Abstract
The development and survival of cancer cells require adaptive mechanisms to stress. Such adaptations can confer intrinsic vulnerabilities, enabling the selective targeting of cancer cells. Through a pooled in vivo short hairpin RNA (shRNA) screen, we identified the ATPase Associated with diverse cellular Activities (AAA-ATPase) Valosin Containing Protein (VCP) as a top stress-related vulnerability in acute myeloid leukemia (AML). We established that AML was the most responsive disease to chemical inhibition of VCP across a panel of 16 cancer types. The sensitivity to VCP inhibition of human AML cell lines, primary patient samples, and syngeneic and xenograft mouse models of AML was validated using VCP-directed shRNAs, overexpression of a dominant negative VCP mutant, and chemical inhibition. By combining mass spectrometry-based analysis of the VCP interactome and phospho-signaling studies, we determined that VCP is important for Ataxia Telangiectasia Mutated (ATM) kinase activation and subsequent DNA repair through homologous recombination in AML. A second-generation VCP inhibitor, CB-5339 was then developed and characterized. Efficacy and safety of CB-5339 were validated in multiple AML models, including syngeneic and patient-derived xenograft murine models. We further demonstrated that combining DNA damaging agents, such as anthracyclines, with CB-5339 treatment synergizes to impair leukemic growth in an MLL-AF9-driven AML murine model. These studies support the clinical testing of CB-5339 as a single agent or in combination with standard of care DNA-damaging chemotherapy for the treatment of AML.
One sentence summary:
Acute myeloid leukemia cells are dependent on the DNA repair function of the AAA-ATPase VCP, opening up a therapeutic avenue.
INTRODUCTION
The past two decades of basic and translational cancer research have brought into the clinic a myriad of small molecules targeting oncogenic dependencies driven by oncogene mutations. The treatment of acute myeloid leukemia (AML) has seen recent progress with the Food and Drug Administration (FDA) approval of drugs for patients with mutations in FLT3 (midostaurin and gilteritinib), IDH2 (enasidenib) and IDH1 (ivosidenib) (1–3). Although conceptually attractive and clinically effective in a subset of patients, targeting mutated oncogenes is not always feasible, for reasons such as loss-of-function mutations, difficult-to-drug oncogenic drivers, or lack of identified mutated drivers. Additionally, clonal evolution under targeted selective pressure frequently results in drug-resistant clonal outgrowth (4).
An emerging alternative therapeutic approach relies on targeting non-oncogenic driver pathways essential for tumor survival, such as cellular stress response (5). Systematic functional genomic screens, using short hairpin RNA (shRNA) or Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR/Cas9) in a number of cancers, have recently revealed new therapeutic targets not suggested by the cancer’s mutational profiles (6). In some cases, in vitro screens are sufficient to identify high fidelity dependencies subsequently validated both in vitro and in vivo. In other cases, particularly in the context of metabolic, immunologic, proteotoxic, or DNA damage stress response targets, an in vivo approach may enhance the identification of true positives.
These evolutionarily conserved stress response pathways are rewired in cancers, such as AML, to permit tumorigenesis and cancer cell survival in the face of oncogenic stress. Indeed, these escape mechanisms allow the clonal selection of cells which can proliferate despite accumulation of damage, increasing the risk of full transformation in a premalignant cell (7). Although necessary for leukemogenesis, these adaptive mechanisms also confer an intrinsic vulnerability and an attractive therapeutic opportunity to selectively target AML cells. Indeed, cellular survival under such unfavorable conditions becomes highly dependent on proficient stress response pathways. Disrupting this common hallmark of cancer through stress sensitization and/or stress overload should ultimately result in cancer cell death (5). For example, targeting oxidative and metabolic stress through BCL-2 inhibition has been shown to have anti-leukemic activity in multiple preclinical models (8). These studies have been recently validated with the promising efficacy of the selective BCL-2 inhibitor venetoclax, now FDA-approved for a subset of patients with AML, combined with low dose cytarabine or hypomethylating agents (9). Therefore, developing functional approaches to unravel the cellular mechanisms to evade stress surveillance may open new therapeutic avenues for AML treatment. Here, we performed an in vivo pooled shRNA screen in an AML mouse model driven by the MLL-AF9 translocation to identify new druggable targets involved in cellular stress reponse pathways.
RESULTS
An in vivo shRNA screen identifies Valosin Containing Protein (VCP) as an AML dependency.
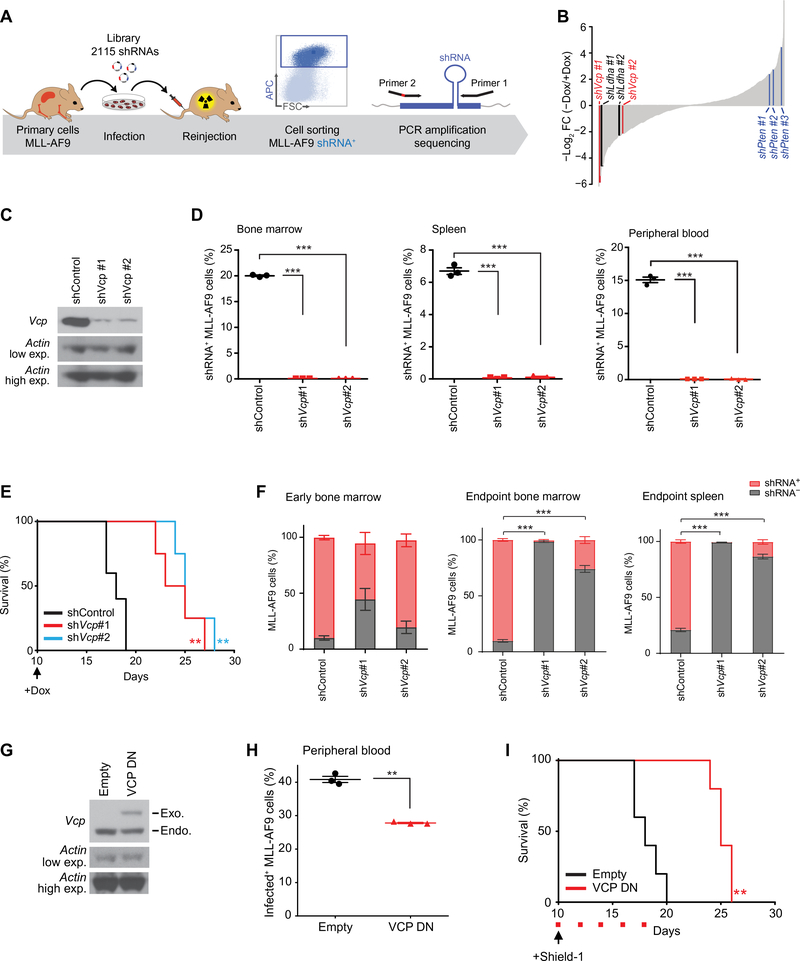
To find therapeutic gene candidates in AML, we designed a custom shRNA library of 2,115 shRNAs targeting 429 genes implicated in various adaptive stress response pathways. This library, cloned into a doxycycline-inducible vector, was transduced as a pool into L-GMP primary MLL-AF9 cells reinjected into sub-lethally irradiated secondary syngeneic recipient mice (10) (Fig. 1A). Using an EdgeR/RIGER comparative analysis of the relative representation of the library hairpins between the two groups of mice treated with and without doxycycline, we identified Vcp, an AAA-ATPase family member, as a top scoring hit (Fig. 1B and table S1). As expected, multiple hairpins directed against the tumor suppressor gene, Pten, as well as a previously described metabolic cancer dependency gene, Ldha scored among the top enriched and depleted genes respectively, providing support for the overall screen quality. We first validated the dependency of the MLL-AF9 blasts on Vcp using two Vcp-directed shRNAs whose efficiency was validated by western blot (Fig. 1C). Doxycycline-induced Vcp knockdown reduced the MLL-AF9-driven disease burden in mouse bone marrow, spleen and peripheral blood as evaluated by flow cytometry (Fig. 1D) and prolonged survival, thereby confirming Vcp dependency in this AML model (Fig. 1E). Because mice ultimately succumbed to AML despite the initial disease burden clearance following Vcp knockdown, we further evaluated disease relapse characteristics in this murine model. Interestingly, at time of death, Vcp knockdown mice harbored a leukemic burden mainly composed of non shRNA-expressing cells, suggesting that disease relapse may result from a positive selection of leukemic cells that have escaped the doxycycline-induced shRNA knock-down (Fig. 1F and fig. S1, A–C). These results thus further validate AML cell dependency on Vcp.
Figure 1. An in vivo shRNA screen identifies VCP as an AML dependency.
(A) Model of the doxycycline-inducible shRNA screening. (B) Waterfall plot of all screened hairpins analyzed using the EdgeR/RIGER method. Top scoring Vcp, Ldha (positive control), and Pten (negative control) hairpins are highlighted in red, black and blue respectively. (C) Western blot for Vcp and Actin in MLL-AF9 cells expressing one control (shControl) and two Vcp-directed shRNAs (shVcp #1 and #2). (D) Proportion of shRNA+ MLL-AF9 cells in bone marrow, spleen and peripheral blood respectively 19, 19 and 16 days post-injection of MLL-AF9 cells expressing either shControl, shVcp#1 or shVcp#2 (n=3 mice per condition). Welch’s t-test in comparison with control condition. Error bars represent mean ± SEM. *** p < 0.001. (E) Kaplan-Meier curves showing overall survival of mice (n=5 for shControl and n=4 for each shVcp group) transplanted with cells expressing shControl, shVcp#1 or shVcp#2. Arrow indicates the beginning of doxycycline treatment. Log-rank (Mantel-Cox) test. ** p < 0.01 by comparison with shControl within the shVcp#1 or the shVcp#2 group. (F) Percentage of shRNA+ and shRNA− MLL-AF9 cells in mice bone marrow 14 days post-injection (Early Bone Marrow) of MLL-AF9 cells expressing either shControl, shVcp#1 or shVcp#2, and in bone marrow and spleen at time of death (n=7 mice per condition). Mann-Whitney test in comparison with the shControl condition. Error bars represent mean ± SEM. *** p < 0.001. (G) Western blot for Vcp and Actin indicating exogenous (Exo) dominant negative (DN) VCP in MLL-AF9 cells treated with Shield-1 for 24 hours. (H) Percentage in peripheral blood of MLL-AF9 cells expressing either an empty or a VCP DN-encoding vector 16 days post-transplantation (n=3 mice per condition). Welch’s t-test in comparison with empty condition. Error bars represent mean ± SEM. ** p < 0.01. (I) Kaplan-Meier curves showing overall survival of mice (n=5 per condition) transplanted with MLL-AF9 cells expressing either an empty vector or VCP DN. Arrow indicates beginning of Shield-1 and red squares indicate days of Shield-1 injection. Log-rank (Mantel-Cox) test. ** p < 0.01 by comparison with empty vector.
VCP is an ATPase protein chaperone which adopts a homo-hexameric conformation comprising six identical subunits arranged in a ring. Each monomeric subunit contains two tandem ATPase Associated with diverse cellular Activities (AAA-ATPase) domains responsible for ATP binding and hydrolysis (D1 and D2). The main ATPase activity is contained in the D2 domain. We further evaluated the VCP dependency in the MLL-AF9 cells using a previously reported ATPase-deficient, mutant form of VCP (E305Q, E578Q). This mutant VCP is unable to hydrolyze ATP but still incorporates into wild type (WT) hexamers, and thus exhibits a dominant negative (DN) function on the complex by abrogating its ATPase activity (11, 12). VCP DN was tagged with a ligand-dependent destabilization domain whose function was to constitutively destabilize VCP DN. Treatment with the membrane-permeable destabilization domain ligand, Shield-1, was then used to block the destabilization domain function and stabilize VCP DN in MLL-AF9 cells (Fig. 1G). Shield-1-induced VCP DN overexpression was sufficient to impair the MLL-AF9-driven disease expansion (Fig. 1H) in vivo and prolong mice survival, thereby establishing that the ATPase activity of VCP is required for AML progression (Fig. 1I).
AML is preferentially sensitive to VCP inhibition.
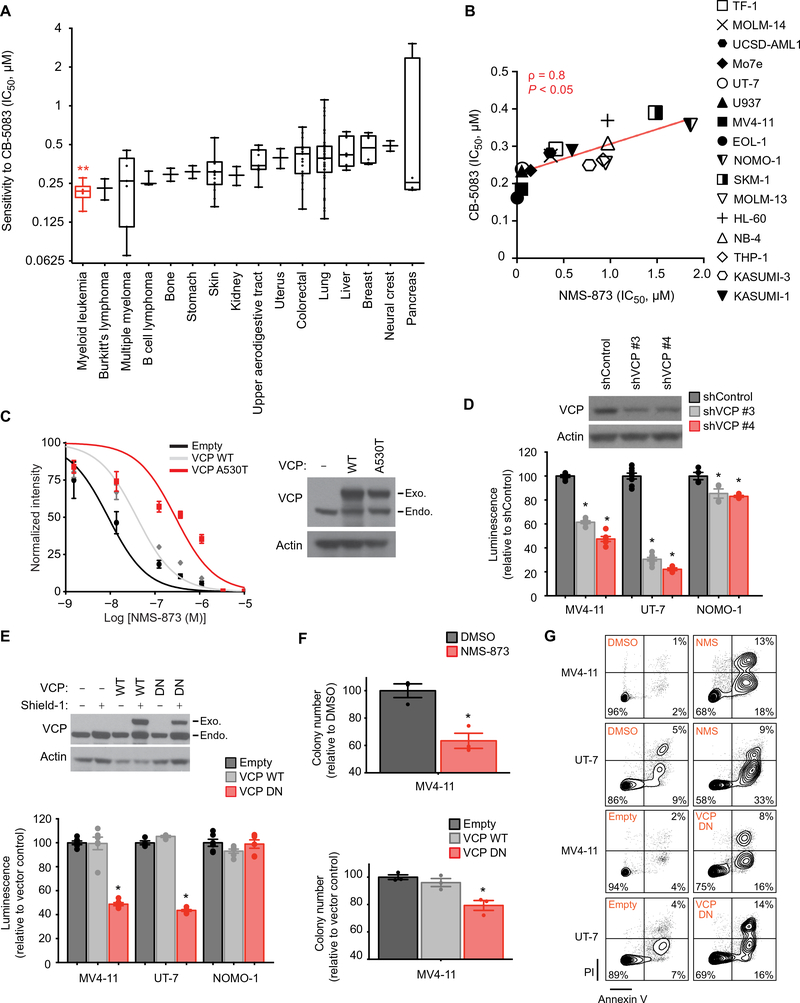
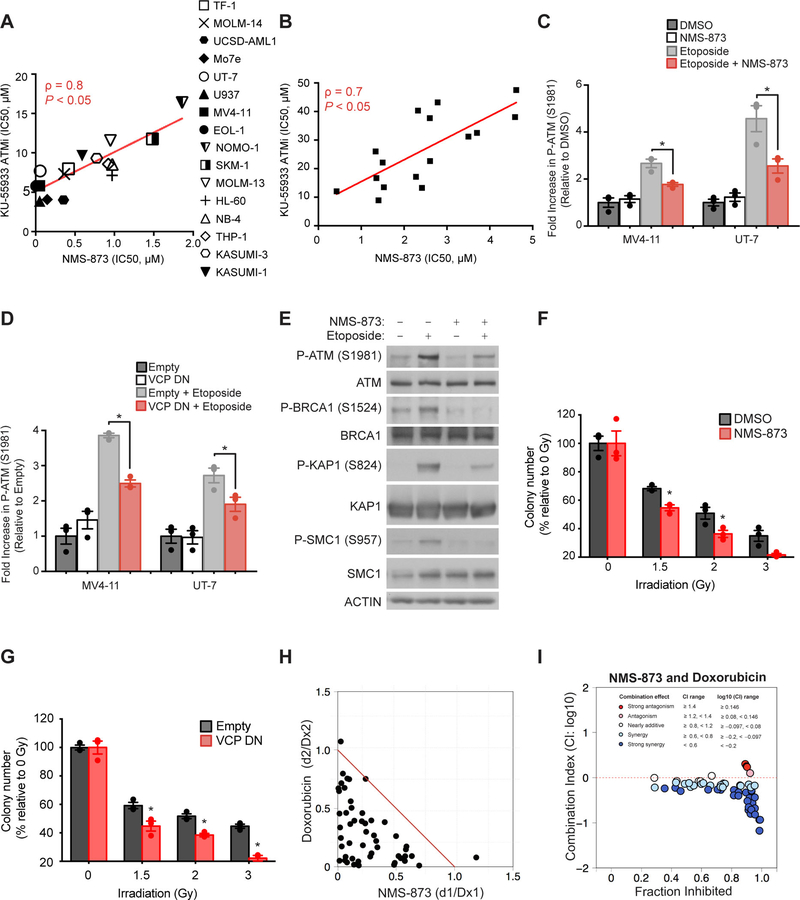
To evaluate whether VCP inhibition elicited a preferential response in AML, we expanded our investigation to a panel of 131 human cancer cell lines, representing 16 cancer types, with the ATP-competitive, small-molecule VCP inhibitor CB-5083 (13, 14). AML cell lines were more sensitive to CB-5083 compared to other cancer subtypes, suggesting that AML is highly dependent on VCP (Fig. 2A and table S2). To further validate these results, we treated a panel of 16 AML cell lines with CB-5083 and a highly-selective allosteric VCP inhibitor NMS-873 (15). The sensitivity of the AML cell lines to these two VCP inhibitors was strongly correlated (Spearman score = 0.8), supporting their on-target effect (Fig. 2B). Moreover, expression of a mutated form of VCP (A530T), which confers resistance to NMS-873 (16), in UT-7 cells rescued the loss of viability induced by NMS-873, thereby confirming the selectivity of NMS-873 for VCP in an AML context (Fig. 2C).
Figure 2. AML cell lines are preferentially sensitive to VCP inhibition.
(A) Distribution of IC50 (concentrations of CB-5083 at which viability was reduced by 50%) in a panel of 131 cancer cell lines treated with CB-5083 for three days in duplicate. Cell lines derived from non-oncogenic tissues and cancer types represented by only one cell line were excluded from this analysis. Error bars represent mean ± SEM of all cell lines within each cancer subtype. Kruskal-Wallis Anova test. ** p < 0.01. (B) Linear regression analysis of the distribution of IC50 of a panel of 16 AML cell lines treated with various concentrations of NMS-873 or CB-5083 for four and five days respectively, with four replicates for each condition. Non-parametric Spearman correlation coefficients (ρ) and associated P-value. (C) Growth inhibition of UT-7 AML cells infected with the indicated Shield-1-inducible constructs and treated with increasing concentrations of NMS-873 for four days. Western blot for VCP and actin indicating VCP WT and a NMS-873-resistant mutant form of VCP (A530T) after Shield-1 treatment for 24 hours. Error bars represent mean of 4 replicates ± SEM. (D) Growth inhibition of indicated AML cell lines infected with either a control or two VCP-directed shRNAs. Western blot for VCP and actin, 6 days post-doxycycline. Data are normalized to the control shRNA and shown relative to day two of doxycycline induction (time of seeding). Welch’s t-test in comparison with control condition. Error bars represent mean of 4 to 8 replicates ± SEM. *p<0.05. (E) Growth inhibition of indicated human AML cell lines infected with a Shield-1-inducible overexpression vector either empty or encoding WT or dominant negative (DN) VCP. Western blot for VCP and actin indicating VCP WT or DN overexpression, in MV4–11 cells 24 hours post-Shield-1. Growth inhibition after four days of Shield-1 treatment is normalized to the empty vector and shown relative to non-induced conditions. Welch’s t-test in comparison with empty condition. Error bars represent mean of 4 to 6 replicates ± SEM. *p<0.05. (F) Colony-forming assay for MV4–11 AML cell line either treated with DMSO or NMS-873 (NMS) at 0.4 μM, or infected with an empty, a VCP WT or DN overexpression construct and treated with Shield-1. Welch’s t-test in comparison with control conditions (DMSO or empty). Error bars represent mean of 3 replicates ± SEM. *p<0.05. (G) Representative FACS plots for annexin V (AV) and propidium iodide (PI) staining of indicated AML cell lines either treated with DMSO or NMS-873 (NMS) at 0.2 μM for 8 days, or infected with an empty or a VCP DN overexpression construct and treated two days with Shield-1. Data representative of two independent experiments.
The anti-leukemic effect of reduced VCP expression and activity was then investigated in three human AML cell lines expressing VCP-directed shRNAs (Fig. 2D) or overexpressing WT or DN VCP (Fig. 2E). Impairment of VCP expression or function markedly decreased viability of the AML cell lines MV4–11 and UT-7, whereas the NOMO-1 cell line was relatively resistant to VCP inhibition, suggesting that all AML subtypes are not equally dependent on VCP (Fig. 2, D and E). The decreased cell viability observed was specific to the loss of VCP function, as overexpression of WT VCP did not impair cell growth. Of note, the profile of sensitivity of these cell lines to VCP knockdown and VCP DN overexpression was similar to their sensitivity to VCP chemical inhibition, supporting on-target activity of both the genetic and chemical approaches used to impair VCP.
Additionally, VCP impairment through either chemical inhibition or VCP DN overexpression resulted in decreased colony forming ability (Fig. 2F). Using Annexin V/propidium iodide staining and flow cytometry analysis of the two most VCP-dependent AML cell lines, MV4–11 and UT-7, we determined that inhibition of VCP ATPase activity using either NMS-873 or overexpression of exogenous VCP DN induced Annexin-positive cell death consistent with apoptosis (Fig. 2G).
VCP inhibition efficiently targets leukemic cells in multiple in vivo AML mouse models and primary AML patient samples.
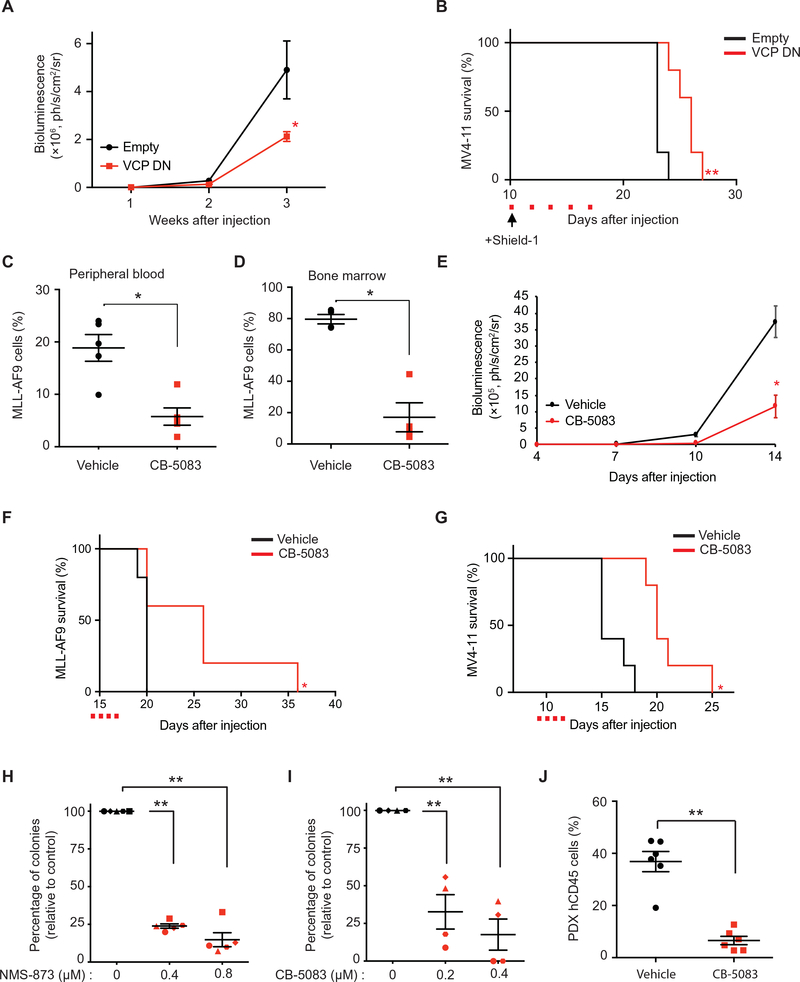
To confirm the anti-leukemic effect of VCP impairment in human AML in vivo, we induced VCP DN overexpression using Shield-1 in MV4–11 cells injected into immunocompromised NSG mice. As we observed in Fig. 1, G–H in the MLL-AF9 syngeneic mouse model, VCP DN overexpression delayed disease progression and improved survival in this MV4–11 orthotopic xenograft model (Fig. 3, A and B). Because NMS-873 does not possess pharmacokinetic properties suitable for in vivo testing, we then evaluated the efficacy of the orally-bioavailable drug candidate recently tested in a phase I clinical trial in patients with advanced solid tumors, CB-5083 (NCT02243917), in both the MLL-AF9 syngeneic and MV4–11 xenograft mouse models. CB-5083 treatment decreased disease burden (Fig. 3, C–E) and prolonged mice survival in both models (Fig. 3, F and G). Importantly, no major weight loss or hematopoietic toxicity based on complete blood counts was observed upon CB-5083 treatment (fig. S2, A–D).
Figure 3. VCP is a dependency in in vivo AML models and primary AML patient samples.
(A) Bioluminescence measurements of mice transplanted with MV4–11-luc cells infected with either an empty or a VCP DN-encoding vector (n=5 mice per condition). Mann-Whitney test in comparison with empty condition. Error bars represent mean ± SEM. * p < 0.05. (B) Kaplan-Meier curves showing overall survival of mice (n=5 per condition) transplanted with MV4–11-luc cells expressing either an empty or a VCP DN-encoding vector. Arrow indicates beginning of Shield-1 and red squares indicate days of Shield-1 injection. Log-rank (Mantel-Cox) test. ** p < 0.01 by comparison with empty vector. (C-D) Proportion of MLL-AF9 cells in peripheral blood (n=5 mice per condition) (C) and bone marrow (n=4 mice per condition) (D), respectively 18 and 20 days post-injection of MLL-AF9 cells. CB-5083 treatment was started at day 14. Mann-Whitney test in comparison with vehicle. Error bars represent mean ± SEM. * p < 0.05. (E) Bioluminescence measurements of mice transplanted with MV4–11-luc cells and treated with CB-5083 or vehicle (n=5 mice per condition). CB-5083 treatment was started at day 9. Welch’s t-test in comparison with vehicle. Error bars represent mean ± SEM. * p < 0.05. (F) Kaplan-Meier curves showing overall survival of mice (n=5 per condition) transplanted with MLL-AF9 and treated with CB-5083. Red squares indicate days of CB-5083 treatment. Log-rank (Mantel-Cox) test. * p < 0.05 by comparison with vehicle. (G) Kaplan-Meier curves showing overall survival of mice (n=5 per condition) transplanted with MV4–11-luc cells and treated with CB-5083 or vehicle. Red squares indicate days of CB-5083 treatment. Log-rank (Mantel-Cox) test. * p < 0.05 by comparison with vehicle. (H-I) Colony-forming assay for primary patient AML samples treated with NMS-873 (n=5) (H) or CB-5083 (n=4) (I). Results represent mean of three replicates for each patient. Welch’s t-test in comparison with the control condition. Error bars represent mean ± SEM. ** p < 0.01. (J) Proportion of hCD45+ leukemic cells in mice peripheral blood 24 days post-injection of patient derived primary AML cells (n=6 mice per condition). CB-5083 treatment was started 20 days post-injection, after engraftment validation. Mann-Whitney test in comparison with vehicle. Error bars represent mean ± SEM. ** p < 0.01.
To further demonstrate the clinical potential of VCP inhibition as an anti-AML therapy, we then treated leukemia cells from five patients with AML and established that the blockade of VCP activity, using either NMS-873 or CB-5083, impaired the capacity of AML blasts to form colonies in methylcellulose (Fig. 3, H–I and table S3). In line with this data, CB-5083 also substantially decreased disease burden in an orthotopic patient-derived xenograft (PDX) model of therapy-related MLL-AF9 positive AML (Fig. 3J and table S3).
Inhibition of the nuclear function of VCP alters AML cell line viability through impairment of DNA repair.
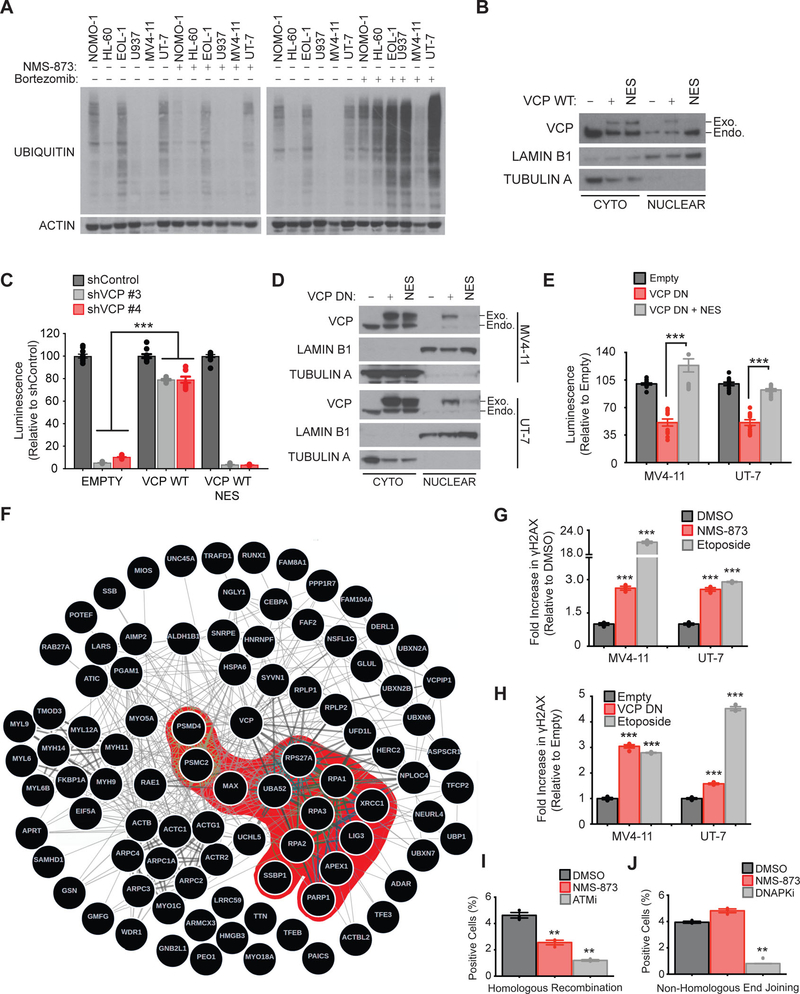
VCP is reported to have many functions in the cell, but the mechanism promoting the dependency in AML was unclear. Previous studies demonstrated that VCP inhibition triggers the endoplasmic reticulum (ER)-associated protein degradation pathway, promoting accumulation of poly-ubiquitinated proteins and generation of proteotoxic stress associated with activation of the unfolded protein response pathway (17, 18). In contrast to these reports, neither NMS-873 treatment at a concentration observed to impair viability in AML nor overexpression of VCP DN induced accumulation of poly-ubiquitinated proteins (Fig. 4A and fig. S3A) or promoted accumulation of the ER stress marker GRP78 (fig. S3B). Poly-ubiquitinated proteins started to accumulate when AML cells were exposed to 5 μM (100-fold the IC50) of NMS-873 (fig. S3C). Moreover, in the panel of 16 AML cell lines tested, sensitivity to NMS-873 did not correlate with sensitivity to bortezomib, a proteasome inhibitor (fig. S3D), and VCP inhibition did not result in any marked changes in the caspase-, trypsin-, or chymotrypsin-like enzymatic activities of the proteasome at concentrations altering cellular viability (fig. S3E).
Figure 4. Inhibition of the nuclear function of VCP alters AML cell line viability through impairment of DNA repair.
(A) Western blot for ubiquitin and actin in the indicated AML cell lines treated with 0.4 μM NMS-873 or 10 nM bortezomib for 24 hours. (B) Western blot for VCP, lamin B1 (nucleus loading control) and tubulin A (cytoplasm loading control) after nuclear/cytoplasmic fractionation of the MV4–11 cell line expressing either a codon optimized “wild-type” VCP (WT) or a NES (nuclear export signal) flanked-WT VCP. (C) Growth inhibition of the indicated MV4–11 cell lines treated with Shield-1 for six days, and five days post-doxycycline supplementation. Data are normalized to shControl for each condition. Mann-Whitney test in comparison to empty vector condition. Error bars represent mean of 10 replicates ± SEM. *** p < 0.001. (D) Western blot for VCP, lamin B1 and tubulin A after nuclear/cytoplasmic fractionation of MV4–11 and UT-7 cell lines expressing either a DN or a NES (nuclear export signal) flanked-DN VCP. (E) Growth inhibition of indicated AML cell lines treated with Shield-1 for four days. Data are normalized to the empty vector and are shown relative to non-induced conditions. Mann-Whitney test in comparison to VCP DN condition. Error bars represent mean of two independent experiments with 6 replicates each ± SEM. *** p < 0.001. (F) Network depicting the VCP interactome established by quantitative mass spectrometry-based analysis of MV4–11 cells stably expressing V5-tagged WT VCP. Results achieving statistical significance (log2FC>0.5 and P-value<0.05) in two biological replicates are shown. VCP interactome was interrogated in a functional enrichment overlapping analysis across the MSigDB database (C2 collection). Protein partners involved in DNA repair, synthesis and cell cycle checkpoints pathways are highlighted in red. −log10FDR calculated through overlapping analysis >1 is defined as threshold of significance. (G-H) FACS analysis of the intracellular expression of γH2AX in MV4–11 and UT-7 AML cells treated for 72 hours with 0.4 μM NMS-873 (G) or expressing either an empty or a VCP DN vector and treated with Shield-1 for 48 hours (H). Etoposide treatment was used as a positive control. Error bars represent mean ± SEM of 3 to 4 replicates. 10,000 cellular events were measured for each replicate condition. Welch’s t-test in comparison with control condition. *** p<0.001. (I-J) FACS analysis of GFP and CD4 expression in RG37-DR-GFP (I) and GC92-NHEJ-CD4 (J) cells respectively at 48 hours post-transfection with I-SceI endonuclease and treated with either 0.4 μM NMS-873, 5 μM KU-55933 (ATMi), or 2.5 μM KU-57788 (DNAPKi). Error bars represent mean ± SEM of 3 replicates. 10,000 cellular events were measured for each replicate. Welch’s t-test in comparison with control condition. ** p<0.01.
To further investigate the mechanism of action by which VCP inhibition may alter AML growth, we exploited the fact that VCP is present in both the nucleus and cytoplasm of AML cells in order to ask whether AML cell survival relies more on the nuclear or the cytoplasmic pool of VCP. Therefore, we depleted endogenous VCP concomitantly with overexpression of a codon-optimized non-shRNA-targetable exogenous VCP WT or VCP WT fused to a Nuclear Export Signal (NES), in the MV4–11 AML cell line. We validated by subcellular fractionation that this NES-mutant VCP WT remained largely absent from the nucleus of MV4–11 AML cell lines (Fig. 4B). As expected, VCP knockdown decreased cellular viability in the MV4–11 AML cell line. Although this effect was significantly abolished by codon-optimized VCP WT overexpression (p<0.0001), VCP WT NES failed to restore MV4–11 cell viability to the same extent as its WT counterpart, thus suggesting an essential role for VCP in nuclear signaling pathways in AML (Fig. 4C). To further support the nuclear essentiality of VCP in AML, we fused a Nuclear Export Signal (NES) to the VCP DN mutant to drive its constitutive export from the nucleus (Fig. 4D). Restricting VCP DN localization to the cytoplasm abrogated its cytotoxic effects on these cells, again suggesting a role for the nuclear fraction of VCP in AML cells survival (Fig. 4E).
To investigate the nuclear functions of VCP in AML, we performed an unbiased mass spectrometry-based characterization of the VCP interactome in the MV4–11 AML cell line, one of the most sensitive cell lines to VCP inhibition. MV4–11 cells were infected with either a V5-tagged VCP or an empty control vector before V5 immunoprecipitation and mass-spectrometry analysis. A total of 735 VCP protein partners were identified (table S4). Two previously described critical VCP substrate adapters, NPLOC4 and UFD1L, scored as top VCP interactors in our screen, thereby acting as internal positive controls and validating our mass-spectrometry approach (fig. S4A). The 92 top scoring hits (p-value < 0.05 and log2 (FC) > 0.5) are represented in the interactome network in Fig. 4F. We interrogated this list of top scoring hits with published, validated gene signatures for enrichment by pathway analysis (table S5) and established that VCP interacts with 41 nuclear proteins markedly enriched among DNA damage recognition and repair pathways (Fig. 4F and table S5). To better explore the VCP interactome in an AML context, we compared our data with previously reported VCP interactome data in HEK-293T cells (19). The overlap and differences between both cellular contexts are highlighted in fig. S4, B and C and table S6. Eighty high confidence VCP-interacting proteins were only identified in the MV4–11 AML cell line (fig. S4C). Interestingly, this AML interactors were markedly enriched among DNA repair and replication pathways in an unbiased enrichment overlapping analysis (fig. S4D). In a similar enrichment analysis, VCP-interacting proteins only detected in the HEK-293T cellular context were mainly enriched among mRNA processing and not DNA repair pathways (fig. S4E).
Based on these results, we then assessed phosphorylation of the histone H2AX (termed as γH2AX), which reflects accumulation of double-strand breaks (DSB) in DNA. Inhibition of VCP using either NMS-873 treatment or VCP DN overexpression increased γH2AX in MV4–11 and UT-7 AML cell lines as was observed in cells treated with etoposide, a topoisomerase inhibitor that induces DSBs (Fig. 4,G and H).
Given that the two major DNA repair pathways in response to DSB are homologous recombination (HR) and non-homologous end-joining (NHEJ), we then explored the effects of VCP impairment on these pathways. We used two reporter systems to evaluate induction of either HR or NHEJ in response to I-SceI overexpression. This endonuclease creates a targeted DSB in an eGFP or CD4 reporter sequence. Successful repair of the DSB by either HR or NHEJ results in the reconstitution of functional eGFP or CD4, which can be quantified using flow cytometry (Fig. 4, I and J). As expected, we observed that pharmacological inhibition of the serine/threonine kinase ATM, a key component of the HR repair pathway (20), impaired HR-mediated DSB repair. Inhibition of VCP with NMS-873 also led to an impairment of HR, whereas it did not impair NHEJ, suggesting that VCP plays an important role in HR DSB repair.
VCP inhibition impairs ATM phosphorylation and downstream signaling resulting in increased sensitivity to DNA-damaging agents.
Many key mammalian DNA damage response signaling pathways stem from activation of the canonical serine/threonine kinase ATM (20). Interestingly, we observed that the pattern of sensitivity to pharmacological inhibition of this key genome guardian using the ATM inhibitor KU-55933, in a panel of 16 AML cell lines (Fig. 5A) and 16 primary AML patient samples (Fig. 5B and table S3), was highly correlated with their sensitivity to NMS-873 (Spearman score = 0.8 and 0.7 respectively). To evaluate the impact of VCP impairment on the DNA damage signaling response, we used etoposide to induce DSBs. We then evaluated the downstream DNA repair signaling cascade after either genetic (VCP DN overexpression) or chemical inhibition of VCP. These experiments were performed using short-term VCP impairment, to allow the evaluation of early effects on DNA repair signaling, before high cellular DNA damage accumulation. Consistent with our previous results, we established that VCP inhibition blocked DSB-induced activation of ATM signaling (Fig. 5, C and D) and ATM’s well-reported protein substrates, BRCA1, SMC1 and KAP1 (Fig. 5E), suggesting that VCP plays a central role in DNA damage sensing and repair in AML. Moreover, in line with a role for VCP at the crosstalk between DNA damage response pathways and AML growth, we established that VCP inhibition sensitizes AML cells to DNA damaging agents, such as irradiation (Fig. 5, F and G) and the anthracycline doxorubicin (Fig. 5, H and I). These findings nominated VCP inhibition as a potential therapeutic avenue to treat patients with AML and supported further preclinical studies to test it in combination with standard chemotherapy regimens using anthracyclines.
Figure 5. VCP inhibition impairs ATM phosphorylation and downstream signaling resulting in increased sensitivity to DNA damaging agents.
(A-B) Linear regression analysis of the distribution of IC50 of a panel of 16 AML cell lines (A) and 16 primary patient AML samples (B) treated with various concentrations of NMS-873 or KU-55933 (ATMi) for four and three days respectively, four replicates for each condition. Non-parametric Spearman correlation coefficient (ρ) and associated P-value are calculated. (C-D) FACS analysis of the intracellular expression of P-ATM (S1981) in MV4–11 and UT-7 AML cells treated with NMS-873 for two hours (C) or with Shield-1 for 12 hours to stabilize VCP DN (D). Etoposide treatment was used to induce DNA damage in order to evaluate the DNA repair signaling response under VCP impairment. Error bars represent mean ± SEM of three replicates. 10,000 cellular events were measured for each replicate. Welch’s t-test in comparison with etoposide conditions for each cell line. * p<0.05. (E) Western blot for P-ATM (S1981), ATM, P-BRCA1 (S1524), BRCA1, P-KAP1 (S824), KAP1, P-SMC1 (S957), SMC1 and actin, from MV4–11 cells treated with NMS-873 for two hours. Etoposide treatment was used to induce DNA damage, to evaluate the DNA repair signaling response under VCP impairment. (F-G) Colony-forming assay for MV4–11 AML cell line treated with 0.2 μM NMS-873 for 24H (F) or infected with a Shield-1 inducible empty or VCP DN overexpression construct (G), and then irradiated with the indicated doses. Welch’s t-test in comparison with each control condition. Error bars represent mean of 3 replicates ± SEM. * p<0.05. (H-I) Isobologram representation (H) or Combination Index (CI) plots (I) for the combination of NMS-873 with doxorubicin in MV4–11 cell line after four days of treatment. Doxorubicin treatment was added after 24 hours of NMS-873 pre-treatment. Results represent the average of four replicates for each dose combination. Dx denotes the drug concentration required to produce x percentage effect alone, and d denotes the drug concentration required to produce the same x percentage effect in combination with the second drug.
CB-5339, a second-generation, potent and selective VCP small-molecule inhibitor offers a promising therapeutic avenue for AML.
CB-5083 is a first-in-class ATP-competitive VCP inhibitor that was recently tested in two phase I clinical trials in solid tumors and multiple myeloma (13, 14) (NCT02243917, NCT0223598). The clinical development of this drug candidate was halted due to an unexpected off-target inhibitory action on phosphodiesterase (PDE) 6, a key regulator of the retinal phototransduction cascade (21), thus resulting in ophthalmological side effects. This finding is reminiscent of the vision disturbances caused by PDE5 inhibitors, which were developed for the treatment of erectile dysfunction and had cross-reactivity with PDE6 (22).
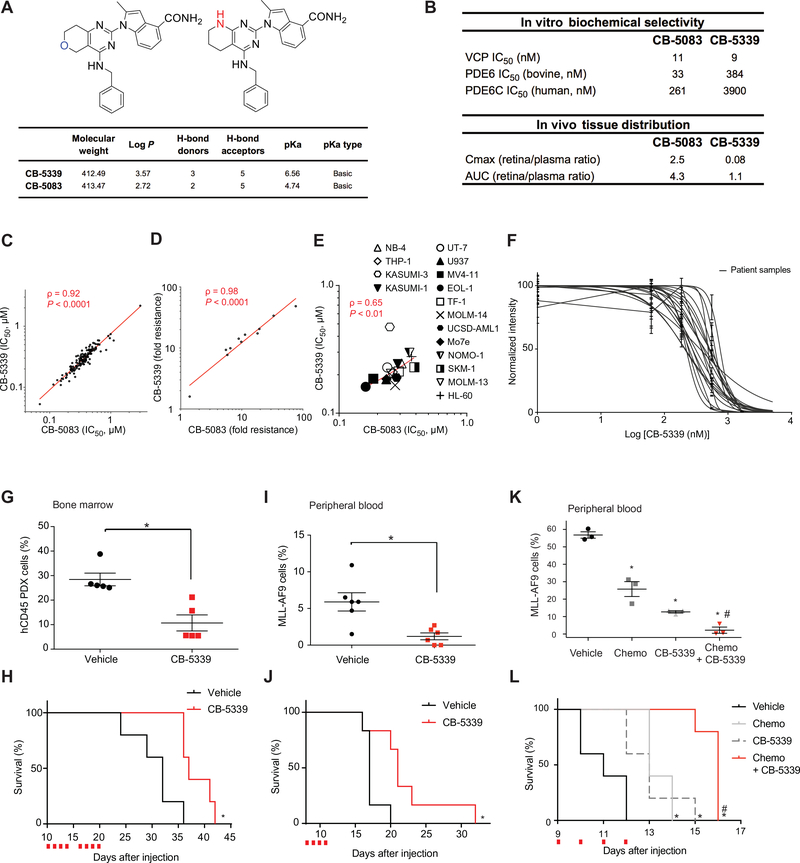
In order to translate our findings into the clinic, a second-generation, highly selective and potent ATP-competitive, oral, small molecule inhibitor of VCP, CB-5339, was developed (Fig. 6A). CB-5339 was 15-fold less active on the human cone PDE6 (PDE6C) in in vitro biochemical assays, and displayed a 31-times lower Cmax in vivo in rat retina compared to CB-5083, thus addressing the off-target limitations of the first-generation compound (Fig. 6B). Importantly, CB-5339 retained the high on-target potency of CB-5083 and displayed a similar sensitivity profile across a panel of 138 cell lines (Fig. 6C). To further characterize the molecular inhibition of VCP using this second-generation drug candidate, the previously established HCT116 colon cancer CB-5083 resistant cell lines were treated with either CB-5083 or CB-5339 (13). These resistant cell lines harbor VCP mutations mainly located in or adjacent to the D2 ATPase domain of VCP. Both compounds showed a comparable biochemical efficacy profile, suggesting a similar molecular inhibitory mechanism and retention of the on-target effect of the first-generation compound on VCP (Fig. 6D). The efficacy of CB-5339 was then assessed in our panel of 16 AML cell lines and was accordingly correlated to the first-generation inhibitor CB-5083 (Fig. 6E). CB-5339 efficacy was also validated on a panel of 16 primary AML patient samples harboring diverse genetic backgrounds (table S3). The median IC50 was 375 nM among these samples, thus supporting the translational potential of CB-5339 (Fig. 6F).
Figure 6. Targeting VCP in AML through a second-generation VCP inhibitor: CB-5339.
(A) Chemical structure and properties of CB-5339 compared to CB-5083. (B) CB-5339 in vitro biochemical selectivity towards PDE6 and retina/plasma tissue distribution compared to CB-5083. (C) Linear regression analysis of IC50 distribution of a panel of 138 cell lines treated with CB-5339 or CB-5083 for three days in duplicates. Non-parametric Spearman correlation coefficient (ρ) and associated P-value. (D) Linear regression analysis of IC50 distribution of a panel of 11 HCT116 cell lines carrying resistance mutations to CB-5083, treated with CB-5339 or CB-5083 for three days in duplicates. Results are presented as fold resistance compared to the parental HCT116 cell line. Non-parametric Spearman correlation coefficient (ρ) and associated P-value. (E) Linear regression analysis of IC50 distribution of a panel of 16 AML cell lines treated with CB-5339 or CB-5083 (four replicates for each condition). Non-parametric Spearman correlation coefficient (ρ) and associated P-value. (F) Growth inhibition of 16 primary patient AML samples treated with increasing concentrations of CB-5339 for six days. Error bars represent mean of two replicates ± SEM. (G) Proportion of hCD45+ leukemic cells in mice bone marrow (n=5 mice per condition) 21 days post-injection of patient-derived primary AML cells. CB-5339 treatment was started 10 days post-injection, after engraftment validation. Mann-Whitney test in comparison with vehicle. Error bars represent mean ± SEM. * p<0.05. (H) Kaplan-Meier curves showing overall survival of mice (n=5 mice per condition) transplanted with patient-derived primary AML cells and treated with CB-5339 at 90 mg/kg. Red squares indicate days of CB-5339 treatment. Log-rank (Mantel-Cox) test. * p < 0.05 by comparison with vehicle. (I) Proportion of MLL-AF9 cells in peripheral blood (n=6 mice per condition) 12 days post-injection of MLL-AF9 cells. CB-5339 treatment was started at day 8. Mann-Whitney test in comparison with vehicle. Error bars represent mean ± SEM. * p<0.05. (J) Kaplan-Meier curves showing overall survival of mice (n=6 per condition) transplanted with MLL-AF9 and treated with CB-5339 at 90 mg/kg. Red squares indicate days of CB-5339 treatment. Log-rank (Mantel-Cox) test. * p < 0.05 by comparison with vehicle. (K) Proportion of MLL-AF9 cells in peripheral blood (n=3 mice per condition) 11 days post-injection of MLL-AF9 cells. Treatment was started at day 9 (CB-5339 at 50 mg/kg for 4 days, Chemo : Doxorubicin at 0.5 mg/kg for 3 days and cytarabine at 75 mg/kg for 5 days). Welch’s t-test. Error bars represent mean ± SEM. * p < 0.05 by comparison with vehicle. # p < 0.05 by comparison with CB-5339 or Chemo groups. (L) Kaplan-Meier curves showing overall survival of mice (n=5 per condition) transplanted with MLL-AF9 and treated as indicated. Red squares indicate days of CB-5339 treatment. Log-rank (Mantel-Cox) test. * p < 0.05 by comparison with vehicle. # p < 0.05 by comparison with CB-5339 or Chemo groups.
To further validate our mechanistic hypothesis using CB-5339, we first studied its effects on polyubiquitin protein accumulation and the activation of the unfolded protein response (UPR) in an AML context. CB-5339 treatment induced dose-dependent polyubiquitin protein accumulation at concentrations ≥ 0.4 μM (fig. S5A). Similarly, the ER stress marker GRP78 accumulated at concentrations ≥ 0.4 μM, whereas spliced XBP-1 and ATF-4 accumulated after treatment with CB-5339 at concentrations ≥ 1.6 μM and 0.8 μM respectively (fig. S5B) arguing for a concentration-dependent increase in proteotoxic stress. We also explored DNA damage response signaling after CB-5339 treatement to evaluate whether CB-5339 also impaired DNA repair at concentrations impacting cellular viability in AML (MV4–11 IC50 = 0.186 μM). Importantly, CB-5339 treatment impaired ATM phosphorylation upon etoposide DSBs induction (fig. S5C) and increased γH2AX accumulation at 0.2 μM (fig. S5D). Accordingly, CB-5339 also synergized with doxorubicin in the MV4–11 AML cell line (fig. S5E). CB-5339 thus displayed both a nuclear and a cytoplasmic effect on AML cell lines. Lower concentrations impaired DNA repair whereas higher concentrations resulted in proteotoxic stress, with both potentially contributing to the viability effect observed with CB-5339 treatment of AML cells. To further evaluate the relative importance of each effect on AML cellular viability, we studied the pattern of sensitivity of 16 AML cell lines to CB-5339 compared to either the proteasome inhibitor bortezomib or the ATM inhibitor KU-55933. Cellular sensitivity to CB-5339 across this panel of AML cell lines was not correlated with bortezomib response whereas it was strongly correlated with KU-55933 sensitivity (Spearman score = 0.67) (fig. S5, F and G). These results are similar to those observed with the allosteric inhibitor of VCP; NMS-873 and are in line with our rescue experiments arguing for a strong dependency of AML on the nuclear function of VCP.
Compared to CB-5083, CB-5339 had a lower clearance in preclinical studies in multiple species, thus providing for a higher bioavailability (fig. S6A). In order to confirm that the highly conserved on-target activity of CB-5339 against VCP translated into a similar in vivo efficacy, we used a MLL-AF9-driven patient-derived xenograft (PDX) AML mouse model. As observed with CB-5083, CB-5339 treatment resulted in decreased bone marrow leukemic infiltration and prolonged mice survival (Fig. 6,G and H).
Similar to the results obtained with this PDX model, CB-5339 treatment decreased circulating leukemic cells and prolonged survival in the MLL-AF9 syngeneic mouse model (Fig. 6, I and J). Moreover, CB-5339 synergized in vivo with standard of care AML chemotherapy, a combination of an anthracycline and cytarabine. This triple combination regimen resulted in decreased leukemia burden and prolonged mouse survival compared to each regimen alone (Fig. 6, K and L). Importantly, CB-5339 was well tolerated as evidenced by stable weight curves and absence of substantial myelosuppression (fig. S6, B–E).
At time of disease relapse, no marked difference in disease burden or cell surface myeloid differentiation markers were observed (fig. S6, F and G). Consistently, differentiation markers were not substantially increased at time of initial disease burden evaluation post-CB-5339 treatment (fig. S6H). These results suggested that CB-5339 did not induce phenotypic differentiation of myeloid leukemia cells and were in agreement with our mechanistic model based on leukemic cell apoptosis rather than cell differentiation.
Co-occurrence of RAS oncogene activation and TP53 deficiency is associated with impaired response to VCP inhibition.
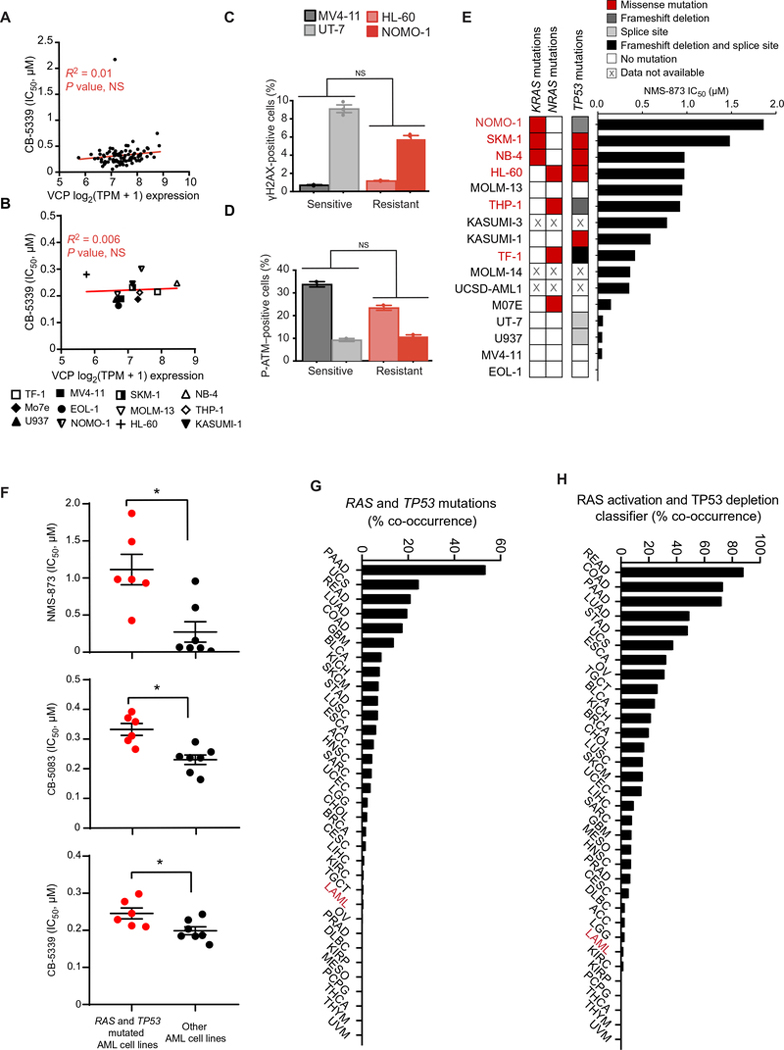
One fundamental step to accelerate the clinical translation of our findings relies on the identification of the most or least responsive AML subgroups to VCP inhibition. To address this question, we first evaluated VCP mRNA expression based on the publicly available Cancer Cell Line Encyclopedia (CCLE) database (23, 24). No correlation was observed between VCP expression and cancer cell line sensitivity to CB-5339 in our panel of AML cell lines (12 available in CCLE) nor in our pan-cancer sensitivity screen (91 cell lines available in CCLE) (Fig. 7, A and B). VCP is thus ubiquitously expressed with very low variation among different cancer cellular contexts and does not explain the differential sensitivity to VCP inhibition.
Figure 7. Co-occurrence of RAS oncogene activation and TP53 deficiency is associated with decreased response to VCP inhibition.
(A-B) Linear regression analysis of IC50 distributions of a panel of 91 cancer cell lines (A) or 12 AML cell lines (B) treated with CB-5339 for three days compared to VCP mRNA expression data extracted from the CCLE database. Cell lines with no available transcriptional data were excluded from this analysis. R-square coefficient and associated P-value. NS=not significant. TPM=Transcripts per Million. (C-D) FACS analysis of the intracellular expression of γH2AX (C) or P-ATM (S1981) (D) in the indicated AML cell lines. Error bars represent mean of 2 to 3 replicates ± SEM. 10,000 cellular events were measured for each replicate. Welch’s t-test. NS=not significant. (E) KRAS, NRAS and TP53 mutational profiles (extracted from CCLE database) of the 16 AML cell lines screened for sensitivity to NMS-873. Cell lines harboring both RAS (KRAS or NRAS) and TP53 mutations are highlighted in red. (F) Distribution of IC50 in a panel of 13 AML cell lines treated with various concentrations of NMS-873, CB-5083 or CB-5339 (four replicates for each condition). The 3 AML cell lines with no available RAS-TP53 mutational status were excluded from this analysis. Mann-Whitney test. * p<0.05. (G-H) RAS and TP53 mutational (G) or RAS activation and TP53 deficiency (H) co-occurrence frequency across pan-cancer TCGA database (10, 294 patient samples, 33 tumor lineages). Functional RAS activation and TP53 deficiency scores were determined based on two previously validated classifiers. AML subtype is highlighted in red. TCGA acronyms are detailed in Table S8.
Because of the role of DNA repair impairment in the underlying mechanism of AML growth blockade by VCP inhibition, we next evaluated both γ-H2AX and pATM basal expression using a quantitative flow cytometry approach in two sensitive and two resistant AML cell lines. Neither of these DNA damage-related markers was correlated with VCP sensitivity (Fig. 7, C and D).
To further assess for biomarkers of VCP sensitivity or resistance in AML, we then analyzed the mutational profiles of the 16 AML cell lines screened for sensitivity to the VCP inhibitor NMS-873 using the CCLE database. Because our initial screen was performed in an MLL-driven mouse model, we first evaluated the impact of MLL rearrangement on VCP dependency. The MLL status was not associated with increased sensitivity to NMS-873. No specific single mutation was associated with VCP sensitivity nor with VCP resistance.
Given the poor prognosis of TP53 and RAS mutations in AML and their potential effect on the DNA repair pathways, we then explored whether the combination of these mutations was associated with resistance to VCP inhibition. We found that harboring co-occurring activating mutations in one of the RAS family genes (KRAS or NRAS) and deleterious mutations in the TP53 gene is associated with decreased response to VCP inhibition (two tailed Fisher exact test odds ratio = 2.67, P-value = 0.002) (Fig. 7E and table S7). Consistently, the top 4 sensitive cell lines EOL-1, MV4–11, U937 and UT-7 harbor at most a single splice site mutation for TP53 and no mutation for KRAS or NRAS. KRAS/NRAS and TP53 mutational co-occurrence was associated with decreased response to VCP inhibition using NMS-873, CB-5083 and CB-5339 (Fig. 7F).
Using the pan-cancer TCGA database (https://cancergenome.nih.gov/publications; (25)), we observed that the frequency of TP53 and RAS mutations co-occurrence in AML was among the lowest across the 33 TCGA cancer cohorts (Fig. 7G and table S8), potentially generating a preferential VCP vulnerability in AML. This observation was validated using recently developed functional classifiers allowing identification of TP53 deficiency (26) and RAS activation (27) (Fig. 7H and table S8). Together, this data suggests that co-occurrence of TP53 deficiency and RAS pathway activation renders cancer cells less sensitive to VCP inhibition and highlights a useful biomarker that could inform future clinical trials.
DISCUSSION
Our studies revealed a strong dependency of AML on the multifunctional AAA-ATPase VCP. VCP has been recently reported to play a role in solid tumors and multiple myeloma progression through regulation of protein homeostasis and ER stress (13, 15, 28). In contrast to these prior studies focused on the role of VCP in proteotoxic stress, our data provided evidence that VCP impairing in an AML context also targets another key cancer hallmark: genomic instability (5, 29).
Previous studies have determined that the yeast homolog of VCP - Cdc48 - and VCP play a key role in regulation of DNA damage tolerance and repair pathways (30)(31–35). Here, we demonstrated that VCP inhibition plays an upstream function in DNA repair in an AML context, through the impairment of the canonical DNA repair kinase ATM. Using mass spectrometry-based interactome analysis, we highlighted the broad role of VCP in AML as a key interactor at the crosstalk of many nuclear signaling cascades, including DNA repair, synthesis and replication.
Targeting non-oncogene addictions such as DNA damage tolerance and repair pathways has recently emerged as a promising cancer therapy as exemplified by the success of Poly(ADP-ribose) polymerase inhibitors in the treatment of cancers deficient for homologous recombination (36). In AML, despite the fact that germ-line mutations in DNA damage response (DDR) genes are very rare, recent reports demonstrated that defective DDR can result from replication and oxidative stress, gene polymorphisms and transcriptional deregulation of DDR players (37, 38) (39). Recent studies accordingly established that pharmacological inhibition of the two serine/threonine kinases ATR and ATM represent potential therapeutic strategies for AML treatment (40). Our results uncovered the role of VCP impairment as another approach to inhibiting DDR in AML. Although complete Vcp knockout (KO) is embryonic lethal in mice, Vcp heterozygous KO is viable and no specific dysfunctional phenotype has been reported (41), suggesting a dosage effect, and offering a therapeutic window for the selective targeting of cancer cells particularly addicted to VCP-driven cellular processes. Given the heightened sensitivity of AML cells to VCP inhibition, we hypothesize that impairment of DDR through VCP inhibition could offer a therapeutic window to selectively target AML cells while sparing normal tissues. In line with this hypothesis, CB-5083 and CB-5339 treatments did not markedly alter normal peripheral blood counts and weight curves in C57/BL6 mice. Accordingly, CB-5083 was not myelosuppressive in the recently conducted phase I trial (NCT02243917).
Cancer cells’ rewiring of DNA repair pathways can both promote cancer development and influence response to chemotherapy (37) (42, 43). Impairment of DDR can be beneficial to cancer cells in the initial steps of tumorigenesis and in tumor evolution, leading to genomic rearrangements and accumulation of somatic mutations (44). Whereas the molecular basis of the DNA damage and repair coordination that enables a tolerable mutational burden in cancer cells is only partially understood, DNA-damaging agents, such as chemotherapy or radiation, are widely used in the clinic and tend to exploit the inability of cancer cells to correctly repair DNA. DDR upregulation can represent an escape mechanism and explain chemotherapy resistance(45). Thus, the combination of standard AML chemotherapy with inhibitors of DDR is a therapeutic strategy of interest. Our data suggests that targeting the DDR through VCP inhibition might also sensitize AML cells to DNA damaging agents, such as anthracyclines. Such a synergistic combination strategy should thus simultaneously help to avoid compensatory pathways activated in response to either chemotherapy regimens alone or single agent VCP inhibitors.
To more effectively target cancer cells vulnerability to DNA damage, one of the major challenges is the identification of biomarkers predicting response to DDR targeted cancer therapy. In our study, we did not identify any specific mutational susceptibility to VCP impairment, suggesting that VCP dependency relies on a broader DDR-mediated vulnerability. However, we did observe a milder response to VCP inhibition in the context of RAS and TP53 co-occurring mutations. RAS/MAPK pathway activating mutations induce oncogenic replicative stress and can result in DDR pathways modulation (46–48)(49). TP53 inactivating mutations result in delayed DNA DSB resolution and a better DNA damage tolerability (50). Oncogenic RAS activates a TP53 dependent DDR and checkpoints resulting in cellular senescence (51). TP53 deficiency could thus cooperate with RAS through overcoming TP53-mediated senescence to promote tumorigenesis (52, 53). Taken together, this data suggests that such a double mutated context results in a modulated DDR and a better DNA damage tolerability potentially explaining resistance to VCP inhibition. Association of both TP53 and RAS mutations is only described in a small number of patients with AML (0.6 to 3.1%) (53) suggesting that a large majority of patients may benefit from VCP inhibition.
Translating preclinical studies into the clinic is challenging and relies on the ability to develop tractable drug candidates (54). Many small molecule inhibitors have been reported to target VCP (14, 15, 55, 56). Among these, the most selective reported to date are two mechanistically divergent small molecules: NMS-873 and CB-5083. NMS-873 is a highly selective allosteric inhibitor that lacks appropriate pharmacological and bioavailability properties and thus is not expected to move forward into the clinic (15). CB-5083 is an ATP-competitive VCP inhibitor that has been recently tested into a phase I trial in solid tumors(14) (NCT02243917). This small molecule’s drug development journey was curtailed by an unanticipated off-target ophthalmological toxicity. A comprehensive set of biochemical studies pointed to a PDE6 inhibitory off-target action as responsible for this side effect. We report here the structure and biochemical properties of a second-generation VCP inhibitor, CB-5339, the lead drug candidate for future clinical trials. Our study offers an opportunity to repurpose this unique class of VCP inhibitors, potentially at a lower dose, for AML, a disease highly dependent on VCP and with a strong unmet clinical need. Moreover, future studies will evaluate CB-5339’s therapeutic potential in solid tumors and neurodegenerative disorders where VCP represents a promising therapeutic target (57).
Limitations of our study include a potentially narrow therapeutic window given the essential and ubiquitous role of VCP in various biological processes. Careful pharmacokinetic and pharmacodynamic studies will be needed to account for inter-individual variability and ensure exposure to an effective and non-toxic dose of CB-5339. Additionally, redundance in the DDR pathways may be responsible for resistance mechanisms that will need to be carefully monitored and mechanistically dissected during future clinical trials.
In conclusion, we identified and validated VCP as a therapeutic target in AML using multiple in vivo and in vitro models. Dissecting the molecular mechanisms underpinning this dependency in AML led to the identification of a nuclear DNA repair function of VCP in leukemia, thereby providing a therapeutic avenue for AML. Our findings thus provide the preclinical and pathophysiological basis for a CB-5339 AML phase I clinical trial as a single agent and in combination with standard of care AML chemotherapy regimens.
MATERIALS AND METHODS
Study design
This study sought to identify stress-related AML dependencies and validate the use of a second-generation clinical candidate VCP inhibitor as a targeted therapy in AML. A pooled shRNA screen was performed in vivo and analyzed using the EdgeR/RIGER method as developed in the supplementary materials and methods. A series of biochemical and functional studies, in murine models, human AML cell lines, and primary patient samples, using genetic and chemical tools, were developed as detailed in the supplementary materials and methods, to validate and understand the critical role of this ATPase in leukemia. To further translate these results to the clinic, a selective ATP-competitive VCP inhibitor was identified. Effects of first-generation (CB-5083) and second-generation (CB-5339) VCP inhibitors in AML were studied in vitro in AML cell lines and primary patient AML samples and in vivo in human cell line or patient-derived orthotopic xenograft and syngeneic mouse models. Sample size was chosen in light of the fact that these in vivo models were historically highly penetrant, aggressive, and consistent. Blinded observers visually inspected mice for obvious signs of illness, such as loss of appetite, hunched posture, and lethargy. Mice were randomly assigned to each treatment group. The number of experimental replicates is specified in each figure legend.
Statistical analysis
Statistical analysis was performed using PRISM 8.0.1 (GraphPad), or the indicated software for more dedicated analysis. Data were analyzed using a nonparametric Mann-Whitney test (with the assumption of no Gaussian distribution of the group) unless otherwise specified, and the threshold of significance (α) was always set at 0.05.
Supplementary Material
Table S1. List of the top depleted genes identified by the in vivo shRNA screening.
Table S2. CB-5083 sensitivity profile across 131 cancer cell lines.
Table S3. Patient characteristics.
Table S4. List of VCP-interacting partners identified by mass spectrometry-based interactome analysis in the MV4–11 AML cell line.
Table S5. List of enriched pathways among VCP interactome.
Table S6. List of VCP-interacting partners identified by mass spectrometry-based interactome analysis in the MV4–11 AML cell line compared to HEK-293T cell line.
Table S7. RAS and TP53 mutational profiles of AML cell lines according to the CCLE database.
Table S8. Mutational and functional co-occurrence of RAS activation and TP53 deficiency in pan-cancer TCGA cohorts.
Table S9: List of antibodies used for western blotting, immunoflourescence and flow cytometry.
Fig. S1. MLL-AF9 AML disease burden evolution after VCP knock-down.
Fig. S2. CB-5083 tolerability profile in mice.
Fig. S3. VCP inhibition does not primarily impair AML cell growth through induction of proteotoxic/ER stress.
Fig. S4. Comparative proteomic analysis identifies VCP-interacting proteins in an AML context.
Fig. S5. CB-5339 impairs DNA repair and induces proteotoxic stress in AML.
Fig. S6. CB-5339 pharmacokinetic properties, tolerability profile and post-treatment relapse characteristics.
ACKNOWLEDGEMENTS
We thank Timur Yusufzai, Raphael Ceccaldi, Sebastian Oeck, Benjamin Manning, Neil Umbreit and Peter Bruno for advice and discussions. We also thank David Wustrow for his insightful contribution to VCP inhibitors molecular structure evolution, selection and screening. We are indebted to Veronique Montcuquet, Nicolas Setterblad, Christelle Doliger, and Sophie Duchez from the Saint-Louis Research Institute Core Facility for their technical support.
FUNDING: This research was supported with grants from the US National Cancer Institute (NCI) (NIH R35 CA210030 to KS, and NIH K08 CA222684 to YP), the Children’s Leukemia Research Foundation (CLRF) (to KS), the Bettencourt Schueller foundation (CCA-INSERM-Bettencourt to LB) and the Bridge Project, a collaboration between the Koch Institute for Integrative Cancer Research at MIT and the Dana-Farber–Harvard Cancer Center (DF-HCC) (to KS and MTH). KS was a Leukemia and Lymphoma Society Scholar. AP is a recipient of support from the starting grant form ERC (Horizon 2020), the ATIP–AVENIR and LNCC French research programs, the EHA research grant for a non-clinical advanced fellow, the FSER foundation grant, the Emergence ville de Paris grant and is supported by the St. Louis Association for leukemia research. LB is a CCA-INSERM-Bettencourt laureate and a recipient of Philippe Foundation and GPM fellowships, and research funding from Association Laurette Fugain, LNCC (Comité de Paris) and Gilead Sciences as a “Gilead Research Scholars award” recipient. JME is a recipient of a Swiss National Science Foundation, a Lady Tata Memorial Trust and a Pediatric Cancer Research Foundation fellowships.
COMPETING INTERESTS: JDV is an employee of Cleave Therapeutics and YT, TW, DA, RLM and HJZ are former employees of Cleave Therapeutics, a biopharmaceutical company developing VCP inhibitors for therapeutic use in oncology. KS has consulted for Rigel Pharmaceuticals, Kronos Bio, and Auron Therapeutics on unrelated topics, receives grant funding from Novartis which did not fund this project, and holds stock options with Auron Therapeutics. LB receives grant support from Gilead Sciences as a “Gilead Research Scholars award recipient”. The other authors declare no competing interests. The following patents are held by Cleave Therapeutics (formerly Cleave Biosciences): (U.S. Patent Nos.: 9,828,363, B2 fused pyrimidines as inhibitors of p97 complex and 10,174,005, B2 fused pyrimidines as inhibitors of p97 complex); HJZ is a listed inventor on these patents. Cleave Therapeutics has filed an additional provisional patent application concerning CB-5339 and its uses (application No: 63/114,435, VCP/p97 inhibitor for the treatment of cancer). JDV, DJA, RLM and HJZ are listed as inventors on the provisional patent.
Footnotes
DATA AND MATERIALS AVAILABILITY: All data associated with this study are available in the paper or the supplementary materials. CB-5339 can be obtained by request to Cleave Therapeutics via materials transfer agreement (contact email: jadvargas@cleavetherapeutics.com).
REFERENCES
- 1.Stone RM, Mandrekar SJ, Sanford BL, Laumann K, Geyer S, Bloomfield CD, Thiede C, Prior TW, Dohner K, Marcucci G, Lo-Coco F, Klisovic RB, Wei A, Sierra J, Sanz MA, Brandwein JM, de Witte T, Niederwieser D, Appelbaum FR, Medeiros BC, Tallman MS, Krauter J, Schlenk RF, Ganser A, Serve H, Ehninger G, Amadori S, Larson RA, Dohner H, Midostaurin plus Chemotherapy for Acute Myeloid Leukemia with a FLT3 Mutation. N Engl J Med 377, 454–464 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DiNardo CD, Stein EM, de Botton S, Roboz GJ, Altman JK, Mims AS, Swords R, Collins RH, Mannis GN, Pollyea DA, Donnellan W, Fathi AT, Pigneux A, Erba HP, Prince GT, Stein AS, Uy GL, Foran JM, Traer E, Stuart RK, Arellano ML, Slack JL, Sekeres MA, Willekens C, Choe S, Wang H, Zhang V, Yen KE, Kapsalis SM, Yang H, Dai D, Fan B, Goldwasser M, Liu H, Agresta S, Wu B, Attar EC, Tallman MS, Stone RM, Kantarjian HM, Durable Remissions with Ivosidenib in IDH1-Mutated Relapsed or Refractory AML. N Engl J Med 378, 2386–2398 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Stein EM, DiNardo CD, Pollyea DA, Fathi AT, Roboz GJ, Altman JK, Stone RM, DeAngelo DJ, Levine RL, Flinn IW, Kantarjian HM, Collins R, Patel MR, Frankel AE, Stein A, Sekeres MA, Swords RT, Medeiros BC, Willekens C, Vyas P, Tosolini A, Xu Q, Knight RD, Yen KE, Agresta S, de Botton S, Tallman MS, Enasidenib in mutant IDH2 relapsed or refractory acute myeloid leukemia. Blood 130, 722–731 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ellis LM, Hicklin DJ, Resistance to Targeted Therapies: Refining Anticancer Therapy in the Era of Molecular Oncology. Clin Cancer Res 15, 7471–7478 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Luo J, Solimini NL, Elledge SJ, Principles of cancer therapy: oncogene and non-oncogene addiction. Cell 136, 823–837 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, Taylor MJ, Johns C, Chicas A, Mulloy JC, Kogan SC, Brown P, Valent P, Bradner JE, Lowe SW, Vakoc CR, RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 478, 524–528 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greaves M, Evolutionary determinants of cancer. Cancer Discov 5, 806–820 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lagadinou ED, Sach A, Callahan K, Rossi RM, Neering SJ, Minhajuddin M, Ashton JM, Pei S, Grose V, O’Dwyer KM, Liesveld JL, Brookes PS, Becker MW, Jordan CT, BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell Stem Cell 12, 329–341 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Konopleva M, Pollyea DA, Potluri J, Chyla B, Hogdal L, Busman T, McKeegan E, Salem AH, Zhu M, Ricker JL, Blum W, DiNardo CD, Kadia T, Dunbar M, Kirby R, Falotico N, Leverson J, Humerickhouse R, Mabry M, Stone R, Kantarjian H, Letai A, Efficacy and Biological Correlates of Response in a Phase II Study of Venetoclax Monotherapy in Patients with Acute Myelogenous Leukemia. Cancer Discov 6, 1106–1117 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Puissant A, Fenouille N, Alexe G, Pikman Y, Bassil CF, Mehta S, Du J, Kazi JU, Luciano F, Ronnstrand L, Kung AL, Aster JC, Galinsky I, Stone RM, DeAngelo DJ, Hemann MT, Stegmaier K, SYK is a critical regulator of FLT3 in acute myeloid leukemia. Cancer Cell 25, 226–242 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Q, Song C, Li CC, Hexamerization of p97-VCP is promoted by ATP binding to the D1 domain and required for ATPase and biological activities. Biochem Biophys Res Commun 300, 253–260 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Dalal S, Rosser MF, Cyr DM, Hanson PI, Distinct roles for the AAA ATPases NSF and p97 in the secretory pathway. Mol Biol Cell 15, 637–648 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson DJ, Le Moigne R, Djakovic S, Kumar B, Rice J, Wong S, Wang J, Yao B, Valle E, Kiss von Soly S, Madriaga A, Soriano F, Menon MK, Wu ZY, Kampmann M, Chen Y, Weissman JS, Aftab BT, Yakes FM, Shawver L, Zhou HJ, Wustrow D, Rolfe M, Targeting the AAA ATPase p97 as an Approach to Treat Cancer through Disruption of Protein Homeostasis. Cancer Cell 28, 653–665 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou HJ, Wang J, Yao B, Wong S, Djakovic S, Kumar B, Rice J, Valle E, Soriano F, Menon MK, Madriaga A, Kiss von Soly S, Kumar A, Parlati F, Yakes FM, Shawver L, Le Moigne R, Anderson DJ, Rolfe M, Wustrow D, Discovery of a First-in-Class, Potent, Selective, and Orally Bioavailable Inhibitor of the p97 AAA ATPase (CB-5083). J Med Chem 58, 9480–9497 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Magnaghi P, D’Alessio R, Valsasina B, Avanzi N, Rizzi S, Asa D, Gasparri F, Cozzi L, Cucchi U, Orrenius C, Polucci P, Ballinari D, Perrera C, Leone A, Cervi G, Casale E, Xiao Y, Wong C, Anderson DJ, Galvani A, Donati D, O’Brien T, Jackson PK, Isacchi A, Covalent and allosteric inhibitors of the ATPase VCP/p97 induce cancer cell death. Nat Chem Biol 9, 548–556 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Her NG, Toth JI, Ma CT, Wei Y, Motamedchaboki K, Sergienko E, Petroski MD, p97 Composition Changes Caused by Allosteric Inhibition Are Suppressed by an On-Target Mechanism that Increases the Enzyme’s ATPase Activity. Cell Chem Biol 23, 517–528 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wojcik C, Rowicka M, Kudlicki A, Nowis D, McConnell E, Kujawa M, DeMartino GN, Valosin-containing protein (p97) is a regulator of endoplasmic reticulum stress and of the degradation of N-end rule and ubiquitin-fusion degradation pathway substrates in mammalian cells. Mol Biol Cell 17, 4606–4618 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dai RM, Li CC, Valosin-containing protein is a multi-ubiquitin chain-targeting factor required in ubiquitin-proteasome degradation. Nat Cell Biol 3, 740–744 (2001). [DOI] [PubMed] [Google Scholar]
- 19.Raman M, Sergeev M, Garnaas M, Lydeard JR, Huttlin EL, Goessling W, Shah JV, Harper JW, Systematic proteomics of the VCP-UBXD adaptor network identifies a role for UBXN10 in regulating ciliogenesis. Nat Cell Biol 17, 1356–1369 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shiloh Y, Ziv Y, The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol 14, 197–210 (2013). [PubMed] [Google Scholar]
- 21.Arshavsky VY, Lamb TD, Pugh EN Jr., G proteins and phototransduction. Annu Rev Physiol 64, 153–187 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Andersson KE, PDE5 inhibitors - pharmacology and clinical applications 20 years after sildenafil discovery. Br J Pharmacol 175, 2554–2565 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, Reddy A, Liu M, Murray L, Berger MF, Monahan JE, Morais P, Meltzer J, Korejwa A, Jane-Valbuena J, Mapa FA, Thibault J, Bric-Furlong E, Raman P, Shipway A, Engels IH, Cheng J, Yu GK, Yu J, Aspesi P Jr., de Silva M, Jagtap K, Jones MD, Wang L, Hatton C, Palescandolo E, Gupta S, Mahan S, Sougnez C, Onofrio RC, Liefeld T, MacConaill L, Winckler W, Reich M, Li N, Mesirov JP, Gabriel SB, Getz G, Ardlie K, Chan V, Myer VE, Weber BL, Porter J, Warmuth M, Finan P, Harris JL, Meyerson M, Golub TR, Morrissey MP, Sellers WR, Schlegel R, Garraway LA, The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.C. Cancer Cell Line Encyclopedia, C. Genomics of Drug Sensitivity in Cancer, Pharmacogenomic agreement between two cancer cell line data sets. Nature 528, 84–87 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bailey MH, Tokheim C, Porta-Pardo E, Sengupta S, Bertrand D, Weerasinghe A, Colaprico A, Wendl MC, Kim J, Reardon B, Ng PK, Jeong KJ, Cao S, Wang Z, Gao J, Gao Q, Wang F, Liu EM, Mularoni L, Rubio-Perez C, Nagarajan N, Cortes-Ciriano I, Zhou DC, Liang WW, Hess JM, Yellapantula VD, Tamborero D, Gonzalez-Perez A, Suphavilai C, Ko JY, Khurana E, Park PJ, Van Allen EM, Liang H, M. C. W. Group, N. Cancer Genome Atlas Research, Lawrence MS, Godzik A, Lopez-Bigas N, Stuart J, Wheeler D, Getz G, Chen K, Lazar AJ, Mills GB, Karchin R, Ding L, Comprehensive Characterization of Cancer Driver Genes and Mutations. Cell 173, 371–385 e318 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knijnenburg TA, Wang L, Zimmermann MT, Chambwe N, Gao GF, Cherniack AD, Fan H, Shen H, Way GP, Greene CS, Liu Y, Akbani R, Feng B, Donehower LA, Miller C, Shen Y, Karimi M, Chen H, Kim P, Jia P, Shinbrot E, Zhang S, Liu J, Hu H, Bailey MH, Yau C, Wolf D, Zhao Z, Weinstein JN, Li L, Ding L, Mills GB, Laird PW, Wheeler DA, Shmulevich I, N. Cancer Genome Atlas Research, Monnat RJ Jr., Xiao Y, Wang C, Genomic and Molecular Landscape of DNA Damage Repair Deficiency across The Cancer Genome Atlas. Cell Rep 23, 239–254 e236 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Way GP, Sanchez-Vega F, La K, Armenia J, Chatila WK, Luna A, Sander C, Cherniack AD, Mina M, Ciriello G, Schultz N, N. Cancer Genome Atlas Research, Sanchez Y, Greene CS, Machine Learning Detects Pan-cancer Ras Pathway Activation in The Cancer Genome Atlas. Cell Rep 23, 172–180 e173 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Moigne R, Aftab BT, Djakovic S, Dhimolea E, Valle E, Murnane M, King EM, Soriano F, Menon MK, Wu ZY, Wong ST, Lee GJ, Yao B, Wiita AP, Lam C, Rice J, Wang J, Chesi M, Bergsagel PL, Kraus M, Driessen C, Kiss von Soly S, Yakes FM, Wustrow D, Shawver L, Zhou HJ, Martin TG 3rd, Wolf JL, Mitsiades CS, Anderson DJ, Rolfe M, The p97 Inhibitor CB-5083 Is a Unique Disrupter of Protein Homeostasis in Models of Multiple Myeloma. Mol Cancer Ther 16, 2375–2386 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Negrini S, Gorgoulis VG, Halazonetis TD, Genomic instability--an evolving hallmark of cancer. Nat Rev Mol Cell Biol 11, 220–228 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Gibbs-Seymour I, Oka Y, Rajendra E, Weinert BT, Passmore LA, Patel KJ, Olsen JV, Choudhary C, Bekker-Jensen S, Mailand N, Ubiquitin-SUMO circuitry controls activated fanconi anemia ID complex dosage in response to DNA damage. Mol Cell 57, 150–164 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vaz B, Halder S, Ramadan K, Role of p97/VCP (Cdc48) in genome stability. Front Genet 4, 60 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bergink S, Ammon T, Kern M, Schermelleh L, Leonhardt H, Jentsch S, Role of Cdc48/p97 as a SUMO-targeted segregase curbing Rad51-Rad52 interaction. Nat Cell Biol 15, 526–532 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Acs K, Luijsterburg MS, Ackermann L, Salomons FA, Hoppe T, Dantuma NP, The AAA-ATPase VCP/p97 promotes 53BP1 recruitment by removing L3MBTL1 from DNA double-strand breaks. Nat Struct Mol Biol 18, 1345–1350 (2011). [DOI] [PubMed] [Google Scholar]
- 34.van den Boom J, Wolf M, Weimann L, Schulze N, Li F, Kaschani F, Riemer A, Zierhut C, Kaiser M, Iliakis G, Funabiki H, Meyer H, VCP/p97 Extracts Sterically Trapped Ku70/80 Rings from DNA in Double-Strand Break Repair. Mol Cell 64, 189–198 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davis EJ, Lachaud C, Appleton P, Macartney TJ, Nathke I, Rouse J, DVC1 (C1orf124) recruits the p97 protein segregase to sites of DNA damage. Nat Struct Mol Biol 19, 1093–1100 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O’Connor MJ, Ashworth A, Carmichael J, Kaye SB, Schellens JH, de Bono JS, Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med 361, 123–134 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Esposito MT, So CW, DNA damage accumulation and repair defects in acute myeloid leukemia: implications for pathogenesis, disease progression, and chemotherapy resistance. Chromosoma 123, 545–561 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Esposito MT, Zhao L, Fung TK, Rane JK, Wilson A, Martin N, Gil J, Leung AY, Ashworth A, So CW, Synthetic lethal targeting of oncogenic transcription factors in acute leukemia by PARP inhibitors. Nat Med 21, 1481–1490 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Inoue S, Li WY, Tseng A, Beerman I, Elia AJ, Bendall SC, Lemonnier F, Kron KJ, Cescon DW, Hao Z, Lind EF, Takayama N, Planello AC, Shen SY, Shih AH, Larsen DM, Li Q, Snow BE, Wakeham A, Haight J, Gorrini C, Bassi C, Thu KL, Murakami K, Elford AR, Ueda T, Straley K, Yen KE, Melino G, Cimmino L, Aifantis I, Levine RL, De Carvalho DD, Lupien M, Rossi DJ, Nolan GP, Cairns RA, Mak TW, Mutant IDH1 Downregulates ATM and Alters DNA Repair and Sensitivity to DNA Damage Independent of TET2. Cancer Cell 30, 337–348 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morgado-Palacin I, Day A, Murga M, Lafarga V, Anton ME, Tubbs A, Chen HT, Ergan A, Anderson R, Bhandoola A, Pike KG, Barlaam B, Cadogan E, Wang X, Pierce AJ, Hubbard C, Armstrong SA, Nussenzweig A, Fernandez-Capetillo O, Targeting the kinase activities of ATR and ATM exhibits antitumoral activity in mouse models of MLL-rearranged AML. Sci Signal 9, ra91 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muller JM, Deinhardt K, Rosewell I, Warren G, Shima DT, Targeted deletion of p97 (VCP/CDC48) in mouse results in early embryonic lethality. Biochem Biophys Res Commun 354, 459–465 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Curtin NJ, DNA repair dysregulation from cancer driver to therapeutic target. Nat Rev Cancer 12, 801–817 (2012). [DOI] [PubMed] [Google Scholar]
- 43.Dobbelstein M, Sorensen CS, Exploiting replicative stress to treat cancer. Nat Rev Drug Discov 14, 405–423 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Libura J, Slater DJ, Felix CA, Richardson C, Therapy-related acute myeloid leukemia-like MLL rearrangements are induced by etoposide in primary human CD34+ cells and remain stable after clonal expansion. Blood 105, 2124–2131 (2005). [DOI] [PubMed] [Google Scholar]
- 45.David L, Fernandez-Vidal A, Bertoli S, Grgurevic S, Lepage B, Deshaies D, Prade N, Cartel M, Larrue C, Sarry JE, Delabesse E, Cazaux C, Didier C, Recher C, Manenti S, Hoffmann JS, CHK1 as a therapeutic target to bypass chemoresistance in AML. Sci Signal 9, ra90 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Kotsantis P, Petermann E, Boulton SJ, Mechanisms of Oncogene-Induced Replication Stress: Jigsaw Falling into Place. Cancer Discov 8, 537–555 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grabocka E, Pylayeva-Gupta Y, Jones MJ, Lubkov V, Yemanaberhan E, Taylor L, Jeng HH, Bar-Sagi D, Wild-type H- and N-Ras promote mutant K-Ras-driven tumorigenesis by modulating the DNA damage response. Cancer Cell 25, 243–256 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tu Z, Aird KM, Bitler BG, Nicodemus JP, Beeharry N, Xia B, Yen TJ, Zhang R, Oncogenic RAS regulates BRIP1 expression to induce dissociation of BRCA1 from chromatin, inhibit DNA repair, and promote senescence. Dev Cell 21, 1077–1091 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hahnel PS, Enders B, Sasca D, Roos WP, Kaina B, Bullinger L, Theobald M, Kindler T, Targeting components of the alternative NHEJ pathway sensitizes KRAS mutant leukemic cells to chemotherapy. Blood 123, 2355–2366 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Jacoby MA, De Jesus Pizarro RE, Shao J, Koboldt DC, Fulton RS, Zhou G, Wilson RK, Walter MJ, The DNA double-strand break response is abnormal in myeloblasts from patients with therapy-related acute myeloid leukemia. Leukemia 28, 1242–1251 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ferbeyre G, de Stanchina E, Lin AW, Querido E, McCurrach ME, Hannon GJ, Lowe SW, Oncogenic ras and p53 cooperate to induce cellular senescence. Mol Cell Biol 22, 3497–3508 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW, Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88, 593–602 (1997). [DOI] [PubMed] [Google Scholar]
- 53.Zhang J, Kong G, Rajagopalan A, Lu L, Song J, Hussaini M, Zhang X, Ranheim EA, Liu Y, Wang J, Gao X, Chang YI, Johnson KD, Zhou Y, Yang D, Bhatnagar B, Lucas DM, Bresnick EH, Zhong X, Padron E, Zhang J, p53−/− synergizes with enhanced NrasG12D signaling to transform megakaryocyte-erythroid progenitors in acute myeloid leukemia. Blood 129, 358–370 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Munos B, Lessons from 60 years of pharmaceutical innovation. Nat Rev Drug Discov 8, 959–968 (2009). [DOI] [PubMed] [Google Scholar]
- 55.Chou TF, Brown SJ, Minond D, Nordin BE, Li K, Jones AC, Chase P, Porubsky PR, Stoltz BM, Schoenen FJ, Patricelli MP, Hodder P, Rosen H, Deshaies RJ, Reversible inhibitor of p97, DBeQ, impairs both ubiquitin-dependent and autophagic protein clearance pathways. Proc Natl Acad Sci U S A 108, 4834–4839 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pohler R, Krahn JH, van den Boom J, Dobrynin G, Kaschani F, Eggenweiler HM, Zenke FT, Kaiser M, Meyer H, A Non-Competitive Inhibitor of VCP/p97 and VPS4 Reveals Conserved Allosteric Circuits in Type I and II AAA ATPases. Angew Chem Int Ed Engl 57, 1576–1580 (2018). [DOI] [PubMed] [Google Scholar]
- 57.Kakizuka A, Roles of VCP in human neurodegenerative disorders. Biochem Soc Trans 36, 105–108 (2008). [DOI] [PubMed] [Google Scholar]
- 58.Dai Z, Sheridan JM, Gearing LJ, Moore DL, Su S, Wormald S, Wilcox S, O’Connor L, Dickins RA, Blewitt ME, Ritchie ME, edgeR: a versatile tool for the analysis of shRNA-seq and CRISPR-Cas9 genetic screens. F1000Res 3, 95 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Robinson MD, McCarthy DJ, Smyth GK, edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Luo B, Cheung HW, Subramanian A, Sharifnia T, Okamoto M, Yang X, Hinkle G, Boehm JS, Beroukhim R, Weir BA, Mermel C, Barbie DA, Awad T, Zhou X, Nguyen T, Piqani B, Li C, Golub TR, Meyerson M, Hacohen N, Hahn WC, Lander ES, Sabatini DM, Root DE, Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci U S A 105, 20380–20385 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fellmann C, Hoffmann T, Sridhar V, Hopfgartner B, Muhar M, Roth M, Lai DY, Barbosa IA, Kwon JS, Guan Y, Sinha N, Zuber J, An optimized microRNA backbone for effective single-copy RNAi. Cell Rep 5, 1704–1713 (2013). [DOI] [PubMed] [Google Scholar]
- 62.Banerji V, Frumm SM, Ross KN, Li LS, Schinzel AC, Hahn CK, Kakoza RM, Chow KT, Ross L, Alexe G, Tolliday N, Inguilizian H, Galinsky I, Stone RM, DeAngelo DJ, Roti G, Aster JC, Hahn WC, Kung AL, Stegmaier K, The intersection of genetic and chemical genomic screens identifies GSK-3alpha as a target in human acute myeloid leukemia. J Clin Invest 122, 935–947 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rappsilber J, Mann M, Ishihama Y, Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc 2, 1896–1906 (2007). [DOI] [PubMed] [Google Scholar]
- 64.Udeshi ND, Mani DR, Eisenhaure T, Mertins P, Jaffe JD, Clauser KR, Hacohen N, Carr SA, Methods for quantification of in vivo changes in protein ubiquitination following proteasome and deubiquitinase inhibition. Mol Cell Proteomics 11, 148–159 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wickham H, ggplot2: Elegant Graphics for Data Analysis.. (Springer-Verlag; New York., 2009). [Google Scholar]
- 66.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP, Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma’ayan A, Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, Monteiro CD, Gundersen GW, Ma’ayan A, Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44, W90–97 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Muslimovic A, Ismail IH, Gao Y, Hammarsten O, An optimized method for measurement of gamma-H2AX in blood mononuclear and cultured cells. Nat Protoc 3, 1187–1193 (2008). [DOI] [PubMed] [Google Scholar]
- 70.Pierce AJ, Johnson RD, Thompson LH, Jasin M, XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev 13, 2633–2638 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guirouilh-Barbat J, Huck S, Bertrand P, Pirzio L, Desmaze C, Sabatier L, Lopez BS, Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Mol Cell 14, 611–623 (2004). [DOI] [PubMed] [Google Scholar]
- 72.Chou TC, Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res 70, 440–446 (2010). [DOI] [PubMed] [Google Scholar]
- 73.Chou TC, Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev 58, 621–681 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. List of the top depleted genes identified by the in vivo shRNA screening.
Table S2. CB-5083 sensitivity profile across 131 cancer cell lines.
Table S3. Patient characteristics.
Table S4. List of VCP-interacting partners identified by mass spectrometry-based interactome analysis in the MV4–11 AML cell line.
Table S5. List of enriched pathways among VCP interactome.
Table S6. List of VCP-interacting partners identified by mass spectrometry-based interactome analysis in the MV4–11 AML cell line compared to HEK-293T cell line.
Table S7. RAS and TP53 mutational profiles of AML cell lines according to the CCLE database.
Table S8. Mutational and functional co-occurrence of RAS activation and TP53 deficiency in pan-cancer TCGA cohorts.
Table S9: List of antibodies used for western blotting, immunoflourescence and flow cytometry.
Fig. S1. MLL-AF9 AML disease burden evolution after VCP knock-down.
Fig. S2. CB-5083 tolerability profile in mice.
Fig. S3. VCP inhibition does not primarily impair AML cell growth through induction of proteotoxic/ER stress.
Fig. S4. Comparative proteomic analysis identifies VCP-interacting proteins in an AML context.
Fig. S5. CB-5339 impairs DNA repair and induces proteotoxic stress in AML.
Fig. S6. CB-5339 pharmacokinetic properties, tolerability profile and post-treatment relapse characteristics.