between viral AA variants and host genome-wide SNPs resulted in a total of 12 HIV AA variants that were associated with a range of 1–512 human SNPs (Bonferroni threshold P value < 4.28 × 10–11; Table 2). Most associations were in the genes gag (n = 5) and nef (n = 5) with the strongest association between Gag242N HIV AA variant and the human rs1248395773 SNP (P value = 1.22 × 10–79). All but 1 association were in the HLA class I gene region (chr6:29 910 247–31 324 939). The association between Gag242N and a host SNP outside the HLA class I gene region (chr15:50785016) was in high linkage disequilibrium (LD; R2 ≥ 0.90) with several associated SNPs from the HLA gene region (Supplementary Table 3).
Table 2.
Twelve HIV AA Variants Significantly Associated With Varying Numbers of Host SNPs
| HIV AA Variant | Lowest P Value | Number of Associated Host SNPs | Region of Associated Host SNPs |
|---|---|---|---|
| GAG242N | 1.22 × 10–79 | 512 | chr6:29818568–32626272; chr15:50785016 |
| GAG302K | 1.35 × 10–16 | 8 | chr6:30993440–31326410 |
| GAG357S | 3.17 × 10–16 | 48 | chr6:31005726–31812038 |
| GAG397R | 1.13 × 10–32 | 186 | chr6:29607476–30222020 |
| GAG403K | 2.78 × 10–19 | 4 | chr6:29830074–29924779 |
| POL432R (RT277R) | 6.26 × 10–39 | 134 | chr6:29611885–30711805 |
| POL837I (INT122I) | 1.85 × 10–11 | 1 | chr6:31316609 |
| NEF071K | 1.38 × 10–31 | 150 | chr6:30861729–31879158 |
| NEF092R | 4.70 × 10–19 | 8 | chr6:29785827–30026290 |
| NEF102H | 8.79 × 10–16 | 1 | chr6:29785827–30026290 |
| NEF105R | 9.38 × 10–12 | 3 | chr6:31242649–31245821 |
| NEF135F | 6.36 × 10–21 | 8 | ch6:29735280–29992261 |
The information on host and viral variant location in the genome together with the lowest P value are reported.
Abbreviations: AA, amino acid; HIV, human immunodeficiency virus; INT, integrase; RT, reverse transcriptase; SNP, single nucleotide polymorphism.
Association of HIV Amino Acid Variants With HLA Alleles
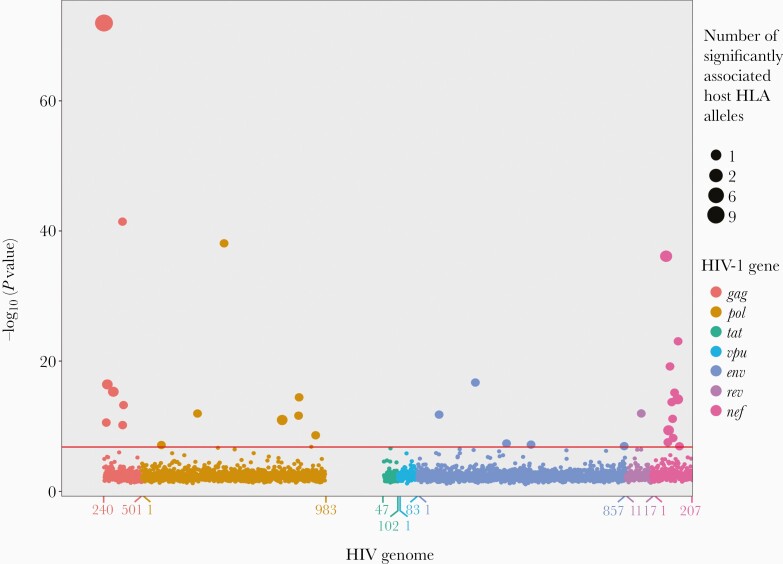
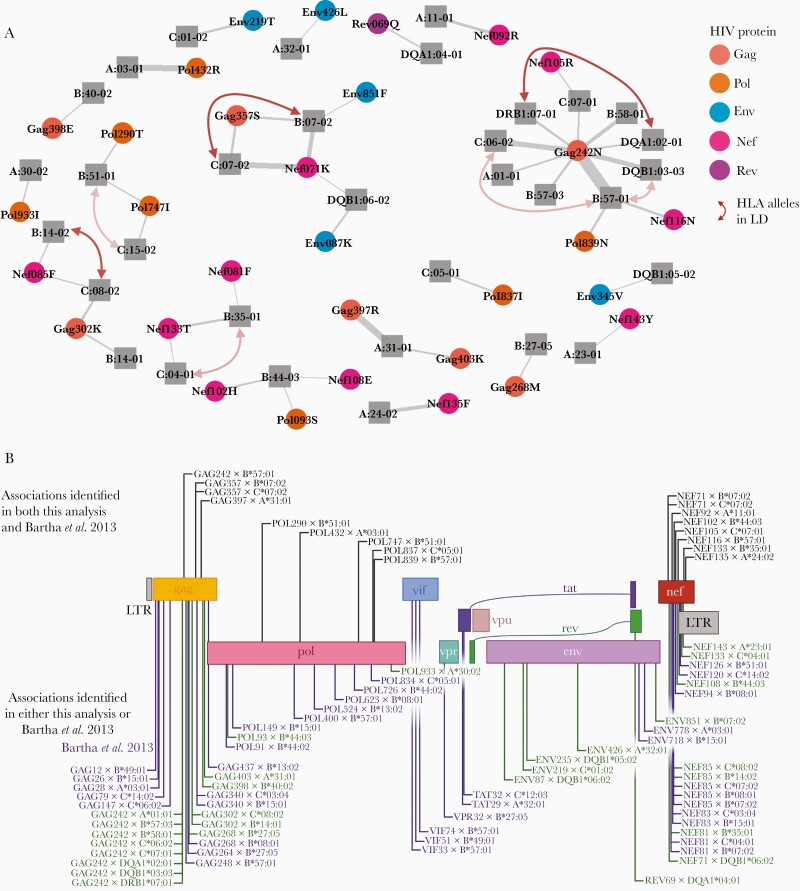
As all the associations in the previous analysis were in the HLA gene region, we next tested the associations between the HIV AA variants and the imputed HLA alleles. We identified 46 associations (Bonferroni threshold P value < 1.29 × 10–7) between 33 different HLA alleles (27 class I and 6 class II) and 31 HIV AA variants (Figure 2). The 12 previously identified associated HIV AA variants were also associated with HLA alleles. As in the previous analysis, most associations were in the genes gag (n = 12) and nef (n = 17). Furthermore, several HLA alleles were in LD while no HIV AA variants were in LD (Figure 3A and Supplementary Table 4). Associated HLA alleles showed different prevalence across participants’ race; however, all of the associated alleles were present in the major subpopulations (white, black, and Hispanic participants) (Supplementary Table 5).
Figure 2.
Associations between HIV amino acid (AA) variants and HLA alleles projected on HIV genome with the strongest association for each HIV AA variant included. Red horizontal line marks the Bonferroni P value threshold (P = 1.29 × 10–7). The x-axis denotes gene AA positions. Circle size denotes the number of significantly associated host HLA alleles.
Figure 3.
A, Network of the associated HLA alleles and HIV AA variants where line thickness represents the association’s strength and red arrows mark HLA alleles in LD. The opacity of the arrows corresponds to squared correlation coefficient (R2). B, Comparison between the HLA type and HIV AA associations identified in this analysis and the Bartha et al study [19]. Abbreviations: AA, amino acid; HIV, human immunodeficiency virus; LD, linkage disequilibrium; LTR, long terminal repeat.
We further compared our findings to a study by Bartha et al [19] that performed a similar analysis although not including HLA class II alleles. Of the 39 associations between HLA class I alleles and HIV AA variants identified in this study, 17 associations overlapped with the 53 associations between HLA class I alleles and HIV AA variants identified by Bartha et al (Figure 3B). Like our analysis, Bartha et al also identified most associations to gag and nef; nonetheless, the largest relative overlap between studies was for pol (5 of 14 associations overlapped). Moreover, out of a total 34 HLA class I alleles associated with HIV AA variants in at least 1 study, 17 HLA alleles were associated with HIV AA variants in both studies.
Viral Load Association With the Interaction Between Viral and Host Genetics
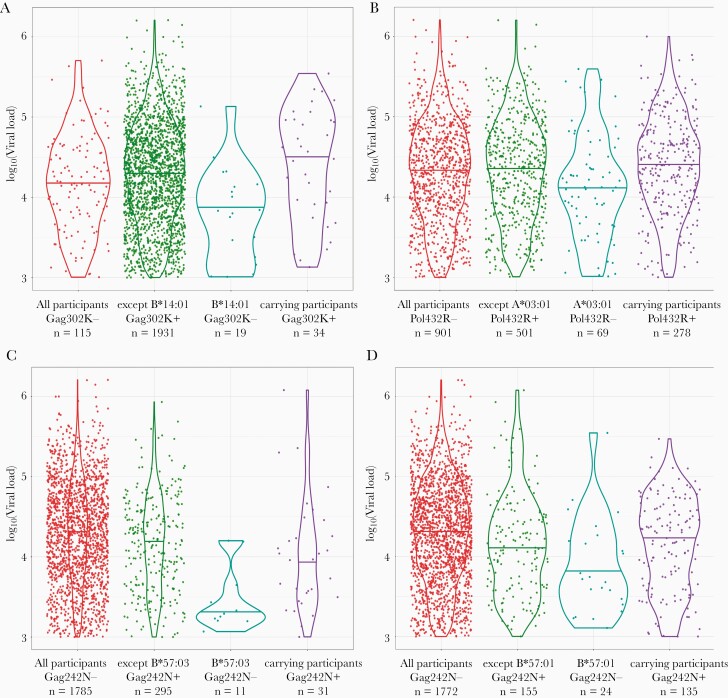
To disentangle the effect of the HLA allele presence together with the associated HIV AA variants, we performed an interaction analysis for the 46 previously identified HLA-HIV associations with VL. The effect sizes with 95% confidence intervals together with the P values of the estimates are reported in Supplementary Table 6. In 3 cases the associated HLA allele and HIV AA variant interaction association with VL was below our P value threshold (P value < 5 × 10–2; Table 3). In all 3 cases, the individual effects of the HLA alleles resulted in lower VL (P value < 5 × 10–2), Gag302K and Pol432R had no effect on VL, and Gag242N showed lower VL (P value < 5 × 10–2). However, the interaction between the HLA alleles and the associated HIV AA variant in all 3 cases showed higher VL (P value < 5 × 10–2; Figure 4). Moreover, the interaction between B*57:01 and Gag242N AA variant did not meet our P value threshold (P value = 8.5 × 10–2); however, it showed the same interaction tendency as B*57:03 allele interaction with Gag242N AA variant (Supplementary Table 8). Both B*57:01 and B*57:03 HLA alleles are known to belong to the same functional group. Furthermore, we predicted the binding affinities in these 4 cases (Supplementary Tables 7 and 8). Epitopes with the associated HIV AA variants were weak or nonbinders; vice versa, epitopes with other than the HIV AAs on the same position were strong binders.
Table 3.
Summary of HLA Alleles, the Associated HIV Amino Acid Variants, and Their Interaction Effect on Viral Load That Passed the P Value Threshold
| Variable | Effect Size (95% CI) | P Value |
|---|---|---|
| Gag242N | −0.136 (−.220 to −.053) | 1.4 × 10 −3 |
| B*57:03 | −0.591 (−.945 to −.237) | 1.1 × 10 −3 |
| Gag242N×B*57:03 | 0.577 (.163 to .991) | 6.4 × 10 −3 |
| Gag302K | 0.005 (−.124 to .135) | 9.3 × 10−1 |
| B*14:01 | −0.419 (−.712 to −.125) | 5.2 × 10 −3 |
| Gag302K×B*14:01 | 0.540 (.185 to .896) | 3.0 × 10 −3 |
| Pol432R | −0.015 (−.082 to .051) | 6.5 × 10−1 |
| A*03:01 | −0.188 (−.324 to −.051) | 7.3 × 10 −3 |
| Pol432R × A*03:01 | 0. 226 (.068 to .384) | 5.1 × 10 −3 |
The variables in bold represent association tests below the P value threshold.
Abbreviations: CI, confidence interval; HIV, human immunodeficiency virus.
Figure 4.
Violin plots of the HIV amino acid (AA) variant and HLA alleles show the 3 associations that had a strong (P < .05) interaction effect on viral load (A–C), and the Gag242N and B*57:01 association that did not have a strong interaction effect on viral load but shows the same tendency as the Gag242N and B*57:03 association (D).
The 3 associations with interaction effects on VL were used to investigate the explained variance in log10 VL. We first applied the model of VL with the 3 HLA alleles and the associated HIV AA variants as covariates, and HLA alleles together with HIV AA variants explained 2.0% of the total variance. When the interactions were added as covariates to the model, the explained variance was 3.6%.
DISCUSSION
The host and HIV viral genome interaction and its effect on VL in demographically diverse populations is poorly understood. In this study, we performed 4 analyses using baseline data from antiretroviral treatment-naive START trial participants to explore the interaction of viral and host genetics together with the impact it has on the VL, which is a proxy for disease progression. While we found little signal in the associations between HIV AA variants and VL, we identified both host SNPs and HLA alleles that were associated with HIV AA variants. Finally, we showed that the interaction of several of these associations was affecting the VL.
The first association analysis with VL against HIV AA variants showed a weak signal, with only 4 associations below the P value threshold. Similar results were observed in a study on persons infected with subtype B by Bartha et al [19]. Only the HIV AA variant Env571W is located in a region previously described as being associated with the replicative fitness of HIV [30, 31]. However, while these associations pass the stringent P value threshold requirements, further functional laboratory validation is necessary to confirm any biological function. From the low number of HIV AA variants associated with VL, it is apparent that single variants in the HIV genome cannot sufficiently explain differences in VL between individuals. The analysis of associations between VL and HIV AA variants is inherently limited, as VL correlates with the ability to detect HIV AA variants: low sequencing coverage occurred more frequently in low VL samples.
The second association analysis with HIV AA variants against human SNPs showed strong associations between HIV AA variants and SNPs in the HLA region. In particular, Gag242N was associated with 511 SNPs in the class I HLA region. Interestingly, none of the associations were observed in the gene env, which might be due to the hypervariable nature of some regions of this gene and that adaptation to the host immune system happens shortly after the initial infection, making it difficult to capture both adapted and nonadapted viral genotypes in the data set [32]. Ultimately, the associations in the host genome were only observed in the HLA gene region. The same observation of the associated SNPs being in the HLA gene region was made by Bartha et al [19]. To improve the sensitivity and biological interpretability of the analysis, we next tested directly for associations between HLA alleles and HIV AA variants.
Of 66 tested class I HLA alleles, 27 were associated with 1 or several HIV AA variants, which could be corresponding to the CD8+ T-cell mediated response primarily interacting with HIV. Our analysis adds to the previous findings of Bartha et al [19] that were based on a smaller and demographically more homogeneous cohort of primarily European descent, and 17 associations between HIV AA and class I HLA alleles overlap between our study and that of Bartha et al [19]. The overlapping associations were located in gag, pol, and nef genes, which suggests these HIV genome regions as likely being under high host pressure. Several factors might explain differences in the associations identified across studies: (1) the START cohort is larger and more demographically heterogeneous than the cohort from Bartha et al [19], where only participants with Western European ancestry and carrying subtype B HIV were included; (2) this analysis included both class I and class II HLA alleles while class II HLA alleles were not part of the Bartha et al study; and (3) Bartha et al did not include a part of the env gene (coding for GP120) while in this analysis the beginning of the gag gene, the end of the pol gene, the beginning of the tat gene, and the genes vif and vpr were not sequenced. Our study supports the findings from Kinloch et al [33], that many escape pathways are not subtype B specific, but rather shared among different subtypes.
Only 6 out of 64 tested HLA class II alleles had associations with P values below the Bonferroni threshold. Furthermore, several associated HLA class II alleles were in LD with other associated HLA alleles, which could indicate that HLA class II alleles have less effect on HIV diversity as the primary immune response is mediated through CD8+ T-cells and CD4+ cells expressing HLA class II molecules play a lesser role during the infection [34]. However, from the 46 identified associations between HLA alleles and HIV AA variants (Figure 3A) we show that a heterogeneous group of HLA alleles is associated with a diverse group of HIV AA variants, which might be essential during the HIV infection for viral adaptation to the host.
The interaction analysis between HIV AA variants with the associated HLA alleles and their influence on VL revealed that, while several HLA alleles are linked to lower VL and the HIV AA variants associate with no or even a slightly lower VL, the interaction between them associated with higher VL; thus the protective effect of the HLA allele is diluted when analyzed with the corresponding viral genotype. Arora et al [35] recently showed that B*57:01 and B*57:03 HLA alleles share similar clusters of HIV-associated epitopes. While predominantly found in different races, our analysis shows that these HLA alleles and their interaction with the associated HIV AA variants show the same impact on VL, which further supports the observed interaction effect as a selected biological trait. This observation is further supported by the predicted epitope binding affinity of the 4 associations where the associated HLA allele had a lower binding affinity to the epitope containing the associated HIV AA variant than the other observed AA variants in the position of interest.
While only a fraction of variance is explained using the 3 associated HLA alleles and HIV AA variants together with their interaction, the observation that addition of the interaction to the model explained almost twice as much variance indicates that the interaction between the viral and host genomes is essential when analyzing viral evolution and adaptation.
Our study has several limitations. First, while it is a demographically diverse and large cohort, we did not have consent for genotyping from Asian START clinical trial participants. Furthermore, larger cohorts that expand on the demographic diversity are needed for the more sensitive association identification in the association of VL to HIV AA variants analyzed in concert with interactive HLA alleles as the analysis of participants with less common HLA alleles did not have enough power. Moreover, because of the study design no follow-up HIV genomes were included in the study, which could have provided further support for the identified associations as the phenomena of the viral adaptation to the host immunotype. SNP-array genotyping of the host genome and HIV amplicon sequencing instead of whole-genome sequencing limited the amount of genetic variants that could be identified for association analysis; however, our SNP arrays were enriched for immune genes of the human genome [15] and the majority of the coding region in the HIV genome was covered. Similarly, the HLA allele information was imputed with high accuracy and not based on conventional HLA typing, thus slightly increasing the uncertainty in our study. In addition, we analyzed only HIV consensus sequences, and accordingly we did not address the intrahost diversity, which would provide more detailed insights at the individual level. Finally, because of the nature of the cohort, no participants with low CD4+ cell count and low VL were included in the analysis and, therefore, HLA alleles from HIV controllers will not be well represented in our analysis.
In conclusion, by using a demographically diverse cohort of treatment-naive HIV-positive persons with different HIV subtypes, we were able to identify and quantify the influence that viral and host genetics have on VL. We observed that HIV AA variants alone explained little to no variation in VL. Instead, we found that a diverse group of HLA class I alleles, and to a lesser extent also HLA class II, impose selection on HIV genomes for variants primarily in gag and nef genes, and that VL was influenced by the interplay of viral and human genetics. Accordingly, HIV AA variants and the host HLA alleles should be analyzed and interpreted together, highlighting that HIV virulence factors are best studied in the context of the host.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank all participants in the START trial and all trial investigators (see Lundgren et al [22] for the complete list of START investigators).
Author contributions. R. L. M. and J. D. L. conceived the study and supervised the project. M. G., M. B., A. G. Z., R. L. M., and J. D. L. designed the analysis plan. M. G. and M. B. conducted the analysis and wrote the manuscript. M. G., M. B., A. G. Z., C. E., D. D. M., V. L. K., G. T., L. V., D. T., J. N., H. C. L., S. S., A. A. P., M. N. P., H. F. G., J. D. L., and R. L. M. revised the analysis plan, and reviewed and edited the manuscript.
Financial support. This work was supported by the Danish National Research Foundation (grant number 126). The START trial was supported by the National Institutes of Health National Institute of Allergy and Infectious Diseases, Division of AIDS (United States) (grant numbers UM1-AI068641, UM1-AI120197, and 1U01-AI36780), National Institutes of Health Clinical Center, National Cancer Institute, National Heart, Lung, and Blood Institute, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institute of Mental Health, National Institute of Neurological Disorders and Stroke, National Institute of Arthritis and Musculoskeletal and Skin Diseases, Agence Nationale de Recherches sur le SIDA et les Hépatites Virales (France), National Health and Medical Research Council (Australia), National Research Foundation (Denmark), Bundes Ministerium für Bildung und Forschung (Germany), European AIDS Treatment Network, Medical Research Council (United Kingdom), National Institute for Health Research, National Health Service (United Kingdom), and University of Minnesota. Antiretroviral drugs for the START clinical trial were donated to the central drug repository by AbbVie, Bristol-Myers Squibb, Gilead Sciences, GlaxoSmithKline/ViiV Healthcare, Janssen Scientific Affairs, and Merck.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Presented in part: virtual Conference on Retroviruses and Opportunistic Infections (CROI) 8–11 March 2020, poster number 00272.
References
- 1. Prugnolle F, Manica A, Charpentier M, Guégan JF, Guernier V, Balloux F. Pathogen-driven selection and worldwide HLA class I diversity. Curr Biol 2005; 15:1022–7. [DOI] [PubMed] [Google Scholar]
- 2. Hertz T, Nolan D, James I, et al. Mapping the landscape of host-pathogen coevolution: HLA class I binding and its relationship with evolutionary conservation in human and viral proteins. J Virol 2011; 85:1310–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fraser C, Lythgoe K, Leventhal GE, et al. Virulence and pathogenesis of HIV-1 infection: an evolutionary perspective. Science 2014; 343:1243727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zanini F, Puller V, Brodin J, Albert J, Neher RA. In vivo mutation rates and the landscape of fitness costs of HIV-1. Virus Evol 2017; 3:vex003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Voronin Y, Holte S, Overbaugh J, Emerman M. Genetic drift of HIV populations in culture. PLoS Genet 2009; 5:e1000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McLaren PJ, Carrington M. The impact of host genetic variation on infection with HIV-1. Nat Immunol 2015; 16:577–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Naranbhai V, Carrington M. Host genetic variation and HIV disease: from mapping to mechanism. Immunogenetics 2017; 69:489–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zucco AG, Bennedbaek M, Ekenberg C, et al. Functional clustering and association of HLA class I alleles to viral load in HIV-positive and ART-naive participants from the INSIGHT START study. HIV Med 2019; 20:181. [Google Scholar]
- 9. Fellay J, Pedergnana V. Exploring the interactions between the human and viral genomes. Hum Genet 2020; 139:777–81. [DOI] [PubMed] [Google Scholar]
- 10. Bartha I, McLaren PJ, Brumme C, Harrigan R, Telenti A, Fellay J. Estimating the respective contributions of human and viral genetic variation to HIV control. PLoS Comput Biol 2017; 13:e1005339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Alizon S, von Wyl V, Stadler T, et al. ; Swiss HIV Cohort Study . Phylogenetic approach reveals that virus genotype largely determines HIV set-point viral load. PLoS Pathog 2010; 6:e1001123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bertels F, Marzel A, Leventhal G, et al. ; Swiss HIV Cohort Study . Dissecting HIV virulence: heritability of setpoint viral load, CD4+ T-cell decline, and per-parasite pathogenicity. Mol Biol Evol 2018; 35:27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Blanquart F, Wymant C, Cornelissen M, et al. ; BEEHIVE Collaboration . Viral genetic variation accounts for a third of variability in HIV-1 set-point viral load in Europe. PLoS Biol 2017; 15:e2001855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bachmann N, Turk T, Kadelka C, et al. ; Swiss HIV Cohort Study . Parent-offspring regression to estimate the heritability of an HIV-1 trait in a realistic setup. Retrovirology 2017; 14:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ekenberg C, Tang MH, Zucco AG, et al. Association between single-nucleotide polymorphisms in HLA alleles and human immunodeficiency virus type 1 viral load in demographically diverse, antiretroviral therapy-naive participants from the strategic timing of antiretroviral treatment trial. J Infect Dis 2019; 220:1325–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fellay J, Shianna KV, Ge D, et al. A whole-genome association study of major determinants for host control of HIV-1. Science 2007; 317:944–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. International HIV Controllers Study, Pereyra F, Jia X, McLaren PJ, et al. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 2010; 330:1551–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fellay J, Ge D, Shianna KV, et al. ; NIAID Center for HIV/AIDS Vaccine Immunology (CHAVI) . Common genetic variation and the control of HIV-1 in humans. PLoS Genet 2009; 5:e1000791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bartha I, Carlson JM, Brumme CJ, et al. A genome-to-genome analysis of associations between human genetic variation, HIV-1 sequence diversity, and viral control. Elife 2013; 2:e01123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Power RA, Davaniah S, Derache A, et al. Genome-wide association study of HIV whole genome sequences validated using drug resistance. PLoS One 2016; 11:e0163746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tough RH, McLaren PJ. Interaction of the host and viral genome and their influence on HIV disease. Front Genet 2018; 9:720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lundgren JD, Babiker AG, Gordin F, et al. ; INSIGHT START Study Group . Initiation of antiretroviral therapy in early asymptomatic HIV infection. N Engl J Med 2015; 373:795–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Verbist BM, Thys K, Reumers J, et al. VirVarSeq: a low-frequency virus variant detection pipeline for Illumina sequencing using adaptive base-calling accuracy filtering. Bioinformatics 2015; 31:94–101. [DOI] [PubMed] [Google Scholar]
- 24. Marees AT, de Kluiver H, Stringer S, et al. A tutorial on conducting genome-wide association studies: quality control and statistical analysis. Int J Methods Psychiatr Res 2018; 27:e1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 2015; 4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet 2006; 2:e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006; 38:904–9. [DOI] [PubMed] [Google Scholar]
- 28. The R Project for Statistical Computing. R. https://www.r-project.org/. Accessed 10 February 2020.
- 29. Jurtz V, Paul S, Andreatta M, Marcatili P, Peters B, Nielsen M. NetMHCpan-4.0: improved peptide-MHC class I interaction predictions integrating eluted ligand and peptide binding affinity data. J Immunol 2017; 199:3360–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sloan EA, Kearney MF, Gray LR, et al. Limited nucleotide changes in the Rev response element (RRE) during HIV-1 infection alter overall Rev-RRE activity and Rev multimerization. J Virol 2013; 87:11173–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sherpa C, Rausch JW, Le Grice SF, Hammarskjold ML, Rekosh D. The HIV-1 Rev response element (RRE) adopts alternative conformations that promote different rates of virus replication. Nucleic Acids Res 2015; 43:4676–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Andrews SM, Zhang Y, Dong T, Rowland-Jones SL, Gupta S, Esbjörnsson J. Analysis of HIV-1 envelope evolution suggests antibody-mediated selection of common epitopes among Chinese former plasma donors from a narrow-source outbreak. Sci Rep 2018; 8:5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kinloch NN, Lee GQ, Carlson JM, et al. Genotypic and mechanistic characterization of subtype-specific HIV adaptation to host cellular immunity. J Virol 2018; 93:e01502-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Blankson JN, Siliciano RF. MHC class II genotype and the control of viremia in HIV-1-infected individuals on highly active antiretroviral therapy. J Clin Invest 2001; 107:549–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Arora J, McLaren PJ, Chaturvedi N, Carrington M, Fellay J, Lenz TL. HIV peptidome-wide association study reveals patient-specific epitope repertoires associated with HIV control. Proc Natl Acad Sci U S A 2019; 116:944–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.