Abstract
Tomato heat stress transcription factor HsfA2 is a shuttling protein with dominant cytoplasmic localization as a result of a nuclear import combined with an efficient export. Besides the nuclear localization signal (NLS) adjacent to the oligomerization domain, a C-terminal leucine-rich motif functions as a nuclear export signal (NES). Mutant forms of HsfA2 with a defective or an absent NES are nuclear proteins. The same is true for the wild-type HsfA2 if coexpressed with HsfA1 or in the presence of export inhibitor leptomycin B (LMB). Fusion of the NES domain of HsfA2 to HsfB1, which is a nuclear protein, caused export of the HsfB1-A2NES hybrid protein, and this effect was reversed by the addition of LMB. Due to the lack of background problems, Chinese hamster ovary (CHO) cells represent an excellent system for expression and functional analysis of tomato Hsfs. The results faithfully reflect the situation found in plant cells (tobacco protoplasts). The intriguing role of NLS and NES accessibility for the intracellular distribution of HsfA2 is underlined by the results of heat stress treatments of CHO cells (41°C). Despite the fact that nuclear import and export are not markedly affected, HsfA2 remains completely cytoplasmic at 41°C even in the presence of LMB. The temperature-dependent conformational transition of HsfA2 with shielding of the NLS evidently needs intramolecular interaction between the internal HR-A/B and the C-terminal HR-C regions. It is not observed with the HR oligomerization domain (HR-A/B region) deletion form of HsfA2 or in HsfA2-HsfA1 hetero-oligomers.
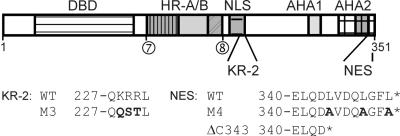
Key regulators of the heat stress (HS) response are the HS transcription factors (Hsfs), which belong to a family of proteins conserved throughout the eukaryotic kingdom (24, 26, 35, 46). Hsfs have a modular structure with an N-terminal DNA-binding domain characterized by a helix-turn-helix motif, an adjacent domain with heptad hydrophobic repeats (HR-A/B) involved in oligomerization, a cluster of basic amino acid residues essential for nuclear import (the nuclear localization signal, or NLS) and a C-terminal activation domain (Fig. 1).
FIG. 1.
Block diagram with functional motifs of HsfA2. The positions of the functional domains and/or motifs mentioned in the text are indicated (from N to C terminus): DNA-binding domain (DBD), oligomerization domain (HR-A/B), the bipartite NLS, activator motifs (AHA1 and AHA2), and the C-terminal heptad hydrophobic repeat region (cross-hatched), including the NES. Sequence details and amino acid exchanges in mutant forms of HsfA2 are given for the NLS and NES, respectively. Deletion points 7 (aa 169 to 170) and 8 (aa 213 to 214) mark the portion of the protein lacking in HsfA2Δ7/8. Sequence details for the NLS mutant M3 and the NES mutant M4 as well as the NES deletion mutant HsfA2ΔC343 are indicated below.
The high degree of structural and functional conservation of Hsfs was documented repeatedly by using heterologous systems for Hsf expression in combination with appropriate reporter assays. Thus, Drosophila melanogaster and human Hsfs were tested in plant cells, Xenopus oocytes, and Saccharomyces cerevisiae (5, 6, 22, 43, 50), and plant Hsfs were tested in yeast, Drosophila, and human cells (4, 5, 8, 17). Using yeast strains with disruption of the endogenous Hsf1 gene, it was shown that many of these heterologous Hsfs were able to replace the yeast Hsf1 in most of its functions, i.e., in Hsf-dependent reporter assays, in the survival function both at 25 and 37°C, and in the generation of the thermotolerant state (5, 12, 22, 48).
In plants, the Hsf system is more complex than in any other organisms investigated so far (26, 28, 35). (i) Besides the constitutively expressed members of the Hsf family, many Hsfs themselves are HS-inducible proteins. (ii) Two classes of Hsfs (class A versus class B) are discriminated by a 21-amino-acid (21-aa) insertion found in the oligomerization domain of class A Hsfs. This insertion is lacking not only in the plant class B Hsfs but also in all Hsfs from other organisms. In addition, the CTAD of class A Hsfs is acidic with two or more short peptide motifs (AHA motifs), which are essential for the activator function (4, 8). In contrast to this, the CTAD of class B Hsfs is neutral or basic, and there is evidence for clear functional differences between class A and class B Hsfs (5, 7).
Tomato HsfA2, a strongly HS-inducible protein, has two remarkable properties. (i) Despite a functional NLS, it does not localize in the nucleus unless coexpressed with the constitutively expressed HsfA1. Evidently, the formation of hetero-oligomers between both Hsfs markedly influences the intracellular distribution and thus activator function of HsfA2 (36). (ii) The ongoing accumulation of HsfA2 and other HS-inducible proteins in the course of an HS period results in a unique storage form in cytoplasmic chaperone complexes composed of 40-nm particles identified earlier as HS granules (HSG) (27). No other Hsf so far identified in tomato plant cells (HsfA1, HsfA3, and HsfB1) is found in the HSG complexes (4, 36).
To investigate the question of whether the nuclear cotransport phenomenon is the result of intrinsic properties of the Hsfs involved or whether other plant proteins are needed, we expressed tomato Hsfs A1 and A2 alone and together in Chinese hamster ovary (CHO) cells and tobacco mesophyll protoplasts. As shown in this paper, the mammalian expression system has great advantages when studying protein interactions contributing to the dynamically changing intracellular distribution of tomato Hsfs. By the application of leptomycin B (LMB) as a covalent inhibitor of the nuclear export receptor exportin 1 (10, 12, 19, 20), we noticed that the dominant cytoplasmic localization of HsfA2 is in fact the result of a strong C-terminal nuclear export signal (NES) combined with a weak or inaccessible NLS. Depending on the expression conditions, HS markedly influences the intracellular distribution of HsfA2 in the presence of LMB.
MATERIALS AND METHODS
General materials and methods.
The use of tobacco (Nicotiana plumbaginifolia) mesophyll protoplasts for the transient expression of Hsfs was described previously (23, 36). After polyethylene glycol-mediated transformation with the indicated expression plasmids, protoplasts were incubated for 15 h at 25°C under dim light. Protoplasts were processed for immunofluorescence as previously described (18, 36).
The rabbit antisera against tomato HsfA1, HsfA2, and HsfB1 were described before (23, 36). Secondary antibodies against rabbit immunoglobulins conjugated with fluorescent dyes CY2 or CY3 were obtained from Dianova (Hamburg, Germany). Fluorescein isothiocyanate (FITC)-phalloidin for actin staining was from Sigma-Aldrich Chemie GmbH (Deisenhofen, Germany).
LMB was kindly provided by Minoru Yoshida, Tokyo, Japan, and used at a final concentration of 2 to 20 ng/ml.
Culture and transfection of CHO-K1 cells.
CHO-K1 cells were maintained in Ham's F12 nutrient mixture supplemented with 10% fetal bovine serum and penicillin-streptomycin (all from Life Technologies, Eggenstein, Germany). Cells were grown in 25-cm2 culture flasks (Nunc, Wiesbaden, Germany) at 37°C under a 5% CO2 atmosphere. Twenty-four hours before transfection, cells were seeded on chamber slides (Nunc). DNA for transfection was prepared using the Plasmid Midi kit (Qiagen, Hilden, Germany). Transfection was performed according to the manufacturer's protocol using the FUGENE 6 Transfection kit (Roche Diagnostics, Mannheim, Germany). Cells were harvested 24 to 36 h after transfection. For luciferase assays, cells were cultured in 24-well plates (Nunc) and transfected as described above.
Protein analysis and Western blotting.
About 2 × 105 CHO cells were washed twice with phosphate-buffered saline (PBS) buffer (137 mM NaCl, 10 mM Na2HPO4, 1.7 mM KH2PO4, 2.6 mM KCl [pH 7.4]) and lysed in 50 μl of hot sodium dodecylsulfate sample buffer. Protein analysis by sodium dodecylsulfate-polyacrylamide gel electrophoresis and Western blotting was performed as described previously (8, 36).
Luciferase reporter assay.
Thirty-six hours after transfection, cells were washed two times in PBS and lysed in 70 μl of cell culture lysis reagent (100 mM potassium phosphate [pH 7.8], 1 mM EDTA, 7mM 2-mercaptoethanol, 1% Triton X-100, 10% glycerol). For all experiments, four cell samples were transformed and processed independently. Experiments were repeated at least three times. Cell lysates were stored at −80°C until usage. Luciferase activity was measured with the luciferase assay reagent (20mM Tricine [pH 7.8], 5 mM MgCl2, 0.1 mM EDTA, 3.3 mM dithiothreitol, 270 μM coenzyme A, 50 μM luciferin, 500 μM ATP) using a Mikrolumat LB 96P luminometer (Eg&G Berthold, Bad Wildbad, Germany). Two seconds after injection of the substrate cocktail, light emission was measured for 30 s. Hsf expression was monitored by Western analysis.
Expression plasmids for plant and mammalian cells.
Standard procedures were used for cloning (2, 34). PCR was done using the Taq Plus Precision system (Stratagene, Amsterdam, The Netherlands). Plant expression vectors are based on the pRT series of vectors (42). Except for three plasmids given below, details of the constructions were described before (5, 23, 43). The NES mutant of HsfA2 (HsfA2M4) was generated by PCR mutagenesis of a C-terminal fragment of HsfA2 with primers 1 and 2, followed by insertion of a BglII/XbaI fragment into pRTHsfA2ΔC343 (22). For construction of the vector encoding the HsfB1-A2NES fusion protein (HsfB1aa1-296xHsfA2aa329-351), the C-terminal part of pRTHsfA2 was amplified by PCR using primers 1 and 3, and a BglII/XbaI fragment was inserted into pRTHsfB1. In the encoded fusion protein, the last 5-aa residues of HsfB1 were replaced by the last 23-aa residues of HsfA2. For generation of the new vector encoding the green fluorescent protein (GFP)-tagged HsfA1 (pCKHsfA1-GFP), the HsfA1 cassette of pRTHsfA1 (43) was amplified by PCR using primers 6 and 7, and the corresponding SacI/XbaI fragment was inserted into pCK-GFP S65C (31).
All mammalian expression vectors were constructed by subcloning the whole Hsf coding regions as XhoI/XbaI fragments from the pRT vectors into pcDNA3 (Invitrogen, Groningen, The Netherlands). The HsfA1-GFP expression plasmid was obtained by subcloning an EcoRl/XbaI fragment from pCK-HsfA1-GFP into pcDNA3.
For construction of the hsp17xluc reporter plasmid for the Hsf-dependent luciferase reporter assays in CHO cells, a promoter-leader fragment of the plant plasmid phsp17xgus (43) was amplified by PCR using primers 4 and 5. It was inserted as an Asp718I/SmaI fragment in front of the luciferase gene of the pGL3-basic vector (Promega, Mannheim, Germany). The soybean hsp17 promoter part of the inserted fragment (nucleotides −321 to −12) (38) is evidently decisive for the remarkably low level of basal luciferase expression in mammalian cells.
Primers.
Primers (F, forward; R, reverse; restriction sites are underlined) are as follows: primer 1 (HsfA2F), 5′AATCAGATCATTGCCATGGGAGAAAAAATCGAAACACAGGAGAGG; primer 2 [HsfA2R(M4) and XbaI], 5′CTTAATGTTCTGCGACATCTAGTTCGACCAAAGCGAATCAGATCTGAGAACACA; primer 3 (HsfA2R and XbaI), 5′GTTGAACCAAAGGAAATCTCAGATCTCGCGCGCG; primer 4 (GusR), 5′TTCGCGATCCAGACTGAATGCC; primer 5 (Hsp17F and Asp718), 5′GGCCTGGTACCCCAATAATAACC; primer 6 (HsfA1-GFP-F and SacI), 5′GAGCTGAGCTCTTACAGCCGGCGC; and primer 7 (HsfA1-GFP-R and XbaI), 5′GAATAGGGCCCTCTAGAAACTACC.
Indirect immunofluorescence of CHO cells.
After being briefly washed in PBS containing 1 mM MgCl2 and 1 mM CaCl2 CHO-K1 cells on chamber slides were fixed for 30 min with freshly prepared 3.7% paraformaldehyde in microtubule-stabilizing buffer (MTSB) (100 mM PIPES [piperazine-N,N′9-bis(2-ethanesulfonic acid] [pH 6.9], 1 mM MgSO4, 2 mM EGTA). After being washed in MTSB, cells were permeabilized by a 15-min treatment with 0.5% Triton X-100 in MTSB, and residual aldehyde groups were blocked by 15-min incubation in PBS-100 mM glycine. After 30-min blocking in PBS–1% bovine serum albumin (Sigma-Aldrich Chemie GmbH), cells were incubated for 3 h with the indicated antisera diluted 1:500 in PBS–1% bovine serum albumin. After being washed twice with MTSB and stained with fluorochrome-coupled goat anti-rabbit secondary antibodies, samples were mounted for microscopic inspection in mounting solution (100 mM Tris-HCl [pH 8.5], 24% glycerol, 9.6% mowiol 4-88) (Calbiochem-Novabiochem, Bad Soden, Germany) and 2.5% 1,4-diazabicyclo(2.2.2)octane. Cells transfected with plasmids encoding GFP-tagged Hsfs were fixed and washed in PBS-100 mM glycine before mounting.
For microscopic analysis, an Axiophot microscope (Zeiss, Oberkochen, Germany) combined with a DP10 Photo System (Olympus, Hamburg, Germany) was used. Captured images were resized and combined using Photoshop 5.5 software (Adobe Systems, La Jolla, Calif.). Confocal laser scan micrographs were captured using a TCS4 microscope (Leica, Bensheim, Germany) and Imaris software (Bitplane, Zürich, Switzerland).
RESULTS
Cytoplasmic localization of tomato HsfA2 is the result of nuclear export.
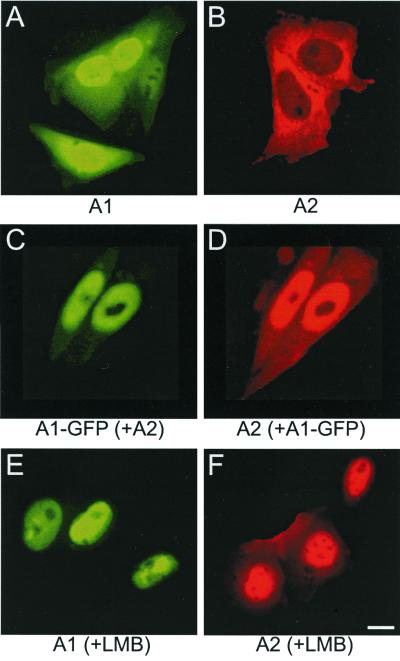
Our earlier experiments, demonstrating the intriguing interaction of HsfA1 and HsfA2 for nuclear localization of the latter, were always complicated by the unknown and probably changing contributions of members of the endogenous Hsf system in plant cells. To investigate the phenomenon in more detail without this experimental background, we decided to use CHO cells for the expression of tomato HsfA2 alone and in combination with HsfA1, followed by immune fluorescence analysis of the intracellular distribution (Fig. 2). Similar to the situation in plant cells, HsfA1 was found distributed between the nucleus and cytoplasm (Fig. 2A), whereas HsfA2 was exclusively cytosolic (Fig. 2B). Following earlier observations (36), the nuclear exclusion of HsfA2 could be overcome by coexpression with HsfA1. Both proteins colocalize with a distribution between the nucleus and cytoplasm (Fig. 2C and D). The results show that the characteristic interaction of the tomato Hsfs A1 and A2 and its consequences for the intracellular localization of HsfA2 reflect intrinsic properties of the two proteins and do not need the presence of other plant-specific proteins.
FIG. 2.
Nuclear localization of tomato HsfA2 in CHO cells depends on physical interaction with HsfA1 or the presence of LMB. CHO cells were transfected with the indicated Hsf expression plasmids. After 20 h of incubation at 37°C, cells were processed for immunofluorescence. (A, C, and E) Detection of HsfA1 with anti-HsfA1 (green channel). (A and E) or of HsfA1-GFP (C); (B, D, and F) detection of HsfA2 with anti-HsfA2 (red channel). (A and B) Cells expressing HsfA1 or HsfA2 alone; (C and D) cells coexpressing HsfA1-GFP and HsfA2; (E and F) same as shown in panels A and B but with LMB (2 ng/ml) added 60 min before cells were harvested. In Fig. 2 to 6, bars represent 10 μm; A1, HsfA1; A2, HsfA2; B1, HsfB1.
An earlier model explaining the nuclear translocation of HsfA2 as a result of unmasking its NLS, either by binding of HsfA1 or by deletion of a C-terminal shielding domain (23, 36), evidently needs an important modification. Adding LMB, an inhibitor of the nuclear exportin 1 dependent export pathway (10, 12, 20), led to an almost complete nuclear localization of HsfA2 without the need of HsfA1 as a helper protein (Fig. 2F). The recompartmentalization was clearly visible after 15 min of incubation in the presence of LMB (2 ng/ml) and was complete after 60 min (data not shown). Although the change in intracellular distribution in the presence of LMB was most pronounced for HsfA2 (Fig. 2B versus F), addition of the drug also led to an exclusive nuclear localization of HsfA1 (Fig. 2A versus E).
Structural determinants of nuclear import and export of HsfA2.
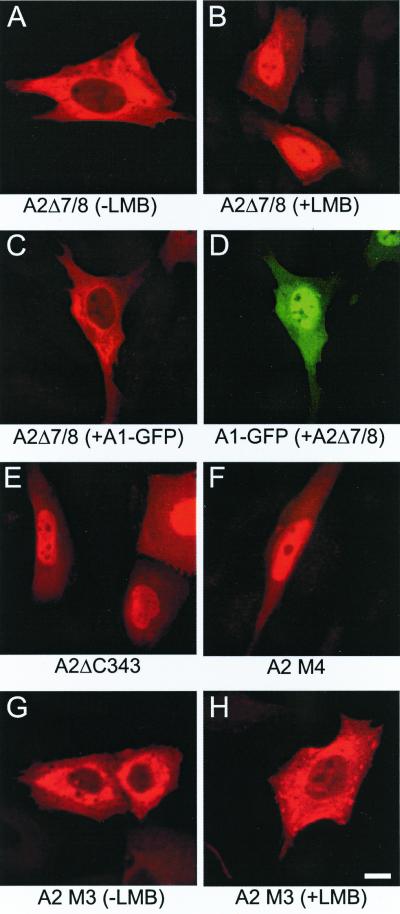
It was shown before that formation of hetero-oligomers of HsfA1 and HsfA2 is a prerequisite for nuclear colocalization of HsfA2 in the presence of HsfA1 (36). To study this effect in more detail, we expressed a deletion form of HsfA2 lacking the oligomerization domain (HsfA2Δ7/8) (Fig. 1). Its intracellular distribution was indistinguishable from that of the wild-type HsfA2; i.e., it was found in the cytoplasm in the absence (Fig. 3A) and in the nucleus in the presence (Fig. 3B) of LMB. As expected for this mutant protein, there was no influence of coexpression with HsfA1. Due to the lack of formation of HsfA1-HsfA2 hetero-oligomers, HsfA2Δ7/8 remained cytoplasmic (Fig. 3C), whereas HsfA1 was detected in the nucleus (Fig. 3D).
FIG. 3.
Structural determinants of HsfA2 influencing its intracellular distribution. For transfection, incubation, and detection conditions see the legend to Fig. 2. HsfA2 and its derivatives were detected in the red channel (A to C and E to H), and HsfA1-GFP was detected in the green channel (D). (A to D) CHO cells were transfected with the expression vector encoding HsfA2Δ7/8 without LMB (A) or with LMB (2 ng/ml) added 60 min before harvesting (B). (C and D) Cells coexpress HsfA2Δ7/8 together with HsfA1-GFP (D). In contrast to the wild-type HsfA2 (Fig. 2), there is no nuclear retention of the HR-A/B deletion form HsfA2Δ7/8 in the presence of HsfA1 (C). (E and F) Nuclear localization of HsfA2 was observed in the absence of LMB if the putative NES was deleted (A2ΔC343 [E]) or made defective by alanine substitutions of the leucine residues (A2M4 [F]). (G and H) The cytoplasmic localization of HsfA2 and its redistribution in the presence of LMB require a functional NLS. The NLS mutant (A2M3) (see Fig. 1) was permanently cytoplasmic irrespective of the absence (G) or presence (H) of LMB.
Upon inspection of the C-terminal part of HsfA2, we noticed a peptide motif rich in leucine residues (340 ELQDLVDQLGFL∗) (Fig. 1). To test the role of this motif as a potential NES, we used a deletion form lacking the last eight amino acid residues (HsfA2ΔC343) and a mutant form of the full-length protein (alanine substitutions of the three leucine residues are underlined) (HsfA2M4). As predicted, both forms had a predominant nuclear localization (Fig. 3E and F), indicating that this C-terminal peptide motif in the HR-C region represents the nuclear export signal of HsfA2. Evidently, the negative effect of the HR-C domain on the activator function of HsfA2 (23) results from the lack of nuclear accumulation due to the presence of the NES in this domain. The pronounced effect of LMB indicates that HsfA2 rapidly shuttles between nucleus and cytoplasm. In keeping with this, no nuclear import could be detected with the NLS mutant of HsfA2 (HsfA2M3), irrespective of the presence or absence of LMB (Fig. 3G and H).
Nuclear export of HsfB1 by fusion with the NES of HsfA2.
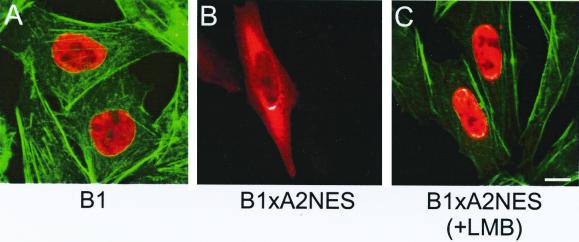
The only member of the class B Hsfs so far identified in tomato cells is HsfB1 (37). This protein is always localized in the nucleus, irrespective of control or HS conditions (36). To support the evidence for the role of the C-terminal NES of HsfA2, we expressed a fusion protein of HsfB1 with the C-terminal 23-aa residues of HsfA2 (HsfB1-A2NES) and compared its intracellular distribution with that of the wild-type protein (Fig. 4). Similar to the findings in plant cells, HsfB1 is also found entirely in the nucleus in CHO cells (Fig. 4A). For better orientation, actin fibers were stained with FITC-phalloidin. As expected, most of the HsfB1-A2NES fusion protein was cytoplasmic, indicating an efficient nuclear export due to the attached NES (Fig. 4B), and this effect was reversible by incubation in the presence of LMB (Fig. 4C).
FIG. 4.
Intracellular localization of HsfB1 and the HsfB1-A2NES fusion protein. CHO cells were transfected with the indicated expression plasmids encoding HsfB1 (aa 1 to 301) or the fusion protein HsfB1 (aa 1 to 296)-HsfA2 (aa 339 to 351) containing the NES domain of HsfA2 (B1-A2 NES). Detection was anti-HsfB1 (red channel). For better orientation, actin fibers were stained with FITC-phalloidin (green channel). (A) The HsfB1 wild type was found exclusively in the nucleus, whereas HsfB1-A2NES was cytoplasmic in the absence (B) and nuclear in the presence (C) of LMB.
Nuclear export is also the dominant effect on the intracellular distribution of HsfA2 in plant cells.
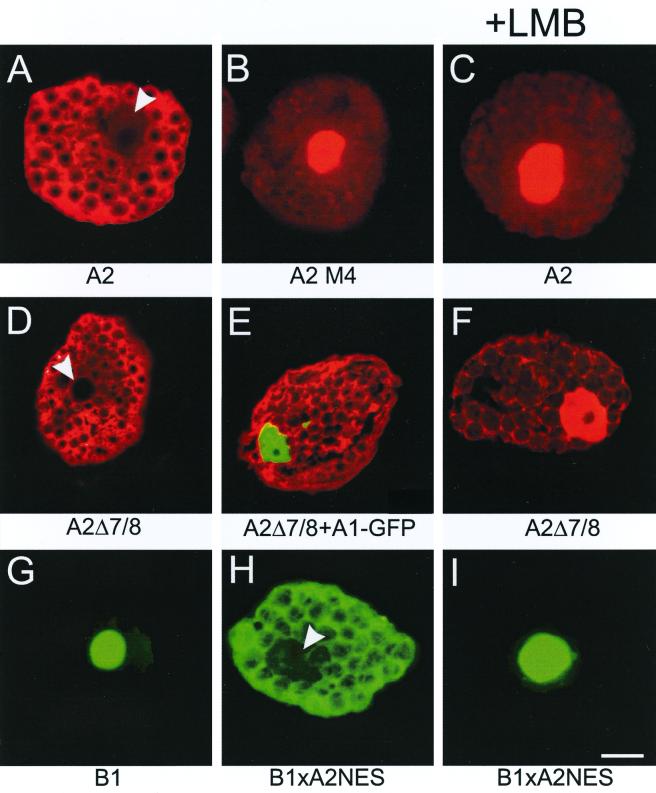
To verify the role of the NES of HsfA2 in plant cells, we also investigated essential aspects of intracellular distribution by transient expression in tobacco protoplasts. As shown earlier (36), expression of HsfA2 alone leads to a dominant cytoplasmic localization (Fig. 5A). However, similar to the results in CHO cells, nuclear retention was observed after the addition of LMB to protoplasts expressing HsfA2 alone (Fig. 5C) or with cells expressing the NES mutant form HsfA2M4 with alanine substitutions in the C-terminal leucine-rich motif (Fig. 5B). As with the results obtained with CHO cells, the intracellular distribution of the deletion form HsfA2Δ7/8, lacking its oligomerization domain, was not influenced by coexpression with HsfA1 (Fig. 5D versus E) but by the addition of LMB (Fig. 5F). Finally, we tested HsfB1 and its mutant form with a C-terminal NES. The results were very similar to those shown in Fig. 4. The exclusive nuclear localization of the wild-type HsfB1 (Fig. 5G) contrasts with the dominant cytoplasmic staining of cells expressing the HsfB1-A2NES fusion protein. Nuclear export of the latter was blocked in the presence of LMB added 60 min before protoplasts were processed for immunostaining (Fig. 5I).
FIG. 5.
Immunofluorescence analysis of the NES function of HsfA2 in tobacco protoplasts. At 16 h after transformation with the indicated Hsf expression plasmids, protoplasts were processed for immunofluorescence. (A and C) Protoplasts expressing HsfA2 alone; (B) cells expressing the NES mutant HsfA2M4; (D to F) protoplasts expressing HsfA2Δ7/8 alone (D and F) or together with HsfA1-GFP with A1-GFP detected in the green channel (E); (G to I) protoplasts expressing the HsfB1 wild type (G) or the HsfB1-A2NES fusion protein (H and I). Samples shown in panels C, F, and I contained LMB (2 ng/ml) added 1 h before fixation. Arrowheads in panels A, D, and H indicate the positions of nuclei.
Effect of HS on the nucleocytoplasmic distribution of HsfA2.
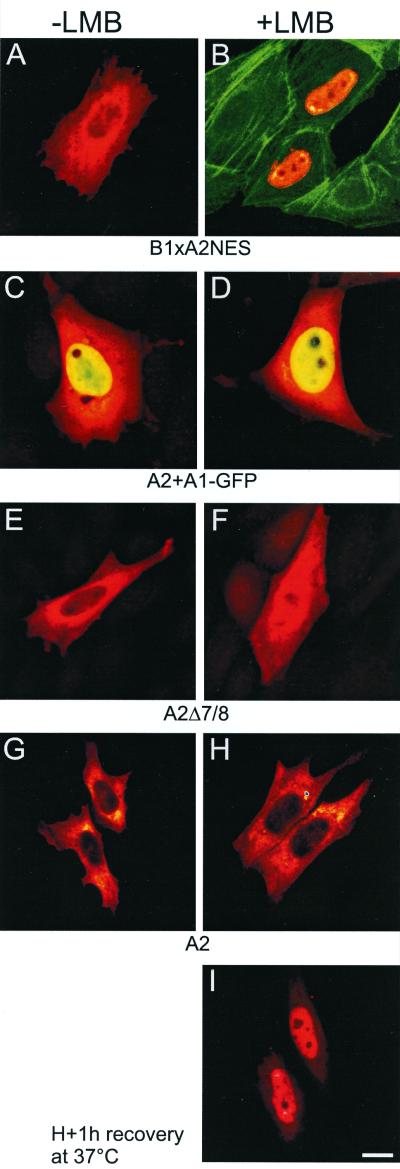
The particularities of intracellular distribution of HsfA2 in CHO cells and tobacco protoplasts are evidently caused by the dynamic balance of nuclear import and export. Considering the predominant incorporation of HsfA2 in cytoplasmic chaperone complexes formed in tomato cells under HS conditions (4, 35), we wanted to know whether the HS-dependent selective binding of HsfA2 to the HSG complexes reflects a conformational transition of the transcription factor and whether the presence of HsfA1 has any effect on such a transition. To this end, we analyzed the localization of HsfA2 alone and/or in combination with HsfA1 in CHO cells under mild HS conditions (41°C), which are close to the HS regime also used with thermotolerant tomato cell cultures (36). As a control, the HsfB1-A2NES was included in the analysis. Twenty-four hours after transfection, cells were shifted for 1 h to 41°C with LMB added during the HS treatment to the indicated samples 30 min before harvesting (Fig. 6).
FIG. 6.
HS effects on the intracellular distribution of tomato Hsfs in CHO cells and on the effect of LMB. CHO cells were incubated for 24 h at 37°C and then shifted for 1 h to 41°C. LMB (2 ng/ml) was added to samples shown in panels B, D, F, H, and I 30 min after the onset of the HS. HsfB1 (A and B) and HsfA2 (C to H) were detected in the red channel (C to H), and HsfA1-GFP was detected in the green channel (C and D). (A and B) Cells transfected with HsfB1-A2NES, with actin detected with FITC-phalloidin in the green channel (B); (C and D) merged pictures of cells coexpressing HsfA1-GFP (green channel) and HsfA2 (red channel); (E and F) cells expressing HsfA2Δ7/8; (G to I) cells expressing HsfA2. LMB-treated cells in sample I were allowed to recover at 37°C for 1 h after HS.
Results with the HsfB1-A2NES fusion protein showed that, similar to the results at 37°C (Fig. 4), most of the protein is also cytoplasmic at 41°C but nuclear in the presence of LMB (Fig. 6A and B). For better orientation, actin was stained with FITC-phalloidin (Fig. 6B). The results with HsfB1-A2NES indicated that neither nuclear import nor export nor the efficiency of LMB as an inhibitor was affected by the 1-h HS at 41°C. The same is evidently true for the results found with CHO cells coexpressing HsfA1 and HsfA2 (Fig. 6C and D) and with those expressing HsfA2Δ7/8 (Fig. 6E and F). Hence, it was surprising that results with HsfA2 were totally different (Fig. 6G to I). As expected, HsfA2 was also cytosolic at 41°C. However, no nuclear import could be found in LMB-containing samples, when LMB was added to cells at 41°C. Even increasing the LMB concentration to 20 ng/ml did not change the outcome of the experiment (data not shown). It is important to recall that a prerequisite for the rapid and remarkable effects of LMB on the intracellular recompartmentalization of HsfA2 from the cytoplasm to the nucleus at 37°C is the constant shuttling of HsfA2 between both compartments. From the abnormal behaviour of HsfA2 at 41°C, we conclude that there is a temperature-dependent conformational transition with intramolecular shielding of the NLS, which was not observed in the mutant form lacking the oligomerization domain or with HsfA2 as part of the hetero-oligomeric complexes with HsfA1. The conformational change of HsfA2 with shielding of the NLS is readily reversible after shifting the LMB-containing cultures back to 37°C. After 15 min of recovery, a considerable portion of HsfA2 was again detected in the nucleus, and recompartmentalization was complete after 60 min of recovery (Fig. 6I).
Nuclear translocation of HsfA2 results in increased expression of an Hsf-dependent reporter.
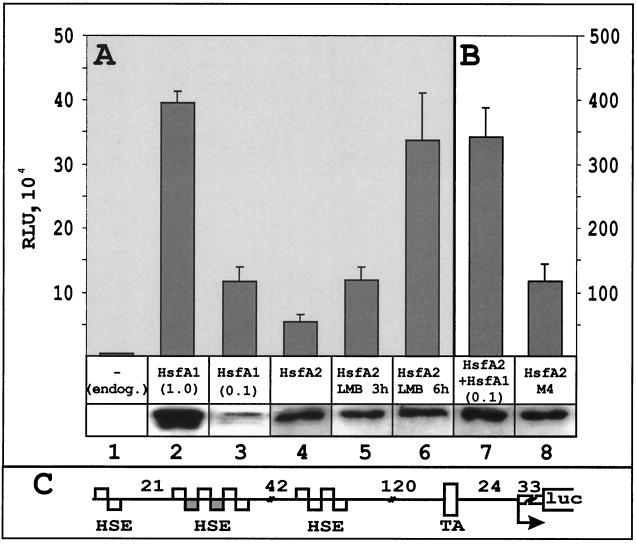
To investigate the consequences of the intracellular redistribution of HsfA2 as a balance of the function of its NLS and NES, we used a luciferase reporter assay. First, we tested a luciferase reporter construct with the human hsp70 promoter (kindly provided by R. Morimoto). However, as a result of the endogenous Hsfs and other activator proteins of CHO cells binding to this complex promoter (45), there was a considerable level of basal activity, which was increased only two- to threefold in the presence of tomato HsfA1 or HsfA2 (data not shown). Interestingly, a solution to the problem was achieved by introducing a plant-specific promoter fragment with the TATA box and scattered Hsf binding motifs (HSE) into the pGL3xluc basal vector (Promega). This promoter fragment was derived from the soybean hsp17.3B gene (38). Compared to the hsp70xluc reporter, this new hsp17xluc reporter had a 60-fold-lower level of basal activity, indicating that interaction with the endogenous Hsfs or other mammalian transcription-activating proteins was very inefficient. However, there was a good response in the presence of various tomato Hsfs (Fig. 7), in some cases resulting in a >800-fold stimulation of luciferase expression, e.g., in sample 7. To avoid problems with luciferase as a temperature-sensitive enzyme, we only used samples incubated at 37°C.
FIG. 7.
Activator assay with the hsp17xluc reporter. Expression of luciferase was measured in extracts of CHO cells incubated at 37°C for 36 h after cotransformation with 1 μg of the reporter plasmid hsp17xluc plus the indicated Hsf expression plasmid (1 and 0.1 μg of HsfA1 and 1 μg of HsfA2 expression plasmid, respectively). LMB (2 ng/ml) was added to samples 5 and 6 at 3 and 6 h before harvesting. Note that the scale shown in panel B is extended 10-fold. Expression of the Hsfs was monitored by Western blotting (see signals shown at the bottom of panel A, samples 2 to 8). In coexpression sample 7, only the signal for HsfA2 is indicated. The hsp17xluc reporter construct (C) contains a promoter fragment of the soybean hsp17.5B gene (see Materials and Methods). TA, TATA box; HSE, AGAAn or nTTCT motifs; shaded boxes, defective HSE (nTTmT); numbers indicate the number of nucleotides between the functional elements of the construct. RLU, relative light units.
The results clearly demonstrate that the improved nuclear localization of HsfA2 is indeed connected with considerably increased luciferase expression. To include all data in one diagram, the scale for the enzyme activity (relative light units, 104) had to be extended 10-fold (Fig. 7B). Compared to the background activity of samples transformed with the reporter construct alone (Fig. 7, sample 1), the luciferase activity was increased 10-fold in cells expressing HsfA2 (sample 4) and 30- to 100-fold in samples expressing different amounts of HsfA1 (samples 2 and 3). Reporter activity was increased 300-fold with the NES mutant of HsfA2 (M4) as activator (sample 8) and 850-fold in cells coexpressing HsfA2 with HsfA1 (sample 7). To optimize the stimulating effect of the coexpression of HsfA2 and HsfA1 on the activator function of the former, we used 1 μg of HsfA2 and 0.1 μg of HsfA1 expression plasmid for cotransformation (see results with samples 3 and 4 as a reference for sample 7). As was observed earlier with plant cells, a relatively small amount of HsfA1 is sufficient to promote the translocation of HsfA2 to the nucleus (36).
Because nuclear retention of HsfA2 was most prominent with LMB (2 ng/ml) in the culture medium, we tested the effect of the drug on reporter activity if LMB was added 3 and 6 h before harvesting. Considering the relatively short time of incubation with LMB, the effect is striking. Compared to activity in sample 4, luciferase activity increased about 2-fold after 3 h and 6.5-fold after 6 h of incubation with LMB (Fig. 7, samples 5 and 6).
DISCUSSION
CHO cells as test system for tomato Hsfs.
As outlined in the introduction, the high degree of evolutionary conservation of the HS response throughout the eukaryotic kingdom and the conserved modular structure of the Hsfs were repeatedly used as a basis for investigations of Hsf functions in heterologous systems (4, 5, 8, 43). Following this line of inquiry, the results in this paper demonstrate that CHO cells represent an efficient system for the expression and analysis of tomato Hsfs. Compared to tobacco mesophyll protoplasts used so far, the advantage of CHO cells is the size of the cells, the lack of chloroplasts filling most of the cell (Fig. 5), and the lack of background problems by interfering endogenous Hsfs (Fig. 7). This situation considerably facilitates detailed studies of the intriguing network of protein interactions influencing the intracellular distribution and function of tomato Hsfs.
As a starting point and to test the use of CHO cells as model system, we studied the intracellular distribution of tomato Hsfs. The results faithfully reflect results reported earlier for tobacco protoplasts and tomato cells (35, 36). (i) HsfB1 is always nuclear (Fig. 4A). (ii) HsfA1 distributes between the cytoplasm and the nucleus (Fig. 2A). (iii) HsfA2 is exclusively found in the cytoplasm unless coexpressed with HsfA1 (Fig. 2B versus D). Colocalization of HsfA1 and HsfA2 requires formation of heterooligomers mediated by the oligomerization domain (HR-A/B region). As expected, a deletion form of HsfA2 lacking the oligomerization domain (HsfA2Δ7/8) is localized in the cytoplasm and cannot be influenced by coexpression with HsfA1 (Fig. 3A and C).
The nuclear export signal of HsfA2.
Our earlier concept that the cytoplasmic localization of HsfA2 results from an intramolecular shielding of the NLS by interaction of the C-terminal HR-C with the HR-A/B region (36) is confirmed but evidently needs an important modification. Addition of LMB to CHO cells expressing HsfA2 alone led to a rapid intracellular redistribution of HsfA2 to the nucleus (Fig. 2F). As demonstrated earlier, LMB is an inhibitor of the nuclear export receptor exportin 1 (10, 19, 20, 39) recognizing Leu-rich motifs in the target proteins (NES). When comparing an NES consensus motif LXXXLXXLXL (9, 12, 25, 29, 30) with the HR-C region of HsfA2, the very C-terminal peptide motif LQDLVDQL GFL* fits such a nuclear export signal. In support of this, mutant forms of HsfA2 with alanine substitutions of the three leucine residues (underlined above) or with a deletion of the last 8-aa residues (HsfA2ΔC343) were both nuclear proteins even in the absence of LMB or HsfA1 as a mediator for nuclear retention (Fig. 3E and F). The role of the C-terminal NES of HsfA2 was confirmed by investigating the intracellular distribution of a fusion protein of HsfB1 with the last 20-aa residues of HsfA2 containing the NES (Fig. 4 and 5).
In contrast to HsfA2, a considerable portion of HsfA1 was always localized in the nucleus (Fig. 2A). As a consequence, the LMB effect was less striking (Fig. 2E) but still detectable. Because a leucine-rich motif with clear homology to the consensus NES was not detectable, we tested a number of C-terminal deletion forms available from earlier experiments aimed at the identification of the activator motifs of HsfA1 (8, 43). All of them were found exclusively in the nucleus (data not shown). In the mutant form with deletion of the C-terminal 26-aa residues (HsfA1ΔC491), the only peptide motif with a weak homology to an NES is 505 TQNMEHLTEQM 515. The significance of this motif remains to be analyzed, e.g., by alanine substitutions of the hydrophobic residues underlined. However, Askjaer et al. (1) reported a similar peptide motif derived from the Ns2 protein of minute virus of mice (DEMTKKFGTLTIHD) with a particularly strong interaction with the exportin 1 receptor. In this context, it is important to notice that the classical leucine-rich NES is only one type of motif recognized by exportin 1. Recently, Hoshino et al. (16) identified a totally unrelated cytoplasmic localization signal in the mammalian Bach 2 repressor. In this case, the exportin 1 recognition motif represents a stretch of mostly hydrophilic amino acid residues with two essential Cys residues.
The intracellular distribution of HsfA2 between the nucleus and cytoplasm is evidently the result of an inefficient nuclear import and an efficient nuclear export. It needs the intact NLS adjacent to the oligomerization domain (see results with the NLS mutant HsfA2M3 [Fig. 3G and H]) and the exposed C-terminal NES. But oligomerization is not required as shown by the results with the deletion form (HsfA2Δ7/8) lacking the HR-A/B region. It is important to notice that the results with the intracellular distribution of HsfA2 are very similar or identical, irrespective of the expression system used, i.e., CHO cells or tobacco protoplasts. This is true not only for the nuclear retention of HsfA2 by coexpression with HsfA1 (36) but also for the effects of LMB and the NES mutations, as well as for the nuclear export of the HsfB1-A2NES hybrid (Fig. 5). Evidently, the characteristic pattern of intracellular localization of tomato Hsfs results from the intrinsic properties of the three proteins involved. Despite some differences in details reported for the plant nuclear import system (15), the components of the nuclear-cytoplasmic transport machineries are functionally conserved between plants and mammals (12, 13, 44), and no additional, plant-specific proteins are needed for the effects described in this paper.
Interestingly, the close similarity between plant and animal systems in many aspects of molecular cell biology does not hold true for one important detail of the reporter assay, i.e., the promoter context of the reporter construct with the pattern of HSE. The high level of basal activity of the human hsp70xluc reporter probably results from the complex pattern of potential binding sites for Hsfs as well as for other activator proteins, e.g., CTF, Sp1, and AP2 (45). It could be reduced more than 60-fold by introducing a soybean hsp17.3B promoter fragment (38). The new hsp17xluc reporter for animal cells provides the basis for a sensitive activity assay for tomato Hsfs in CHO cells (Fig. 7). The dominant nuclear localization of HsfA2 as a result of coexpression with HsfA1, mutation of the NES, or the addition of LMB is nicely documented by a considerable increase of Hsf-dependent increase of luciferase expression.
Nucleocytoplasmic shuttling as controlled balance of import and export.
Nucleocytoplasmic redistribution of proteins involved in signal transduction and transcription regulation is evidently an important process in many systems. The balance of NLS and NES and changes in the accessibility caused by interaction with other proteins or by protein modification are decisive for the intracellular localization of these proteins. Four examples may illustrate this point. (i) The RelA subunit (p65) of the NF-κB transcription factor is a cytoplasmic protein unless it is complexed with the second subunit (p50). Shielding of the NES of p65 or the balance of two NLSs (one each in p65 and p50) with one NES of p65 may be responsible for the effect (14). In addition, the newly synthesized inhibitor protein I-κB with an NES in its C-terminal domain mediates the reexport of NF-κB and thus terminates the response (33, 41). (ii) Intracellular distribution of the transcription factor NFAT is controlled by the accessibility of two NESs in the N terminus and two NLSs in the C-terminal part of the molecule. Binding of the Ca2+-calcineurin phosphatase to the NES domain triggers nuclear import by shielding the NES and unmasking the NLS by dephosphorylation (3, 49). (iii) The tumor suppressor protein p53 contains two NLSs and one NES, which is part of the oligomerization domain. Because of NES shielding, the p53 tetramers are nuclear, whereas monomers or dimers are cytoplasmic (40). Nuclear retention and the stability of the tetrameric state are increased by stress-induced phosphorylation of Ser392 in the C-terminal NLS domain. (iv) In G2-arrested Xenopus oocytes, maturation is triggered by progesteron. The effect depends on the cytoplasmic release and subsequent nuclear import of the dual-specificity phophatase Cdc25 activating the Cdc2-cyclinB1 complex. Nuclear export of Cdc25 and binding to cytoplasmic 14-3-3 proteins are characteristic for the G2-arrested state (21, 32, 47).
The nucleocytoplasmic distribution of the tomato HsfA2 is markedly influenced by heterooligomerization with HsfA1 (Fig. 2 and 5). The molecular basis for this influence is unclear. It is tempting to speculate about masking of the NES of HsfA2 or about the balance of NLS and NES in the HsfA1-HsfA2 hetero-oligomer. Additional insights into mechanisms influencing the intracellular distribution of HsfA2 came from the experiments with CHO cells subjected to a mild HS of 41°C in the absence and presence of LMB (Fig. 6). In contrast to the results seen at 37°C, HS led to a complete exclusion of HsfA2 from the nucleus even in the presence of LMB. Because there is no general effect on nuclear import or export or on the inhibitory effect of LMB at 41°C, at least not under our experimental conditions, the simplest explanation for this remarkable behavior of HsfA2 is a reversible conformational transition with shielding of its NLS, probably mediated by interaction of the HR-A/B with the HR-C region. In support of this assumption, the intracellular redistribution at 41°C in the presence of LMB was normal for CHO cells expressing HsfA2Δ7/8, the HR-A/B deletion form of HsfA2 (Fig. 6). Since the temperature of 41°C is also in the range of the physiological HS response for tomato, it is tempting to speculate that this HS-induced conformational change of HsfA2 may contribute to its efficient storage in the cytoplasmic chaperone complexes of HSG formed in HS tomato cells (36). The faithful reconstruction of many aspects of protein interaction in the tomato Hsf system in CHO cells represents a good basis for experiments with coexpression of HsfA2 with the components of the HSG, in particular class I and II small Hsps of tomato, to reconstruct the HSG or related complexes in a non-plant system and to study the evident competition between HsfA1 and the small Hsps for interaction with HsfA2.
ACKNOWLEDGMENTS
We thank Claudia Tietjen for excellent technical assistance and Kapil Bharti and Markus Fauth for helpful discussions and comments during the preparation of the manuscript.
This work was supported by grants of the Deutsche Forschungsgemeinschaft Bonn (SFB 474) and by the Fonds der Chemischen Industrie.
REFERENCES
- 1.Askjaer P, Bachi A, Wilm M, Bischoff F R, Weeks D L, Ogniewski V, Ohno M, Niehrs C, Kjems J, Mattaj I W, Fornerod M. RanGTP-regulated interaction of CRM1 with nucleoporins and a shuttling DEAD-box helicase. Mol Cell Biol. 1999;19:6276–6285. doi: 10.1128/mcb.19.9.6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons, Inc.; 1993. [Google Scholar]
- 3.Beals C R, Sheridan C M, Turck C W, Gardner P, Crabtree G R. Nuclear export of NF-ATc enhanced by glycogen synthase kinase-3. Science. 1997;275:1930–1933. doi: 10.1126/science.275.5308.1930. [DOI] [PubMed] [Google Scholar]
- 4.Bharti K, Schmidt E, Lyck R, Heerklotz D, Bublak D, Scharf K-D. Isolation and characterization of HsfA3, a new heat stress transcription factor of Lycopersicon peruvianum. Plant J. 2000;22:355–365. doi: 10.1046/j.1365-313x.2000.00746.x. [DOI] [PubMed] [Google Scholar]
- 5.Boscheinen O, Lyck R, Queitsch C, Treuter E, Zimarino V, Scharf K D. Heat stress transcription factors from tomato can functionally replace HSF1 in the yeast Saccharomyces cerevisiae. Mol Gen Genet. 1997;255:322–331. doi: 10.1007/s004380050503. [DOI] [PubMed] [Google Scholar]
- 6.Clos J, Westwood J T, Becker P B, Wilson S, Lambert U, Wu C. Molecular cloning and expression of a heaxameric Drosophila heat shock factor subject to negative regulation. Cell. 1990;63:1085–1097. doi: 10.1016/0092-8674(90)90511-c. [DOI] [PubMed] [Google Scholar]
- 7.Czarnecka-Verner E, Yuan C-X, Scharf K D, English G, Gurley W B. Plants contain a novel multi-member family of heat shock factors without transcriptional activator potential. Plant Mol Biol. 2000;43:459–471. doi: 10.1023/a:1006448607740. [DOI] [PubMed] [Google Scholar]
- 8.Döring P, Treuter E, Kistner C, Lyck R, Chen A, Nover L. Role of AHA motifs for the activator function of tomato heat stress transcription factors HsfA1 and HsfA2. Plant Cell. 2000;12:265–278. [PMC free article] [PubMed] [Google Scholar]
- 9.Fischer U, Michael M, Lührmann R, Dreyfuss G. Signal-mediated nuclear export pathways of proteins and RNAs. Trends Cell Biol. 1996;6:290–293. doi: 10.1016/0962-8924(96)20030-3. [DOI] [PubMed] [Google Scholar]
- 10.Fornerod M, Ohno M, Yoshida M, Mattaj I W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 11.Gallo G J, Prentice H, Kingston R E. Heat shock factor is required for growth at normal temperatures in the fission yeast Schizosaccharomyces pombe. Mol Cell Biol. 1993;13:749–761. doi: 10.1128/mcb.13.2.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Görlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 13.Haasen D, Köhler C, Neuhaus G, Merkle T. Nuclear export of proteins in plants: AtXPO1 is the export receptor for leucine-rich nuclear export signals in Arabidopsis thaliana. Plant J. 1999;20:695–705. doi: 10.1046/j.1365-313x.1999.00644.x. [DOI] [PubMed] [Google Scholar]
- 14.Harhaj E W, Sun S C. Regulation of RelA subcellular localization by a putative nuclear export signal and p50. Mol Cell Biol. 1999;19:7088–7095. doi: 10.1128/mcb.19.10.7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hicks G R, Smith H M S, Lobreaux S, Raikhel N. Nuclear import in permeabilized protoplasts from higher plants has unique features. Plant Cell. 1996;8:1337–1352. doi: 10.1105/tpc.8.8.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoshino H, Kobayashi A, Yoshida M, Kudo N, Oyake T, Motohashi H, Hayashi N, Yamamoto M, Igarashi K. Oxidative stress abolishes leptomycin B-sensitive nuclear export of transcription repressor Bach2 that counteracts activation of Maf recognition element. J Biol Chem. 2000;275:15370–15376. doi: 10.1074/jbc.275.20.15370. [DOI] [PubMed] [Google Scholar]
- 17.Hübel A, Lee J H, Wu C, Schöffl F. Arabidopsis heat shock factor is constitutively active in Drosophila and human cells. Mol Gen Genet. 1995;248:136–141. doi: 10.1007/BF02190794. [DOI] [PubMed] [Google Scholar]
- 18.Kirschner M, Winkelhaus S, Thierfelder J, Nover L. Transient expression and heat stress induced co-aggregation of endogenous and heterologous small heat stress proteins in tobacco protoplasts. Plant J. 2000;24:397–412. doi: 10.1046/j.1365-313x.2000.00887.x. [DOI] [PubMed] [Google Scholar]
- 19.Kudo N, Matsumori N, Taoka H, Fujiwara D, Schreiner E P, Wolff B, Yoshida M, Horinouchi S. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc Natl Acad Sci USA. 1999;96:9112–9117. doi: 10.1073/pnas.96.16.9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kudo N, Wolff B, Sekimoto T, Schreiner E P, Yoneda Y, Yanagida M, Horinouchi S, Yoshida M. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp Cell Res. 1998;242:540–547. doi: 10.1006/excr.1998.4136. [DOI] [PubMed] [Google Scholar]
- 21.Kumagai A, Dunphy W G. Binding of 14-3-3 proteins and nuclear export control the intracellular localization of the mitotic inducer Cdc25. Genes Dev. 1999;13:1067–1072. doi: 10.1101/gad.13.9.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu X D, Liu P C C, Santoro N, Thiele D J. Conservation of a stress response: human heat shock transcription factors functionally substitute for yeast Hsf. EMBO J. 1997;16:6466–6477. doi: 10.1093/emboj/16.21.6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lyck R, Harmening U, Höhfeld I, Scharf K D, Nover L. Intracellular distribution and identification of the nuclear localization signals of two tomato heat stress transcription factors. Planta. 1997;202:117–125. doi: 10.1007/s004250050110. [DOI] [PubMed] [Google Scholar]
- 24.Morimoto R I. Regulation of the heat shock transcriptional response: cross talk between family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 1998;12:3788–3796. doi: 10.1101/gad.12.24.3788. [DOI] [PubMed] [Google Scholar]
- 25.Nakielmy S, Dreyfuss G. Nuclear export of proteins and RNAs. Curr Opin Cell Biol. 1997;9:420–429. doi: 10.1016/s0955-0674(97)80016-6. [DOI] [PubMed] [Google Scholar]
- 26.Nover L, Scharf K-D. Heat stress proteins and transcription factors. Cell Mol Life Sci. 1997;53:80–103. doi: 10.1007/PL00000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nover L, Scharf K-D, Neumann D. Cytoplasmic heat shock granules are formed from precursor particles and are associated with a specific set of mRNAs. Mol Cell Biol. 1989;9:1298–1308. doi: 10.1128/mcb.9.3.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nover L, Scharf K D, Gagliardi D, Vergne P, Czarnecka-Verner E, Gurley W B. The Hsf world: classification and properties of plant heat stress transcription factors. Cell Stress Chaperones. 1996;1:215–223. doi: 10.1379/1466-1268(1996)001<0215:thwcap>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohno M, Fornerod M, Mattaj I W. Nucleocytoplasmic transport: the last 200 nanometers. Cell. 1998;92:327–336. doi: 10.1016/s0092-8674(00)80926-5. [DOI] [PubMed] [Google Scholar]
- 30.Pollard V W, Malim M H. The HIV-1 Rev protein. Annu Rev Microbiol. 1998;52:491–532. doi: 10.1146/annurev.micro.52.1.491. [DOI] [PubMed] [Google Scholar]
- 31.Reichel C, Mathur J, Eckes P, Langenkemper K, Koncz C, Schell J, Reiss B, Maas C. Enhanced green fluorescence by the expression of an Aequora victoria green fluorescent protein mutant in mono- and dicotyledonous plant cells. Proc Natl Acad Sci USA. 1996;95:1495–1499. doi: 10.1073/pnas.93.12.5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rittinger K, Budman J, Xu J A, Volinia S, Cantley L C, Smerdon S J, Gamblin S J, Yaffe M B. Structural analysis of 14-3-3 phosphopeptide complexes identifies a dual role for the nuclear export signal of 14-3-3 in ligand binding. Mol Cell. 1999;4:153–166. doi: 10.1016/s1097-2765(00)80363-9. [DOI] [PubMed] [Google Scholar]
- 33.Rodriguez M S, Thompson J, Hay R T, Dargemont C. Nuclear retention of IκBα protects it from signal-induced degradation and inhibits NFκB transcriptional activation. J Biol Chem. 1999;274:9108–9115. doi: 10.1074/jbc.274.13.9108. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Scharf K-D, Höhfeld I, Nover L. Heat stress response and heat stress transcription factors. J Biosci. 1998;23:313–329. [Google Scholar]
- 36.Scharf K-D, Heider H, Höhfeld I, Lyck R, Schmidt E, Nover L. The tomato Hsf system: HsfA2 needs interaction with HsfA1 for efficient nuclear import and may be localized in cytoplasmic heat stress granules. Mol Cell Biol. 1998;18:2240–2251. doi: 10.1128/mcb.18.4.2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scharf K-D, Rose S, Zott W, Schöffl F, Nover L. Three tomato genes code for heat stress transcription factors with a region of remarkable homology to the DNA-binding domain of the yeast HSF. EMBO J. 1990;9:4495–4501. doi: 10.1002/j.1460-2075.1990.tb07900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schöffl F, Rieping M, Baumann G, Bevan M W, Angermüller S. The function of plant heat shock promoter elements in the regulated expression of chimaeric genes in transgenic tobacco. Mol Gen Genet. 1989;217:246–253. doi: 10.1007/BF02464888. [DOI] [PubMed] [Google Scholar]
- 39.Stade K, Ford C S, Guthrie C, Weis K. Exportin 1 (CRM1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 40.Stommel J M, Marchenko N D, Jimenez G S, Moll U M, Hope T J, Wahl G M. A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J. 1999;18:1660–1672. doi: 10.1093/emboj/18.6.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tam W F, Lee L H, Davis L, Sen R. Cytoplasmic sequestration of Rel proteins by IκBα requires CRM1-dependent nuclear export. Mol Cell Biol. 2000;20:2269–2284. doi: 10.1128/mcb.20.6.2269-2284.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Töpfer R, Schell J, Steinbiss H H. Versatile cloning vectors for transient gene expression and direct gene transfer in plant cells. Nucleic Acids Res. 1988;16:8725. doi: 10.1093/nar/16.17.8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Treuter E, Nover L, Ohme K, Scharf K-D. Promoter specificity and deletion analysis of three heat stress transcription factors of tomato. Mol Gen Genet. 1993;240:113–125. doi: 10.1007/BF00276890. [DOI] [PubMed] [Google Scholar]
- 44.Ward B M, Lazarowitz S G. Nuclear export in plants: use of geminivirus movement proteins for a cell-based export assay. Plant Cell. 1999;11:1267–1276. doi: 10.1105/tpc.11.7.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Williams G T, McClanahan T K, Morimoto R I. E1a transactivation of the human HSP70 promoter is mediated through the basal transcriptional complex. Mol Cell Biol. 1989;9:2574–2587. doi: 10.1128/mcb.9.6.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu C. Heat stress transcription factors. Annu Rev Cell Biol. 1995;11:441–469. doi: 10.1146/annurev.cb.11.110195.002301. [DOI] [PubMed] [Google Scholar]
- 47.Yang J, Winkler K, Yoshida M, Kornbluth S. Maintenance of G(2) arrest in the Xenopus oocyte: a role for 14-3-3-mediated inhibition of Cdc25 nuclear import. EMBO J. 1999;18:2174–2183. doi: 10.1093/emboj/18.8.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yuan C-X, Czarnecka-Verner E, Gurley W B. Expression of human heat shock transcription factors 1 and 2 in HeLa cells and yeast. Cell Stress Chaperones. 1997;2:263–275. doi: 10.1379/1466-1268(1997)002<0263:eohhst>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu J Y, McKeon F. NF-AT activation requires suppression of Crm1-dependent export by calcineurin. Nature. 1999;398:256–260. doi: 10.1038/18473. [DOI] [PubMed] [Google Scholar]
- 50.Zuo J, Baler R, Dahl G, Voellmy R. Activation of the DNA-binding ability of human heat shock transcription factor 1 may involve the transition from an intramolecular to an intermolecular triple-stranded coiled-coil structure. Mol Cell Biol. 1994;14:7557–7568. doi: 10.1128/mcb.14.11.7557. [DOI] [PMC free article] [PubMed] [Google Scholar]