Significance
The search for genetic variants associated with resistance or susceptibility to HIV/AIDS already yielded insights that allowed developing therapeutics that contribute to dramatic reduction in AIDS-related comorbidities. Unfortunately, nearly all studies focused on European-descent populations, even though most infections happen in Africa. In this genome-wide association study we explored the genetic background of several hundred individuals from Botswana in search of associations with HIV infection. We discovered several genetic variant associations in genes implicated in HIV/AIDS pathogenesis. We also confirmed several associations reported in previous research. This study provides valuable data on the influence of human genetic variation on HIV-1C infection and pathogenesis in southern Africa, a region with the world’s largest number of HIV-1 infections.
Keywords: AIDS, HIV-C, GWAS, Botswana, GWATCH
Abstract
Although there have been many studies of gene variant association with different stages of HIV/AIDS progression in United States and European cohorts, few gene-association studies have assessed genic determinants in sub-Saharan African populations, which have the highest density of HIV infections worldwide. We carried out genome-wide association studies on 766 study participants at risk for HIV-1 subtype C (HIV-1C) infection in Botswana. Three gene associations (AP3B1, PTPRA, and NEO1) were shown to have significant association with HIV-1C acquisition. Each gene association was replicated within Botswana or in the United States–African American or United States–European American AIDS cohorts or in both. Each associated gene has a prior reported influence on HIV/AIDS pathogenesis. Thirteen previously discovered AIDS restriction genes were further replicated in the Botswana cohorts, extending our confidence in these prior AIDS restriction gene reports. This work presents an early step toward the identification of genetic variants associated with and affecting HIV acquisition or AIDS progression in the understudied HIV-1C afflicted Botswana population.
HIV infection is a fatal chronic disease that has spread across all continents since its emergence in the early 1980s. Important progress has been made against the epidemic in the last two decades. The global incidence–prevalence ratio has declined from 11.2% in 2000 to 4.6% in 2018. Despite this, the world is not yet on track to end HIV/AIDS as a public health threat (1). There are 38 million (31.6 million to 44.5 million) people in the world living with HIV; 32.7 million (24.8 million to 42.2 million) people who died from AIDS-related conditions since the onset of the pandemic. An estimated 690,000 (500,000 to 970,000) died of HIV/AIDS in 2019 (1). Despite the important breakthroughs in therapeutic approaches, as well as efforts and resources invested in the fight against the HIV epidemic for about four decades, no efficient vaccine or cure exists.
Sub-Saharan Africa is the region most affected by the HIV pandemic. Botswana falls within the top three countries with the highest HIV prevalence in the world, with an HIV prevalence rate of 20.3% (17.3 to 21.8) among adults (15 to 49 y of age) (1).
Currently HIV-1 subtype C (HIV-1C), which is widespread in the Botswana population, accounts for more than half of all global HIV-1 infections and several times more than any other HIV-1 subtype. Despite this fact, HIV-1C infection remains understudied as prior research largely focused on HIV-1 subtypes (mostly B) more prevalent in Europe and in the United States (2–8).
HIV/AIDS exhibits considerable epidemiological heterogeneity (strength of the innate, humoral, and cell-mediated immune responses, variation in response to antiretroviral therapy, ART), much of which may be attributed to host genetic factors (3–8). African populations in general have the highest overall genetic variation, and it has been hypothesized that treatment-naive Botswana people exhibit noticeable heterogeneity in natural disease progression (varying ability to maintain high CD4 counts and low viral load) in part due to the variable host genetic makeup (2, 9). Complex gene and gene–environment interactions affect HIV-1 infection and AIDS progression in major ways (6, 10). Much remains to be discovered about the genetic basis of individual host genetic differences in progression to AIDS in this geographic region.
Identification of genetic factors is important for several reasons. For example, the mechanism of HIV/AIDS restriction by genetic variants has previously encouraged the development of salvage pathway antiretroviral medications (e.g., enfuvirtide, maraviroc, which block HIV-1 binding to CCR-5 receptors and membrane fusion), while predicting progression to AIDS for a given genotype can be used to inform clinical trials for new drugs (4, 11–14). In this study, we searched for genetic factors associated with HIV-1C infection/acquisition and disease progression in Botswana.
Genome-wide association studies (GWAS) are a standard approach to scan for novel genetic variants that are associated with resistance or susceptibility to a particular disease (15–18). Here, we employed a GWAS approach to discover gene variants that regulate HIV-1C acquisition and are also related to HIV-1 pathogenesis. In this GWAS, we identified several novel genetic variants that demonstrated statistically significant association with HIV-1C infection in a Botswana cohort using single nucleotide polymorphisms (SNPs) determined by Illumina microarray (MA; n = 809) and by whole-genome sequencing (WGS; n = 364). We report gene associations for three HIV-1C associated loci: AP3B1, PTPRA, and NEO1. The discoveries were verified by independent cohort replication and by looking for previously reported implications in HIV-1 host-pathogenesis. We further replicated prior AIDS restriction gene (ARG) association signals from analyses with Botswana HIV-1C cohorts, adding confidence to the existing knowledge about gene influences in HIV/AIDS.
Results
We developed a cohort of 838 young African women living in Botswana who were exposed to or infected with HIV-1C. Study participants, each enrolled in one or more Botswana-AIDS Partnership studies (Table 1), were examined in a GWAS to search for genetic variants that influenced HIV-1C acquisition. The cohort was parsed into a case-controlled design with 447 HIV infected individuals and 315 HIV exposed but uninfected controls (Table 2). We genotyped these samples using two methods. First, DNA from 476 individuals were genotyped with the Illumina Human Omni 2.5 M BeadChip array (MA cohort), which resulted in genotypes for 1,374,256 SNPs after filtering, as described in Table 1. Second, the complete genomes of 364 individuals (nonoverlapping with the MA study participants) were sequenced (WGS cohort). SNPs annotated from the WGS samples were genotyped as described in Methods for a total of 8,636,400 SNPs that passed filters for discordant SNPs, missing genotypes, failure to conform to Hardy–Weinberg equilibrium (HWE), and minor allele frequency (MAF) < 0.05 (Table 1).
Table 1.
Botswana study participants and SNP filtering
| Cohort | Step | Dropped | Included |
| Study participant filtering steps | |||
| WGS | Total subjects | 364 | |
| Discordant genotypes (WGS/MA) | 3 | 361 | |
| No phenotype | 1 | 360 | |
| low genotyping rate | 2 | 358 | |
| IBD | 2 | 356 | |
| PCA outliers | 0 | 356 | |
| Males | 20 | 336 | |
| Total used | 336 | ||
| MA | Total subjects | 809 | |
| MA/WGS duplicates | 333 | 476 | |
| PCA outliers | 6 | 470 | |
| Males | 40 | 430 | |
| Total used | 430 | ||
| MA-WGS (combined- shared SNPs) | Total subjects | 1,173 | |
| MA/WGS duplicates | 335 | 838 | |
| Discordant genotypes (WGS/MA) | 1 | 837 | |
| No phenotype | 1 | 836 | |
| low genotyping rate | 1 | 835 | |
| IBD | 2 | 833 | |
| PCA outliers | 10 | 823 | |
| Males | 61 | 762 | |
| Total used | 762 | ||
| Annotated SNPs and filtering steps | |||
| WGS | Loaded from .bim file | 19,715,426 | |
| Discordant SNPs (>5%) | 66,218 | 19,649,208 | |
| Missing genotype data (0.05) | 869,548 | 18,779,660 | |
| HWE | 164,425 | 18,615,235 | |
| Minor allele threshold (0.05) | 9,978,835 | 8,636,400 | |
| Total used | 8,636,400 | ||
| MA | Loaded from .bim file | 1,822,601 | |
| Discordant SNPs (>5%) | 66,218 | 1,756,383 | |
| Missing genotype data (0.05) | 8,250 | 1,748,133 | |
| HWE | 134 | 1,747,999 | |
| Minor allele threshold (0.05) | 373,743 | 1,374,256 | |
| Total used | 1,374,256 | ||
| MA-WGS (combined-shared SNPs) | Loaded from .bim file | 1,705,582 | |
| Missing genotype data (0.05) | 5,981 | 1,699,601 | |
| HWE | 2,066 | 1,697,535 | |
| Minor allele threshold (0.05) | 363,591 | 1,333,944 | |
| Total used | 1,333,944 |
In the top half are steps for filtering of study participants as described in Methods. Each row includes the number of individuals dropped (filtered) and retained in each step. Total indicates the final count of individuals with genotypes actually used in the analyses of MA, WGS, and combined MA-WGS cohorts from Botswana. In the bottom half are the same for filtering steps for SNPs. IBD, identity by descent test for close relative; PCS, principal component analysis; HWE, Hardy–Weinberg departue at P > 0.0001.
Table 2.
Cohorts interrogated in this study
| Application | Cohort | Genotyping platform | n individuals | HIV+ | HIV− | Phenotype | SNPs after filtering (see Table 1) | Source (if published) |
| Discovery | Botswana | WGS | 336 | 256 | 80 | HIV acquisition | 8,636,400 | Present study |
| Discovery | Botswana | MA | 430 | 194 | 236 | HIV acquisition | 1,374,256 | Present study |
| Combined cohort | Botswana | MA+WGS | 762 | 447 | 315 | HIV acquisition | 1,333,944 | Present study |
| Replication | United States-EA | MA | 1,527 | HIV acquisition; AIDS progression; sequelae; ART | 700,022 | (8, 18) | ||
| Replication | United States-AA | MA | 1,460 | HIV acquisition; AIDS progression; sequelae; ART | 700,022 | (8, 18) |
AA, African Americans; EA, European Americans.
Genetic-association analysis was carried out with filtered study participants of three datasets, allowing us to perform both a discovery and a replication study. Three discovery datasets (Table 2) were considered: 1) the MA group of 430 females (194 HIV+ and 236 HIV−, but HIV-exposed); 2) the WGS group of 336 females (256 HIV+ and 80 of HIV−, but HIV-exposed); 3) shared SNPs from the MA and WGS study participants were then combined and analyzed for 1,333,944 shared SNP variants determined for a MA-WGS study group of 762 females (447 HIV+ and 315 HIV−, but HIV-exposed).
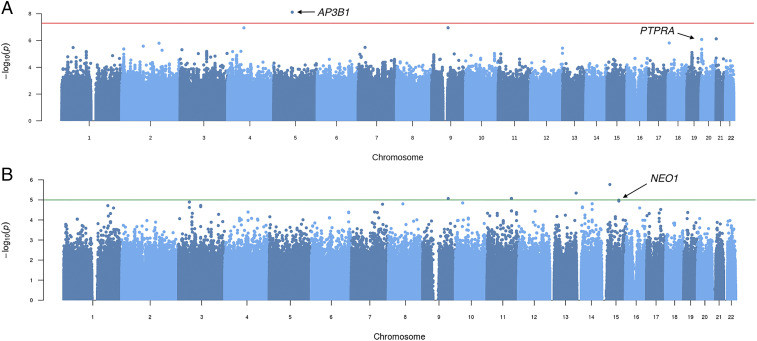
We performed a GWAS using logistic regression in PLINK on the MA, WGS, and combined MA-WGS groups, all exposed to HIV-1C, looking for genetic variants that influence HIV-1C acquisition. Manhattan plots for each group were obtained (Fig. 1 and Table 3). WGS Manhattan plots for HIV infection, using tests for allele and genotype frequency distribution in HIV+ vs. HIV− people, revealed significant associations of multiple SNPs with HIV acquisition (Fig. 1, Table 3, SI Appendix, Table S1, and Dataset S1), including a single SNP, rs572880838 on chromosome 5 (position 77,590,422; GRCh37) within the AP3B1 (Adaptor Related Protein Complex 3 Subunit Beta 1) gene, that reached the standard GWAS threshold for genome-wide significance after Bonferroni correction (Methods) for both the allele frequency and dominant genotype models (P = 7.6 × 10−9; odds ratio [OR] = 6.711) (Figs. 1A and 2A and Table 3). The AP3B1 gene product, AP-3, is well known to influence HIV-1 gag assembly and AIDS pathogenesis (Discussion)
Fig. 1.
Manhattan plots for associations with HIV infection using tests for allele and genotype frequency distribution in HIV+ vs. HIV− people. (A) WGS dataset of 336 people under the allelic model revealed departure of 8,636,400 SNPs from expectation, including associations within the AP3B1 (rs572880838; position 5:77,590,422; P = 7.6 × 10−9; OR = 6.711) and PTPRA (rs6076463; position 20:3012815; P = 8.33E-07; OR = 0.172) genes. (B) MA dataset: 430 people (1,374,256 SNP) under the allelic model revealed 1,374,256 SNPs. NEO1 (rs9920504; position 15:73284181; P = 1.03 × 10−5; OR = 0.465) (Table 3). Manhattan plots for the combined MA-WGS cohort (n = 762) did not reveal any SNPs that exceeded genome-wide significance (P < 3.75 × 10−8).
Table 3.
Gene variants associated with HIV-1C infection
| Cohort | CHR | SNP | Coordinates | Allele (minor) | Test | NMISS | OR | STAT | P value | Gene | Distance | RR | AR | EF |
| WGS | 5 | rs572880838 | 77590422 | G | ALLELIC | 333 | 6.711 | 5.78 | 7.62E-09 | AP3B1 | 0 | 1.210 | 0.074 | 0.029 |
| 5 | rs572880838 | 77590422 | G | DOM | 333 | 6.711 | 5.78 | 7.62E-09 | AP3B1 | 0 | NaN | NaN | NaN | |
| 20 | rs6076463 | 3012815 | T | ALLELIC | 336 | 0.172 | −4.93 | 8.33E-07 | PTPRA | 0 | 0.564 | −0.029 | 0.030 | |
| 20 | rs6076463 | 3012815 | T | DOM | 336 | 0.172 | −4.93 | 8.33E-07 | PTPRA | 0 | NaN | NaN | NaN | |
| 20 | rs3787480 | 3016895 | A | ALLELIC | 336 | 0.179 | −4.59 | 4.46E-06 | PTPRA | 0 | 0.573 | −0.025 | 0.025 | |
| 20 | rs3787480 | 3016895 | A | DOM | 336 | 0.179 | −4.59 | 4.46E-06 | PTPRA | 0 | NaN | NaN | NaN | |
| MA | 15 | rs9920504 | 73284181 | C | ALLELIC | 422 | 0.465 | −4.41 | 1.03E-05 | NEO1 | 59870 | 0.648 | −0.102 | 0.018 |
| 15 | rs7172316 | 73287152 | G | ALLELIC | 430 | 0.470 | −4.39 | 1.14E-05 | NEO1 | 56899 | 0.654 | −0.100 | 0.017 | |
| 15 | rs9920504 | 73284181 | C | DOM | 422 | 0.437 | −4.06 | 4.94E-05 | NEO1 | 59870 | 0.398 | −0.040 | 0.015 | |
| 15 | rs7172316 | 73287152 | G | DOM | 430 | 0.445 | −4.01 | 6.05E-05 | NEO1 | 56899 | 0.395 | −0.039 | 0.015 |
AR, attributable risk; EF, explained fraction; Distance, distance to the closest gene (see column Gene); NaN, not a valid number; NMISS, number of nonmissing individuals included; RR, relative risk; STAT, coefficient t-statistic.
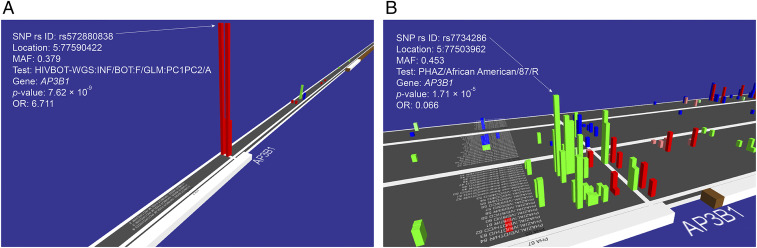
Fig. 2.
GWATCH snapshots of associations in the AP3B1 gene region for HIV infection and AIDS progression. GWATCH is a web-based dynamic real-time genome browser designed to discover, view, and assess hits from GWAS and WGS association studies (https://botswana.gen-watch.org/) (9, 18). The platform displays a three-dimensional viewer above and moving along human chromosomes with baseline x axis listing all association tests performed in the GWAS. Adjacent SNPs genotyped along each chromosome are the indices of the y axis. The rising blocks illustrate the resulting log of P value obtained (blocks are shown only for P < 10−3 to avoid clutter of noise) for each SNP–test combination, with color showing direction of association (red for susceptible vs. green for protective) and color intensity indicating the hazard ratio/OR (unified under the term QAS for Quantitative Association Statistic) (18) of each association. The combination of nonindependent proxy SNPs (which track the operative SNP by LD) and the nonindependent SNP association tests produce clusters of P value peaks in associated regions. Nonassociated regions are generally flat with random false signals appearing infrequently. (A) HIV infection tests in the Botswana WGS discovery dataset (n = 336 study participants [256 HIV+ and 80 HIV−] genotyped with 8,636,400 SNPs). (B) AIDS progression tests in the replication AA cohort (n = 1,460 study participants genotyped with 700,022 SNPs) (Table 2). The American replication cohorts consist of multiple tests with different numbers of individuals in each. The individuals’ numbers for each test are listed in GWATCH (https://gen-watch.org/#/option0) under the “List of tests & Manhattan plots” tab for each cohort. The elevated P values reflect a series of different case-control hypothesis tests for AIDS progression among HIV infected AA study participants (https://botswana.gen-watch.org/) (9, 18).
Replication and validation of associated variants were performed for the exact implicated SNP association in the nondiscovery cohorts as well as in previously interrogated American AIDS cohorts that included actively gay male participants, recipients of HIV-1 contaminated clotting factors (hemophiliacs), contaminated donor blood transfusions, and persons who inject drugs and who exchanged HIV-contaminated syringes in urban settings (Table 2). In cases where a discovered associated SNP was not present in the replication cohort, we tested proxy SNPs with r2 > 0.8 when feasible. The AP3B1 SNP-rs572880838 had but one proxy to in the WGS SNP in the Botswana WGS dataset (rs777424597; r2 > 0.8) (SI Appendix, Fig. S1); however, neither AP3B1-SNP was included among the genotyped MA SNPs, making exact SNP replication (or nonreplication) impossible.
A less stringent but informative “gene region” replication approach involved the loading of the GWAS results (WGS and MA) for discovery and replication cohorts into the GWATCH program (9, 18). GWATCH includes a suite of GWAS analytical programs that allow discovered SNP association signals to be quantified, viewed, and compared for gene region associations with multiple AIDS association test hypotheses and statistical models (i.e., HIV infection, Cox regression for survival analysis, case-control categorical analyses of AIDS progression, various AIDS defining pathologies, and progression to AIDS after ART treatment) (Methods). We used GWATCH to explore and replicate AIDS associations in AIDS progression tests in the replication African American (AA) cohort-ALIVE (n = 1,460 study participants genotyped with 700,022 SNPs) (Fig. 2B and Table 2) (https://botswana.gen-watch.org/). In addition, the AP3B1 gene showed a series of SNPs that were highly associated with AIDS progression (P = 1.7 × 10−5 for Cox regression for survival analysis), including various AIDS defining end point analyses of the United States-European American (EA) cohort (n = 1,527 study participants genotyped with 700,022 SNPs; P = 2.1 × 10−3) (Table 2 and SI Appendix, Fig. S2) (https://botswana.gen-watch.org/).
The associated SNP (rs572880838) is located in the promoter (regulatory 5′UTR region) of the AP3B1 gene, which comprises 27 exons encoding a 140-kD, 194-amino acid protein AP3B1. This promoter region is a binding site for multiple different transcription factors (19); rs572880838 could potentially influence the regulation of AP3B1 gene transcription or affect the secondary structure of the transcript itself (SI Appendix, Fig. S3). The rs572880838 has a noticeably high MAF (MAF = 0.379) in the Botswana population (based on our dataset of 336 WGS), where HIV-1C infection is prevalent. This Botswana MAF is higher than in any of the human populations studied to date. In the 1000 Genomes Project the highest detected population MAF of an AP3B1 SNP (rs572880838) is 0.2654 (Gambian) (20). Two (rs572880838) SNP alleles are distributed in TT and TG genotypes in the Botswana population, while the expected homozygous GG genotype is remarkably absent. The same AP3B1 SNP (rs572880838) GG genotype is also completely absent among all (African, European, and Asian) populations sequenced in the 1000 Genomes Project (n = 2,504 people), potentially indicating an adverse (perhaps lethal) phenotype for the AP3B1 SNP (rs572880838) GG genotypes worldwide. Several mutations, including a 63-bp deletion of AP3B1 Exon 15, can cause Hermansky–Pudlak syndrome, a rare inherited disorder characterized by decreased pigmentation (albinism), visual impairment, and blood platelet dysfunction that is sometimes fatal (21–24). To date there are no clinical pathologies reported in association with the SNP rs572880838 in the AP3B1 gene, here associated with HIV-1C acquisition in Botswana.
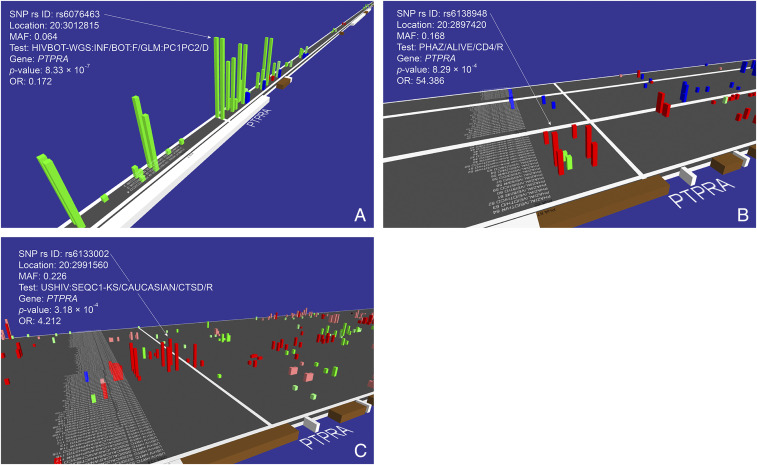
A second gene-association signal that approached (but did not exceed) genome-wide significance for the WGS dataset involves SNPs within the PTPRA (protein tyrosine phosphatase receptor type A) locus on chromosome 20 (rs6076463; P = 8.3 × 10−7; OR = 0.172 for allele and dominant genotype models) (Figs. 1A and 3A, Table 3, SI Appendix, Table S1, and Dataset S1). This SNP has multiple proxy SNPs that are all associated with HIV acquisition (Fig. 3A). The strongest PTPRA SNP (rs6076463; P = 8.3 × 10−7; OR = 0.172) was not genotyped in the MA cohort nor was any informative linkage disequilibrium (LD) proxy SNP included in the MA genotyped SNPs. The PTPRA gene region showed multiple SNP associations with progression to AIDS endpoints revealed in the GWATCH snapshot of AAs (Fig. 3B) (P = 8.3 × 10−4; https://botswana.gen-watch.org/). PTPRA gene variants were also significantly associated with the development of certain AIDS defining conditions (sequelae; namely Kaposi’s sarcoma, PJP [Pneumocytis jiroveci pneumonia], B cell lymphoma) in the EA cohorts screened by GWATCH (Fig. 3C) (P = 3.2 × 10−4). The PTPRA gene has also been reported as having a role in HIV/AIDS pathogenesis (25–27) (Discussion)
Fig. 3.
GWATCH snapshots of associations in the PTPRA gene region. (A) HIV infection tests in the Botswana WGS discovery cohort (n = 336 study participants [256 HIV+ and 80 HIV−] genotyped with 8,636,400 SNPs). (B) AIDS progression tests in the replication AA cohort (also called ALIVE cohort, n = 1,460 study participants genotyped with 700,022 SNPs). (C) HIV/AIDS progression (sequelae) tests in the Botswana replication EA Mod A cohort (n = 1,527 study participants genotyped with 700,022 SNPs) (https://botswana.gen-watch.org/) (9, 18).
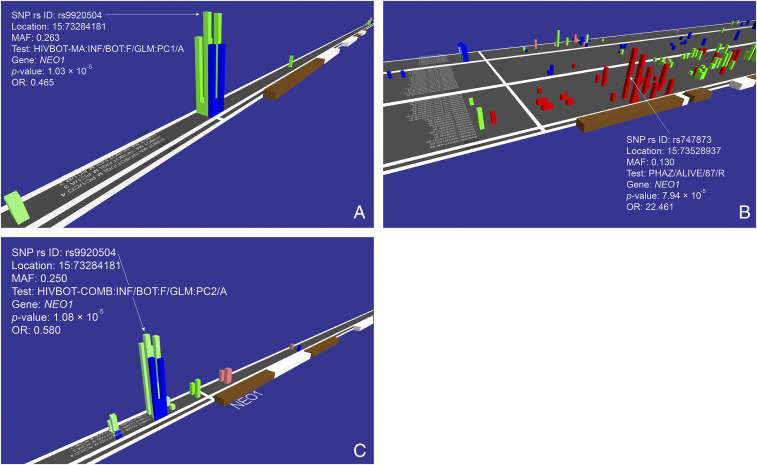
A third genetic association, initially detected as genome-wide significant in the GWAS on MA (Fig. 1B), involves multiple SNPs within the promoter region of the NEO1 gene associated with HIV infection (chromosome band 15q24.1; rs9920504; P = 1.03 × 10−5; OR = 0.465) (Fig. 4A, Table 3, SI Appendix, Table S1, and Dataset S1). A GWATCH screen shot showed multiple SNPs within the NEO1 gene in the AA cohort that were associated with AIDS progression using Cox regression for survival analysis (P = 7.94 × 10−5) (Fig. 4B). Furthermore, NEO1 gene variants were also associated with the development of specific AIDS-defining conditions (sequelae: Kaposi’s sarcoma, PCP, B cell lymphoma) in the EA cohorts screened by GWATCH (SI Appendix, Fig. S4) (P = 9.59 × 10−5). Finally, the identical NEO1 SNP of the discovery cohort analysis (rs9920504) (Fig. 4A and Table 3) was also significantly associated in an HIV infection GWAS using the MA-WGS Botswana combined cohort (P = 1.09 × 10−5) (Fig. 4C). The NEO1 gene encodes a cell surface protein member of the immunoglobulin superfamily that functions in multiple tissues and in chronic inflammation of HIV infected patients (28–30) (Discussion). The combined MA-WGS cohort analyses for allele and genotyping effect did not yield any SNPs that exceeded genome-wide significance in allele or genotype association tests (https://botswana.gen-watch.org/).
Fig. 4.
GWATCH snapshots of associations in the NEO1 gene region. (A) HIV infection tests in the discovery MA cohort (n = 430 study participants genotyped [194 HIV+ and 236 HIV−] with 1,374,256 SNPs). (B) AIDS progression tests in the replication United States-AA cohort (n = 1,460 study participants genotyped with 700,022 SNPs). (C) HIV infection tests in the replication combined MA-WGS cohort (n = 762 study participants genotyped with 1,333,944 SNPs) (https://botswana.gen-watch.org/) (9, 18).
We further searched across the three Botswana discovery cohorts (Table 2) for possible SNP and gene replications in HIV-1 acquisition of previously reported ARGs. We found 13 genes of the 41 tested that showed P < 0.005 for one or more stages of HIV infection and AIDS pathogenesis (Table 4 and SI Appendix, Table S2) (4, 7, 8). These include several loci within the HLA gene complex, as seen previously in EA-based GWAS but not reported to date in African cohorts. The CCR2-64I allele was associated with HIV infection in a Kenya cohort of sex workers (41), while PROX1 has not been replicated to our knowledge prior to this study (32). The failure to replicate some of the additional associated ARGs might be explained in some cases by differences in ethnic populations used for GWAS and in HIV subtypes. For example, CCR5-∆32 is not present among African ethnic groups. Despite these differences, we were able to replicate the loci in such genes as NCOR2, TRIM5, CXCL12, and several HLA genes (Table 4). The present replication increases our confidence in the validity of these prior genetic associations.
Table 4.
Replications of previously discovered AIDS restriction genes in the cohorts of this study
| Dataset | Gene | Chr | SNP | Position | test | P value | Odds ratio | Citation (original source) |
| WGS | NCOR2 | 12 | rs701025 | 125042336 | DOM | 8.0E-05 | 3.48 | (31) |
| PROX1 | 1 | rs366684 | 214187262 | DOM | 5.1E-04 | 0.33 | (32) | |
| PROX1-AS1 | 1 | rs853757 | 214051639 | REC | 5.6E-04 | 0.24 | (32) | |
| TRIM5 | 11 | rs71490145 | 5691528 | DOM | 5.9E-04 | 0.38 | (33) | |
| CXCL12 (SDF1) | 10 | rs1898318 | 44825430 | DOM | 6.6E-04 | 0.33 | (34) | |
| HCP5 | 6 | rs7746061 | 31419216 | ALLELIC | 1.2E-03 | 0.31 | (35) | |
| MYH9 | 22 | rs28478212 | 36734593 | ALLELIC | 2.1E-03 | 0.38 | (36) | |
| HLA-DRB1 | 6 | rs9270209 | 32556443 | ALLELIC | 2.5E-03 | 2.19 | (37) | |
| TSG101 | 11 | rs867590536 | 18508258 | ALLELIC | 2.8E-03 | 0.39 | (38) | |
| HLA-DMB | 6 | 6:32908833 | 32908833 | ALLELIC | 3.4E-03 | 0.36 | (39) | |
| HLA-C | 6 | rs1049709 | 31236854 | DOM | 5.5E-03 | 0.37 | (35) | |
| HLA-A | 6 | rs1059514 | 29911190 | REC | 5.8E-03 | 0.34 | (40) | |
| CCR2 * | 3 | rs62242985 | 46385638 | ALLELIC | 8.3E-03 | 1.79 | (41) | |
| MA | TRIM5 | 11 | rs73404271 | 5739483 | ALLELIC | 4.2E-04 | 2.17 | (33) |
| CXCL12 (SDF1) | 10 | rs800323 | 44801673 | REC | 1.8E-03 | 0.39 | (34) | |
| NCOR2 | 12 | rs12814949 | 124976612 | DOM | 5.3E-03 | 0.47 | (31) | |
| HLA-DQB1-AS1 | 6 | rs6689 | 32627700 | ALLELIC | 8.7E-03 | 0.54 | (37) |
See SI Appendix, Table S2 for more information on the original associations.
In LD; 9,587 bp.
Discussion
In this study we performed a GWAS analysis using a combination of WGS and MA data obtained from 766 individuals (after filtering) (Table 1) from Botswana, either infected or exposed to HIV-1C, in order to identify genetic factors associated with HIV-1C acquisition in a Botswana population. The results presented reveal candidate gene associations with HIV-1C acquisition and were replicated in independent AIDS cohorts (Table 2). This is one of the few studies of the impact of human genetic variation on HIV-1C infection and pathogenesis in Botswana or in southern Africa, a locale with the world’s majority of HIV-1 infections. We report here several genes with significant association with HIV-1C acquisition: AP3B1, PTPRA, and NEO1 (Table 3). Evidence of replication in independent datasets (listed in Table 2) including AA AIDS cohorts is presented for each associated locus (Results). We researched the recently discovered loci associated with HIV-1C infection and found that each gene plays an important functional role in HIV/AIDS disease processes.
AP3B1 encodes a clathrin-associated protein complex required for HIV-1 assembly and release (42–44). The matrix region of HIV-1 Gag interacts directly with the δ-subunit of the AP-3 complex (coded by the AP3B1 gene), and this interaction plays an important functional role in viral particle assembly. Disruption of this interaction eliminates Gag trafficking to multivesicular bodies and diminishes HIV particle formation (43). In cultures of AP-3–deficient fibroblasts, induced by mutations in the AP3B1 gene, HIV-1 particle assembly and release are diminished, while both transient and stable expression of the full-length wild-type β3A subunit in these cells restores the impaired virus assembly and release (44). Intact and stable AP-3 complex is required for HIV-1 assembly and release, and the involvement of the AP-3 complex in late stages of the HIV-1 replication cycle is independent of clathrin-mediated endocytosis (44). The β-subunit of AP-3 is a target of IP7-mediated pyrophosphorylation, which modulates its interaction with Kif3A (a motor protein of the kinesin superfamily, which is also involved in an intracellular process required for HIV-1 Gag release) and, as a consequence, affects the release of HIV-1 virus-like particles (42).
PTPRA encodes a membrane receptor, member of the protein tyrosine phosphatase (PTP) family, which regulates a variety of cellular processes, including cell growth, differentiation, mitotic cycle, and oncogenic transformation. The PTPRA gene was shown to be up-regulated in CD4+ cells from HIV-1 patients (25, 26). It has also been detected among exosomal proteins found in urinary extracellular vesicles from HIV-1+ patients (27).
The human Neogenin 1 (NEO1) gene encodes a multifunctional cell surface protein that is a member of the immunoglobulin superfamily, implicated in tissue morphogenesis, angiogenesis, myoblast differentiation, and axon guidance (28). Plasma membrane expression of NEO1 is down-regulated in CEMT4 T cells infected with VSV-G pseudotyped HIV-1 (29). Neogenin also demonstrates a greater than twofold increased relative abundance in HIV-1+ blood plasma and some studies suggest that it may play an important role in chronic inflammation induced by HIV infection. It was proposed that functional regulation of neogenin plasma level may have a therapeutic value in HIV+ patients (30).
It seems notable that all three important associations discovered in the African Botswana population showed gene replications in the United States-AA AIDS cohort (Figs. 2B, 3B, and 4B and Table 2). By contrast, over 30 ARGs were discovered in the United States Caucasian cohort (4, 7, 8) with very few previously described ARGs found in the same United States-AA cohort (Table 2). This difference may actually be attributable to ethnic genetic background. Furthermore, we were able to replicate the reported associations for 13 previously found ARGs, such as NCOR2, TRIM5, CXCL12, and several HLA genes (Table 4 and SI Appendix, Table S2). These replications provide additional evidence in support of the previously reported associations and strengthen our confidence in the newly detected ones.
Our study was limited by rather small sample sizes and a gender emphasis on females, though previous ARG studies in AIDS cohorts from Western countries were biased toward male participants. More expansive studies in the future are certainly needed in this population, as well as in others, particularly in other African regions, ethnic groups, in men and in children in order to assess the findings more thoroughly. Multiple kinds of genome-level data (e.g., genomic DNA, epigenetic data, transcriptome data, siRNA screens) used in conjunction with resequencing strategies identifying rare causal variants are necessary to improve our comprehension of the role of host genome variability on HIV-1 acquisition, progression, and transmission at regional and global scales (6, 17). Finally, nongenetic factors are unaccounted for in present-day GWAS studies, such as host microbiome, epigenome, environmental, and social factors, which surely contribute to HIV-1 transmission and progression. These components should be assessed by the future study designs in this important search for the host genetic determinants of HIV/AIDS transmission and pathogenesis (6).
Methods
Populations Studied.
Participants from two earlier Botswana Harvard AIDS Institute Partnership studies, Mashi and Tshedimoso, were enrolled in GWAS study. The Mashi study (clinicaltrials.gov identifier NCT00197587; NIH R01 HD37793) was a mother-to-child prevention 2 × 2 factorial randomized clinical trial with peripartum (single-dose nevirapine vs. placebo) and postpartum infant feeding formula vs. breastfeeding with infant zidovudine prophylaxis) interventions presented elsewhere (45–49). A subset of 436 individuals who participated in Mashi study were enrolled in this study. The Tshedimoso study (NIH AI057027 Markers of Viral Set Point in Primary HIV-1C Infection) was presented elsewhere (50–70).
A subset of 373 individuals who were screened for the Tshedimoso study were enrolled in the GWAS study. The samples all came from the two cohorts that were collected/enrolled for the original clinical studies of largely Tswana ethnicity study participants.
The largest cohort was recruited only for HIV+ pregnant women to test interventions to prevent mother-to-child transmission. Thus, no males were ever involved in that cohort. As this study was performed in Botswana, a country with one of the highest HIV-1 prevalence (20.3%), a high proportion of citizens could be considered as likely exposed to HIV-1 infection. Individual exposure was not quantified. The data were obtained in collaboration with Harvard T. H. Chan School of Public Health AIDS Initiative (Boston, MA) and the Botswana/Harvard AIDS Institute (Gaborone, Botswana).
This study was conducted by the Botswana Harvard AIDS Institute Partnership and approved according to The Declaration of Helsinki. All participants consented to participate in the study. Institutional Review Board (IRB) approval was from Botswana Ministry of Health and Wellness–Health Research Development Committee and Harvard School of Public Health IRB (reference no.: HPDME 13/18/1).
Whole-Genome Sequencing.
WGS was performed for 364 samples at 30× coverage (Table 1). Quality control of raw WGS reads was performed using FastQC v0.11.5 (71) and 23-mer counts distribution with KraTER (72). Trimmomatic (73) with default parameters was used to remove adapters and trim low-quality read ends. Trimmed reads were mapped to the human reference genome GRCh38 (top-level main chromosomes) using BWA-MEM v0.7.15 (74) with default parameters and the read alignments were sorted with SAMtools sort v0.1.19 (75, 76). These alignments were then used for genotyping by SAMtools and BCFtools toolchain (SAMtools mpileup with -Q 30 -q 30 -m, and BCFtools call) (76). After checking the F1-score between genotypes in WGS and microarray data with the DP (the variant read depth) range from 1 to 10 and for QUAL (the Phred-scaled quality score of alternative alleles of a variable site) from the set 15, 20, 25, 30, 35, and 40 variants with DP < 5 and QUAL < 20, and singletons with single called genotype were filtered out. The resulting variants were annotated using Ensembl (19). VEP WGS genotypes were lifted down to genome build hg19 using UCSC liftOver tool to make them compatible with MA data.
WGS Sample Filtering.
All genotype quality control was performed using PLINK 1.9 (77) and custom scripts. First, for the samples with both Illumina MA genotypes and WGS genotypes, we calculated the fraction of concordant genotypes between the two methods. WGS samples with genotype concordance below 90% were removed. All males were removed, as there were no HIV-infected samples among them. In addition, we removed principal component analysis (PCA) outliers based on visual inspection of the plot of PC1 and PC2, samples with missing genotype rate >5% and cryptic relatives (PI_HAT > 0.25) (Table 1 and SI Appendix, Fig. S5A). These steps resulted in 336 study participants independent of the microarray individuals (Tables 1 and 2).
MA Sample Filtering.
For MA sample filtering, 809 individual samples were genotyped using the Illumina Infinium Omni2.5-8 BeadChip genotyping MA [333 samples which overlap with WGS were excluded from the MA dataset (Table 1)]. All males were removed, as were PCA outliers, samples with missing genotype rate > 1%, and cryptic relatives (PI_HAT > 0.01) based on PLINK 1.9 variant statistics (Table 1). Sample filtering resulted in 430 individuals genotyped by microarray that were independent of the WGS cohort (Tables 1 and 2 and SI Appendix, Fig. S5B). Recently, additional MA chips have been developed for specifically evaluating African populations, namely the Infinium H3Africa Consortium Array (v2) and African Diaspora Power Chip, but these were unavailable at the time of our genotyping.
Association Tests.
Prior to testing genetic variants for association with HIV, we performed SNP filtering and excluded SNPs with call rate < 95%, MAF < 5% or HWE P value < 1e-4 (Table 1). The resulting genotype data were used for the association tests. Association tests were performed using PLINK v1.9 logistic regression: we applied additive allelic (–logistic) models for alleles, dominant (–logistic dominant), recessive (–logistic recessive), and codominant (–logistic genotypic) models for genotypes. PCs 1 and 2 were used as covariates (–covar) in the WGS dataset, and PC1 was used as a covariate in the MA dataset to account for potential batch effects based on visual inspection of PCA plots. To avoid inadequate P values in the codominant model, we calculated the P value only for the variants having enough observations (≥3) for each sample group, thus excluding those variants for which it is not possible to test the codominant model.
The GWAS P value threshold for genome-wide significance for WGS and MA-based GWAS follows the corrected P value threshold convention (P < 5 × 10−8 and P < 10−5, respectively) (78). These thresholds are corrected from the standard Bonferroni application that assumes every genetic variant tested is independent of the others (i.e., ignoring LD) and considering false-discovery rate procedures, permutation-based approaches, and Bayesian approaches (78). Relative risk, attributable risk, and explained fraction were calculated as defined in refs. 8 and 79.
The numbers of HIV+ and HIV− study participants used for each test for Botswana analyses are presented in Table 2 and also in Figs. 2–4. The American replication cohorts consist of multiple tests with different numbers of individuals in each. The individuals’ numbers for each test are listed in GWATCH (https://gen-watch.org/#/option0) under the “List of tests & Manhattan plots” tab for each cohort.
Visualization of the Results Using GWATCH.
Genotyping and clinical data were loaded into the GWATCH analytical suite (9, 18) for further analysis and visualization. GWATCH is a web-based dynamic real-time genome browser designed to discover, view, and assess genetic association hits and genomic environment using multiple association test designs from GWASs (botswana.gen-watch.org/). GWATCH displays a moving three-dimensional viewer above along human chromosomes with baseline x axis listing all association tests performed in the GWAS (https://www.youtube.com/watch?v=vFHRCb4bUGs). Adjacent SNPs genotyped along each chromosome are the indices of the y axis. The rising blocks (Figs. 2–4) illustrate the resulting negative logarithm with base 10 of P values obtained for each SNP–test combination, with color showing the direction of association (green indicates allele or genotype resistance to infection or disease OR < 1.0; red indicates susceptibility OR > 1.0). Color intensity of the value bars indicate the hazard ratio/OR of each association listed in Table 3. The P values for the gene-associated SNPS in replication cohorts were corrected by the number of SNPS in each gene interrogated in SI Appendix, Table S3.
Acknowledgments
We thank Lada Antonova (ITMO University) for technical support in manuscript preparation. This work was supported, in part, by Russian Ministry of Science Mega-grant 11.G34.31.0068 and St. Petersburg State University (Genome Russia Grant 1.52.1647.2016 and St. Petersburg State University project no. 51148284). P.K.T. is funded by the Sub-Saharan African Network for TB/HIV Research Excellence (SANTHE), a DELTAS Africa Initiative (Grant DEL-15-006), through the Wellcome Trust (Grant 107752/Z/15/Z).
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2107830118/-/DCSupplemental.
Data Availability
SNP genotype data have been deposited in https://botswana.gen-watch.org/ (Botswana GWATCH database database). All other study data are included in the article and supporting information. Previously published data were used for this work (18).
References
- 1.UN Joint Programme on HIV/AIDS (UNAIDS), UNAIDS Data 2019 (2020). Available at https://www.unaids.org/sites/default/files/media_asset/2019-UNAIDS-data_en.pdf. Accessed 6 October 2021.
- 2.Essex M., Mboup S., Kanki P. J., Marlink R. G., Tlou S. D., AIDS in Africa (Kluwer Academic/Plenum Publishers, New York, ed. 2, 2002). [Google Scholar]
- 3.Fellay J., Shianna K. V., Telenti A., Goldstein D. B., Host genetics and HIV-1: The final phase? PLoS Pathog. 6, e1001033 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Brien S. J., Hendrickson S. L., Host genomic influences on HIV/AIDS. Genome Biol. 14, 201 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McLaren P. J., Carrington M., The impact of host genetic variation on infection with HIV-1. Nat. Immunol. 16, 577–583 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thami P. K., Chimusa E. R., Population structure and implications on the genetic architecture of HIV-1 phenotypes within Southern Africa. Front. Genet. 10, 905 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.An P., Winkler C. A., Host genes associated with HIV/AIDS: Advances in gene discovery. Trends Genet. 26, 119–131 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Brien S. J., Nelson G. W., Human genes that limit AIDS. Nat. Genet. 36, 565–574 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Xie W., et al., Genome-wide analyses reveal gene influence on HIV disease progression and HIV-1C acquisition in Southern Africa. AIDS Res. Hum. Retroviruses 33, 597–609 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fellay J., Host genetics influences on HIV type-1 disease. Antivir. Ther. 14, 731–738 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hughes A., Barber T., Nelson M., New treatment options for HIV salvage patients: An overview of second generation PIs, NNRTIs, integrase inhibitors and CCR5 antagonists. J. Infect. 57, 1–10 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Manfredi R., Sabbatani S., A novel antiretroviral class (fusion inhibitors) in the management of HIV infection. Present features and future perspectives of enfuvirtide (T-20). Curr. Med. Chem. 13, 2369–2384 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Aquaro S., et al., Specific mutations in HIV-1 gp41 are associated with immunological success in HIV-1-infected patients receiving enfuvirtide treatment. J. Antimicrob. Chemother. 58, 714–722 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Dorr P., et al., Maraviroc (UK-427,857), a potent, orally bioavailable, and selective small-molecule inhibitor of chemokine receptor CCR5 with broad-spectrum anti-human immunodeficiency virus type 1 activity. Antimicrob. Agents Chemother. 49, 4721–4732 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirschhorn J. N., Daly M. J., Genome-wide association studies for common diseases and complex traits. Nat. Rev. Genet. 6, 95–108 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Hutcheson H. B., et al., Detecting AIDS restriction genes: From candidate genes to genome-wide association discovery. Vaccine 26, 2951–2965 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Telenti A., Goldstein D. B., Genomics meets HIV-1. Nat. Rev. Microbiol. 4, 865–873 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Svitin A., et al., GWATCH: A web platform for automated gene association discovery analysis. Gigascience 3, 18 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yates A. D., et al., Ensembl 2020. Nucleic Acids Res. 48, D682–D688 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.1000 Genomes Project Consortium; Auton A.et al., A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gahl W. A., et al., Genetic defects and clinical characteristics of patients with a form of oculocutaneous albinism (Hermansky-Pudlak syndrome). N. Engl. J. Med. 338, 1258–1264 (1998). [DOI] [PubMed] [Google Scholar]
- 22.Dell’Angelica E. C., Shotelersuk V., Aguilar R. C., Gahl W. A., Bonifacino J. S., Altered trafficking of lysosomal proteins in Hermansky-Pudlak syndrome due to mutations in the beta 3A subunit of the AP-3 adaptor. Mol. Cell 3, 11–21 (1999). [DOI] [PubMed] [Google Scholar]
- 23.Jung J., et al., Identification of a homozygous deletion in the AP3B1 gene causing Hermansky-Pudlak syndrome, type 2. Blood 108, 362–369 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones M. L., et al., Disruption of AP3B1 by a chromosome 5 inversion: A new disease mechanism in Hermansky-Pudlak syndrome type 2. BMC Med. Genet. 14, 42 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hyrcza M. D., “Gene expression changes in immune cells during human immunodeficiency virus 1 (HIV-1) infection,” PhD thesis, University of Toronto, ON, Canada (2009).
- 26.van ’t Wout A. B., et al., Cellular gene expression upon human immunodeficiency virus type 1 infection of CD4(+)-T-cell lines. J. Virol. 77, 1392–1402 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anyanwu S. I., et al., Detection of HIV-1 and human proteins in urinary extracellular vesicles from HIV+ patients. Adv. Virol. 2018, 7863412 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson N. H., Key B., Neogenin: One receptor, many functions. Int. J. Biochem. Cell Biol. 39, 874–878 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Matheson N. J., et al., Cell surface proteomic map of HIV infection reveals antagonism of amino acid metabolism by Vpu and Nef. Cell Host Microbe 18, 409–423 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang W., et al., Glycoproteomic analysis identifies human glycoproteins secreted from HIV latently infected T cells and reveals their presence in HIV+ plasma. Clin. Proteomics 11, 9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chinn L. W., et al., Genetic associations of variants in genes encoding HIV-dependency factors required for HIV-1 infection. J. Infect. Dis. 202, 1836–1845 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herbeck J. T., et al., Multistage genomewide association study identifies a locus at 1q41 associated with rate of HIV-1 disease progression to clinical AIDS. J. Infect. Dis. 201, 618–626 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Javanbakht H., et al., Effects of human TRIM5alpha polymorphisms on antiretroviral function and susceptibility to human immunodeficiency virus infection. Virology 354, 15–27 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Winkler C., et al., Genetic restriction of AIDS pathogenesis by an SDF-1 chemokine gene variant. ALIVE Study, Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort (SFCC). Science 279, 389–393 (1998). [DOI] [PubMed] [Google Scholar]
- 35.Fellay J., et al., A whole-genome association study of major determinants for host control of HIV-1. Science 317, 944–947 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kopp J. B., et al., MYH9 is a major-effect risk gene for focal segmental glomerulosclerosis. Nat. Genet. 40, 1175–1184 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang J., et al., Human leukocyte antigens and HIV type 1 viral load in early and chronic infection: Predominance of evolving relationships. PLoS One 5, e9629 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bleiber G.et al.; Swiss HIV Cohort Study , Use of a combined ex vivo/in vivo population approach for screening of human genes involved in the human immunodeficiency virus type 1 life cycle for variants influencing disease progression. J. Virol. 79, 12674–12680 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aissani B., et al., SNP screening of central MHC-identified HLA-DMB as a candidate susceptibility gene for HIV-related Kaposi’s sarcoma. Genes Immun. 15, 424–429 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramírez de Arellano E., et al., Novel association of five HLA alleles with HIV-1 progression in Spanish long-term non progressor patients. PLoS One 14, e0220459 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anzala O. A., et al., Rapid progression to disease in African sex workers with human immunodeficiency virus type 1 infection. J. Infect. Dis. 171, 686–689 (1995). [DOI] [PubMed] [Google Scholar]
- 42.Azevedo C., Burton A., Ruiz-Mateos E., Marsh M., Saiardi A., Inositol pyrophosphate mediated pyrophosphorylation of AP3B1 regulates HIV-1 Gag release. Proc. Natl. Acad. Sci. U.S.A. 106, 21161–21166 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dong X., et al., AP-3 directs the intracellular trafficking of HIV-1 Gag and plays a key role in particle assembly. Cell 120, 663–674 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Liu L., et al., Defective HIV-1 particle assembly in AP-3-deficient cells derived from patients with Hermansky-Pudlak syndrome type 2. J. Virol. 86, 11242–11253 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shapiro R. L., et al., Maternal single-dose nevirapine versus placebo as part of an antiretroviral strategy to prevent mother-to-child HIV transmission in Botswana. AIDS 20, 1281–1288 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Thior I.et al.; Mashi Study Team , Breastfeeding plus infant zidovudine prophylaxis for 6 months vs formula feeding plus infant zidovudine for 1 month to reduce mother-to-child HIV transmission in Botswana: A randomized trial: The Mashi Study. JAMA 296, 794–805 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Rossenkhan R., et al., Infant feeding practices were not associated with breast milk HIV-1 RNA levels in a randomized clinical trial in Botswana. AIDS Behav. 16, 1260–1264 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Powis K. M., et al., Effects of in utero antiretroviral exposure on longitudinal growth of HIV-exposed uninfected infants in Botswana. J. Acquir. Immune Defic. Syndr. 56, 131–138 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dryden-Peterson S., et al., Increased risk of severe infant anemia after exposure to maternal HAART, Botswana. J. Acquir. Immune Defic. Syndr. 56, 428–436 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mann J. K., et al., Nef-mediated down-regulation of CD4 and HLA class I in HIV-1 subtype C infection: Association with disease progression and influence of immune pressure. Virology 468-470, 214–225 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Novitsky V., et al., Magnitude and frequency of cytotoxic T-lymphocyte responses: Identification of immunodominant regions of human immunodeficiency virus type 1 subtype C. J. Virol. 76, 10155–10168 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Novitsky V., et al., Association between virus-specific T-cell responses and plasma viral load in HIV-1 subtype C infection. J. Virol. 77, 882–890 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Novitsky V. A., et al., Interactive association of proviral load and IFN-gamma-secreting T cell responses in HIV-1C infection. Virology 349, 142–155 (2006). [DOI] [PubMed] [Google Scholar]
- 54.Novitsky V., Gaolathe T., Woldegabriel E., Makhema J., Essex M., A seronegative case of HIV-1 subtype C infection in Botswana. Clin. Infect. Dis. 45, e68–e71 (2007). [DOI] [PubMed] [Google Scholar]
- 55.Novitsky V., et al., Identification of primary HIV-1C infection in Botswana. AIDS Care 20, 806–811 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Novitsky V., et al., Evolution of proviral gp120 over the first year of HIV-1 subtype C infection. Virology 383, 47–59 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Novitsky V., et al., Better control of early viral replication is associated with slower rate of elicited antiviral antibodies in the detuned enzyme immunoassay during primary HIV-1C infection. J. Acquir. Immune Defic. Syndr. 52, 265–272 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Novitsky V., et al., Timing constraints of in vivo gag mutations during primary HIV-1 subtype C infection. PLoS One 4, e7727 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Novitsky V., et al., Viral load and CD4+ T-cell dynamics in primary HIV-1 subtype C infection. J. Acquir. Immune Defic. Syndr. 50, 65–76 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Novitsky V., et al., HIV-1 subtype C-infected individuals maintaining high viral load as potential targets for the “test-and-treat” approach to reduce HIV transmission. PLoS One 5, e10148 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Novitsky V., et al., Dynamics and timing of in vivo mutations at Gag residue 242 during primary HIV-1 subtype C infection. Virology 403, 37–46 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Novitsky V., et al., Extended high viremics: A substantial fraction of individuals maintain high plasma viral RNA levels after acute HIV-1 subtype C infection. AIDS 25, 1515–1522 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Novitsky V., et al., Evolutionary gamut of in vivo Gag substitutions during early HIV-1 subtype C infection. Virology 421, 119–128 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Novitsky V., et al., Transmission of single and multiple viral variants in primary HIV-1 subtype C infection. PLoS One 6, e16714 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Novitsky V., Essex M., Using HIV viral load to guide treatment-for-prevention interventions. Curr. Opin. HIV AIDS 7, 117–124 (2012). [DOI] [PubMed] [Google Scholar]
- 66.Novitsky V., Wang R., Rossenkhan R., Moyo S., Essex M., Intra-host evolutionary rates in HIV-1C env and gag during primary infection. Infect. Genet. Evol. 19, 361–368 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Novitsky V., Moyo S., Wang R., Gaseitsiwe S., Essex M., Deciphering multiplicity of HIV-1C infection: Transmission of closely related multiple viral lineages. PLoS One 11, e0166746 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rossenkhan R., et al., tat Exon 1 exhibits functional diversity during HIV-1 subtype C primary infection. J. Virol. 87, 5732–5745 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rossenkhan R., et al., Transmitted/founder HIV-1 subtype C viruses show distinctive signature patterns in Vif, Vpr, and vpu that are under subsequent immune pressure during early infection. AIDS Res. Hum. Retroviruses 32, 1031–1045 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wright J. K., et al., Influence of Gag-protease-mediated replication capacity on disease progression in individuals recently infected with HIV-1 subtype C. J. Virol. 85, 3996–4006 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Andrews S., FastQC: A Quality Control Tool for High Throughput Sequence Data. Babraham Bioinformatics (Babraham Institute, Cambridge, UK, 2010). [Google Scholar]
- 72.Kliver S., Tamazian G., Brukhin V., O’Brien S., Kommisarov A., KrATER: K-mer analysis tool easy to run. MCCMB 128 (2017). [Google Scholar]
- 73.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li H., Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv [Preprint] (2013). https://arxiv.org/abs/1303.3997 (Accessed 1 April 2021).
- 75.Li H.et al.; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Danecek P., Schiffels S., Durbin R., Multiallelic calling model in bcftools (-m) (2014). https://samtools.github.io/bcftools/call-m.pdf. Accessed 6 October 2021.
- 77.Chang C. C., et al., Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 4, 10.1186/s13742-015-0047-8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fadista J., Manning A. K., Florez J. C., Groop L., The (in)famous GWAS P-value threshold revisited and updated for low-frequency variants. Eur. J. Hum. Genet. 24, 1202–1205 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nelson G. W., O’Brien S. J., Using mutual information to measure the impact of multiple genetic factors on AIDS. J. Acquir. Immune Defic. Syndr. 42, 347–354 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
SNP genotype data have been deposited in https://botswana.gen-watch.org/ (Botswana GWATCH database database). All other study data are included in the article and supporting information. Previously published data were used for this work (18).