Abstract
We recently developed a highly effective immunization procedure for the generation of monoclonal antibodies (MAbs) directed against the porcine reproductive and respiratory syndrome virus (E. Weiland, M. Wieczorek-Krohmer, D. Kohl, K. K. Conzelmann, and F. Weiland, Vet. Microbiol. 66:171–186, 1999). The same method was used to produce a panel of 16 MAbs specific for the equine arteritis virus (EAV). Ten MAbs were directed against the EAV nucleocapsid (N) protein, and five MAbs recognized the major viral envelope glycoprotein (GL). Two of the EAV GL-specific MAbs and one antibody of unknown specificity neutralized virus infectivity. A comparison of the reactivities of the MAbs with 1 U.S. and 22 newly obtained European field isolates of EAV demonstrated that all N-specific MAbs, the three nonneutralizing anti-GL MAbs, and the weakest neutralizing MAb (MAb E7/d15-c9) recognized conserved epitopes. In contrast, the two MAbs with the highest neutralization titers bound to 17 of 23 (MAb E6/A3) and 10 of 23 (MAb E7/d15-c1) of the field isolates. Ten of the virus isolates reacted with only one of these two MAbs, indicating that they recognized different epitopes. The GL-specific MAbs and the strongly neutralizing MAb of unknown specificity (MAb E6/A3) were used for the selection of neutralization-resistant (NR) virus variants. The observation that the E6/A3-specific NR virus variants were neutralized by MAb E7/d15-c1 and that MAb E6/A3 blocked the infectivity of the E7/d15-c1-specific NR escape mutant confirmed that these antibodies reacted with distinct antigenic sites. Immunoelectron microscopy revealed for the first time that the antigenic determinants recognized by the anti-GL MAbs were localized on the virion surface. Surprisingly, although the immunofluorescence signal obtained with the neutralizing antibodies was relatively weak, they mediated binding of about three times as much gold granules to the viral envelope than the nonneutralizing anti-GL MAbs.
Equine arteritis virus (EAV) is the etiological agent of equine viral arteritis (EVA), a respiratory and reproductive disease that affects horses throughout the world (18). EAV usually causes subclinical infections but may also produce clinical illness with symptoms resembling those of equine influenza (45). Infection of pregnant mares frequently results in abortion, and the virus has occasionally been associated with foal death (10). EAV can establish a persistent infection in the genital tracts of peripubertal colts and stallions (25). Although only one serotype of the virus has been recognized, EAV isolates differ in genomic sequence, antigenic properties, and pathogenic qualities (18). Virus-neutralizing (VN) antibodies are first detected 1 to 2 weeks after natural or artificial infection with EAV. These antibodies generally persist for many years and help to protect horses against EVA but rarely prevent reinfection (32). Accordingly, VN antibody titers have proven to be reliable indicators of the effectiveness of EAV vaccination protocols. The observation that colostrum from immune mares but not colostrum from nonimmune dams moderates or prevents EVA in young foals (33) emphasizes the importance of VN antibodies in the protection against EAV.
EAV is a spherical, enveloped, positive-stranded RNA virus that belongs to the genus Arterivirus within the monogeneric family Arteriviridae. Other members of this virus group are lactate dehydrogenase-elevating virus, porcine reproductive and respiratory syndrome virus (PRRSV), and simian hemorrhagic fever virus. Arteriviruses are the smallest representatives of the order Nidovirales, which further includes the bigeneric family Coronaviridae (15, 40). Negatively stained arterivirus particles have diameters of 55 to 75 nm (51) and contain a relatively smooth surface without prominent spikes. The nucleocapsids of the arterivirus possess an icosahedral structure and accommodate single copies of the viral genomic RNA. The genome of EAV has a length of 12.7 kb and contains eight genes (11, 41). The largest gene consists of two open reading frames (ORFs; ORFs 1a and 1b) which occupy nearly three-quarters of the viral genome. ORFs 1a and 1b are directly translated from the viral genomic RNA and direct the synthesis of nonstructural proteins. The other genes (ORFs 2a, 2b, and 3 through 7) are expressed from a 3′ coterminal nested set of six leader-containing subgenomic mRNAs (13) and code for the structural proteins of the virus with the possible exception of EAV ORF 3. EAV particles contain three major virion proteins: (i) a phosphorylated nucleocapsid protein (N) of 14 kDa encoded by ORF 7, (ii) a 16-kDa unglycosylated membrane protein (M) specified by ORF 6, and (iii) a heterogeneously N-glycosylated membrane protein (GL) of 30 to 42 kDa encoded by ORF 5 (14, 26, 55). The GL and M proteins are present in virus particles as covalently linked heterodimers (16). In addition, three minor structural proteins have been identified in EAV particles: (i) the poorly characterized translation product of ORF 4, (ii) an N-glycosylated membrane protein (GS) of 25 kDa encoded by ORF 2b, and (iii) an 8-kDa unglycosylated envelope protein (E) specified by ORF 2a (14, 17, 41).
The humoral immune response of horses to EAV is mainly directed against the three major protein components of the virus (7, 8, 9, 24, 28, 31). By pepscan analysis, an immunodominant epitope was localized between amino acids 70 and 99 of the EAV GL protein (30). Immunization of mice with EAV particles yielded both VN-positive (VN+) and VN-negative (VN−) monoclonal antibodies (MAbs). The neutralizing MAbs whose antigen specificities could be determined were all directed against the EAV GL protein of the virus, whereas the nonneutralizing MAbs recognized either the EAV N or GL protein (2, 3, 4, 6, 12, 23, 29, 52). Consistently, peptides derived from the GL ectodomain induced VN antibodies in mice, rabbits, and/or horses (2, 7). Different MAbs directed against the same virus displayed differential reactivities with laboratory isolates and field strains of EAV, indicating that the GL protein contains multiple antigenic determinants (2, 3, 23, 29, 52, 54). Nucleotide sequence analysis of MAb neutralization-resistant (NR) escape mutants and field isolates of EAV showed that the GL protein is composed of constant and variable domains and led to the identification of at least four neutralization sites within the second half of the hydrophilic amino-terminal ectodomain (2, 3, 5, 23, 42, 43, 54). Furthermore, the observation that only 4 of 11 MAbs that showed strong competition for binding to virions in an enzyme-linked immunosorbent assay (ELISA) reacted with the GL protein in an immunoblot assay (3) implied that some GL-specific MAbs are directed against linear epitopes, whereas others recognize conformational epitopes.
In the study described here, we have established and characterized a new panel of 16 EAV-specific MAbs. The protein specificities of 15 of these MAbs were determined, and the binding of each of the antibodies to a unique collection of 23 field isolates of EAV was investigated. Our experiments revealed that 14 of 16 of the MAbs recognized each of the virus isolates that were tested. These MAbs may thus be good candidates for the development of novel diagnostic assays for the detection of EAV-specific antigens or antibodies. We also investigated the usefulness of the anti-EAV MAbs for immunofluorescence studies and tested their ability to neutralize viral infectivity and to interact with the virus exterior. Unexpectedly, three of the GL-specific MAbs bound to the surfaces of the virions but could not prevent virus entry.
MATERIALS AND METHODS
Cells and viruses.
OST-7.1 (21), rabbit kidney (RK-13), and Vero cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco-BRL Life Technologies) supplemented with 10% fetal calf serum (FCS), 100 IU of penicillin per ml, and 100 μg of streptomycin per ml (DMEM–10% FCS). BHK-21 cells were cultured in Glasgow minimal essential medium (Gibco-BRL Life Technologies) containing 10% FCS, and Sp2/0-Ag14 cells (39) and hybridomas were propagated in RPMI 1640 medium supplemented with 12% FCS. Virus stocks of the Bucyrus strain of EAV (19) and a plaque derivative thereof (EAV B15) were grown in Vero cells, whereas the Utrecht variant of the Bucyrus strain of EAV (EAV Utr) was propagated in BHK-21 cells (14). The virions were concentrated by centrifugation through a 20% (wt/wt) sucrose cushion in Tris-buffered saline (TBS) for 5 h at 25,000 rpm and 4°C in an SW27 rotor (Beckman). The virus pellet was resuspended in TBS and was further purified in a 10 to 50% (wt/wt) continuous sucrose gradient prepared in TBS by spinning for 24 h at 32,000 rpm and 4°C in an SW41 Ti rotor (Beckman). Virus particles were harvested by puncturing the bottom of the centrifuge tube and were directly used for immunoelectron microscopy. Alternatively, the virus suspension was diluted with TBS and was centrifuged for 1 h at 45,000 rpm and 4°C in an SW55 Ti rotor (Beckman) to produce a highly concentrated virus preparation for immunizations and Western blotting.
The field isolates of EAV were collected in Germany between 1991 and 1997 from aborted foals or horse semen; S1112 virus, however, was isolated in the United States (see Table 3). The viruses were passaged two to three times in Vero cells before preparation of stocks in these cells.
TABLE 3.
Reactivities of EAV-specific MAbs with EAV field isolates by IPMA
| Virus isolate/ yr of isolation/ passage no.a | Source | Reaction with neutralizing anti-GL MAbs
|
||
|---|---|---|---|---|
| E6/A3 | E7/d15-c1 | E7/d15-c9 | ||
| V2416/1991/4 | Aborted foal | +b | + | + |
| V2716/1991/4 | Aborted foal | +b | + | + |
| S1112/1992/4 | Semen | + | − | + |
| V8633/1993/3 | Semen | + | − | + |
| V4987/1994/3 | Semen | + | − | + |
| V979/1994/4 | Semen | +b | − | + |
| V8062/1995/4 | Semen | + | − | + |
| V2592/1995/3 | Aborted foal | − | + | + |
| V2715/1995/3 | Aborted foal | − | + | + |
| V4202/1995/3 | Semen | + | + | + |
| V4675/1995/3 | Semen | + | − | + |
| V8908/1995/3 | Semen | +b | + | + |
| V1790/1995/4 | Semen | + | + | + |
| V1791/1995/3 | Semen | +b | − | + |
| V1792/1995/3 | Semen | + | −c | + |
| V273/1996/4 | Semen | − | − | + |
| V1763/1996/4 | Semen | − | − | + |
| V1764/1996/4 | Semend | − | − | + |
| V4137/1996/4 | Semen | − | − | + |
| V1206/1996/4 | Semen | + | − | + |
| V5915/1996/3 | Semen | + | + | + |
| V152/1997/3 | Semene | + | + | + |
| V200/1997/4 | Aborted foal | +b | + | + |
Passage in Vero cells.
A major part of the infected cells were reactive.
A minor part of the infected cells were reactive.
Obtained 3 months later from the same stallion that provided isolate V273/1996/4.
Obtained 1 month later from the same stallion that provided isolate V5915/1996/3.
The recombinant vaccinia viruses vAVE07, vAVE16, and vAVE25, which express the EAV N, M, and GL proteins, respectively (2, 14), were propagated in RK-13 cells and their titers were determined. Transient expression experiments were carried out in OST-7.1 cells as described by Balasuriya et al. (2).
Plasmid construction.
Recombinant DNA techniques were adopted from Sambrook et al. (37) and Ausubel et al. (1). To produce a fusion product of the EAV N protein and the amino terminus of the bacteriophage MS2 polymerase, the 0.5-kb HindIII fragment from pAVI07 (14) was inserted into the HindIII site of the bacterial expression vector pEX30A (44). The resulting construct carrying the EAV-specific part in the proper orientation was denoted pAVP07. ORF 4 was cloned behind the T7 promoter of pBluescript KS(+) by insertion of a filled-in 0.6-kb DdeI fragment from cDNA clone PB106 (14) into the SmaI site of the vector to generate plasmid pAVI04. Subsequently, pAVI04 was digested with SpeI and BglII, and the large vector-containing fragment was ligated to an adapter molecule consisting of oligonucleotides 830 (5′-CTAGGATCCACCACCATGAA-3′) and 831 (5′-GATCTTCATGGTGGTGGATC-3′). The resulting construct was designated pMRI14. The cloning results were verified by restriction enzyme digestions and nucleotide sequence analysis of alkali-denatured plasmid DNA with a T7 DNA polymerase sequencing kit (Amersham Pharmacia Biotech) and [α-33P]dATP (≥2,500 Ci per mmol; 10 mCi per ml; Amersham Pharmacia Biotech). Recombinant plasmids were transfected into Escherichia coli strain PC2495 (Phabagen). In the case of construct pAVP07, the bacterial acceptor strain already contained plasmid pcI857 (36) which encodes a temperature-sensitive mutant of the bacteriophage lambda cI repressor protein.
Inclusion body purification.
A fresh 20-ml culture of pAVP07- and pcI857-transfected PC2495 cells was mixed with 180 ml of prewarmed Luria-Bertani medium containing 50 mg of ampicillin and kanamycin per ml, and the mixture was incubated overnight at 30°C. Next, 800 ml of similar medium heated to 42°C was added to the bacterial culture. After incubation for 4 h at 42°C, the bacteria were pelleted by centrifugation for 10 min at 5,000 rpm and 4°C in a JA10 rotor (Beckman). The cell pellet was resuspended in 60 ml of ice-cold lysis buffer (50 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA [TNE]) and frozen at −70°C. The suspension was then thawed and supplemented with the protease inhibitors leupeptin, pepstatin A, and phenylmethylsulfonyl fluoride to final concentrations of 1, 1, and 50 μg per ml, respectively. After stirring for 5 min at 4°C, 60 mg of lysozyme was added and the sample was incubated for 15 min at room temperature (RT). The sample was then squeezed three times through a 21-gauge needle, supplemented with 120 μl of Triton X-100, 600 μl of 1 M MgCl2, 60 μl of 1 M MnCl2, and 600 μg of DNase I (grade II; Boehringer Mannheim), and incubated for another quarter of an hour at RT. The cell lysate was centrifuged for 15 min at 10,000 rpm and 4°C in a JA20 rotor (Beckman), and the resulting pellet was resuspended in 30 ml of ice-cold TNE–1% Nonidet P-40–1% sodium deoxycholate. The inclusion bodies were pelleted from the suspension by another round of centrifugation and were washed with 50 mM Tris-HCl (pH 8.0) containing 1, 3, and 5 M urea, respectively. The final protein preparation (named FP07) was taken up in 3 ml of phosphate-buffered saline (PBS), (partially) dissolved by sonication, and analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide (PAA) gel electrophoresis (PAGE) prior to injection into rabbits.
Polyclonal antisera.
The production and characterization of the EAV M protein-specific antipeptide serum has been reported previously (14). To generate an antiserum that recognizes the whole EAV N protein, a rabbit was injected subcutaneously with 2 ml of antigen emulsion containing 1 ml of Freund's complete adjuvant and 1 mg of FP07 in 1 ml of PBS. At 4, 8, and 12 weeks after primary immunization, the animals were boosted with 500 μg of antigen in incomplete Freund's adjuvant. Two weeks later the animal was bled and the serum was stored in aliquots at −20°C.
MAbs.
The properties of EAV GL protein-specific MAb 93B have been documented elsewhere (23). The new panel of anti-EAV MAbs was established by fusion of Sp2/0-Ag14 myeloma cells with spleen cells of STU mice (38) immunized by the intraperitoneal route with the Bucyrus strain of EAV. The virus used for immunization was propagated in Vero cells, purified by density gradient centrifugation as described above, and mixed with an equal volume of aluminum hydroxide adjuvant (Pierce). To diminish the production of antibodies directed against host cell components, immunotolerance against uninfected Vero cells was induced in the mice prior to antigen administration (49). For the primary immunization 109 50% tissue culture infective doses (TCID50s) of virus contained in a total volume of 350 μl was used per mouse. The mice were boosted with 250 μl of the same antigen preparation at 26 days after the first immunization. A final injection with 3 × 108 TCID50s of virus in 200 μl of TBS without adjuvant was given 3 weeks after the second immunization and 3 days before the cell fusion. Splenocytes were fused with myeloma cells essentially as described previously (49). An indirect immunofluorescence test (see below) carried out with EAV- and mock-infected Vero cells was used to screen the hybridoma-spent media for anti-EAV MAbs. Hybridomas that secreted EAV-specific MAbs were cloned by the limiting dilution method. The isotypes of the MAbs were determined by using an IsoStrip Mouse Monoclonal Antibody Isotyping Kit (Boehringer Mannheim). To measure the antibody concentration in the hybridoma culture supernatants, a commercial ELISA for the quantification of murine immunoglobulin G (IgG) (Boehringer Mannheim) was used.
Indirect immunoperoxidase monolayer assay (IPMA).
Mock- and EAV-infected Vero cells in 96-well microtiter plates (Greiner) were fixed for 25 min with ice-cold 96% ethanol at the indicated time points after infection. Next, the cells were washed two times for 3 min each time with PBS and incubated overnight at RT with EAV-specific MAb diluted in PBS containing 5% FCS (PBS–5% FCS) and 0.1% Tween 20. After several 3-min washes with PBS, the cells were incubated for 45 min with a 1:1,000 dilution of a horseradish peroxidase (HRPO)-conjugated goat-anti mouse IgG (heavy and light chains) antibody (Bio-Rad) in PBS–5% FCS. Following three additional washes with PBS, 100 μl of substrate solution (200 mM sodium acetate [pH 5.0] containing 200 μg of aminoethylcarbazole [AEC] per ml and 0.03% H2O2) was added to the cells. The enzymatic reaction was terminated after 20 min by replacing the substrate with water.
ELISA.
The ELISA was identical to the IPMA except that 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) (ABTS) was used as a substrate. After addition of 100 μl of a ready-to-use ABTS solution (Hoechst) to each well, the plates were incubated for 20 min at RT. The HRPO reaction was stopped by adding 50 μl of 1% SDS to each well, and the optical density was measured at 405 nm in an SLT Spectra plate reader.
Metabolic labeling experiments.
For the EAV- and mock-infected BHK-21 cells, the radiolabeling was started at 8 to 10 h postinfection (p.i.), while the OST-7.1 cells infected with recombinant vaccinia viruses were labeled at 5 h after addition of the inoculum. In each case, the radiolabeling was preceded by a starvation period of 30 min. The labeling medium consisted of DMEM without l-cystine, l-glutamine, and l-methionine (BioWhittaker) which was supplemented with 5% dialyzed FCS, 10 mM HEPES (pH 7.4), 2 mM l-alanyl–l-glutamine, and 20 μl of Redivue Pro-mix l-[35S]-in vitro cell labeling mix ([35S]Met plus [35S]Cys; >1,000 Ci per mmol; 10 mCi per mmol; Amersham Pharmacia Biotech) per ml. After labeling for 30 min to 2 h, the culture dishes were placed on ice. True cell lysates or total lysates of cells and media were then prepared by established procedures (17). Whenever indicated the SDS concentration in the postnuclear supernatant was increased to 0.5%.
Immunoprecipitation, gel electrophoresis, and fluorography.
Immunoprecipitations were performed as described by Balasuriya et al. (2). When lysates of EAV-infected cells were used as the antigen, the immunoprecipitations were either carried out in the absence of a reducing agent or in the presence of 1 mM dithiothreitol to disrupt disulfide-linked GL-M heterodimers. Prior to analysis in SDS–15% PAA gels, the immunoprecipitates were resuspended in 30 μl of sample buffer and were either incubated for 5 min at 96°C (N protein) or left at RT for ≥15 min (M and GL proteins). After electrophoresis the gels were fixed, impregnated with scintillant, and dried on filter paper as described previously (41). The gels were exposed at −70°C to Kodak X-OMAT LS films, and molecular weight estimations were based on digital images of the fluorographs obtained with a DC40 camera (Eastman Kodak Company) and were analyzed with Kodak Digital Science 1D image analysis software, version 1.6.
Immunoblotting.
Sucrose gradient-purified virus particles were dissolved in SDS-PAGE sample buffer containing 5% 2-mercaptoethanol and were applied to a discontinuous SDS-PAA gel containing 12% acrylamide. After electrophoresis the proteins were transferred to a nitrocellulose membrane (46) which was cut in strips and blocked with 5% nonfat dry milk powder (Heirler) in TBS containing 0.1% Tween 20 (TBST) for 1 h at RT. Next, the filter pieces were incubated with the culture medium (diluted 1:10 in the case of the N-specific MAbs and 1:2 in the case of the anti-GL antibodies) of the EAV-specific hybridomas for 1 h at ambient temperature, followed by incubation for 16 h at 4°C, and were washed three times for 5 min each time in TBST. The nitrocellulose strips were probed with a biotin-labeled anti-mouse IgG antibody (MaxTag Immunoblotting Kit; Biotrend) diluted 1:200 in TBST for 30 min at RT. After three 10-min washings, HRPO-conjugated streptavidin was added to the filter paper pieces at a dilution of 1:200 in TBST. Following a 30-min incubation, the nitrocellulose strips were washed again, and AEC-containing substrate solution was added (see above). The enzymatic reaction was stopped after 15 min.
VN assays.
To find out whether an MAb displayed any VN activity, 10-fold serial dilutions of EAV in DMEM–10% FCS were mixed with equal volumes of undiluted hybridoma culture supernatant or with fresh culture medium, and the mixtures were incubated for 1 h at 37°C. Next, 100 μl of the virus-antibody mixture was added to 3 wells of a 96-well microtiter plate containing confluent monolayers of Vero cells. Three days later the wells were screened for the development of cytopathic effects (CPEs) and were stained for viral antigen in an IPMA carried out with the N-specific MAb E1/b5. The neutralization titer of the VN MAbs was subsequently determined by incubation of twofold serial dilutions of the MAb-containing culture medium with 100 TCID50s of EAV B15 for 1 h at 37°C. It was expressed as the reciprocal of the maximum dilution at which 100% of the cell monolayers were protected from a CPE and/or stained negative for viral antigen in an IPMA. Finally, a complement-dependent neutralization test was performed to analyze whether the nonneutralizing GL-specific MAbs rendered EAV sensitive to inactivation by complement. To this end, serial 10-fold dilutions of EAV B15 were mixed with an equal volume of a 1:2 dilution of each MAb in DMEM–10% FCS or with an equal volume of culture medium devoid of antibodies. Following incubation for 1 h at 37°C, complement-containing guinea pig serum (Sigma) diluted in DMEM–10% FCS was added to the virus mixtures to final concentrations of between 5 and 20%. After another 1-h incubation at 37°C, the samples were added to 1-day-old monolayers of Vero cells in 96-well microtiter plates. Following an incubation period of 3 days, the cells were subjected to IPMA as described above.
Generation of NR virus variants.
To select NR viral escape mutants, 5 × 104 TCID50s of EAV B15 in 5 μl were incubated with 1 ml of undiluted culture supernatant of hybridomas E3/c16, E3/d10, E6/A3, E6/B8, E7/d15-c1, and E7/d15-c9 for 1 h at 37°C. The virus-antibody mixtures were then added to confluent monolayers of Vero cells and the mixtures were incubated at 37°C until cytopathic changes occurred. A 5-μl portion of the supernatant of the infected cells was incubated for 1 h at 37°C with 1 ml of the selecting MAb and was used to inoculate fresh monolayers of Vero cells. After another round of selection, a plaque assay was performed and the cell cultures were covered with a 0.5% agarose overlay. Individual plaques were collected by aspiration at 96 h p.i. and were used for the preparation of large virus stocks in Vero cells. NR virus variants received the name of the MAb used for their selection followed by the plaque number.
Immunofluorescence assay (IFA).
Vero cells in suspension were either mock infected or infected with EAV B15 at a multiplicity of infection of 0.2 TCID50s per cell and were seeded onto multitest slides. At 26 h after plating the cells were briefly immersed twice in PBS, fixed for 25 min with ice-cold 96% ethanol, air dried, and stored at −70°C until use. After thawing, the cells were incubated for 90 min with EAV-specific MAbs at various dilutions in PBS–5% FCS. Following three 2-min washes with PBS, the cells were stained with a fluorescein-conjugated horse-anti mouse IgG (heavy and light chain) antibody (Camon) diluted 1:150 in PBS–5% FCS for 45 min at ambient temperature. After washing the cells were embedded in glycerol buffer containing 25 mg of 1,4-diazobicyclo-(2,2,2)-octane and 2 μg of propidium iodide per ml. The samples were examined with a Zeiss Axioskop microscope, and pictures were taken with a Zeiss ×40 objective by using Ilford 400 Delta Professional black and white film.
Immunoelectron microscopy.
Sucrose-gradient-purified virus was adsorbed to copper grids (400 mesh) for 10 min at RT. The grids were subsequently washed four times for 3 min each time with PBS–0.5% bovine serum albumin (BSA) and incubated for 45 min with undiluted hybridoma culture supernatant. After further washing steps with PBS–0.5% BSA, the grids were immersed for 45 min in drops of goat anti-mouse IgG plus IgM (heavy and light chains)–5-nm gold conjugate (Biocell) diluted 1:15 in PBS–0.5% BSA. Thereafter, the grids were washed three times with PBS–0.5% BSA, three times with PBS, and once with distilled water. They were then subjected to negative staining with 1% unbuffered uranyl acetate solution for 45 s at ambient temperature. The specimens were examined and photographed with a Zeiss 910 electron microscope.
RESULTS
Production of EAV-specific MAbs.
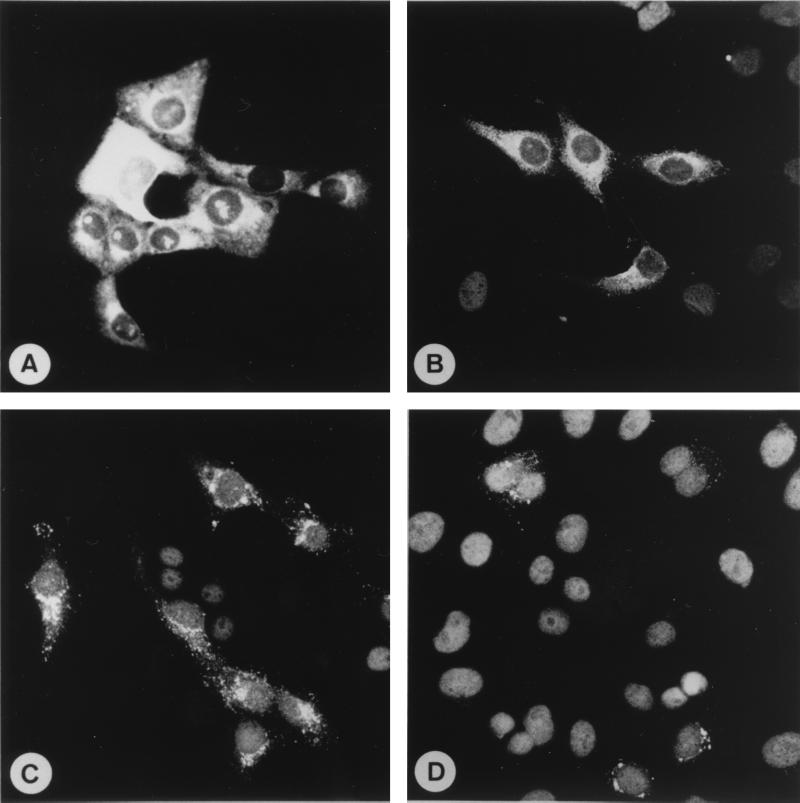
Immunization of mice with PRRSV particles has yielded MAbs directed against five different structural proteins (20, 48, 50, 53). In contrast, all EAV-specific murine MAbs that have been obtained thus far recognize either the N or the GL protein of the virus (2, 3, 4, 6, 12, 23, 29, 52). A possible explanation for this discrepancy is the fact that only BALB/c mice have been immunized for the production of EAV-specific MAbs, while different mouse strains were used to generate PRRSV-specific hybridomas. In an attempt to expand the existing repertoire of EAV-specific MAbs, STU mice were intraperitoneally injected with a sucrose gradient-purified preparation of the B15 isolate of EAV. To increase the likelihood of recovering hybridomas that secrete virus-specific antibodies, the mice were made immunotolerant for the Vero cells used to propagate EAV prior to immunization. Three fusion experiments yielded a total of 780 hybridomas after plating of 20% of the fusion products of each spleen into four 96-well and two 24-well plates. Twenty-eight hybridomas recognized EAV-specific antigen in an indirect IFA carried out with EAV- and mock-infected Vero cells, and 16 of them remained stable after cloning. Interestingly, different staining patterns were observed for the EAV-specific MAbs when applied as undiluted hybridoma culture supernatants. Ten of the MAbs (MAbs E1/A19, E1/b5, E1/c4, E3/a9, E3/a61, E3/d17, E3/d52, E6/A7, E6/a27, and E6/d24) gave a very intense cytoplasmic fluorescence that required only very short exposure times for recording (Fig. 1A). Furthermore, all of these MAbs stained distinct areas in some of the nuclei of EAV-infected cells but not the nuclei of mock-infected cells. A very similar immunofluorescence pattern was previously reported for a panel of six MAbs directed against the N protein of the vaccine strain of EAV (29). Three other MAbs (MAbs E3/c16, E3/d10, and E6/B8) caused a bright labeling of the cytoplasms of EAV-infected Vero cells (Fig. 1B). When used at a higher dilution the distal parts of the cells were no longer labeled by these antibodies. Instead, the immunofluorescence signal was concentrated in juxtanuclear granular structures. The sizes of these granules appeared to decrease with increasing distance from the nucleus. The labeling pattern of MAb E7/d15-c1 was also characterized by fine and coarse dots in the perinuclear region (Fig. 1C). MAb E6/A3 produced the weakest immunofluorescence signal but showed an intracellular distribution comparable to that of MAb E7/d15-c1 (Fig. 1D). The staining pattern of MAb E7/d15-c9 could not be easily deduced, as this antibody cross-reacted with an unidentified host cell component (data not shown).
FIG. 1.
Immunofluorescence staining of EAV B15-infected Vero cells with MAbs E1/A19 (A), E6/B8 (B), E7/d15-c1 (C), and E6/A3 (D). The cells were fixed at 26 h p.i. with ice-cold 96% ethanol. Incubation of mock-infected Vero cells with the same antibodies did not yield a detectable immunofluorescence signal (data not shown).
Isotyping of the anti-EAV MAbs revealed that they belonged to the IgG1, IgG2a, or IgG2b subclass (Table 1). Quantification of the immunoglobulins secreted in the culture supernatants indicated that the antibody concentration varied from 3 μg per ml for hybridoma E1/A19 to 85 μg per ml for hybridoma E6/A7 (Table 2). These hybridoma culture media were subsequently used for an ELISA in which fixed EAV-infected Vero cells served as the specific antigen and fixed mock-infected Vero cells were used as the negative control. As is evident from Table 1, the immunoglobulin concentration in the hybridoma culture media did not correlate with the optical densities (ODs) obtained in the ELISA. For instance, MAbs E3/a9, E3/a61, and E3/d17 produced high net absorbance values in the ELISA but were of low concentration. Likewise, the highly concentrated E7/d15-c1 and E7/d15-c9 antibody preparations gave rise to rather low ELISA titers. Furthermore, MAb E7/d15-c9 showed an exceptionally high background absorbance which is consistent with the IFA results that showed cross-reactivity with a cellular protein. The group of six MAbs that produced a granular intracellular immunofluorescence pattern yielded ODs that were at the lower end of the spectrum. The ELISA readings for these MAbs generally correlated with their IFA signals; e.g., MAb E6/A3 produced the weakest immunofluorescence staining and gave the lowest specific absorbance value in the ELISA. Finally, an IPMA was used to establish the maximum dilution at which the MAbs still gave a clearly detectable coloration of EAV-infected Vero cells. As is shown in Table 1, the highest dilution of the MAbs at which a specific signal was observed was not merely determined by the initial antibody concentration.
TABLE 1.
Characteristics of EAV-specific MAbs
| MAba | Isotype | IgG concn (μg/ml) | ELISA ODb | Antibody titerc | Protein specificity | Neutralization (titer)d |
|---|---|---|---|---|---|---|
| E1/A19 | 2a | 3 | 2.6 (0.21) | 40 | N | − |
| E1/b5 | 1 | 20 | 3.3 (0.18) | 80 | N | − |
| E1/c4 | 1 | 5 | 2.0 (0.19) | 20 | N | − |
| E3/a9 | 1 | 5 | 3.2 (0.18) | 20 | N | − |
| E3/a61 | 1 | 9 | 3.2 (0.24) | 20 | N | − |
| E3/c16 | 2a | 42 | 1.9 (0.32) | 40 | GL | − |
| E3/d10 | 2b | 30 | 1.4 (0.31) | 10 | GL | − |
| E3/d17 | 2b | 9 | 3.3 (0.21) | 20 | N | − |
| E3/d52 | 2b | 12 | 2.4 (0.22) | 20 | N | − |
| E6/A3 | 2a | 22 | 0.6 (0.21) | 3 | GL? | +(80) |
| E6/A7 | 1 | 85 | 2.6 (0.21) | 160 | N | − |
| E6/a27 | 1 | 5 | 1.8 (0.18) | 10 | N | − |
| E6/B8 | 1 | 12 | 1.0 (0.28) | 10 | GL | − |
| E6/d24 | 1 | 4 | 2.8 (0.17) | 40 | N | − |
| E7/d15-c1 | 2b | 50 | 1.2 (0.32) | 20 | GL | +(40) |
| E7/d15-c9 | 2a | 42 | 1.7 (0.71) | 40 | GL | +(5) |
Hybridoma culture supernatant.
OD reading for EAV-infected (mock-infected) Vero cells that were fixed to the ELISA plate.
By IPMA as the reciprocal of the highest dilution of the culture supernatant at which specific cytoplasmic reactivity was clearly observed.
Reciprocal of the highest dilution of the culture supernatant that completely inhibited the CPE of 100 TCID50s of EAV.
TABLE 2.
Reactivities of (putative) GL-specific MAbs with NR virus variants in IPMA
| Escape mutant | Reactivity
|
|||||
|---|---|---|---|---|---|---|
| Nonneutralizing anti-GL MAbs
|
Neutralizing anti-GL MAbs
|
|||||
| E3/c16 | E3/d10 | E6/B8 | E6/A3 | E7/d15-c1 | E7/d15-c9 | |
| NR E6/A3, plaque no. 5 | + | + | + | − | + | + |
| NR E7/d15-c1, plaque no. 7 | + | + | + | + | − | + |
| NR E7/d15-c9,a plaque no. 4 | + | + | + | + | + | + |
Although stainable by the MAb used for selection, this NR was less well neutralized by MAb E7/d15-c9 than the parental virus.
EAV-specific MAbs recognize either the EAV N or GL protein.
To get a first impression of the protein specificity of the EAV-specific MAbs, EAV Utr- and mock-infected BHK-21 cells were metabolically labeled for 2 h starting at 8 h p.i. Total lysates of cells and media were prepared, aliquots of which were incubated with culture supernatants of each of the EAV-specific hybridomas or with monospecific antisera that recognize the three major virion proteins (N, M, and GL) of EAV. The resulting immunoprecipitates were analyzed in an SDS–15% PAA gel. This experiment indicated that nine MAbs (MAbs E1/A19, E1/b5, E1/c4, E3/a9, E3/a61, E3/d17, E3/d52, E6/A7, and E6/a27) recognized the EAV N protein, whereas two MAbs (MAbs E7/d15-c1 and E7/d15-c9) precipitated substantial amounts of both the EAV GL and M proteins. The latter finding can be explained by the fact that the EAV GL and M proteins form disulfide-linked heterodimers in virus-infected cells (16). For MAbs E3/c16, E3/d10, E6/A3, E6/B8, and E6/d24, no decisive conclusion could be drawn as to the EAV proteins that they recognized (data not shown).
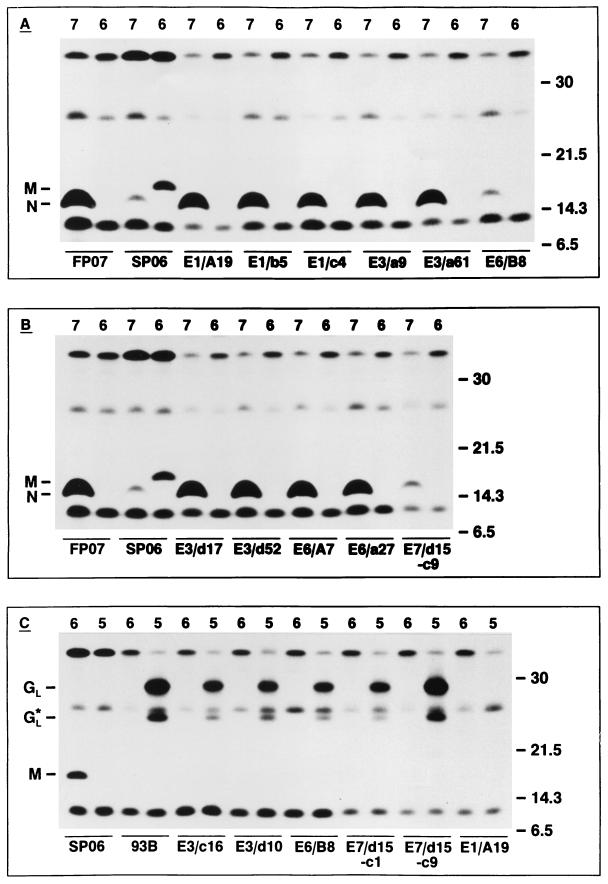
To unequivocally determine the protein specificity of the MAbs directed against EAV, we used vaccinia virus recombinants that express the genes for the GL, M, or N protein of EAV Utr. Immunoprecipitations were performed with each of the MAbs with lysates from vAVE07-, vAVE16-, or vAVE25-infected OST-7.1 cells as the antigen source. As is clear from Fig. 2A and B, MAbs E1/A19, E1/b5, E1/c4, E3/a9, E3/a61, E3/d17, E3/d52, E6/A7, and E6/a27 specifically recognized a 15-kDa protein in lysates of vAVE07-infected cells but not in extracts from cells infected with vAVE06. As the same protein species was recognized by an antiserum directed against an N-specific bacterial fusion protein, we concluded that these MAbs indeed react with the core protein of EAV. To exclude the possibility that the precipitation of the independently expressed EAV N protein was the result of direct binding to the immunosorbent and/or the formation of aggregates, we also incubated the lysate of vAVE07-infected cells with two GL-specific MAbs (MAbs E6/B8 and E7/d15-c9; see below). With either of these MAbs, only minute amounts of the EAV N protein were brought down, which confirms our previous interpretation that MAbs E1/A19, E1/b5, E1/c4, E3/a9, E3/a61, E3/d17, E3/d52, E6/A7, and E6/a27 are N specific. Of the remaining seven MAbs, five reacted with proteins of 29 and 25 kDa in the lysate of the vAVE25-infected OST-7.1 cells (Fig. 2C). Since the same two protein species were recognized by GL-specific MAb 93B (23) and none of these MAbs specifically immunoprecipitated a polypeptide from the lysate of vAVE16-infected cells, our data showed that MAbs E3/c16, E3/d10, E6/B8, E7/d15-c1, and E7/d15-9 are GL specific.
FIG. 2.
(A and B) Immunoprecipitation analysis of N-specific MAbs. OST-7.1 cells were infected with the vaccinia virus recombinants vAVE07 or vAVE16, which express the EAV N and M proteins, respectively, and were labeled for 1 h starting at 5 h p.i. with [35S]Met plus [35S]Cys. Total lysates of cells and media were prepared and incubated with 0.5 to 3 μl of N-specific (FP07) or M-specific (SP06) rabbit antiserum or 100 μl of culture supernatant from mouse hybridoma E1/A19, E1/b5, E1/c4, E3/a9, E3/a61, E3/d17, E3/d52, E6/A7, E6/a27, E6/B8, or E7/d15-c9. (C) Immunoprecipitation analysis of GL-specific MAbs. OST-7.1 cells were infected with the vaccinia virus recombinants vAVE16 (lanes 6) and vAVE25 (lanes 5), which express the EAV M and GL proteins, respectively (12), and were labeled as described above. Total lysates of cells and media were prepared and incubated with 0.5 μl of anti-M rabbit serum (SP06) or ascitic fluid containing the GL-specific MAb 93B (IgG2a subtype) (23) or 100 μl of culture supernatant from mouse hybridoma E3/c16, E3/d10, E6/B8, E7/d15-c1, E7/d15-c9, or E1/A19. The numbers at the right indicate the sizes (in kilodaltons) and positions of marker proteins analyzed in the same SDS–15% PAA gels. The positions of the N and M proteins and the full-length and a truncated form (GL*) (12) of the GL glycoprotein are displayed at the left.
MAbs E6/A3 and E6/d24 did not recognize the independently expressed N, M, or GL protein of EAV Utr, nor did they react with the expression products of ORFs 2b and 4 at an SDS concentration in the undiluted cell lysate of 0.5% (data not shown). The reactivities of these antibodies with the translation products of ORFs 2a and 3 have not been tested.
The failure to determine the protein specificities of MAbs E6/A3 and E6/d24 in immunoprecipitation assays may be explained in three ways: (i) MAbs E6/A3 and E6/d24 are directed against conformational epitopes which are destroyed during the analytical procedures, (ii) MAbs E6/A3 and E6/d24 are of low avidity and/or recognize poorly expressed proteins, and (iii) MAbs E6/A3 and E6/d24 react with epitopes that differ between EAV B15 and EAV Utr. Although Table 1 indicates that MAb E6/A3 may indeed possess poor binding characteristics, this is definitely not the case for MAb E6/d24, which had relatively high IPMA and ELISA titers, despite a low antibody concentration (Table 1). Moreover, the observation that each of the EAV-specific MAbs including E6/A3 and E6/d24 did recognize Vero cells infected with the Utr strain of EAV in an IPMA (data not shown) militates against the third possibility. In addition, MAb E6/A3 efficiently neutralized EAV Utr (data not shown).
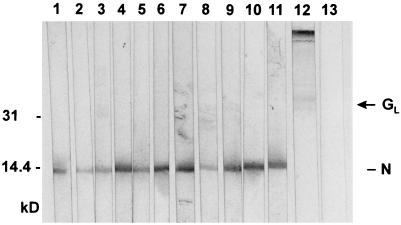
As an alternative approach to establishing the protein specificities of MAbs E6/A3 and E6/d24, we performed immunoblotting experiments with EAV B15 virions that serve as the antigen. Remarkably, in these assays MAb E6/d24 clearly recognized the EAV N protein (Fig. 3, lanes 10 and 11), whereas MAbs E6/A3, E7/d15-c1, and E7/d15-c9 did not specifically bind to any viral protein on the Western blot (data not shown). In contrast, MAbs E3/c16, E3/d10, and E6/B8 weakly bound to a polypeptide of between 30 and 42 kDa (Fig. 3, lane 12) which, on the basis of its size and appearance, is believed to correspond to the EAV GL protein. In agreement with our earlier findings, MAbs E1/A19, E1/b5, E1/c4, E3/a9, E3/a61, E3/d17, E3/d52, E6/A7, and E6/a27 strongly recognized the EAV N protein (Fig. 3, lanes 1 through 9).
FIG. 3.
Immunoblot analysis of EAV-specific MAbs. Nitrocellulose strips containing electrophoretically separated virion proteins of EAV were incubated with MAbs E1/b5 (lane 1), E1/c4 (lane 2), E1/A19 (lane 3), E3/d17 (lane 4), E3/d52 (lane 5), E3/a61 (lane 6), E3/a9 (lane 7), E6/a27 (lane 8), E6/A7 (lane 9), and E6/d24 (lane 10, diluted 1:3; lane 11, diluted 1:8), with a mixture of MAbs E3/c16, E3/d10, and E6/B8 (lane 12), and with the anti-PRRSV MAb P11/d72 (lane 13) (50). The positions of the (putative) N and GL proteins of EAV are shown at the right side; the numbers at the left side indicate positions and sizes (in kilodaltons) of marker proteins analyzed in the same gel.
Detection of neutralizing and nonneutralizing anti-EAV GL MAbs.
Experiments in which constant amounts of antibody were mixed with different doses of virus indicated that only MAbs E6/A3, E7/d15-c1, and E7/d15-c9 displayed VN activity (data not shown). To compare the neutralizing capacities of these MAbs, a constant number of infectious virus particles was mixed with different amounts of each antibody and added to Vero cells. Complete inhibition of the CPE was accomplished by the culture supernatant of hybridoma E6/A3 at dilutions of up to 1:80, by the E7/d15-c1 culture supernatant at a maximum dilution of 1:10, and by the E7/d15-c9 culture supernatant at dilutions of up to 1:5 (Table 1). Protection of half of the cell monolayers was observed after treatment of the virus with 1:320, 1:160, and 1:20 dilutions of MAbs E6/A3, E7/d15-c1, and E7/d15-c9, respectively. However, despite the absence of a visible CPE, single cells and small foci expressing the EAV N protein were detected by IPMA in the cell cultures inoculated with the virus-antibody complexes at 3 days after infection. In concordance with the neutralization titers, the least residual infectivity was observed in the monolayers infected with the E6/A3-treated virus, whereas E7/d15-c9-treated EAV particles yielded the largest number of infectious centers. Thus, only three MAbs showed VN activity.
To investigate whether the nonneutralizing GL-specific MAbs could sensitize EAV for neutralization by complement (see references 27 and 35 and references therein) a microneutralization test in the presence of guinea pig serum was performed. None of the VN− anti-GL MAbs was capable of blocking EAV infectivity in the presence of complement. Furthermore, addition of complement did not affect the neutralization indices of MAbs E6/A3, E7/d15-c1, and E7/d15-c9 (data not shown).
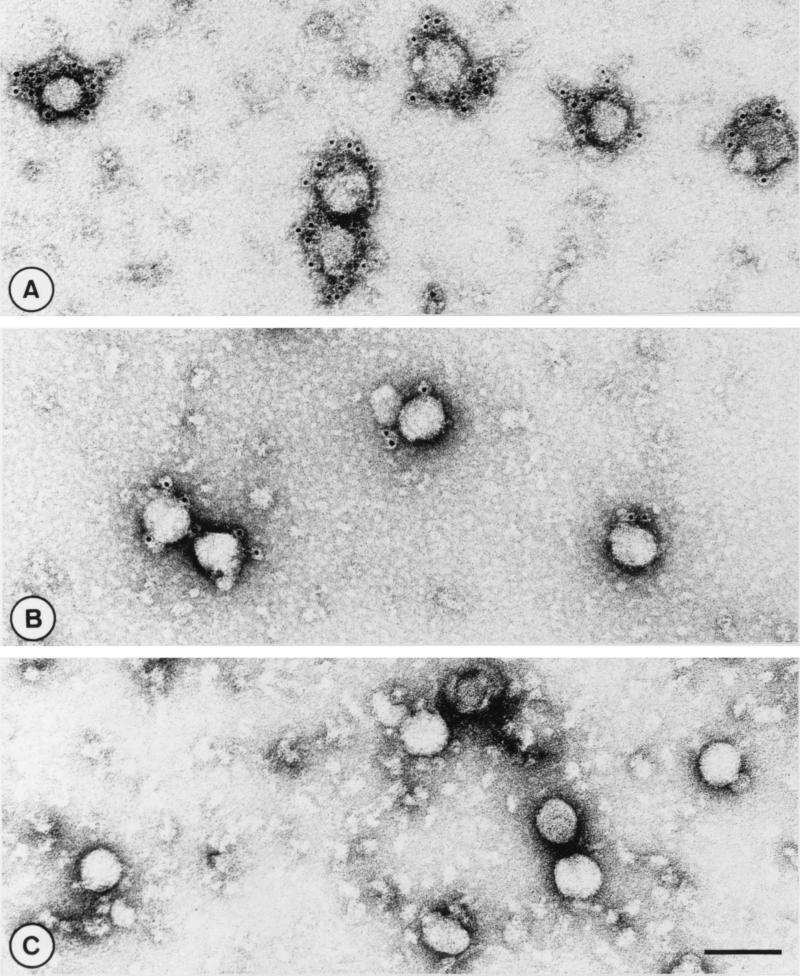
Immunoelectron microscopy.
To investigate whether the different anti-GL MAbs recognized surface components of virions, immunoelectron microscopy was performed. For this purpose, the virus particles were first incubated with diluted hybridoma culture supernatant and were then immersed in a solution of goat anti-mouse antibodies conjugated to 5-nm gold particles. As expected, none of the N-specific MAbs interacted with the viral surface (Fig. 4C). In contrast, all VN+ and GL-specific MAbs mediated binding of immunogold particles to the viral envelope. Interestingly, labeling was more pronounced with the neutralizing MAbs than with the nonneutralizing anti-GL MAbs (compare Fig. 4A and B). Statistical analyses of 100 virions per antibody indicated that the neutralizing MAb E7/d15-c1 on average mediated the binding of 8.8 gold granules per virus particle, whereas the average number of gold particles attached to E3/c16-coated virions was 2.7. A similar threefold difference in the recruitment of gold particles at the viral surface was observed when the binding mediated by VN+ antibodies E6/A3 and E7/d15-c9 and the binding mediated by VN− MAbs E3/d10 and E6/B8 were compared.
FIG. 4.
Immunoelectron microscopy of EAV particles. Sucrose gradient-purified virions were first incubated with MAb E7/d5-c1 (A), E3/c16 (B), or E1/A19 (C) and were then labeled with gold-conjugated secondary antibodies. Bar, 100 nm.
Selection of NR variant viruses.
To find out whether the VN antibodies recognized related or independent epitopes, EAV B15 was repeatedly passaged in the presence of MAbs E6/A3, E7/d15-c1, and E7/d15-c9 in an attempt to generate NR escape mutants of the virus. Individual virus clones were then selected in plaque assays, amplified in Vero cells, and subjected to microneutralization tests. For comparison, the selection procedure was also performed with the nonneutralizing GL-specific MAbs E3/c16, E3/d10, and E6/B8. Eleven NR virus variants of MAb E6/A3 were obtained, whereas treatment of EAV B15 with MAb E7/d15-c1 yielded one NR escape mutant. The E6/A3 NR viruses were recognized by all five GL-specific antibodies in IPMAs (Table 2) and were neutralized with the same efficiency as the parental virus by MAbs E7/d15-c1 and E7/d15-c9 (data not shown). Similarly, the E7/d15-c1-specific viral escape mutant was no longer labeled or neutralized by the selecting MAb, but cells infected with this virus were stained as intensively with the other four anti-GL MAbs and the strongly neutralizing MAb E6/A3 as EAV B15-infected cells. Furthermore, MAbs E6/A3 and E7/d15-c9 were still able to neutralize E7/d15-c1 NR virus infectivity. In contrast, the plaque derivatives of EAV B15 that were obtained after treatment of the virus for three consecutive passages with MAb E7/d15-c9 were still recognized by the selecting MAb in IPMAs (Table 2). However, these virus clones were less well neutralized by MAb E7/d15-c9 than the parental virus. Finally, the properties of EAV B15 were not detectably altered after selection with the nonneutralizing GL-specific MAbs E3/c16, E3/d10, and E6/B8. Taken together these data indicate that the three VN+ MAbs recognize distinct epitopes.
Interaction of MAbs with field isolates of EAV.
To discriminate MAbs directed against conserved epitopes from antibodies that recognize variable protein domains, we compared the binding of each of the 16 EAV-specific MAbs to a panel of 23 field isolates of EAV in IPMAs of virus-infected Vero cells. The 10 N-specific MAbs as well as the 3 nonneutralizing anti-GL MAbs and weakly neutralizing MAb E7/d15-c9 recognized all virus isolates. In contrast, the other two VN+ MAbs reacted only with a subset of the field viruses. Eight EAV isolates were recognized by both of these MAbs, two field viruses reacted with MAb E7/d15-c1 but not with MAb E6/A3, and eight other EAV isolates were recognized by MAb E6/A3 but not by MAb E7/d15-c1. Four field viruses did not react with either of the two strongly neutralizing MAbs. Although the IPMAs were always carried out with monolayers infected with different amounts of virus to compensate for differences in titer and/or growth rate between EAV isolates, it was sometimes hard to decide whether a specific MAb recognized the entire virus population. In particular, MAb E6/A3 showed a very faint staining of infected cells in IPMAs that corresponded to its weak signal in IFA and ELISA, and it seemed to recognize only a fraction of the EAV-infected cells for the virus isolates V200, V979, V1791, V2416, V2716, and V8908 (Table 3). The different reactivities of the VN+ MAbs with the EAV field isolates once again illustrated that they recognize distinct epitopes.
DISCUSSION
In this paper, we have described the production and characterization of a panel of 16 EAV-specific MAbs. Immunoblotting and immunoprecipitation analyses revealed that 10 MAbs were directed against the EAV ORF 7-encoded N protein and five MAbs reacted with the EAV ORF 5-encoded GL protein. Two of the GL-specific MAbs (E7/d15-c1 and E7/d15-c9) displayed VN activity, while the other three anti-GL MAbs did not inhibit the infectivity of EAV. The protein specificity of one MAb (MAb E6/A3) could not be determined, but on the basis of its VN capacity we speculate that it is also directed against the EAV GL protein. The observation that MAb E6/A3 produces similar labeling patterns by immunofluorescence microscopy of infected cells and immunoelectron microscopy of virus particles as MAbs E7/d15-c1 and E7/d15-c9 would be consistent with this notion. Moreover, as mentioned above, the GL protein is the only target of EAV-neutralizing antibodies identified until now (2, 3, 4, 7, 12, 23, 29, 52, 54).
Although the immunization method and mouse strain that we used to produce EAV-specific MAbs differed from those used by others, like in previous studies (2, 3, 4, 6, 12, 23, 29, 52), only antibodies directed against the N and GL proteins of EAV were obtained. This result was unexpected since the same immunization procedure yielded MAbs specific for five different PRRSV polypeptides (50, 53). Moreover, the EAV M protein was recently shown to be more consistently recognized in immunoblots by serum antibodies obtained from naturally or experimentally infected horses than either the N or the GL protein (24, 31). Collectively, these findings indicate that differences exist between the murine and equine humoral immune responses to EAV and between the immunogenicities of the structural proteins of EAV and PRRSV in mice. If one assumes that arteriviruses have similar virion architectures, the antigenic differences between the structural proteins of EAV and PRRSV most likely result from subtle changes in their tertiary or quaternary structures or are a direct consequence of differences in their primary amino acid sequences. In addition, differences in the purity and integrity of the virus preparations used for immunization may have contributed to the observed differences in antibody response following the inoculation of mice with EAV or PRRSV. If indeed the stability of the virus has an effect on the number of structural proteins for which murine antibodies can be obtained, chemical and/or physical disruption of EAV particles may help to expand the existing repertoire of EAV-specific MAbs.
If MAb E6/A3 is indeed directed against the EAV GL protein, it probably recognizes a conformational epitope which is destructed during the preparation of protein samples for immunoblotting or immunoprecipitation. In contrast, the other VN+ GL-specific MAbs seem to be directed against epitopes that are preserved during cell lysis and immunoprecipitation analysis but which do not survive the denaturing conditions that apply to SDS-PAGE and Western blotting. The nonneutralizing GL-specific MAbs most likely interact with (partially) linear epitopes as they weakly bound to the GL protein in an immunoblot. The same may hold true for the N-specific MAbs which strongly recognized the N protein after Western blotting. Interestingly, the N-specific MAb E6/d24 reacted with the EAV N protein in IFA and IPMA and on an immunoblot but not in an immunoprecipitation analysis. A possible explanation for this peculiar result may be that MAb E6/d24 recognizes a linear epitope that is exposed on the Western blot and after fixation of virus-infected cells with 96% ethanol but is not accessible for antibody binding at the low detergent concentrations in the immunoprecipitation buffer. Alternatively, MAb E6/d24 recognizes a conformational epitope that is denatured by the detergents present in the cell lysis and immunoprecipitation buffer but refolds after transfer of the N protein to nitrocellulose and is preserved during ethanol fixation of EAV-infected cells. It thus seems that we have raised N-specific MAbs directed against at least two different epitopes. These binding sites are highly conserved since all 10 N-specific MAbs reacted with each of the EAV isolates tested. Ultimately, competitive binding assays will have to be performed to determine the number of independent epitopes recognized by the anti-N MAbs.
The GL-specific MAbs E3/c16, E3/d10, E6/B8, and E7/d15-c9 also reacted with all 23 field isolates of EAV in IPMAs. Since MAbs E3/c16, E3/d10, and E6/B8 exhibited similar properties in antigen binding assays and VN tests, they presumably recognize identical or related epitopes. MAb E7/d15-c9 most likely binds to a different part of the GL protein as it displayed weak VN activity, in contrast to the other three antibodies. The strongly neutralizing GL-specific MAb E7/d15-c1 reacted with only 10 of the 23 EAV field isolates and therefore recognizes yet another epitope on the major envelope glycoprotein of the virus. Consistently, MAb E7/d15-c1 NR escape mutants were still recognized in IPMAs by the four other GL-specific MAbs and by MAb E6/A3. Moreover, loss of the E7/d15-c1-specific epitope did not influence virus neutralization by MAbs E7/d15-c9 and E6/A3. A possible fourth epitope on the EAV GL protein may be defined by the strongly neutralizing MAb E6/A3 as it reacted with a different subset of the 23 EAV field isolates than MAb E7/d15-c1. In addition, NR virus variants of MAb E6/A3 were still recognized by the GL-specific MAbs E3/c16, E3/d10, E6/B8, E7/d15-c1, and E7/d15-c9. Competitive binding assays between MAb E6/A3 and the GL-specific MAbs produced by others (2, 3, 4, 12, 23, 29, 52, 54) may provide definitive evidence for its protein specificity. The same type of experiments and nucleotide sequence analyses of the E7/d15-c1- and E6/A3-specific NR escape mutants will also be helpful in defining the epitopes recognized by MAbs E3/c16, E3/d10, E6/A3, E6/B8, E7/d15-c1, and E7/d15-c9. Furthermore, it may be of interest to find out whether the viruses selected in the presence of MAbs E3/c16, E3/d10, E6/B8, and E7/d15-c9 have undergone sequence changes in the EAV GL protein.
A general picture that emerged from our studies on the GL-specific antibodies was that the VN+ MAbs weakly reacted with the intracellular GL protein in IFAs but strongly bound to extracellular virus particles, whereas the VN− MAbs poorly labeled secreted virions but produced a bright immunofluorescence in EAV-infected cells. The most plausible explanation for these findings is that there are differences in the accessibilities of the epitopes recognized by VN+ and VN− GL-specific MAbs between intra- and extracellular antigens. The observation that after dilution the immunofluorescence patterns obtained with MAbs E3/c16, E3/d10, and E6/B8 resembled those of undiluted MAbs E6/A3 and E7/d15-c1 may then be a consequence of the fact that the GL protein mainly accumulates at the intracellular side of virus assembly. In this scenario, the heavily labeled perinuclear granules would both contain GL molecules (being) incorporated in virus particles and GL molecules that are not engaged in virus assembly, while the more peripheral cytoplasmic areas which were only stained by the VN− MAbs would mainly contain spillover of the GL protein. The virus-associated GL protein would be most effectively recognized by the VN+ and the VN− MAbs, whereas the free GL molecules would be preferentially bound by the VN− MAbs. Such an interpretation would also fit the results obtained by van der Meer et al. (47), who found that at early times p.i. the GL protein is exclusively localized in the Golgi complex but that at later stages of infection vesicles elsewhere in the cytoplasm are labeled as well.
It may seem surprising that GL-specific MAbs E3/c16, E3/d10, and E6/B8 bound to the surfaces of virus particles but were unable to reduce EAV infectivity or to sensitize the virions for neutralization by complement. The amount of gold granules that decorated the viral surface after immunogold labeling of virions with these MAbs was, however, fairly low. Individual virus particles could thus still contain enough unshielded GL protein to mediate virus entry. In addition, McCollum and Swerczek (34) and Fukunaga et al. (22) previously noticed that complement enhancement of virus neutralization is a strain-dependent phenomenon. The fact that the neutralization titers of the VN+ MAbs were not affected by the presence of complement may indicate that EAV B15 indeed belongs to the virus isolates whose neutralization is insensitive to complement addition.
The relevance of VN− GL-specific antibodies in natural EAV infections remains to be elucidated. The absence of an absolute correlation between the virus neutralization titer of convalescent-phase equine sera and their reactivity with a GL-specific bacterial expression product in an ELISA (7) may indicate that horses do produce nonneutralizing anti-GL antibodies in response to an EAV infection, although this result could also be explained in a number of other ways. Moreover, Chirnside et al. (8) reported induction of nonneutralizing GL-specific antibodies in 9 of 25 horses immunized with a chemically inactivated whole-virus vaccine. The situation in naturally infected horses may, however, differ from that in vaccinated animals. The observation that the VN− anti-GL MAbs recognized each of the EAV isolates tested indicates that their epitopes are not subject to immune selection in vivo. These constant epitopes which have not been previously identified could be potentially useful for the development of a serological test that is less sensitive to the antigenic variation than the VN assay. Furthermore, the N- and GL-specific MAbs that recognize each of the EAV isolates tested may find application in sandwich ELISAs for the detection of anti-EAV antibodies or capture ELISAs for the detection of EAV-specific antigens.
ACKNOWLEDGMENTS
We thank Amy Glaser for supplying us with the EAV GL-specific MAb 93B. We are indebted to Bernard Moss and to Peter Timoney for providing us with the OST-7.1 cell line and EAV field isolate S1112, respectively. We gratefully acknowledge the excellent technical assistance of Britta Kanzok and Karina Mildner and thank Henrik Garoff for general support.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1987. [Google Scholar]
- 2.Balasuriya U B R, MacLachlan N J, de Vries A A F, Rossitto P V, Rottier P J M. Identification of a neutralization site in the major envelope glycoprotein (GL) of equine arteritis virus. Virology. 1995;207:518–527. doi: 10.1006/viro.1995.1112. [DOI] [PubMed] [Google Scholar]
- 3.Balasuriya U B R, Patton J F, Rossitto P V, Timoney P J, McCollum W H, MacLachlan N J. Neutralization determinants of laboratory strains and field isolates of equine arteritis virus: identification of four neutralization sites in the amino-terminal ectodomain of the GL envelope glycoprotein. Virology. 1997;232:114–128. doi: 10.1006/viro.1997.8551. [DOI] [PubMed] [Google Scholar]
- 4.Balasuriya U B R, Rossitto P V, DeMaula C D, MacLachlan N J. A 29K envelope glycoprotein of equine arteritis virus expresses neutralization determinants recognized by murine monoclonal antibodies. J Gen Virol. 1993;74:2525–2529. doi: 10.1099/0022-1317-74-11-2525. [DOI] [PubMed] [Google Scholar]
- 5.Balasuriya U B R, Timoney P J, McCollum W H, MacLachlan N J. Phylogenetic analysis of open reading frame 5 of field isolates of equine arteritis virus and identification of conserved and nonconserved regions in the GL envelope glycoprotein. Virology. 1995;214:690–697. doi: 10.1006/viro.1995.0087. [DOI] [PubMed] [Google Scholar]
- 6.Chirnside E D, Cook R F, Lock M W, Mumford J A. Monoclonal antibodies to equine arteritis virus. In: Powell D G, editor. Proceedings of the 5th International Conference on Equine Infectious Diseases, Lexington, Ky., 1987. Lexington: The University Press of Kentucky; 1988. pp. 262–267. [Google Scholar]
- 7.Chirnside E D, de Vries A A F, Mumford J A, Rottier P J M. Equine arteritis virus-neutralizing antibody in the horse is induced by a determinant on the large envelope glycoprotein GL. J Gen Virol. 1995;76:1989–1998. doi: 10.1099/0022-1317-76-8-1989. [DOI] [PubMed] [Google Scholar]
- 8.Chirnside E D, Francis P M, de Vries A A F, Sinclair R, Mumford J A. Development and evaluation of an ELISA using recombinant fusion protein to detect the presence of host antibody to equine arteritis virus. J Virol Methods. 1995;54:1–13. doi: 10.1016/0166-0934(95)00020-U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chirnside E D, Francis P M, Mumford J A. Expression cloning and antigenic analysis of the nucleocapsid protein of equine arteritis virus. J Virol Methods. 1995;54:277–288. doi: 10.1016/0168-1702(95)00098-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.del Piero F, Wilkins P A, Lopez J W, Glaser A L, Dubovi E J, Schlafer D H, Lein D H. Equine viral arteritis in newborn foals: clinical, pathological, serological, microbiological and immunohistochemical observations. Equine Vet J. 1997;29:178–185. doi: 10.1111/j.2042-3306.1997.tb01666.x. [DOI] [PubMed] [Google Scholar]
- 11.den Boon J A, Snijder E J, Chirnside E D, de Vries A A F, Horzinek M C, Spaan W J M. Equine arteritis virus is not a togavirus but belongs to the coronaviruslike superfamily. J Virol. 1991;65:2910–2920. doi: 10.1128/jvi.65.6.2910-2920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deregt D, de Vries A A F, Raamsman M J B, Elmgren L D, Rottier P J M. Monoclonal antibodies to equine arteritis virus proteins identify the GL protein as a target for virus neutralization. J Gen Virol. 1994;75:2439–2444. doi: 10.1099/0022-1317-75-9-2439. [DOI] [PubMed] [Google Scholar]
- 13.de Vries A A F, Chirnside E D, Bredenbeek P J, Gravestein L A, Horzinek M C, Spaan W J M. All subgenomic mRNAs of equine arteritis virus contain a common leader sequence. Nucleic Acids Res. 1990;18:3241–3247. doi: 10.1093/nar/18.11.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Vries A A F, Chirnside E D, Horzinek M C, Rottier P J M. Structural proteins of equine arteritis virus. J Virol. 1992;66:6294–6303. doi: 10.1128/jvi.66.11.6294-6303.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Vries A A F, Horzinek M C, Rottier P J M, de Groot R J. The genome organization of the Nidovirales: similarities and differences between arteri-, toro-, and coronaviruses. Semin Virol. 1997;8:33–47. doi: 10.1006/smvy.1997.0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Vries A A F, Post S M, Raamsman M J B, Horzinek M C, Rottier P J M. The two major envelope proteins of equine arteritis virus associate into disulfide-linked heterodimers. J Virol. 1995;69:4668–4674. doi: 10.1128/jvi.69.8.4668-4674.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Vries A A F, Raamsman M J B, van Dijk H A, Horzinek M C, Rottier P J M. The small envelope glycoprotein (GS) of equine arteritis virus folds into three distinct monomers and a disulfide-linked dimer. J Virol. 1995;69:3441–3448. doi: 10.1128/jvi.69.6.3441-3448.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Vries A A F, Rottier P J M, Glaser A L, Horzinek M C. Equine viral arteritis. In: Studdert M J, editor. Virus infections of equines. Amsterdam, The Netherlands: Elsevier Science Publishers; 1996. pp. 171–200. [Google Scholar]
- 19.Doll E R, Bryans J T, McCollum W H, Crowe M. Isolation of a filterable agent causing arteritis of horses and abortion by mares. Its differentiation from the equine abortion (influenza) virus. Cornell Vet. 1957;47:3–41. [PubMed] [Google Scholar]
- 20.Drew T W, Meulenberg J J M, Sands J J, Paton D J. Production, characterization and reactivity of monoclonal antibodies to porcine reproductive and respiratory syndrome virus. J Gen Virol. 1995;76:1361–1369. doi: 10.1099/0022-1317-76-6-1361. [DOI] [PubMed] [Google Scholar]
- 21.Elroy-Stein O, Moss B. Cytoplasmic expression system based on constitutive synthesis of bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1990;87:6743–6747. doi: 10.1073/pnas.87.17.6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fukunaga Y, Imagawa H, Kanemaru T, Kamada M. Complement-dependent serum neutralization with virulent and avirulent Bucyrus strains of equine arteritis virus. Vet Microbiol. 1993;36:379–383. doi: 10.1016/0378-1135(93)90104-f. [DOI] [PubMed] [Google Scholar]
- 23.Glaser A L, de Vries A A F, Dubovi E J. Comparison of equine arteritis virus isolates using neutralizing monoclonal antibodies and identification of sequence changes in GL associated with neutralization resistance. J Gen Virol. 1995;76:2223–2233. doi: 10.1099/0022-1317-76-9-2223. [DOI] [PubMed] [Google Scholar]
- 24.Hedges J F, Balasuriya U B R, Ahmad S, Timoney P J, McCollum W H, Yilma T, MacLachlan N J. Detection of antibodies to equine arteritis virus by enzyme linked immunosorbant assays utilizing GL, M and N proteins expressed from recombinant baculoviruses. J Virol Methods. 1998;76:127–137. doi: 10.1016/s0166-0934(98)00131-1. [DOI] [PubMed] [Google Scholar]
- 25.Holyoak G R, Little T V, McCollum W H, Timoney P J. Relationship between onset of puberty and establishment of persistent infection with equine arteritis virus in the experimentally infected colt. J Comp Pathol. 1993;109:26–46. doi: 10.1016/S0021-9975(08)80238-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hyllseth B. Structural proteins of equine arteritis virus. Arch Gesamte Virusforsch. 1973;40:177–188. doi: 10.1007/BF01242536. [DOI] [PubMed] [Google Scholar]
- 27.Hyllseth B, Pettersson U. Neutralization of equine arteritis virus: enhancing effect of guinea pig serum. Arch Gesamte Virusforsch. 1970;32:337–347. doi: 10.1007/BF01250061. [DOI] [PubMed] [Google Scholar]
- 28.Kheyar A, Martin S, St-Laurent G, Timoney P J, McCollum W H, Archambault D. Expression cloning and humoral immune response to the nucleocapsid and membrane proteins of equine arteritis virus. Clin Diagn Lab Immunol. 1997;4:648–652. doi: 10.1128/cdli.4.6.648-652.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kondo T, Akashi H, Fukunaga Y, Sugita S, Sekiguchi K, Wada R, Kamada M. Production and characterisation of monoclonal antibodies against structural proteins of equine arteritis virus. In: Nakajima H, Plowright W, editors. Proceedings of the 7th International Conference on Equine Infectious Diseases, Tokyo, Japan, 1994. Newmarket, England: R & W Publications, Ltd.; 1994. pp. 21–26. [Google Scholar]
- 30.Kondo T, Sugita S, Fukunaga Y, Imagawa H. Identification of the major epitope in the GL protein of equine arteritis virus (EAV) recognized by antibody in EAV-infected horses using synthetic peptides. J Equine Sci. 1998;9:19–23. [Google Scholar]
- 31.MacLachlan N J, Balasuriya U B R, Hedges J F, Schweidler T M, McCollum W H, Timoney P J, Hullinger P J, Patton J F. Serologic response of horses to the structural proteins of equine arteritis virus. J Vet Diagn Invest. 1998;10:229–236. doi: 10.1177/104063879801000302. [DOI] [PubMed] [Google Scholar]
- 32.McCollum W H. Development of a modified virus strain and vaccine for equine viral arteritis. J Am Vet Med Assoc. 1969;155:318–322. [PubMed] [Google Scholar]
- 33.McCollum W H. Studies of passive immunity in foals to equine viral arteritis. Vet Microbiol. 1976;1:45–54. [Google Scholar]
- 34.McCollum W H, Swerczek T W. Studies of an epizootic of equine viral arteritis in racehorses. Equine Vet J. 1978;2:293–299. [Google Scholar]
- 35.Radwan A I, Burger D. The mechanisms of neutralization of sensitized equine arteritis virus by complement components. J Gen Virol. 1974;25:229–237. doi: 10.1099/0022-1317-25-2-229. [DOI] [PubMed] [Google Scholar]
- 36.Remaut E, Tsao H, Fiers W. Improved plasmid vectors with a thermoinducible expression and temperature-regulated runaway replication. Gene. 1983;22:103–113. doi: 10.1016/0378-1119(83)90069-0. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 38.Schäfer W. Der Mäuse-Inzuchtstamm STU. Entwicklung und Eigenschaften. Z Naturforsch Teil C. 1979;34:306–309. [PubMed] [Google Scholar]
- 39.Shulman M, Wilde C D, Köhler G. A better cell line for making hybridomas secreting specific antibodies. Nature. 1978;276:269–270. doi: 10.1038/276269a0. [DOI] [PubMed] [Google Scholar]
- 40.Snijder E J, Meulenberg J J M. The molecular biology of arteriviruses. J Gen Virol. 1998;79:961–979. doi: 10.1099/0022-1317-79-5-961. [DOI] [PubMed] [Google Scholar]
- 41.Snijder E J, van Tol H, Pedersen K W, Raamsman M J B, de Vries A A F. Identification of a novel structural protein of arteriviruses. J Virol. 1999;73:6335–6345. doi: 10.1128/jvi.73.8.6335-6345.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stadejek T, Björklund H, Ros Bascuñana C, Ciabatti I M, Scicluna M T, Amaddeo D, McCollum W H, Autorino G L, Timoney P J, Paton D J, Klingeborn B, Belák S. Genetic diversity of equine arteritis virus. J Gen Virol. 1999;80:691–699. doi: 10.1099/0022-1317-80-3-691. [DOI] [PubMed] [Google Scholar]
- 43.St-Laurent G, Lepage N, Carman S, Archambault D. Genetic and amino acid analysis of the GL protein of Canadian, American and European equine arteritis virus isolates. Can J Vet Res. 1997;61:72–76. [PMC free article] [PubMed] [Google Scholar]
- 44.Strebel K, Beck E, Strohmaier K, Schaller H. Characterization of foot-and-mouth disease virus gene products with antisera against bacterially synthesized fusion proteins. J Virol. 1986;57:983–991. doi: 10.1128/jvi.57.3.983-991.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Timoney P J, McCollum W H. Equine viral arteritis. Vet Clin N Am Equine Pract. 1993;9:295–309. doi: 10.1016/S0749-0739(17)30397-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van der Meer Y, van Tol H, Krijnse Locker J, Snijder E J. ORF1a-encoded replicase subunits are involved in the membrane association of the arterivirus replication complex. J Virol. 1998;72:6689–6698. doi: 10.1128/jvi.72.8.6689-6698.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Nieuwstadt A P, Meulenberg J J M, van Essen-Zandbergen A, Petersen-den Besten A, Bende R J, Moormann R J M, Wensvoort G. Proteins encoded by open reading frames 3 and 4 of the genome of Lelystad virus (Arteriviridae) are structural proteins of the virion. J Virol. 1996;70:4767–4772. doi: 10.1128/jvi.70.7.4767-4772.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weiland E, Thiel H-J, Hess G, Weiland F. Development of monoclonal neutralizing antibodies against bovine viral diarrhea virus after pretreatment of mice with normal bovine cells and cyclophosphamide. J Virol Methods. 1989;24:237–244. doi: 10.1016/0166-0934(89)90026-8. [DOI] [PubMed] [Google Scholar]
- 50.Weiland E, Wieczorek-Krohmer M, Kohl D, Conzelmann K K, Weiland F. Monoclonal antibodies to the GP5 of porcine reproductive and respiratory syndrome virus are more effective in virus neutralization than monoclonal antibodies to the GP4. Vet Microbiol. 1999;66:171–186. doi: 10.1016/s0378-1135(99)00006-1. [DOI] [PubMed] [Google Scholar]
- 51.Weiland F, Granzow H, Wieczorek-Krohmer M, Weiland E. Electron microscopic studies on the morphogenesis of PRRSV in infected cells—comparative studies. In: Schwyzer M, Ackermann M, Bertoni G, Kocherhans R, McCullough K, Engels M, Wittek R, Zanoni R, editors. Immunobiology of viral infections, Proceedings of the 3rd Congress of the European Society for Veterinary Virology, Interlaken, Switzerland. Lyon, France: Fondation Marcel Mérieux; 1995. pp. 499–502. [Google Scholar]
- 52.Westcott D, Lucas M, Paton D. Equine arteritis virus: antigenic analysis of strain variation. In: Schwyzer M, Ackermann M, Bertoni G, Kocherhans R, McCullough K, Engels M, Wittek R, Zanoni R, editors. Immunobiology of viral infections, Proceedings of the 3rd Congress of the European Society for Veterinary Virology, Interlaken, Switzerland. Lyon, France: Fondation Marcel Mérieux; 1995. pp. 479–483. [Google Scholar]
- 53.Wieczorek-Krohmer M, Weiland F, Conzelmann K, Kohl D, Visser N, van Woensel P, Thiel H-J, Weiland E. Porcine reproductive and respiratory syndrome virus (PRRSV): monoclonal antibodies detect common epitopes on two viral proteins of European and U.S. isolates. Vet Microbiol. 1996;51:257–266. doi: 10.1016/0378-1135(96)00047-8. [DOI] [PubMed] [Google Scholar]
- 54.Yamaguchi S, Kanno T, Akashi H, Kondo T. Identification of two neutralization sites in GL protein of equine arteritis virus by means of monoclonal antibodies. J Equine Sci. 1997;8:7–11. [Google Scholar]
- 55.Zeegers J J W, van der Zeijst B A M, Horzinek M C. The structural proteins of equine arteritis virus. Virology. 1976;73:200–205. doi: 10.1016/0042-6822(76)90074-x. [DOI] [PubMed] [Google Scholar]