Abstract
An extremely diverse group of human papillomavirus (HPV) types consisting of epidermodysplasia verruciformis (EV)-associated HPV types and other cutaneous HPV types (e.g., HPV types 2 and 3) is associated with nonmelanoma cancers and benign lesions of the skin. The frequent presence of multiple HPV types in single skin biopsy specimens of renal transplant recipients prompted us to develop PCR techniques for the detection of distinct (sub)groups of genotypically related cutaneous HPV types, i.e., three subgroups of EV-associated HPV types and two groups (A2 and A4) of other cutaneous HPV types. This approach generally allowed a reliable identification of HPV genotypes by direct sequencing of the PCR products, despite the frequent occurrence of multiple infections. The targeted spectrum of HPV types comprises 66 cutaneous HPV types including 21 putative novel HPV types. We also detected 17 putative novel HPV subtypes. We demonstrated that the skin of nearly all renal transplant recipients who developed various benign and (pre)malignant skin lesions was persistently infected with one or more EV-associated HPV types and/or HPV types belonging to groups A2 and A4. The frequency and distribution of EV-associated HPV and HPV types belonging to groups A2 and A4 were similar in biopsy specimens from hyperkeratotic papillomas (77.5%), squamous cell carcinomas (77.8%), and actinic keratoses (67.9%) but appeared to be lower in specimens of basal cell carcinomas (35.7%), benign lesions (38.5%), and clinically normal skin (32.3%). These findings suggest that renal transplant recipients are prone to persistent cutaneous HPV infection. Our data do not support the existence of high-risk cutaneous HPV types.
Human papillomaviruses (HPVs) are small, circular DNA viruses which are confined to the epithelial cells of both mucosa and skin (27). Recent epidemiological studies by PCR techniques demonstrated a strong association of certain high-risk genital HPV types, e.g., HPV type 16 (HPV-16), with cervical dysplasia and carcinoma. Molecular biological studies confirmed their role in the pathogenesis of cervical dysplasia and carcinoma (27). The association of many cutaneous HPV types with specific skin lesions in general and nonmelanoma skin cancer in particular remains enigmatic, partly because of the large number of cutaneous HPV types identified so far.
The earliest evidence for the involvement of specific HPV types in human skin cancer originates from observations for patients suffering from a hereditary disorder, epidermodysplasia verruciformis (EV) (17). About one-third of the EV patients develop multifocal cutaneous squamous cell carcinomas mainly on sun-exposed parts of the body. These patients are commonly infected with a group of genotypically related HPV types (EV-associated HPVs) that induce characteristic, macular skin lesions disseminated over the body (17).
Previously, we and others described broad-spectrum PCR methods, each of which enabled the detection of a large number of genotypically related HPV types, e.g., all EV-associated HPV types (1, 10, 22, 25). The nested PCR approach was shown to be highly specific and sensitive and circumvented the problem that DNA similarity is small even in highly conserved regions of the different HPV genotypes (10). A high prevalence of EV-associated HPV DNA was reported in squamous cell carcinomas and actinic keratoses from both renal transplant recipients (RTRs), and a slightly lower prevalence was reported in immunocompetent patients (1, 6, 7, 10, 22, 24, 25).
We and others used direct sequence analysis of amplified PCR products for HPV typing (1, 10, 22). The presence of multiple HPV types in a single biopsy specimen or smear complicates reliable typing by this method. Therefore, we intended to design PCR techniques which are specific for smaller subgroups of cutaneous HPV types. On the basis of phylogenetic studies, genotypically related HPV types have been united into groups of HPVs (5). For some of these groups a similar tropism has been determined (15, 26). We describe three nested EV-associated HPV (group B1) subgroup-specific PCRs and a PCR targeted to amplification of a subgroup of HPV types belonging to groups A2 and A4 (5). These PCR techniques enabled the characterization of multiple coinfecting HPV types in biopsy specimens from various benign and (pre)malignant skin lesions and normal skin. The value of the newly developed PCR approaches was also established by the identification of a series of new putative novel HPV types and subtypes. Furthermore, it was established that in renal transplant recipients specific cutaneous HPV types do not appear to be confined to specific skin lesions. Notably, no evidence was generated for the association of specific HPV types with skin cancers, in contrast to an earlier report (8). Interestingly, in most of the renal transplant recipients one or two HPV types among the other HPV types infecting the skin were consistently detectable in biopsy specimens which had been collected through the years from different types of skin lesions. This indicates that most RTRs with benign or (pre)malignant skin lesions are persistently infected with cutaneous HPV types.
MATERIALS AND METHODS
Tissue samples.
Biopsy specimens (n = 351) were collected from a group of Dutch RTRs. Half of the biopsy specimens were obtained when the RTR presented at the outpatient dermatology clinic (Leiden University Medical Center [LUMC], Leiden, The Netherlands) with lesions suspect for skin cancer. The other part of the lesions were collected in the same clinic as part of a case-control clinical study (7). Investigations were approved by the local institutional review board (LUMC).
Lesions were characterized according to the histological picture into squamous cell carcinomas, keratoacanthomas, Bowen's disease, basal cell carcinomas, actinic keratoses, hyperkeratotic papillomas, verrucae vulgares, verrucae planae, verrucae seborrheicae, and benign lesions such as dermatitis, cysts, and nevi. All biopsy specimens were divided into two: one half was processed for routine histology, the other half was snap-frozen in liquid nitrogen and stored at −70°C prior to DNA preparation. Clinically normal skin was not assessed for histology, and the complete biopsy specimens were snap-frozen and stored.
DNA extraction.
Tissue specimens were minced and treated overnight with proteinase K (100 μg/ml) and sodium dodecyl sulfate (0.5% [wt/vol]) at 56°C. DNA was extracted after heat inactivation of the proteinase K (10 min at 95°C) by a guanidium isothiocyanate-diatom based method (2).
PCR approaches.
The nested EV-associated HPV PCR approaches (PCR-A, PCR-B, and PCR-C) developed to target the three different subgroups of EV-associated HPV types (group B1 [5]) are indicated in Table 1. The nucleotide sequences and the annealing sites of the common 5′ EV-specific primer (CP62) and the 3′ subgroup-specific primers (CP71A, -B, and -C) are indicated in Table 2. The CP62-CP71 (A, B, and C) primer set amplifies 874- to 908-bp products, depending on the target HPV type and the 3′ primer used. For the second-step PCR, a common nested primer set consisting of two primers (primers CP64 and CP70A) with annealing sites that matched all HPV types mentioned above was designed (Table 1).
TABLE 1.
PCR approaches
| PCR and step (nested primer combination) | HPV (sub)group detecteda | Size (bp) of nested PCR product |
|---|---|---|
| PCR-A | ||
| First step (CP62-CP71A) | Group B1, subgroup A, HPV types 5, 8, 12, 14, 19, 20, 21, 25, 36, and 47 | 545 |
| Second step (CP64-CP70A) | ||
| PCR-B | ||
| First step (CP62-CP71B) | Group B1, subgroup B, HPV types 9, 15, 17, 22, 23, 37, 38, and 49 | 533–536 |
| Second step (CP64-CP70A) | ||
| PCR-C | ||
| First step (CP62-CP71C) | Group B1, subgroup C: HPV type 24 | 545 |
| Second step (CP64-CP70A) | ||
| 2/3-PCR | ||
| First step (CP61M-CP70M) | Group A2, HPV types 3, 10, 28, and 29, and group A4. HPV types 2, 27, and 57 | 707–719 |
| Second step (CP62M-CP69M) |
Groups A2, A4, and B1 as described in reference 5.
TABLE 2.
Primer sequences and annealing sites
| Primer | Sequencea | HPV type, annealing site (position)b |
|---|---|---|
| CP62 | GTW AAT GAA AYT TGY AAA TAT CC | HPV-8, 6520–6542; HPV-15, 6378–6400; HPV-24, 6391–6413 |
| CP71A | AAA CAT TCC TTG AAT ACY GTG | HPV-8, 7432–7452 |
| CP71B | GKY AAA AAT RCT TCA CAT ACC | HPV-15, 7263–7283 |
| CP71C | CAT GAA TAA CAA GCA TTC TTG G | HPV-24, 7294–7315 |
| CP64 | TAY TTY CCW ACW GTH AGT GG | HPV-8, 6754–6773 |
| CP70A | AAY TTT CKA CCY ARA GRA TAY TGA TC | HPV-8, 7273–7298 |
| CP61M | GAT ATG GTG GAY ACH GGS TWY GGT GC | HPV-10, 6414–6439 |
| CP70M | TTT CKT CCC ARR GGR AAY TG | HPV-10, 7203–7222 |
| CP62M | AAT AAG TCW GAY GTK CCY YTD GAT ATT TG | HPV-10, 6465–6493 |
| CP69M | TCC YTW AGA TCY ACW TCC CA | HPV-10, 7161–7180 |
The primers (primers CP61M and CP70M) and the respective annealing sites for the first step of the so-called 2/3-PCR targeted to HPV groups A2 and A4 are also indicated in Table 1. The CP61M-CP70M primer set amplifies 800- to 1,006-bp product depending on the target HPV type. The second step of the 2/3-PCR is performed with primers CP62M and CP69M (Table 1).
PCR amplification.
Amplification reactions in both the first- and second-step PCRs were performed by the method of Saiki et al. (19) in 50 μl of a reaction mixture containing 50 mM KCl, 10 mM Tris-HCl (pH 8.8), 3.6 mM MgCl2, 0.1 mg of bovine serum albumin per ml, each deoxynucleoside triphosphate at a concentration of 0.2 mM (Pharmacia), 1 U of Taq DNA polymerase (Perkin-Elmer Cetus), and 150 ng of both primers. In the first-step PCR, 5 μl of DNA (0.5 to 2% of total DNA extractable from the biopsy specimen) was used as input. The subsequent 40 cycles of amplification were performed for 1 min at 95°C, 1 min at 55°C, and 2 min at 72°C. In the nested PCR, 3 μl of the first-step PCR was used as input. In this PCR, 35 cycles of amplification were performed (1 min at 95°C, 1 min at 55°C, and 2 min at 72°C). All PCRs were performed in a Perkin-Elmer Cetus thermal cycler. For every two samples a negative control (water instead of DNA) was included. These controls were processed in the same way as the tissue specimens throughout the DNA preparation and the first- and second-step PCRs, and they were never found to be positive for HPV. All tissue samples were randomly analyzed. The investigator (R.J.M.B.) who performed the PCR analysis was blinded to the clinical data.
Sequence analysis.
The PCR products of the second-step PCR were reamplified by a PCR with a T7 primer extended nested primer set, T7CP65 (5′-TAA TAC GAC TCA CTA TAG GGC A[A/G]G GGT CA[C/T] AA[C/T] AAT GG[C/T] AT-3′) and CP69A (5′-TC[A/T] GT[C/T] AT[A/G] TCT ACA T[C/T]C CA-3′). Ten to 50 ng of the PCR product was used as input, and the amplification was performed for 30 cycles as described above. Sequencing was done on an ABI 370/373 automated sequencer with the Sequence Navigator computer program (Macintosh). DNA labeling and sampling were performed according to the manufacturer's protocol.
Phylogenetic analysis.
Alignment of the sequences were done by using the Clustal program (11) and was corrected by hand. The phylogenetic analyses were done by the neighbor-joining method as implemented in the MEGA package (20). All data were sorted by use of the Microsoft Office package.
Nucleotide sequence accession numbers.
The following putative novel sequences were submitted to GenBank (available at http://www.hpv-web.lanl.gov): HPV-X13 to HPV-X15 (accession nos. [ACs] AF054873, AF054874, and AF054876, respectively), HPV-X20 to HPV-X27 (ACs AF054877, AF054878, L38914, AF054879, AF054881, AF054882, AF054883, and AF054884, respectively), HPV-X29 (AC AF054885), HPV-X32 to HPV-X35 (ACs AF054886, AF054887, AF055710, and AF055711, respectively), HPV-XS1 to HPV-XS5 (ACs AF091448, AF091449, AF091450, AF091451, and AF091452, respectively), HPV-5c (AC AF091436), HPV-15b (AC AF091457), HPV-17b (AC AF097699), HPV-20b (AC AF091438), HPV-22b (AC AF091439), HPV-23b (AC AF091440), HPV-24b (AC AF091441), HPV-36b (AC AF091442), HPV-38b (AC AF091443), HPV-38c (AC AF091444), HPV-X2b (AC AF097700), HPV-X4b (AC AF091445), HPV-X10b (AC AF091446), HPV-X11b (AC AF091447), HPV-X11c (AC AF097701), HPV-X14b (AC AF154875), and HPV-X23b (AC AF054880).
RESULTS
HPV primer design.
A phylogenetic tree was constructed for all EV-associated HPV types of group B1 belonging to supergroup B, as defined earlier (5) (Fig. 1). Subsequently, three different first-step PCR primer sets were created for three nonoverlapping subgroups that, when combined, comprise the entire spectrum of the EV-associated HPV types. These subgroups were subgroup A (HPV-5, -8, -12, -14, -19, -20, -21, -25, -36, and -47), subgroup B (HPV-9, -15, -17, -22, -23, -37, -38, and -49), and subgroup C (HPV-24). The first-step PCR for the three EV-associated HPV subgroups was performed with one common 5′ primer and three different 3′ primers (see Table 1 and Materials and Methods). In a similar fashion, a nested PCR approach (referred to as 2/3-PCR) was designed for HPV types belonging to groups A2 and A4 (5) (HPV-2, -3, -10, -27, -28, -29, and -57).
FIG. 1.
A neighbor-joining tree based on the 92-amino-acid sequence of the PCR-amplified part of the L1 open reading frame of all EV-associated HPV types including the putative novel (sub)types (see Materials and Methods).
Analysis of clinical specimens.
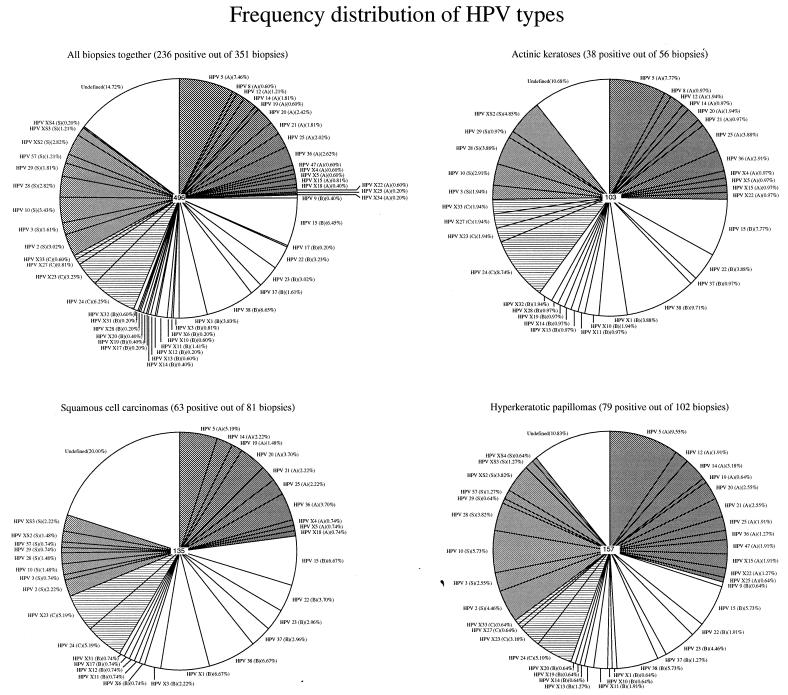
The HPV group- and subgroup-specific PCRs were conducted with a large number (n = 351) of skin biopsy specimens from Dutch RTRs. These skin biopsy specimens were taken from 81 squamous cell carcinomas, 14 basal cell carcinomas, 56 actinic keratoses, 102 hyperkeratotic papillomas, 12 verrucae vulgares, 17 verrucae planae, 16 verrucae seborrheicae, 5 keratoacanthomas, 14 Bowen's disease, and 13 benign lesions (dermatitis, cysts, and nevi); and 31 biopsy specimens were taken from clinically normal skin. Two hundred thirty-six of 351 (67.2%) lesions were found to be positive for HPV. None of the HPV (sub)groups were clearly associated with a particular type of lesion (Table 3). The frequency of HPV DNA in biopsy specimens from squamous cell carcinomas (77.8%) appeared to be higher than that in biopsy specimens from basal cell carcinomas (35.7%). HPV types belonging to EV-specific subgroup B were most frequently found (n = 149; 42.5%). The other subgroups were also frequently encountered: EV-specific subgroup A in 113 (32.5%) biopsy specimens, EV-specific subgroup C in 81 (23.1%) biopsy specimens, and the 2/3 group in 97 (27.6%) biopsy specimens. The frequencies of the HPV types in the different lesions from which less than 80 biopsy specimens were collected are shown in Table 3. The frequencies in squamous cell carcinomas, actinic keratoses, hyperkeratotic papillomas, and all biopsy specimens together are shown in Fig. 2.
TABLE 3.
HPV frequency in skin lesions from RTRs by (sub)group-specific nested PCRs
| Type of lesiona | No. of lesions | No. (%) of lesions positive for HPV
|
HPV typesb | ||||
|---|---|---|---|---|---|---|---|
| PCR-A | PCR-B | PCR-C | 2/3-PCR | All PCRs | |||
| SCC | 81 | 32 (39.5) | 41 (50.6) | 23 (28.4) | 18 (22.2) | 63 (77.8) | See Fig. 2 |
| KA | 5 | 1 (20.0) | 2 (40.0) | 1 (20.0) | 1 (20.0) | 2 (40.0) | Detected once, HPV types 5, 24, 36, X27, and X32; detected two times, undefined |
| MB | 14 | 5 (35.7) | 6 (42.9) | 2 (14.3) | 3 (21.4) | 9 (64.3) | Detected once, HPV types 2, 8, 9, 10, 12, 20, 28, 29, 36, 37, X4, and X53; detected two times, HPV types 24, 38, and X1; detected three times, HPV types 5 and 15 undefined |
| BCC | 14 | 0 (0.0) | 3 (21.4) | 4 (28.6) | 2 (14.3) | 5 (35.7) | Detected once, HPV types 15, 28, 29, 38, X1, and X23; detected two times, HPV type 24 and undefined |
| AK | 56 | 20 (35.7) | 29 (51.8) | 15 (26.8) | 13 (23.2) | 38 (67.9) | See Fig. 2 |
| HP | 102 | 43 (42.2) | 51 (50.0) | 23 (22.6) | 37 (36.3) | 79 (77.5) | See Fig. 2 |
| VV | 12 | 2 (16.7) | 6 (50.0) | 3 (25.0) | 6 (50.0) | 9 (75.0) | Detected once, HPV types 2, 5, 10, 17, 24, 29, 38, X5, X11, and X23; detected two times, HPV type 57 and undefined; detected three times, HPV types 22 and 23 |
| VP | 7 | 0 (0.0) | 1 (14.3) | 2 (28.6) | 6 (85.7) | 6 (85.7) | Detected once, HPV types 22, 23, and 36; detected two times, HPV type 29; detected four times, undefined |
| VS | 16 | 7 (43.8) | 7 (43.8) | 5 (31.3) | 4 (25.0) | 10 (62.5) | Detected once, HPV types 2, 3, 8, 10, 21, 38, X1, X3, X11, and X18; detected two times, HPV type 15; detected 11 times, undefined |
| BL | 13 | 1 (7.7) | 2 (15.4) | 1 (7.7) | 2 (15.4) | 5 (38.5) | Detected once, HPV types X1, X34, and XS2; detected two times, undefined |
| NS | 31 | 2 (6.5) | 1 (3.2) | 2 (6.5) | 5 (16.1) | 10 (32.3) | Detected once, HPV types 24, 29, 57, and X20; detected two times, HPV types 2 and 5 and undefined |
| All | 351 | 113 (32.2) | 149 (42.5) | 81 (23.1) | 97 (27.6) | 236 (67.2) | See Fig. 2 |
SCC, squamous cell carcinoma; KA, keratoacanthoma; MB, Bowen's disease, BCC, basal cell carcinoma; AK, actinic keratosis; HP, hyperkeratotic papilloma; VV, verruca vulgaris; VP, verruca plana; VS, verruca seborrheica; BL, benign lesion (dermatitis, cysts, nevi); NS, clinically normal skin.
The number of lesions with this HPV type are indicated; undefined, HPV type is unknown.
FIG. 2.
The frequency distribution of HPV types in all skin biopsy specimens, actinic keratoses, squamous cell carcinomas, and hyperkeratotic papillomas. The number of HPVs detected is indicated in the middle of the pie charts. Undefined, the HPV type is unknown. The EV-associated HPV subgroups are depicted in different grey scales and by shading.
From twenty-three RTRs, four or more biopsy specimens which had been taken over a period of months to years were analyzed by the four PCR approaches. Only two of these patients did not have any HPV-positive biopsy specimens. All other patients were infected at least at one site (Table 4). In nearly all RTRs one or more HPV types were consistently detected in biopsy specimens taken at different time points.
TABLE 4.
HPV types detected in RTRs from whom four or more biopsy specimens were available, organized by frequency of HPV infection
| Patient no. | Gender (age [yr])a | Type of lesionb | Time of biopsy (mo) relative to first biopsy | Location | HPV detection or HPV type detected byc:
|
|||
|---|---|---|---|---|---|---|---|---|
| PCR-A | PCR-B | PCR-C | 2/3-PCR | |||||
| 1 | M (49) | SCC | 0 | Face | − | − | − | − |
| SCC | 3 | Face | − | X17 | − | ND | ||
| SCC | 27 | Forearm | − | − | − | − | ||
| AK | 43 | Lower leg | − | − | − | − | ||
| AK | 43 | Forearm | − | − | − | − | ||
| AK | 43 | Forearm | − | − | − | − | ||
| NS | 43 | Buttock | − | − | − | − | ||
| SCC | 58 | Hand | − | − | − | ND | ||
| 2 | F (53) | SCC | 0 | Hand | X18 | − | − | − |
| AK | 33 | Upper arm | − | − | − | + | ||
| BCC | 33 | Trunk | − | − | − | − | ||
| NS | 33 | Buttock | − | − | − | 2a | ||
| Chronic inflammation | 44 | Hand | − | − | − | − | ||
| 3 | F (48) | MB | 0 | Upper leg | − | 15b, 38 | − | − |
| AK | 6 | Forearm | − | − | − | 3 | ||
| NS | 6 | Buttock | − | − | − | − | ||
| VV | 6 | Upper leg | − | − | − | − | ||
| 4 | F (58) | HP | 0 | Forearm | − | X11 | − | XS2 |
| HP | 21 | Upper arm | − | − | − | XS2 | ||
| HP | 21 | Lower leg | − | − | − | XS2 | ||
| HP | 21 | Hand | + | − | X27 | − | ||
| 5 | M (64) | SCC | 0 | Hand | − | 23, 38 | − | − |
| HP | 18 | Upper leg | − | − | − | − | ||
| HP | 18 | Fore arm | − | 38 | − | − | ||
| NS | 18 | Buttock | − | − | − | − | ||
| VV | 29 | Hand | − | 23, 38 | − | − | ||
| 6 | M (56) | SCC | 0 | Trunk | − | X1 | − | − |
| Dermal | 2 | Face | − | − | − | − | ||
| Nevus | ||||||||
| SCC | 9 | Forearm | − | 37 | − | − | ||
| BCC | 9 | Face | − | X1 | − | − | ||
| Fibroma molle | 23 | Trunk | − | − | − | − | ||
| SCC | 25 | Hand | − | + | − | − | ||
| SCC | 57 | Forearm | − | − | − | − | ||
| SCC | 64 | Face | + | X1 | + | |||
| SCC | 67 | Leg | − | 37 | − | − | ||
| SCC | 70 | Trunk | + | + | + | ND | ||
| 7 | F (57) | HP | 0 | Forearm | − | − | − | 57 |
| SCC | 3 | Forearm | 5c | − | − | − | ||
| NS | 3 | Forearm | 5c | − | − | − | ||
| VS | 9 | Upper leg | + | − | − | − | ||
| HP | 14 | Trunk | 5c | X10, 22 | − | − | ||
| SCC | 18 | Lower leg | 36 | − | − | − | ||
| SCC | 37 | Lower leg | − | − | − | − | ||
| SCC | 43 | Lower leg | − | + | − | − | ||
| AK | 45 | Forearm | 5c | 22 | − | − | ||
| NS | 45 | Buttock | − | + | − | − | ||
| MB | 45 | Lower leg | 5c | − | − | − | ||
| 8 | M (49) | SCC | 0 | Hand | 25 | 23 | − | 10 |
| SCC | 22 | Hand | 14 | 38b | − | − | ||
| HP | 22 | Hand | + | + | − | − | ||
| SCC | 38 | Upper arm | 25 | − | − | − | ||
| SCC | 44 | Not known | 14, 25 | − | − | − | ||
| 9 | M (57) | HP | 0 | Hand | 21 | + | − | 3 |
| SCC | 30 | Hand | 19, 21 | − | − | 2a | ||
| AK | 30 | Hand | − | − | − | − | ||
| SCC | 30 | Hand | + | − | − | − | ||
| NS | 36 | Buttock | − | − | − | − | ||
| HP | 43 | Forearm | 19, 21 | − | − | 3 | ||
| HP | 43 | Upper arm | + | − | − | − | ||
| NS | 43 | Buttock | − | − | − | − | ||
| 10 | F (45) | MB | 0 | Trunk | − | − | 24 | 29 |
| NS | 5 | Upper arm | − | − | 24 | − | ||
| BCC | 5 | Upper arm | − | − | 24 | 29 | ||
| SCC | 7 | Hand | − | − | 24 | 29 | ||
| SCC | 39 | Upper leg | − | − | − | − | ||
| HP | 39 | Lower leg | − | − | − | 29 | ||
| 11 | F (68) | MB | 0 | Upper arm | 5c | − | − | XS3 |
| SCC | 0 | Hand | 5c | − | − | − | ||
| HP | 15 | Forearm | 5c, X15 | − | − | + | ||
| SCC | 32 | Forearm | 5c | − | − | XS3 | ||
| SCC | 36 | Lower leg | 5c | X31 | − | XS3 | ||
| SCC | 40 | Lower leg | − | X31 | − | XS3 | ||
| HP | 58 | Forearm | X15 | X33 | − | 2a | ||
| HP | 58 | Forearm | X15 | − | − | XS3 | ||
| AK | 58 | Upper arm | X15 | − | − | − | ||
| MB | 66 | Lower leg | − | − | − | − | ||
| SCC | 67 | Hand | − | − | − | − | ||
| SCC | 75 | Face | + | − | + | + | ||
| 12 | M (44) | SCC | 0 | Lower leg | − | − | − | − |
| SCC | 9 | Lower leg | − | − | − | XS2 | ||
| SCC | 14 | Hand | − | 15, 37 | − | 3 | ||
| AK | 21 | Upper leg | − | 15, 37 | − | XS2 | ||
| HP | 24 | Upper leg | − | 15, 37 | − | 3 | ||
| HP | 27 | Lower leg | − | 15 | − | XS2 | ||
| 13 | F (53) | SCC | 0 | Upper leg | + | + | 24 | − |
| SCC | 8 | Hand | + | + | 24 | − | ||
| SCC | 8 | Lower leg | − | 22 | − | − | ||
| MB | 34 | Trunk | 5, 8, 12 | 9, 15c | 24 | − | ||
| NS | 34 | Buttock | − | − | − | − | ||
| AK | 47 | Face | − | − | − | ND | ||
| 14 | M (62) | SCC | 0 | Face | − | X12 | X23 | ND |
| SCC | 15 | Face | − | + | X23 | ND | ||
| VV | 19 | Trunk | X5 | 23 | X23 | ND | ||
| SCC | 23 | Face | 36 | 38c | X23 | ND | ||
| SCC | 29 | Forearm | + | 38c | X23 | ND | ||
| 15 | F (52) | HP | 0 | Lower leg | − | + | − | − |
| SCC | 25 | Forearm | 36b | + | X23, [36b] | ND | ||
| HP | 25 | Hand | + | + | + | ND | ||
| HP | 25 | Lower leg | + | + | − | ND | ||
| NS | 25 | Buttock | − | − | − | ND | ||
| HP | 27 | Hand | 36b, 5a | 23, 15 | X23 | ND | ||
| MB | 34 | Trunk | + | X1, X35 | − | ND | ||
| SCC | 37 | Hand | 36b | X1, 15 | + | ND | ||
| 16 | M (59) | VS | 0 | Trunk | − | 15, X3 | + | − |
| SCC | 24 | Forearm | 20, 21 | + | 24, X23 | − | ||
| SCC | 29 | Lower leg | − | 15, X3 | + | − | ||
| SCC | 59 | Face | − | 15, X3 | + | − | ||
| 17 | F (61) | MB | 0 | Upper leg | − | X1 | − | 10 |
| VS | 0 | Face | 8 | X1 | + | 10 | ||
| HP | 0 | Upper leg | − | − | − | − | ||
| SCC | 3 | Hand | [X1] | X1 | − | − | ||
| 18 | M (60) | AK | 0 | Face | − | 22 | − | − |
| SCC | 14 | Face | − | 23 | 24 | − | ||
| Molluscum contagiosum | 14 | Trunk | − | + | − | − | ||
| VV | 57 | Forearm | 5a | 22b, 23b, 23 | [23b] | 29 | ||
| VP | 57 | Forearm | − | 22b, 23b | + | 29 | ||
| 19 | M (66) | KA | 0 | Lower leg | 5a, 36 | X32 | X27, 24 | − |
| AK | 56 | Upper arm | 25, 36 | X32, 15, 22b | X27, [36] | − | ||
| AK | 56 | Forearm | 25, 36 | 22b | X27, 24 | + | ||
| NS | 56 | Buttock | 5a | − | − | − | ||
| AK | 68 | Hand | 5, 36 | 15b | 24b | − | ||
| 20 | M (64) | BCC | 0 | Trunk | − | 38 | 24 | 28 |
| HP | 29 | Upper arm | 14, 47 | 15, X20 | + | Mix | ||
| HP | 29 | Forearm | 14, 47 | + | 24a, [14] | 2a | ||
| NS | 29 | Buttock | − | X20 | − | |||
| 21 | M (53) | SCC | 0 | Hand | + | − | X23 | XS2 |
| AK | 7 | Face | 20 | X14b, X32 | X33 | + | ||
| Dermatitis | 23 | Hand | X34 | − | + | XS2 | ||
| SCC | 34 | Fore arm | 5b | X6 | − | ND | ||
| VS | 46 | Trunk | − | − | − | ND | ||
| NS | 46 | Buttock | − | − | − | − | ||
| AK | 46 | Forearm | 5a, 20 | X28 | X33 | − | ||
M, male; F, female.
See footnote a of Table 3 for definitions of abbreviations.
−, HPV negative; +, HPV positive, type not determined; brackets, false-positive result; ND, not done.
Specificities of the PCR approaches for subgroups of HPV types.
The different PCR approaches generally proved to be specific for the respective targeted (sub)groups of HPV types, despite the frequent finding of multiple HPV types in a single biopsy specimen. Only infrequently EV-associated HPV types belonging to either subgroup A, B, or C were amplified by a PCR not mediated by the corresponding subgroup-specific primer pair (see, e.g., Table 4, patients 15, 17, 18, 19, and 20). No EV-associated HPV types were detected by the 2/3-PCR designed to detect groups A2 and A4.
Novel HPV types and subtypes.
HPV typing was generally performed by direct sequence analysis of a PCR-amplified 264- to 276-nucleotide fragment and by applying a 10% distance at the nucleotide level for definition of a novel type (5). In those RTRs in whom more than one HPV type was detected by one of the four PCR approaches (Table 4), the PCR product was molecularly cloned before sequence analysis. Sixteen new putative novel EV-associated HPV types were identified in this study (HPV-X13 to HPV-X15, HPV-X20 to HPV-X27, HPV-X29, and HPV-X32 to HPV-X35) and five new putative novel HPV types belonging to the A2 and A4 group (HPV-XS1 to HPV-XS5) were identified (Tables 3 and 4). Also, 17 novel subtypes of 14 EV-associated HPV types were identified, and these differed by 2 to 10% at nucleotide level. The nucleotide sequences of the novel (sub)types have been submitted to GenBank (see Materials and Methods).
All new putative novel EV-associated HPV (sub)types appeared to fit into the phytogenetic tree shown in Fig. 1. This provided evidence that the unknown sequences were amplified from novel EV-associated HPV (sub)types distinct from all known (sub)types.
DISCUSSION
HPV subgroup-specific PCR methods.
In an effort to develop an HPV DNA detection method which allows the efficient identification of multiple HPV types within a single lesion and also of novel HPV types, we designed four PCR approaches mediated by nested primer sets which targeted specific HPV (sub)groups. One PCR approach targeted the HPV types belonging to the A2 and A4 HPV groups (HPV types 2 and 3 and related types) (5). The other three targeted subgroups that have been chosen from within the B1 group of EV-associated HPV types (5). For the detection of the subgroups of EV-associated HPV types, broad-spectrum PCR primers were designed with an annealing site in the L1 open reading frames of all HPV types, including previously identified novel HPV sequences. The 3′ primers were chosen from sequences in the long control region, since the conserved binding sites for transcription regulatory proteins in this region served well as target sites for these subgroup-specific primers.
An advantage of these nested PCR approaches was the potential to identify the different HPV types present in a single lesion by direct sequence analysis of the PCR products in general without the necessity of molecular cloning. The PCR fragment of 264 to 276 nucleotides which was used for EV-associated HPV (sub)typing as described before (1) encompasses variable and conserved genomic segments of the L1 open reading frame. The conserved part of this fragment overlaps the DNA segment amplified by PCR with the MY09-MY11 primer pair (13), which is commonly used for the identification of mucosal HPV types. Putative novel types were identified as sequences with dissimilarities of more than 10% compared to all other known sequences, as defined previously (1, 5, 9, 15). The typing protocol by sequencing resulted in unambiguous signals when one HPV type was present. When direct sequencing indicated that more than one HPV type was present, molecular cloning of the PCR product was performed to identify the HPV types. In general, the nested PCR approaches appeared to be specific for the targeted HPV subgroups, since positive PCR signals were only infrequently obtained from nontargeted HPV types. This problem occurred only when PCR methods were used for the detection of the EV-associated HPV subgroups. This is probably due to the presence of a relatively high EV-associated HPV copy number in some lesions. RTRs with lesions that contain such a high copy number of HPV appear to be uncommon (Table 4, patients 15, 17, 18, 19, and 20).
Different numbers of HPV types were detected by the respective EV-associated HPV subgroup-specific PCR approaches. Subgroup B is very large, with a high continuous diversity throughout the cluster; subgroup A is less diverse, with a characteristic subdivision into clades; and subgroup C started out from only HPV-24 and in this study is found to comprise only one phylogenetically related group of HPVs (Fig. 1).
Novel HPV types, subtypes, and variants.
The usefulness of the nested PCR approaches was also borne out by the detection of about 16 new putative novel EV-associated HPV types, 5 new putative novel HPV types belonging to the A2 and A4 groups (HPV-XS1 to HPV-XS5), and 17 HPV subtypes (see Materials and Methods). Of several HPV types two or three distinct subtypes with dissimilarities of 2 to 10% at the nucleotide level from their reference HPV type could be identified (data not shown). These dissimilar nucleotides were not concentrated within the variable segment but were randomly distributed. Most of the mismatching nucleotides were silent substitutions (data not shown). The occurrence of subtypes was not limited to EV-associated HPV types since subtypes of HPV-57 were also found. Besides subtypes, variants were identified with a low level of sequence diversity (≤2%) (data not shown). This degree of diversity has previously been observed among most HPV types that have been investigated more thoroughly and is generally used to describe variants (5). In our study we found a similar degree of variability within EV-associated HPV types, but this variability was only occasionally observed in HPV-2, HPV-3, or related HPV types (Table 4 and data not shown). A recent study of HPV-2, -27, and -57, however, describes large numbers of variants of these three HPV types (4).
HPV DNA persists in the skin of RTRs.
Epidemiological studies by broad consensus PCRs clearly indicate that EV-associated HPV is commonly found in skin lesions from RTRs and are also frequently detected throughout the population of immunocompetent individuals (for a review, see reference 18). Our previous studies revealed the presence of EV-associated HPV in about 80% of the biopsy specimens from (pre)malignant skin lesions from a group of Dutch RTRs (1, 6, 7). In the study described in this report we demonstrated in most of the RTRs the persistence of one or two HPV types, since we detected these types in biopsy specimens collected over a period of months to years from benign, premalignant, and malignant lesions and also from normal skin (Table 4, patients 4 to 19 and 21). Notably, the frequency and distribution of the detected HPV types in the different lesions, i.e., in hyperkeratotic papillomata, squamous cell carcinomas, and actinic keratoses, were very similar (Table 3; Fig. 2). The frequency of EV-associated HPV detection in basal cell carcinomas appeared to be comparable to the frequency of HPV detection in benign lesions and clinically normal skin but lower than the frequency of HPV detection in cutaneous squamous cell carcinomas and hyperkeratotic lesions (Table 3).
In the present report a persistent and frequent association of HPV types belonging to groups A2 and A4 with benign lesions, (pre)malignant lesions, and normal skin of some individual patients has been found (Table 4, patients 10, 11, 12, and 21). It extends a previous observation that, in particular, HPV-3 and related types were frequently detected in benign lesions from immunocompromised patients (16). These data suggest that RTRs are sensitive to infection with one of these HPV types, probably due to failing immune surveillance. This observation must be corroborated by more extensive epidemiological studies.
Some groups demonstrated the presence of still other HPV types (22; for a review, see reference 18), but in this and previous studies we failed to detect them. These include mucosal HPV types (groups A1, A3, and A5 to A11) that are associated with genital carcinomas (i.e., HPV-16 and -56) and cutaneous HPV types without a known widespread histopathological association (i.e., HPV-41 and -48). In our previous attempts to detect HPV in skin lesions we also failed to detect mucosal HPV types using multiple broad consensus PCR methods (1, 6, 25). The PCR method for the detection of HPV-2 and -3, and related types (groups A2 and A4) described here has been found to detect mucosal HPV types in cervical smears, but these HPV types were not found in skin lesions from RTRs (data not shown). Preliminary PCR studies also targeted other cutaneous HPV types, including HPV-1, -4, and -65, that have been regularly detected in skin warts of immunocompetent patients (12). Also, these HPV types have not been detected in biopsy specimens from skin lesions from RTRs (data not shown).
In conclusion, we report that nearly all RTRs who develop benign or (pre)malignant skin lesions are persistently infected with at least one HPV type, often for long periods. Recently, it was reported that high-risk genital HPV types often persist in immunocompromised human immunodeficiency virus-infected patients (14, 23). An increased HPV persistence may explain the higher prevalence of genital HPV infections as well as the increased risk of cervical neoplasia in human immunodeficiency virus-infected women (21). It will be important to investigate whether cutaneous HPV types also persist more frequently in the skin of immunocompromised patients than in the skin of immunocompetent individuals. Such studies will be very difficult to perform by analyzing multiple lesions from a large number of immunosuppressed and immunocompetent individuals. Our recent detection of EV-associated HPV types in plucked hairs in a considerable part of the population (3, 3a) may facilitate these kinds of studies.
ACKNOWLEDGMENTS
We thank Ingeborg Boxman and Cees Sol for helpful suggestions and critical reading of the manuscript, Wim van Est for preparing the figures, and Anneke Wiersma for typing the manuscript.
The research was supported by the Dutch Kidney Foundation (grant C94-1390).
REFERENCES
- 1.Berkhout R J, Tieben L M, Smits H L, Bouwes Bavinck J N, Vermeer B J, ter Schegget J. Nested PCR approach for detection and typing of epidermodysplasia verruciformis-associated human papillomavirus types in cutaneous cancers from renal transplant recipients. J Clin Microbiol. 1995;33:690–695. doi: 10.1128/jcm.33.3.690-695.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boom R, Sol C J, Salimans M M, Jansen C L, Wertheim-van Dillen P M, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boxman I L, Berkhout R J, Mulder L H, Wolkers M C, Bouwes Bavinck J N, Vermeer B J, ter Schegget J. Detection of human papillomavirus DNA in plucked hairs from renal transplant recipients and healthy volunteers. J Investig Dermatol. 1997;108:712–715. doi: 10.1111/1523-1747.ep12292090. [DOI] [PubMed] [Google Scholar]
- 3a.Boxman I L A, Russell A, Mulder L H C, Bouwes Bavinck J N, ter Schegget J, Green A the Nambour Skin Cancer Prevention Study Group. Case-control study in a subtropical Australian population to assess the relation between non-melanoma skin cancer and epidermodysplasia verruciformis human papillomairus DNA in plucked eyebrow hairs. Int J Cancer. 2000;86:118–121. doi: 10.1002/(sici)1097-0215(20000401)86:1<118::aid-ijc18>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 4.Chan S Y, Chew S H, Egawa K, Grussendorf-Conen E I, Honda Y, Rubben A, Tan K C, Bernard H U. Phylogenetic analysis of the human papillomavirus type 2 (HPV-2), HPV-27, and HPV-57 group, which is associated with common warts. Virology. 1997;239:296–302. doi: 10.1006/viro.1997.8896. [DOI] [PubMed] [Google Scholar]
- 5.Chan S Y, Delius H, Halpern A L, Bernard H U. Analysis of genomic sequences of 95 papillomavirus types: uniting typing, phylogeny, and taxonomy. J Virol. 1995;69:3074–3083. doi: 10.1128/jvi.69.5.3074-3083.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Jong-Tieben L M, Berkhout R J, Smits H L, Bouwes Bavinck J N, Vermeer B J, van der Woude F J, ter Schegget J. High frequency of detection of epidermodysplasia verruciformis-associated human papillomavirus DNA in biopsies from malignant and premalignant skin lesions from renal transplant recipients. J Investig Dermatol. 1995;105:367–371. doi: 10.1111/1523-1747.ep12320803. [DOI] [PubMed] [Google Scholar]
- 7.de Jong-Tieben L M, Berkhout R J, ter Schegget J, Vermeer B J, de Fijter J W, Bruijn J A, Westendorp R G J, Bouwes Bavinck J N. The prevalence of human papillomavirus DNA (EV-HPV) in benign keratotic skin lesions of renal transplant recipients with and without a history of skin cancer is equally high: a clinical study to assess risk factors for keratotic skin lesions and skin cancer. Transplantation. 2000;69:44–49. [PubMed] [Google Scholar]
- 8.de Villiers E M, Lavergne D, McLaren K, Benton E C. Prevailing papillomavirus types in non-melanoma carcinomas of the skin in renal allograft recipients. Int J Cancer. 1997;73:356–361. doi: 10.1002/(sici)1097-0215(19971104)73:3<356::aid-ijc9>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 9.Guerrero E, Shah K V. Polymerase chain reaction in HPV diagnosis. Papillomavirus Rep. 1991;2:115–118. [Google Scholar]
- 10.Harwood C A, Spink P J, Surentheran T, Leigh I M, Hawke J L, Proby C M, Breuer J, McGregor J M. Detection of human papillomavirus DNA in PUVA-associated non-melanoma skin cancers. J Investig Dermatol. 1998;111:123–127. doi: 10.1046/j.1523-1747.1998.00240.x. [DOI] [PubMed] [Google Scholar]
- 11.Higgins D G, Sharp P M. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 12.Jablonska S, Majewski S, Obalek S, Orth G. Cutaneous warts. Clin Dermatol. 1997;15:309–319. doi: 10.1016/s0738-081x(96)00170-8. [DOI] [PubMed] [Google Scholar]
- 13.Manos M M, Ting Y, Wright D K, Lewis A J, Broker T R, Wolinsky S M. The use of polymerase chain reaction amplification for the detection of genital papillomaviruses. Mol Diagn Hum Cancer Cancer Cells. 1989;7:209–214. [Google Scholar]
- 14.Minkoff H, Feldman J, DeHovitz J, Landesman S, Burk R. A longitudinal study of human papillomavirus carriage in human immunodeficiency virus-infected and human immunodeficiency virus-uninfected women. Am J Obstet Gynecol. 1998;178:982–986. doi: 10.1016/s0002-9378(98)70535-6. [DOI] [PubMed] [Google Scholar]
- 15.Myers G, Lu H, Calef C, Leitner T. Heterogeneity of papillomaviruses. Semin Cancer Biol. 1996;7:349–358. doi: 10.1006/scbi.1996.0044. [DOI] [PubMed] [Google Scholar]
- 16.Obalek S, Favre M, Szymanczyk J, Misiewicz J, Jablonska S, Orth G. Human papillomavirus (HPV) types specific of epidermodysplasia verruciformis detected in warts induced by HPV3 or HPV3-related types in immunosuppressed patients. J Investig Dermatol. 1992;98:936–941. doi: 10.1111/1523-1747.ep12460892. [DOI] [PubMed] [Google Scholar]
- 17.Orth G, Jablonska S, Jarzabek-Chorzelska M, Obalek S, Rzesa G, Favre, Croissant O. Characteristics of the lesions and risk of malignant conversion associated with the type of human papillomavirus involved in epidermodysplasia verruciformis. Cancer Res. 1979;39:1074–1082. [PubMed] [Google Scholar]
- 18.Pfister H, ter Schegget J. Role of HPV in cutaneous premalignant and malignant tumors. Clin Dermatol. 1997;15:335–347. doi: 10.1016/s0738-081x(96)00162-9. [DOI] [PubMed] [Google Scholar]
- 19.Saiki R K, Chang C A, Levenson C H, Warren T C, Boehm C D, Kazazian H H, Jr, Erlich H A. Diagnosis of sickle cell anemia and beta-thalassemia with enzymatically amplified DNA and nonradioactive allele-specific oligonucleotide probes. N Engl J Med. 1988;319:537–541. doi: 10.1056/NEJM198809013190903. [DOI] [PubMed] [Google Scholar]
- 20.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 21.Shah K V. Human papillomaviruses and anogenital cancers. N Engl J Med. 1997;337:1386–1388. doi: 10.1056/NEJM199711063371911. [DOI] [PubMed] [Google Scholar]
- 22.Shamanin V, zur Hausen H, Lavergne D, Proby C M, Leigh I M, Neumann C, Hamm H, Goos M, Haustein U F, Jung E G. Human papillomavirus infections in nonmelanoma skin cancers from renal transplant recipients and nonimmunosuppressed patients. J Natl Cancer Inst. 1996;88:802–811. doi: 10.1093/jnci/88.12.802. [DOI] [PubMed] [Google Scholar]
- 23.Sun X W, Kuhn L, Ellerbrock T V, Chiasson M A, Bush T J, Wright T C., Jr Human papillomavirus infection in women infected with the human immunodeficiency virus N. Engl J Med. 1997;337:1343–1349. doi: 10.1056/NEJM199711063371903. [DOI] [PubMed] [Google Scholar]
- 24.Surentheran T, Harwood C A, Spink P J, Sinclair A L, Leigh I M, Proby C M, McGregor J M, Breuer J. Detection and typing of human papillomaviruses in mucosal and cutaneous biopsies from immunosuppressed and immunocompetent patients and patients with epidermodysplasia verruciformis: a unified diagnostic approach. J Clin Pathol. 1998;51:606–610. doi: 10.1136/jcp.51.8.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tieben L M, Berkhout R J, Smits H L, Bouwes Bavinck J N, Vermeer B J, Bruijn J A, van der Woude F J, ter Schegget J. Detection of epidermodysplasia verruciformis-like human papillomavirus types in malignant and premalignant skin lesions of renal transplant recipients. Br J Dermatol. 1994;131:226–230. doi: 10.1111/j.1365-2133.1994.tb08496.x. [DOI] [PubMed] [Google Scholar]
- 26.Van Ranst M, Kaplan J B, Burk R D. Phylogenetic classification of human papillomaviruses: correlation with clinical manifestations. J Gen Virol. 1992;73:2653–2660. doi: 10.1099/0022-1317-73-10-2653. [DOI] [PubMed] [Google Scholar]
- 27.zur Hausen H. Papillomavirus infections—a major cause of human cancers. Biochim Biophys Acta. 1996;1288:F55–F78. doi: 10.1016/0304-419x(96)00020-0. [DOI] [PubMed] [Google Scholar]