Abstract
Pseudoisocytosine (J), a neutral analogue of protonated cytosine, is currently the gold standard modified nucleobase in peptide nucleic acids (PNA) for formation of J•G-C triplets stable at physiological pH. The present study shows that triple helical recognition of RNA and DNA may be significantly improved by using 2-aminopyridine (M) instead of J. The positively charged M forms three-fold stronger M+•G-C triplets than J with uncompromised sequence selectivity. Replacement of six Js with Ms in a PNA 9-mer increased its binding affinity by about two orders of magnitude. M-modified PNAs prefer binding double-stranded RNA over DNA and disfavor off-target binding to single-stranded nucleic acids. Taken together, the results show that M is a promising modified nucleobase that significantly improves triplex-forming PNAs and may provide breakthrough developments for therapeutic and biotechnology applications.
Graphical Abstract
INTRODUCTION
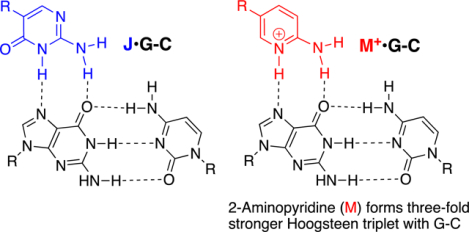
Peptide nucleic acid (PNA) is an artificial biopolymer that combines DNA nucleobases with a neutral and achiral protein-like backbone (Figure 1).1 PNA was originally designed as a DNA mimic to improve the properties of triplex-forming oligonucleotides,1, 2 Because of the lack of electrostatic repulsion between the neutral amide backbone and the negatively charged phosphates, PNA was expected to have superior binding to double-stranded DNA (dsDNA) than negatively charged triplex-forming oligonucleotides.1, 2 However, unfavorable cytosine protonation (due to pKa ~4.5) to form the C+•G-C Hoogsteen triplet (Figure 1) prevented triplex formation at physiological pH. To overcome this limitation, pseudoisocytosine (J), a neutral analogue of protonated cytosine,3, 4 was introduced in PNA.5 J is currently the gold standard modification for development of triplex-forming PNAs as probes and therapeutics.6–10 Herein, we report that 2-aminopyridine (M, pKa ~6.711), a cationic analogue of protonated cytosine,12 is superior to neutral J as a modified nucleobase in PNAs and significantly improves the triple-helical recognition of dsRNA and dsDNA.
Figure 1.
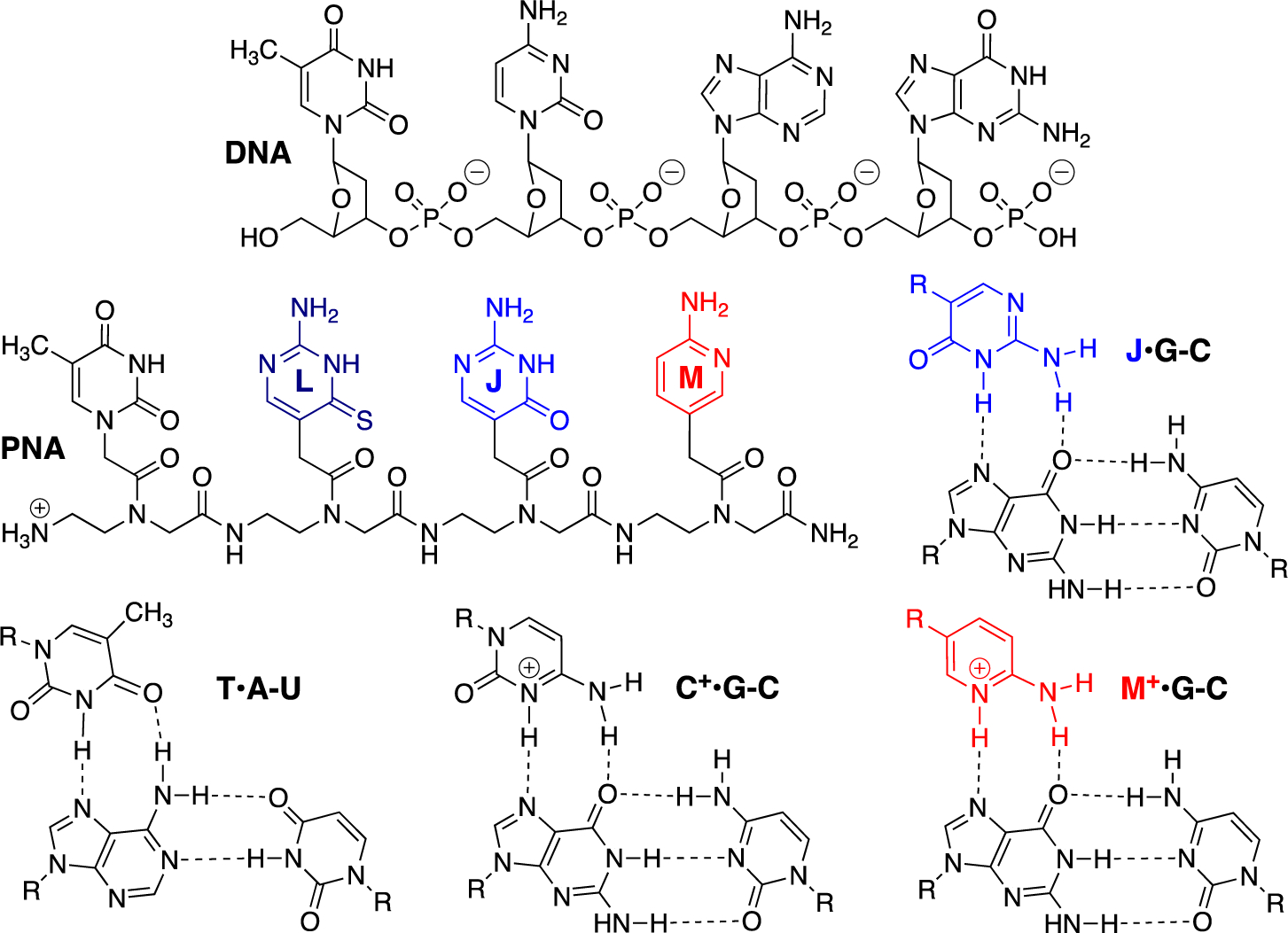
Structures of DNA, PNA and Hoogsteen hydrogen-bonded base triplets.
PNA is a remarkable nucleic acid analogue that binds single-stranded DNA and RNA (ssDNA and ssRNA) with very high affinity and sequence selectivity.13, 14 These favorable binding properties have made PNA the ligand of choice in broad research and diagnostic applications that detect DNA and RNA using Watson-Crick hybridization, such as, molecular beacons for RT-PCR or fluorescence in situ hybridization (FISH) probes.15, 16 Due to the high affinity, PNA is capable of displacing pyrimidine rich strands of dsDNA to form a 2:1 PNA-DNA strand-invasion triplex with the purine rich strand.1 In the strand-invasion triplex, one PNA molecule forms Watson-Crick duplex while the second PNA molecule forms Hoogsteen triplex with the purine rich DNA strand, which collectively displaces the pyrimidine rich strand of DNA as a single-stranded P-loop.1 This remarkable binding mode enabled development of tail-clamp PNAs,17 capable of opening genomic dsDNA through invasion, which has been proposed as a novel gene editing therapeutic approach.6–8 However, despite the impressive molecular recognition properties and success as in vitro probes and diagnostics, to the best of our knowledge, PNA has not yet entered in vivo applications or clinical trials.18, 19 The main problems are related to poor cellular uptake and unfavorable pharmacokinetics.20–22 Hence, development of novel chemical modifications that improve both nucleic acid binding as well as ability to reach the targets in biological systems (e.g., cellular uptake and pharmacokinetics) are critical for closing the gap between PNA and in vivo applications, including drug development. Our present study suggests that 2-aminopyridine nucleobase may be one of such modifications that will help realize PNA’s clinical potential.
2-Aminopyridine (M) and its derivatives were originally developed as modified nucleobases in triplex-forming oligonucleotides to improve their binding affinity at neutral pH.23–26 In our pursuit of triple-helical recognition of dsRNA, we found that M-modified PNAs had high affinity and sequence selectivity for dsRNA at physiological pH and salt conditions.12, 27, 28 Moreover, M-modified PNAs showed improved cellular uptake,27, 28 which suggested that M-modification of PNA might contribute to solving two long-standing challenges for in vivo applications of PNAs – low stability of triplexes and poor cellular uptake in biological systems. In these early studies, we also noted that a J-modified PNA had weaker binding to a dsRNA than an M-modified PNA.12 While this was surprising as J-modification was well established and widely used for recognition of G-C through triplex formation, this very preliminary observation was not followed up until the present study. Herein, we compared binding affinity and sequence selectivity of several M- and J-modified PNAs designed to target dsRNA and dsDNA by triplex formation, as well as ssRNA and ssDNA by duplex and tail-clamp formation. In all cases M-modified PNAs had significantly higher on-target affinity than J-modified PNAs. Despite the cationic nature, M-modified PNAs retained high sequence selectivity. We propose that using M instead of J will provide significant improvement of PNAs ability to enter triplex-binding modes including the stand invasion and tail-clamp formation, which will be critical in the development of improved PNAs as probes and therapeutics.
MATERIALS AND METHODS
PNA synthesis, purification and analysis
PNA synthesis used commercial Fmoc-protected PNA monomers for A,T,C, and G purchased from Link Technologies (part of LGC Genomics Ltd, UK). M monomer12 and J monomer5, 29 were synthesized using previously reported literature procedures. Solid-phase synthesis of PNA was done on an Expedite 8909 synthesizer at 2 μmol scale using NovaSyn TG Sieber resin (Novabiochem) as previously reported by our group.30 PNAs were cleaved from the solid support using 0.6 mL of 20% m-cresol in TFA for 2.0 h using two-syringe pull-push method.30 The cleavage cocktail was distributed into three Eppendorf tubes and precipitated by the addition of chilled diethyl ether (~1.0 mL) followed by the centrifugation (15000 rpm). The crude PNA was dissolved in purified water (~1.0 mL) and analyzed using a Shimadzu LCMS 2020 single quadrupole instrument. For LC conditions and analytical traces of crude PNA synthesis mixtures, see Supplemental Information (Figures S1–S12).
Crude synthetic PNAs were purified using reverse phase HPLC and a gradient of acetonitrile (MeCN) in water containing 0.1% formic acid as a mobile phase modifier. The purity and identity of the final products were confirmed by LCMS (for details, see Table S1 and Figures S1–S12). Pure PNAs were quantified by dissolving in 1.0 mL purified water, and measuring absorbance at 260 nm. For commercially available monomers, extinction coefficients from the manufacturer were used. An extinction coefficient of 2800 M−1cm−1 was used for J and a coefficient of 806 M−1cm−1 was used for M. The PNA solution was lyophilized and the solid diluted with purified water to give a stock solution of 0.24 mM.
DNA and RNA oligonucleotides
DNA oligonucleotides were purchased from Eurofin, checked for purity by HPLC, quantified by absorbance at 260 nm, and used without further purification. RNA oligonucleotides were purchased from Dharmacon. Prior to use, RNA oligonucleotides were deproteced in accordance with manufacturer instructions and purified using reverse phase HPLC and a gradient of MeCN in 50 mM aqueous triethylammonium acetate buffer. The purity of the final products was confirmed by reinjection in HPLC.
Isothermal titration calorimetry (ITC)
ITC experiments were done on a MicroCal iTC200 instrument at 25 °C in 50 mM potassium phosphate buffer (pH 7.4) containing 2 mM MgCl2, 90 mM KCl, 10 mM NaCl. In a typical experiment 2.45 μL aliquots of PNA solution were sequentially injected from a 40 μL rotating syringe (750 rmp) into 200 μL of RNA or DNA hairpin solution. In the case of tail-clamp PNAs (PNA10-PNA12), we used reverse titrations of DNA into PNA to conserve PNA. The specific cell and syringe concentrations employed in different experiments are given in Supplemental Information (Figures S13–S34). Results were analyzed using MicroCal PEAQ-ITC software (Figure S35 and Tables S2–S5, S10, and S15).
UV-melting experiments
UV-melting experiments were performed on Shimadzu UV-2600 or UV-1800 spectrophotometers equipped with TMSPC-8 temperature controllers. A temperature ramp rate of 0.5 °C was used in all cases. Absorbance was monitored at 300 nm for triplex formation involving M-modified PNAs (18 μM), 270 nm for triplex formation involving PNAs containing only J and T nucleobases (18 μM), 260 nm for duplex-forming PNAs (2.5 μM), and 280 nm for tail-clamp-forming PNAs (2.5 μM). Experiments were performed in the ITC buffer containing 50 mM potassium phosphate, 2 mM MgCl2, 90 mM KCl, 10 mM NaCl at pH 7.4. Solutions were prepared as follows. Oligonucleotide was dissolved in buffer, vortexed, centrifuged, and heated to 95°C for 3–5 minutes. The solution was cooled to 4° C and kept at that temperature for an additional 3–5 minutes. PNA was then added to the sample and the mixture was vortexed and centrifuged again. Solutions were prepared to a final volume of 230 μL which was distributed evenly between two cells in an eight cell cuvette. Samples were then analyzed through the heating-cooling cycle three times, typically from 20°C to 95°C. Typical melting curves are shown in Figures S36–S41; the experimental results are listed in Tables S6–S9, S11–S14, and S16. Five of the replicates were then used to determine average and standard deviation in melting temperature (Tm).
RESULTS
M forms three-fold stronger triplets with G-C than J
We started our study by comparing the stability and sequence selectivity of triplex-forming M- and J-modified PNA 9-mers using isothermal titration calorimetry (ITC) in a hairpin model system (Figure 2). In the present study, we replaced the closing U4 tetraloop in hairpins used in our earlier studies12, 27, 28 with the unusually stable C(UUCG)G tetraloop31 in HRP1-HRP4, which improved the overall quality of ITC data most likely, by rigidifying the RNA hairpins. M-modified PNA1 and PNA2, having either M or T nucleobase at the variable position (magenta in Figure 2) had high binding affinity and sequence selectivity for the matched HRP1 and HRP2, respectively (Table 1). The M-modified PNAs had unusually high affinity for dsRNA as binding affinity of PNA1 and PNA2 for the matched DNA hairpins (Table 1, rightmost column) was 41- and 13-fold lower, respectively, and similar to the stability of mismatched PNA-dsRNA triplexes. Higher affinity of PNA for dsRNA was consistent with our earlier studies12, 27, 28, 32 and those by Nielsen and co-workers who reported33 that PNA sequences longer than 9-mers were required to form stable PNA-dsDNA triplexes. These results showed that M-modified PNAs were excellent triplex-forming ligands for recognition of dsRNA.
Figure 2.
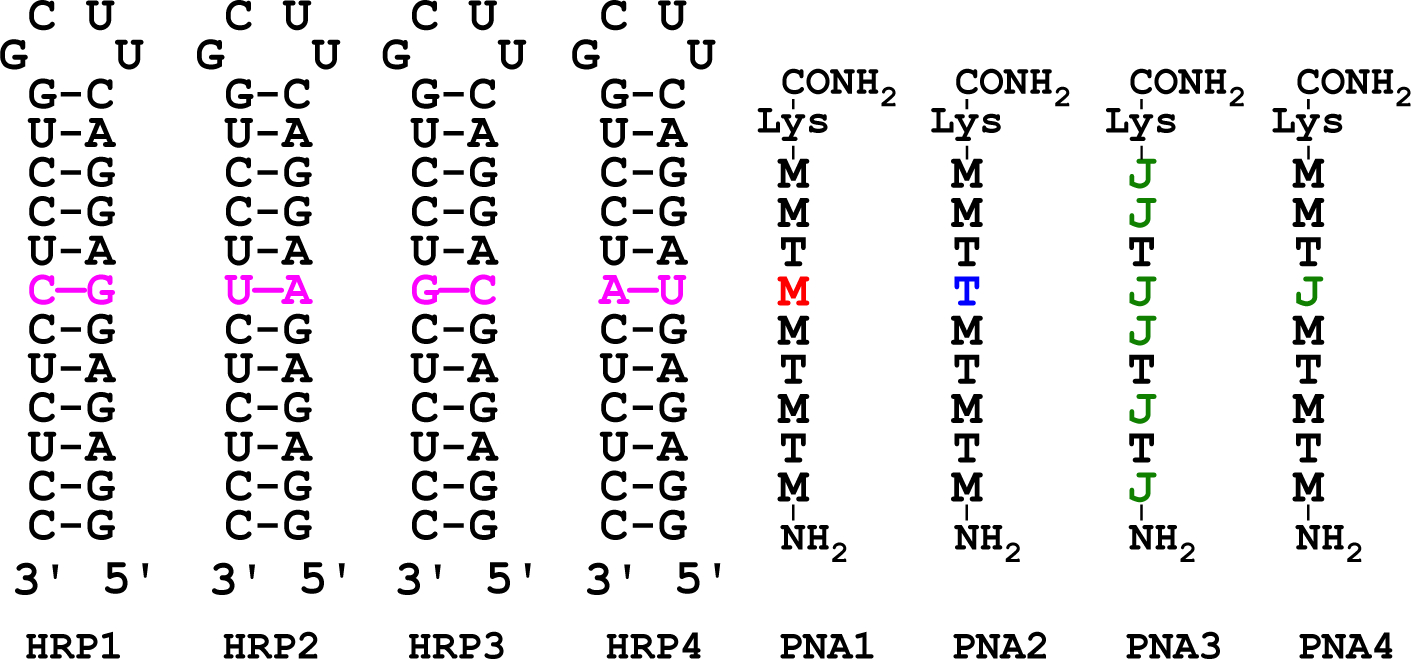
Structures of the RNA hairpins and PNAs to study the affinity and sequence selectivity of triplex formation.
Table 1.
Binding affinity and sequence selectivity of PNAs by ITC and UV thermal melting.
| PNA/dsRNA | Affinitya | HRP1 | HRP2 | HRP3 | HRP4 | dsDNA |
|---|---|---|---|---|---|---|
| PNA1 (M) | Ka × 106 M−1 | 33 ± 1 | 1.3 ± 0.1 | 1.3 ± 0.1 | 1.2 ± 0.1 | 0.8 ± 0.1b |
| Tm °C | 66.5 ± 0.7 | 36.3 ± 0.4 | 36.8 ± 0.4 | 32.6 ± 0.4 | 35.0 ± 0.3b | |
| PNA2 (T) | Ka × 106 M−1 | 1.9 ± 0.1 | 12 ± 1 | 0.9 ± 0.1 | 0.4 ± 0.1 | 0.9 ± 0.1c |
| Tm °C | 46.4 ± 0.5 | 69.6 ± 0.8 | 35.4 ± 0.4 | 34.6 ± 0.2 | 29.4 ± 0.4c | |
| PNA4 (J) | Ka × 106 M−1 | 11 ± 1 | 1.6 ± 0.4 | 0.9 ± 0.1 | 0.4 ± 0.1 | 0.2 ± 0.1b |
| Tm °C | 60.7 ± 0.6 | 43.8 ± 0.6 | 39.1 ± 0.3 | 35.7 ± 0.5 | 29.7 ± 0.3b |
Association constants, Ka × 106 M−1, average of three experiments ± stand. dev., for binding of PNAs to the respective hairpins in 50 mM potassium phosphate buffer (pH 7.4) containing 2 mM MgCl2, 90 mM KCl, 10 mM NaCl at 25 °C. UV thermal melting temperatures, Tm °C, average of five experiments ± stand. dev. measured at 300 nm and 18 μM of each dsRNA (or dsDNA) and PNA in the ITC buffer as above.
dsDNA of the same sequence as HRP1.
dsDNA of the same sequence as HRP2.
In contrast, the affinity of PNA3, built of J and T nucleobases only, was difficult to measure by ITC under the experimental conditions of Table 1 because of low signal strength (due to modest binding enthalpy) and weak binding. When the concentration of hairpins in ITC experiments was increased four-fold to 40 μM, we were able to measure Ka ~0.4 μM−1 for PNA3-HRP1 and ~0.1 μM−1 for PNA3-dsDNA. While the accuracy of these numbers is limited, affinity of PNA1 was about two orders of magnitude higher than that of PNA3, suggesting that M formed significantly stronger triplets with G-C than J. This conclusion was confirmed when the J nucleobase was placed only at the variable position. The binding affinity of PNA4 was reduced three-fold compared to PNA1 having M at the same position (c.f., Ka 33 and 11 μM−1 in Table 1).
The binding affinity and sequence selectivity of PNA4 was similar to that of PNA2 suggesting that J•G-C and T•A-U formed equally stable triplets. This was not surprising as both triplets are stabilized by two similar Hoogsteen hydrogen bonds (Figure 1). The high affinity of M+•G-C triplet compared to either T•A-U or J•G-C triplets was both surprising and remarkable, because our recent structural studies suggested that while the positively charged N-H of M formed a strong hydrogen bond with G-C, the hydrogen bonding from the exocyclic −NH2 of M was relatively weak.34
UV thermal melting confirms the ITC results
UV melting was done at 300 nm where the M heterocycle has unique absorbency, which specifically reports triplex melting with little interference from conformational transitions of native RNA or DNA. Overall, the melting temperature (Tm) values (Table 1) were consistent with Kas obtained using ITC. While the matched triplexes had Tms in the 60–70 °C range, most of the mismatched triplexes melted in the 29–37 °C range. Notable exceptions were PNA2-HRP1 and PNA4-HRP2 triplexes that had higher than expected Tms, ~46 and ~44 °C, respectively. However, these were not out of line with somewhat higher Kas obtained using ITC. It should be noted that ITC and UV melting report on stability of triplexes at different temperatures (25 °C and Tm, respectively). Due to potential differences in dependency of triplex-formation thermodynamics on temperature, these methods do not have to provide the same affinity values.35 The generally good correlation between Kas and Tms confirmed our conclusion that M was superior to J for recognition of G-C base pair.
Given the low affinity of PNA3, we increased the length and decreased the M/J ratio in another attempt to directly compare M and J using PNA5 and PNA6 (Figure 3a). The M-modified PNA5 showed the expected high binding affinity for HRP5r (Ka 28 ± 1 μM−1) that was reduced ten-fold for the DNA target HRP5d (Ka 2.7 ± 0.6 μM−1). Interestingly, the binding affinity of 13-mer PNA5 having six Ms was similar to that of 9-mer PNA1 also having six Ms, which emphasized the importance of the positively charged M for driving the stability of triplexes. In contrast, the J-modified PNA6 produced complex ITC traces showing binding stoichiometry higher than one equivalent of PNA binding to either RNA or DNA target. We were not able to analyze binding of PNA6 using the same 1:1 model as used for PNA5, which suggested that PNA6 might be binding to RNA and DNA hairpins using a more complex mode than triplex, possibly involving strand invasion.
Figure 3.
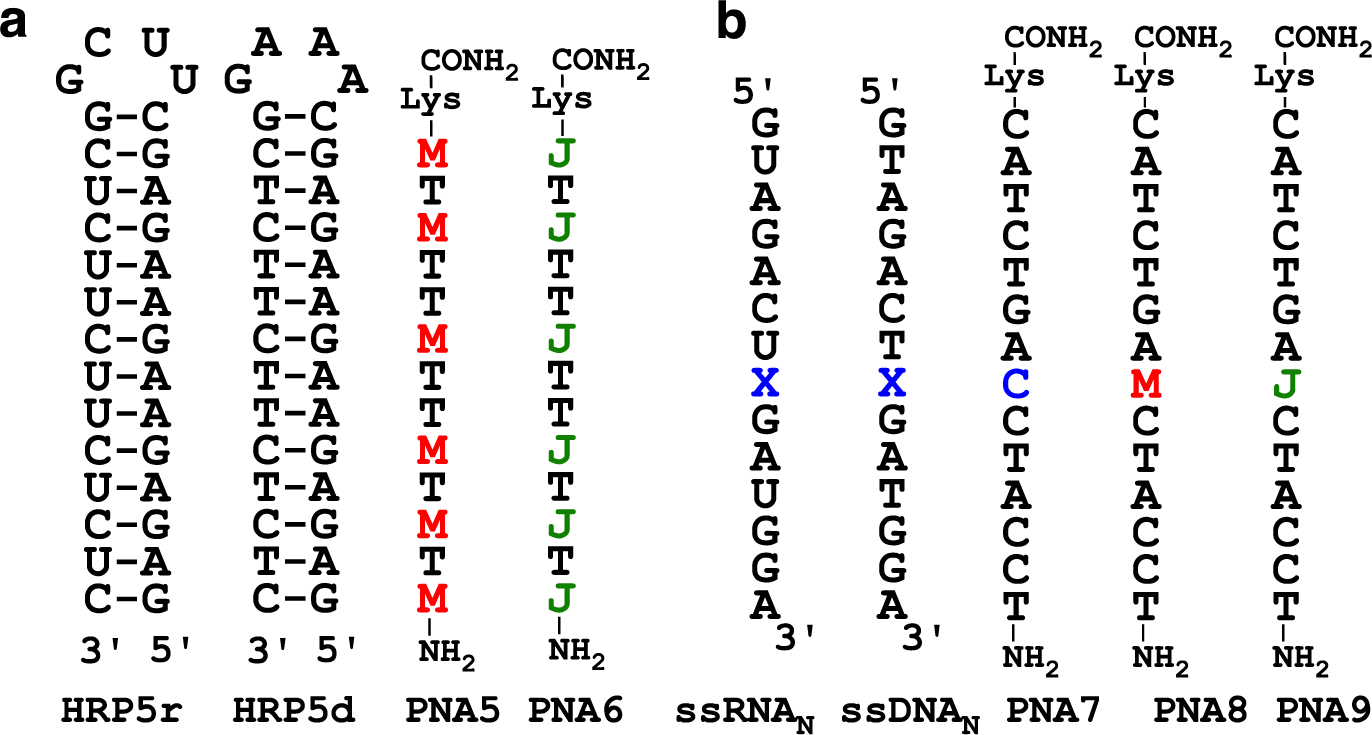
RNA, DNA, and PNA sequences to compare J and M in triplex (a) and duplex (b) contexts.
M disfavors Watson-Crick pairing with native nucleobases
In strand invasion, PNA forms Watson-Crick base pairs with one strand of duplex displacing the other as a P-loop. To evaluate their ability to form Watson-Crick like base pairs, we placed M and J in the middle of a 14-mer PNA designed to form a duplex with complementary ssRNA or ssDNA (Figure 3b). PNA7 having C at the variable position formed the expected stable matched duplexes with ssRNA1 and ssDNA1 (Table 2). The lower stability of duplexes involving PNA8 suggested that M did not form stable Watson-Crick base pairs with any of the natural nucleobases. In contrast, PNA9 having J at the variable position showed similar binding properties as PNA7. Previous studies have indicated that due to tautomeric forms, J may form stable J-G base pairs (Figure 4a).36 We propose that M disfavors Watson-Crick pairing with G because it forms only two hydrogen bonds in the unprotonated state and has repulsive interactions when protonated (Figure 4a).
Table 2.
Stability (Tm °C) of M- and J-modified duplexes.a
| ssRNAN (X) | PNA7 (C) | PNA8 (M) | PNA9 (J) |
|---|---|---|---|
| ssRNA1 (G) | 73.2 ± 0.5 | 59.1 ± 0.7 | 72.2 ± 0.6 |
| ssRNA2 (A) | 57.8 ± 0.4 | 55.4 ± 0.3 | 57.4 ± 0.5 |
| ssRNA3 (C) | 56.4 ± 0.4 | 56.8 ± 0.6 | 58.0 ± 0.4 |
| ssRNA4 (U) | 56.8 ± 0.3 | 55.8 ± 0.6 | 58.5 ± 0.5 |
| ssDNAN (X) | PNA7 (C) | PNA8 (M) | PNA9 (J) |
| ssDNA1 (G) | 66.0 ± 0.7 | 45.3 ± 0.2 | 66.3 ± 0.5 |
| ssDNA2 (A) | 45.6 ± 0.5 | 41.5 ± 0.6 | 45.9 ± 0.4 |
| ssDNA3 (C) | 44.5 ± 0.3 | 46.5 ± 0.4 | 48.9 ± 0.3 |
| ssDNA4 (T) | 47.4 ± 0.3 | 45.6 ± 0.4 | 49.7 ± 0.2 |
Tm °C, average of five experiments ± stand. dev. measured at 260 nm and 2.5 μM of each ssRNAN or ssDNAN and PNA in the ITC buffer as in Table 1.
Figure 4.
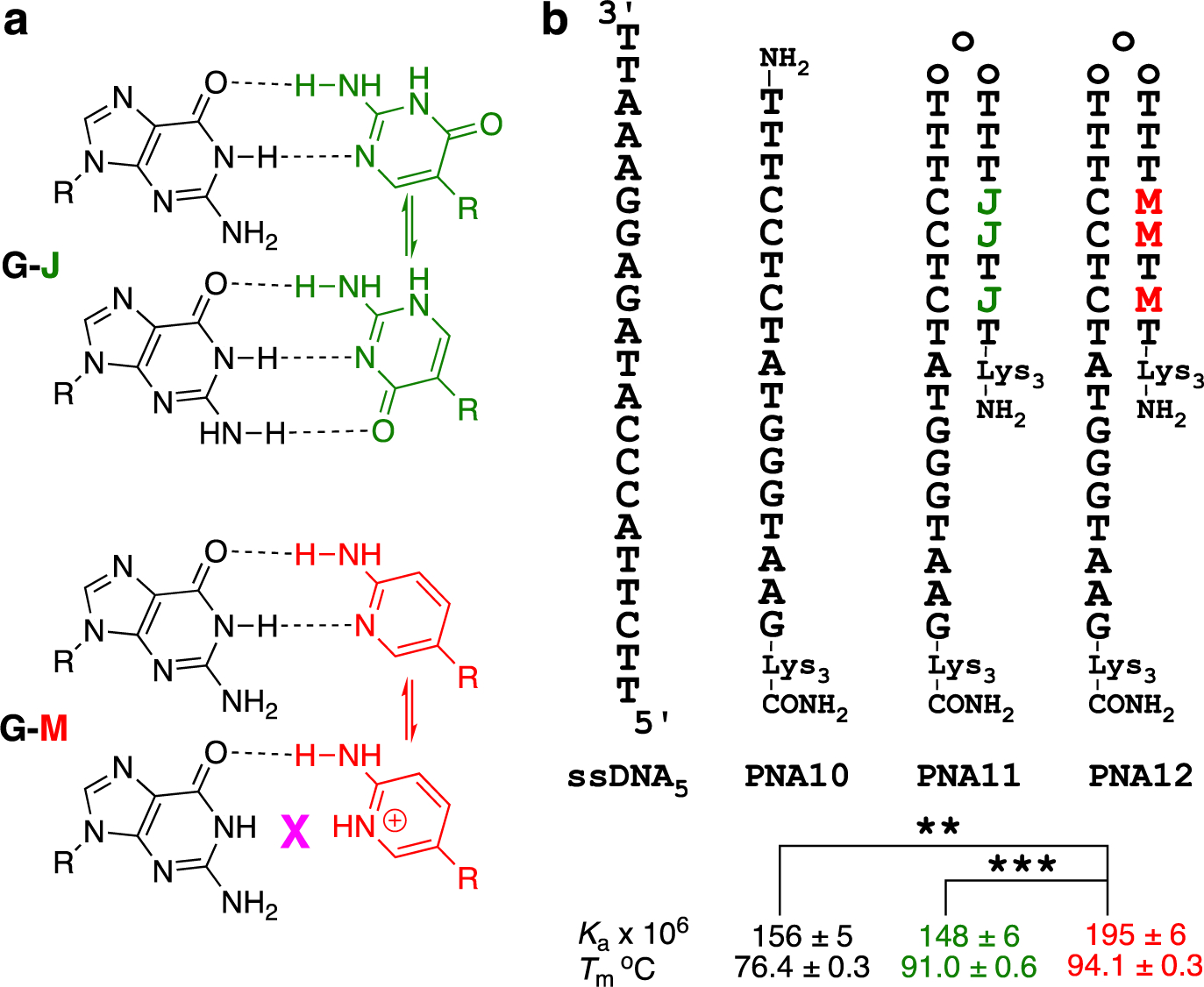
Comparison of J and M Watson-Crick base pairing (a) and in tail-clamp PNA (b); the three o in tail-clamps denote three 2-(2-aminoethoxy)ethoxy acetic acid spacers. Asterisks indicate two-tailed P values: ** denotes P ≤ 0.01, and *** denotes P ≤ 0.001, as calculated using Student’s t-test.
M improves tail-clamp PNA binding affinity
Finally, we compared M and J in the context of a tail-clamp PNA (Figure 4b) reported to target the F508del mutation in the cystic fibrosis transmembrane conductance regulator gene.8 As expected, PNA10 formed a stable duplex with ssDNA5. Adding the J-clamp in PNA11 had small effect on Ka by ITC, but increased the Tm in UV melting (Figure 4b). The M-clamp in PNA12 provided both higher Tm and a statistically significant improvement in Ka. Compared to other PNAs studied, such as PNA1 vs PNA3, the increase in stability comparing PNA11 and PNA12 was relatively modest, which was not entirely unexpected as there were only three Js changed for Ms.
DISCUSSION
As a DNA mimic designed to recognize other nucleic acids, PNA has remarkably favorable biophysical properties – high binding affinity, excellent sequence selectivity, and complete resistance to degradation by nucleases or proteases. Since it’s inception in 1991,1 PNA has become a staple component of many assays and diagnostics.15, 16 However, compared to other nucleic acid derivatives,18, 19 PNA based therapeutics have been remarkably difficult to move to clinical trials. While the reasons behind this lag may be many and complex, unfavorable cellular uptake and pharmacokinetics appear to be the most apparent bottlenecks.20–22 There is an obvious need for novel chemical modifications that would address the remaining problems for in vivo applications of PNA while maintaining and improving its favorable nucleic acid binding properties. From this perspective, our previous preliminary observations that M-modified PNAs showed enhanced cellular uptake27, 28 are especially encouraging. The uptake was directly proportional to the number of Ms – longer and more M rich PNAs were taken up more efficiently.28 Our present study compares M-modified triplex-forming PNAs to the current gold standard J-modified PNAs demonstrating that M-modified PNAs have superior binding affinity and on-target selectivity. We propose that using M instead of J will allow cleaner and stronger triplex-formation improving PNA’s properties as a probe and diagnostic tool and may help to break the barriers for PNA entering clinical applications.
Pseudoisocytosine or J nucleobase (Figure 1) is a neutral analogue of protonated cytosine,3, 4 which was introduced in PNA in 1995.5 Using J instead of C alleviated the problem of unfavorable cytosine protonation (pKa ~4.5), required to form the C+•G-C Hoogsteen triplet, and enabled triplex formation at physiological pH. While J has been widely accepted as the gold standard PNA modification, we12 and others37–39 have noted that the binding affinity and on-target specificity of J may be further improved. Nielsen and co-workers37 reported that 1,8-naphthyridin-2,7-(1,8H)-dione showed improved binding to G-C base pairs at pH 7 compared to J; however, this observation was not followed up with more detailed studies. Chen and co-workers reported that 4-thio-pseudoisocytosine (L in Figure 1) formed a more stable L•G-C Hoogsteen triplet than J, which was proposed to be due to enhanced van der Waals contacts, base stacking, hydrogen bonding, and reduced dehydration energy.38 A PNA 8-mer having three Js replaced with Ls had ~4-fold higher binding affinity than the parent J-modified PNA.38 For comparison, replacement of six Js with Ms in a PNA 9-mer in the present study increased the PNA’s binding affinity by ~100-fold. Later the same group reported that simple guanidinium groups used as nucleobase surrogates showed similar binding to L when placed as isolated modifications; however, consecutive guanidinium nucleobases were not tolerated.39 Not unexpectedly, the cationic guanidinium nucleobases improved cellular uptake of the modified PNAs as judged by fluorescence microscopy.39 In contrast to guanidinium nucleobases, we have used three28, 40, 41 and even four27 consecutive M modifications without any negative effect on binding affinity or selectivity.
Our current study shows that M by far outperforms J as a modified nucleobase to recognize G-C base pairs in triplex-forming PNAs. The stronger binding of M-modified PNAs compared to J-modified PNAs was driven by large favorable enthalpy (c.f., ΔH of PNA1 in Table S2 with ΔH of PNA3 in Table S4). Because of low affinity and enthalpy (weak ITC signal) of fully J-modified PNA, comparison of J and M was done in a model system of 9-mer M-modified PNAs containing an internal variable site each with a different nucleobase, M, T, and J in PNA1, PNA2 and PNA4, respectively. In this system (Table 1), M formed about 3-fold stronger M+•G-C triplet than J while retaining high sequence selectivity. Most remarkably, replacement of six Js with Ms increased the binding affinity of all M-modified PNA1 ~100-fold compared to the all J-modified PNA3. The stabilities and selectivities of J•G-C and T•A-U triplets were similar as expected from similar hydrogen bonding schemes. The superior affinity of M was due to the partial positive charge (pKa ~6.711) resulting in strong, enthalpy-driven binding.
Consistent with our earlier studies12, 27, 28, 32, PNA forms more stable triplexes with dsRNAs than with dsDNAs. Our recent structural studies34 suggest that this is driven by a hydrogen bonding between the amide N-H of PNA backbone and non-bridging phosphate oxygens, which is favored by their matching distances in RNA, but disfavored by mismatching distances in DNA. Our current results suggest that the positively charged M may have additional preference for binding RNA over DNA. Interestingly, the difference between stability of PNA-dsRNA and PNA-dsDNA triplexes decreases with decreasing M/T ratio in the series of PNA1 (6:3, 41-fold), PNA2 (5:4, 13-fold) and PNA5 (6:7, 10-fold). A potential explanation may be that the positively charged M-modified PNAs have more favorable electrostatic attraction (larger kon) for the narrower major groove of RNA (aligned with negatively charged phosphates) than for the wider major groove of dsDNA. Another advantage of M is its weak ability for Watson-Crick base-pairing. This is important for future applications in biological systems where M-modified PNAs will have stronger preference for binding to target dsRNA or dsDNA and less off-target binding to ssRNA or ssDNA.
There has been little follow up after the pioneering studies of M in triplex-forming DNA oligonucleotides.42, 43 The reasons may be related to less than desired gains in stability of DNA triplexes or complicated synthesis of C-nucleosides.26, 42, 43 In contrast, M has been an excellent and relatively easy to prepare modification of triplex-forming PNAs. Our present results conclusively affirm that M is a superior modified nucleobase for triplex-forming PNAs that will improve binding affinity and selectivity in broad range of applications that currently use J to form stable triplets with G-C base pairs at physiologically relevant conditions. Collectively, our present and earlier studies12, 27, 28 demonstrate that M is a unique nucleobase that combines excellent RNA binding properties with improved cellular uptake, which will improve modified PNAs as research and diagnostic tools as well as potential therapeutic agents. To that end we and others have already used M-modified triplex-forming PNAs to inhibit translation of mRNA41 and maturation of pre-miRNA,44 for detection of A-to-I editing,40 and to drive dsRNA-specific templated reactions.45 In this context, our present study demonstrates that using M instead of J will provide significant improvement of PNAs performance in applications requiring triple-helical recognition of complex folded RNA and DNA.
Supplementary Material
ACKNOWLEDGMENT
We thank Vipin Kumar for help with the ITC data analysis and for design of UV melting experiments at 300 nm. This work was supported by National Institutes of Health (R35 GM130207 to E.R.) and National Science Foundation (CHE-1708761 to E.R.).
Footnotes
Supporting Information. General experimental procedures, synthesis, purification, and LC-MS characterization of PNA oligomers; ITC results and representative titration images; UV melting results.
REFERENCES
- [1].Nielsen PE, Egholm M, Berg RH, and Buchardt O (1991) Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide, Science 254, 1497–1500. [DOI] [PubMed] [Google Scholar]
- [2].Buchardt O, Egholm M, Berg RH, and Nielsen PE (1993) Peptide nucleic acids and their potential applications in biotechnology, Trends Biotechnol. 11, 384–386. [DOI] [PubMed] [Google Scholar]
- [3].Ono A, Ts’o POP, and Kan LS (1991) Triplex formation of oligonucleotides containing 2’-O-methylpseudoisocytidine in substitution for 2’-deoxycytidine, J. Am. Chem. Soc 113, 4032–4033. [Google Scholar]
- [4].Ono A, Ts’o POP, and Kan LS (1992) Triplex formation of an oligonucleotide containing 2’-O-methylpseudoisocytidine with a DNA duplex at neutral pH, J. Org. Chem 57, 3225–3230. [Google Scholar]
- [5].Egholm M, Christensen L, Dueholm KL, Buchardt O, Coull J, and Nielsen PE (1995) Efficient pH-independent sequence-specific DNA binding by pseudoisocytosine-containing bis-PNA, Nucleic Acids Res. 23, 217–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Chin JY, Kuan JY, Lonkar PS, Krause DS, Seidman MM, Peterson KR, Nielsen PE, Kole R, and Glazer PM (2008) Correction of a splice-site mutation in the beta -globin gene stimulated by triplex-forming peptide nucleic acids, Proc. Natl. Acad. Sci. U. S. A 105, 13514–13519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Bahal R, Ali McNeer N, Quijano E, Liu Y, Sulkowski P, Turchick A, Lu Y-C, Bhunia DC, Manna A, Greiner DL, Brehm MA, Cheng CJ, Lopez-Giraldez F, Ricciardi A, Beloor J, Krause DS, Kumar P, Gallagher PG, Braddock DT, Mark Saltzman W, Ly DH, and Glazer PM (2016) In vivo correction of anaemia in β-thalassemic mice by γPNA-mediated gene editing with nanoparticle delivery, Nat. Commun 7, 13304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].McNeer NA, Anandalingam K, Fields RJ, Caputo C, Kopic S, Gupta A, Quijano E, Polikoff L, Kong Y, Bahal R, Geibel JP, Glazer PM, Saltzman WM, and Egan ME (2015) Nanoparticles that deliver triplex-forming peptide nucleic acid molecules correct F508del CFTR in airway epithelium, Nat. Commun 6, 6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Li W, Shi H, Dong B, Nie K, Liu Z, and He N (2016) Recognition Mechanisms and Applications of Peptide Nucleic Acids Targeting Double-stranded DNA, Curr. Med. Chem 23, 4681–4705. [DOI] [PubMed] [Google Scholar]
- [10].Saadati A, Hassanpour S, Guardia M. d. l., Mosafer J, Hashemzaei M, Mokhtarzadeh A, and Baradaran B (2019) Recent advances on application of peptide nucleic acids as a bioreceptor in biosensors development, TrAC, Trends Anal. Chem 114, 56–68. [Google Scholar]
- [11].Stewart R, and Harris MG (1978) Comparison of the acidities and basicities of amino-substituted nitrogen heterocycles, J. Org. Chem 43, 3123–3126. [Google Scholar]
- [12].Zengeya T, Gupta P, and Rozners E (2012) Triple Helical Recognition of RNA Using 2-Aminopyridine-Modified PNA at Physiologically Relevant Conditions, Angew. Chem., Int. Ed 51, 12593–12596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Egholm M, Buchardt O, Nielsen PE, and Berg RH (1992) Peptide nucleic acids (PNA). Oligonucleotide analogs with an achiral peptide backbone, J. Am. Chem. Soc 114, 1895–1897. [Google Scholar]
- [14].Egholm M, Nielsen PE, Buchardt O, and Berg RH (1992) Recognition of guanine and adenine in DNA by cytosine and thymine containing peptide nucleic acids (PNA), J. Am. Chem. Soc 114, 9677–9678. [Google Scholar]
- [15].Saarbach J, Sabale PM, and Winssinger N (2019) Peptide nucleic acid (PNA) and its applications in chemical biology, diagnostics, and therapeutics, Curr. Opin. Chem. Biol 52, 112–124. [DOI] [PubMed] [Google Scholar]
- [16].Frickmann H, Zautner AE, Moter A, Kikhney J, Hagen RM, Stender H, and Poppert S (2017) Fluorescence in situ hybridization (FISH) in the microbiological diagnostic routine laboratory: a review, Crit. Rev. Microbiol 43, 263–293. [DOI] [PubMed] [Google Scholar]
- [17].Bentin T, Larsen HJ, and Nielsen PE (2003) Combined Triplex/Duplex Invasion of Double-Stranded DNA by “Tail-Clamp” Peptide Nucleic Acid, Biochemistry 42, 13987–13995. [DOI] [PubMed] [Google Scholar]
- [18].Shen X, and Corey DR (2018) Chemistry, mechanism and clinical status of antisense oligonucleotides and duplex RNAs, Nucleic Acids Res. 46, 1584–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Setten RL, Rossi JJ, and Han S. p. (2019) The current state and future directions of RNAi-based therapeutics, Nat. Rev. Drug Discovery 18, 421–446. [DOI] [PubMed] [Google Scholar]
- [20].Nielsen PE (2010) Sequence-selective targeting of duplex DNA by peptide nucleic acids, Curr. Opin. Mol. Ther 12, 184–191. [PubMed] [Google Scholar]
- [21].Nielsen PE (2005) Addressing the challenges of cellular delivery and bioavailability of peptide nucleic acids (PNA), Q. Rev. Biotech 38, 345–350. [DOI] [PubMed] [Google Scholar]
- [22].Roberts TC, Langer R, and Wood MJA (2020) Advances in oligonucleotide drug delivery, Nat. Rev. Drug Discovery 19, 673–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hildbrand S, and Leumann C (1996) Enhancing DNA triple helix stability at neutral pH by the use of oligonucleotides containing a more basic deoxycytidine analog, Angew. Chem., Int. Ed. Engl 35, 1968–1970. [Google Scholar]
- [24].Bates PJ, Laughton CA, Jenkins TC, Capaldi DC, Roselt PD, Reese CB, and Neidle S (1996) Efficient triple helix formation by oligodeoxyribonucleotides containing α- or β−2-amino-5-(2-deoxy-D-ribofuranosyl) pyridine residues, Nucleic Acids Res. 24, 4176–4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Cassidy SA, Slickers P, Trent JO, Capaldi DC, Roselt PD, Reese CB, Neidle S, and Fox KR (1997) Recognition of GC base pairs by triplex forming oligonucleotides containing nucleosides derived from 2-aminopyridine, Nucleic Acids Res. 25, 4891–4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hildbrand S, Blaser A, Parel SP, and Leumann CJ (1997) 5-Substituted 2-Aminopyridine C-Nucleosides as Protonated Cytidine Equivalents: Increasing Efficiency and Specificity in DNA Triple-Helix Formation, J. Am. Chem. Soc 119, 5499–5511. [Google Scholar]
- [27].Muse O, Zengeya T, Mwaura J, Hnedzko D, McGee DW, Grewer CT, and Rozners E (2013) Sequence Selective Recognition of Double-Stranded RNA at Physiologically Relevant Conditions Using PNA-Peptide Conjugates, ACS Chem. Biol 8, 1683–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Hnedzko D, McGee DW, Karamitas YA, and Rozners E (2017) Sequence-selective recognition of double-stranded RNA and enhanced cellular uptake of cationic nucleobase and backbone-modified peptide nucleic acids, RNA 23, 58–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Hudson RHE, and Wojciechowski F (2008) A Fmoc/Boc pseudoisocytosine monomer for peptide nucleic acid synthesis, Can. J. Chem 86, 1026–1029. [Google Scholar]
- [30].Brodyagin N, Hnedzko D, MacKay JA, and Rozners E (2020) Nucleobase-Modified Triplex-Forming Peptide Nucleic Acids for Sequence-Specific Recognition of Double-Stranded RNA, Methods Mol. Biol. (Totowa, NJ, U. S.) 2105, 157–172. [DOI] [PubMed] [Google Scholar]
- [31].A ntao VP, Lai SY, and Tinoco I Jr. (1991) A thermodynamic study of unusually stable RNA and DNA hairpins, Nucleic Acids Res. 19, 5901–5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ryan CA, and Rozners E (2020) Impact of Chirality and Position of Lysine Conjugation in Triplex-Forming Peptide Nucleic Acids, ACS Omega 5, 28722–28729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hansen ME, Bentin T, and Nielsen PE (2009) High-affinity triplex targeting of double stranded DNA using chemically modified peptide nucleic acid oligomers, Nucleic Acids Res. 37, 4498–4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kotikam V, Kennedy SD, MacKay JA, and Rozners E (2019) Synthetic, Structural, and RNA Binding Studies on 2-Aminopyridine-Modified Triplex-Forming Peptide Nucleic Acids, Chem. Eur. J 25, 4367–4372. [DOI] [PubMed] [Google Scholar]
- [35].Mergny J-L, and Lacroix L (2003) Analysis of thermal melting curves, Oligonucleotides 13, 515–537. [DOI] [PubMed] [Google Scholar]
- [36].Hartono YD, Pabon-Martinez YV, Uyar A, Wengel J, Lundin KE, Zain R, Smith CIE, Nilsson L, and Villa A (2017) Role of Pseudoisocytidine Tautomerization in Triplex-Forming Oligonucleotides: In Silico and in Vitro Studies, ACS Omega 2, 2165–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Christensen C, Eldrup AB, Haaima G, and Nielsen PE (2002) 1,8-Naphthyridin-2,7-(1,8H)-dione is an effective mimic of protonated cytosine in peptide nucleic acid triplex recognition systems, Bioorg. Med. Chem. Lett 12, 3121–3124. [DOI] [PubMed] [Google Scholar]
- [38].Devi G, Yuan Z, Lu Y, Zhao Y, and Chen G (2014) Incorporation of thio-pseudoisocytosine into triplex-forming peptide nucleic acids for enhanced recognition of RNA duplexes, Nucleic Acids Res. 42, 4008–4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Krishna MS, Wang Z, Zheng L, Bowry J, Ong AAL, Mu Y, Prabakaran M, and Chen G (2019) Incorporating G-C Pair-Recognizing Guanidinium into PNAs for Sequence and Structure Specific Recognition of dsRNAs over dsDNAs and ssRNAs, Biochemistry 58, 3777–3788. [DOI] [PubMed] [Google Scholar]
- [40].Annoni C, Endoh T, Hnedzko D, Rozners E, and Sugimoto N (2016) Triplex-forming peptide nucleic acid modified with 2-aminopyridine as a new tool for detection of A-to-I editing, Chem. Commun 52, 7935–7938. [DOI] [PubMed] [Google Scholar]
- [41].Endoh T, Hnedzko D, Rozners E, and Sugimoto N (2016) Nucleobase-Modified PNA Suppresses Translation by Forming a Triple Helix with a Hairpin Structure in mRNA In Vitro and in Cells, Angew. Chem., Int. Ed 55, 899–903. [DOI] [PubMed] [Google Scholar]
- [42].Rusling DA, Le Strat L, Powers VEC, Broughton-Head VJ, Booth J, Lack O, Brown T, and Fox KR (2005) Combining nucleoside analogues to achieve recognition of oligopurine tracts by triplex-forming oligonucleotides at physiological pH, FEBS Letters 579, 6616–6620. [DOI] [PubMed] [Google Scholar]
- [43].Rusling DA, Powers VEC, Ranasinghe RT, Wang Y, Osborne SD, Brown T, and Fox KR (2005) Four base recognition by triplex-forming oligonucleotides at physiological pH, Nucleic Acids Res. 33, 3025–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Endoh T, Brodyagin N, Hnedzko D, Sugimoto N, and Rozners E (2021) Triple-Helical Binding of Peptide Nucleic Acid Inhibits Maturation of Endogenous MicroRNA-197, unpublished data. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Kim KT, Chang D, and Winssinger N (2018) Double-Stranded RNA-Specific Templated Reaction with Triplex Forming PNA, Helv. Chim. Acta 101, e1700295. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


