Abstract
Background
Genome-wide DNA methylation profiling has recently been developed into a tool that allows tumor classification in central nervous system tumors. Extracellular vesicles (EVs) are released by tumor cells and contain high molecular weight DNA, rendering EVs a potential biomarker source to identify tumor subgroups, stratify patients and monitor therapy by liquid biopsy. We investigated whether the DNA in glioblastoma cell-derived EVs reflects genome-wide tumor methylation and mutational profiles and allows noninvasive tumor subtype classification.
Methods
DNA was isolated from EVs secreted by glioblastoma cells as well as from matching cultured cells and tumors. EV-DNA was localized and quantified by direct stochastic optical reconstruction microscopy. Methylation and copy number profiling was performed using 850k arrays. Mutations were identified by targeted gene panel sequencing. Proteins were differentially quantified by mass spectrometric proteomics.
Results
Genome-wide methylation profiling of glioblastoma-derived EVs correctly identified the methylation class of the parental cells and original tumors, including the MGMT promoter methylation status. Tumor-specific mutations and copy number variations (CNV) were detected in EV-DNA with high accuracy. Different EV isolation techniques did not affect the methylation profiling and CNV results. DNA was present inside EVs and on the EV surface. Proteome analysis did not allow specific tumor identification or classification but identified tumor-associated proteins that could potentially be useful for enriching tumor-derived circulating EVs from biofluids.
Conclusions
This study provides proof of principle that EV-DNA reflects the genome-wide methylation, CNV, and mutational status of glioblastoma cells and enables their molecular classification.
Keywords: exosome, glioma, methylome, mutation, proteomics
Key Points.
The extracellular vesicle (EV) DNA methylome enables noninvasive brain tumor classification.
Glioma-associated mutations and copy number variations are present in EV-DNA.
Glioma-associated EV proteins may facilitate the enrichment of tumor-derived EVs.
Importance of the Study.
Glioma patients have increased levels of extracellular vesicles (EVs) in their bloodstream. Circulating tumor-derived EVs has been recognized as promising tumor surrogates that carry important biomarkers, such as tumor-derived RNA, proteins, and DNA. While previous studies largely focused on identifying tumor-specific RNA and protein signatures, EV-associated DNA represents a relatively unexplored source of information. Here, we demonstrate that EV-DNA faithfully reflects the DNA methylation class of glioblastoma cells and tumors and thereby allows their molecular classification. Furthermore, MGMT promoter methylation, genome-wide copy number variations, and driver mutations present in original tumors are detectable in EV-DNA with high accuracy. Proteomic profiling identifies glioma-associated proteins that may become valuable for tumor-derived EV enrichment from biofluids to enhance the detection sensitivity for tumor-specific genetic alterations. Our findings provide a step toward diagnostic and therapeutic monitoring of glioma patients via analysis of EV-DNA.
Cancer cells release EVs carrying complex biologically active molecules into the tumor microenvironment and bloodstream. Thus, EVs are being distributed throughout the body, rendering them attractive targets for liquid biopsies in cancer patients. We previously showed that patients suffering from glioblastoma and other central nervous system (CNS) tumors have increased levels of circulating EVs and that tumor-specific EVs are present in the circulation of glioma-bearing mice.1 Furthermore, elevated plasma EV concentrations in glioblastoma patients were found to drop after surgery but rise again at tumor relapse, suggesting that EV dynamics can reflect the disease state.2
EVs comprise different entities, including exosomes, microvesicles, and large oncosomes.3 The molecular cargo inside EVs consists of proteins, RNA, DNA, and lipids and is protected from fragmentation and degradation by the surrounding EV membrane. Much of the recent interest in glioma EVs was triggered by the discovery that EV-encapsulated RNA and proteins in patient blood can provide information on the tumor of origin. Studies in glioblastoma patients suggested that miRNA signatures and tumor-associated proteins identified in EVs from cerebrospinal fluid (CSF) or blood could serve as diagnostic biomarkers.2,4–6 Furthermore, tumor-specific molecules, such as mutant epidermal growth factor receptor EGFRvIII protein and mRNA, as well as mutant IDH mRNA and DNA were detected in EVs obtained from glioma cell cultures or liquid biopsies of glioblastoma patients.7–12 While these results highlight the diagnostic potential of EVs, the informative value of mutations in few selected genes affected by recurrent hotspot mutations such as IDH1 or EGFR is limited to only a subset of patients bearing these alterations. A more comprehensive profiling is necessary, in order to classify tumors with unknown genetic alterations and to monitor changes in the genetic or epigenetic tumor make-up over the course of disease treatment and progression.
Studies in other types of cancer have shown that high molecular weight double-stranded DNA from all chromosomes is present in EVs and can reflect the genome-wide mutational status of parental tumor cells.13–17 We therefore surmised that EV-DNA could be leveraged to obtain comprehensive information on the mutational status of gliomas and could also permit DNA methylation profiling, which is a powerful tool for the classification of brain tumors.18
Here we demonstrate that DNA is contained and protected in EVs secreted by glioblastoma cells, by using super-resolution single-EV microscopy. Methylation array analysis and targeted next-generation sequencing (NGS) reveal that EV-associated DNA mirrors the complete landscape of mutations and copy number variations (CNV) present in parental glioma cells as well as original tumors. The tumor methylation class, as well as MGMT promoter methylation status, can be correctly determined by analyzing glioblastoma cell-derived EV-DNA, and different EV isolation techniques yield consistent results.
Materials and Methods
Human Specimens
Glioma tissue and non-tumorous temporal lobe tissue from patients undergoing epilepsy surgery were obtained as approved by the medical ethics committee of the Hamburg chamber of physicians (PV4904, PV5034). Informed written consent was obtained from all patients.
Cell Culture
Glioblastoma stem-like (GS) cell cultures were established and cultured as described previously19 and detailed in the Supplementary Methods. Cell lines NCH1681 (anaplastic astrocytoma) and NCH551b (glioblastoma) were kindly provided by Dr. Christel Herold-Mende (Heidelberg University Hospital, Heidelberg, Germany).20
Isolation and Size Analysis of EVs
EVs were isolated from conditioned medium by differential centrifugation as described previously21 or by size exclusion chromatography (SEC), and analyzed by nanoparticle tracking analysis (NTA) as detailed in the Supplementary Methods.
Imaging Flow Cytometry (IFCM)
EVs were analyzed by IFCM as reported1 and as detailed in the Supplementary Methods.
Transmission Electron Microscopy (TEM)
The morphology of EVs was analyzed by TEM as detailed in the Supplementary Methods.
Super-Resolution Imaging of Single EVs
EV-associated DNA was visualized and quantified using Direct Stochastic Optical Reconstruction Microscopy (dSTORM) as detailed in the Supplementary Methods.
Methylation Array Analysis
Infinium MethylationEPIC arrays (850k) were used to obtain genome-wide DNA methylation and CNV profiles as described in the Supplementary Methods.
Mutation Analysis by Gene Panel NGS and Digital Droplet PCR (ddPCR)
Coding sequences or mutational hot spot regions were analyzed by NGS and ddPCR as detailed in the Supplementary Methods.
EV Surface Epitope Analysis
Multiplex profiling of EV surface epitopes was performed using the MACSPlex assay (Miltenyi Biotec) as detailed in the Supplementary Methods.
Differential Quantitative Proteomics
Samples were analyzed by differential quantitative proteomics, using a LC-MS/MS system as described in the Supplementary Methods.
Immunoblot Analysis
Western blot analysis was performed as reported22 and as detailed in the Supplementary Methods.
Statistical Analysis
One-way ANOVA with post-hoc Bonferroni or Kruskal-Wallis test with Dunn’s correction was conducted to compare multiple groups with normal or non-Gaussian sample distribution. Statistical analyses were performed using Microsoft Office Excel 2016 or GraphPad Prism 8.
Results
Characterization of Glioma EVs
To obtain tumor cell-derived EVs, we established primary cell cultures from freshly resected isocitrate dehydrogenase wild-type (IDHwt) glioblastomas, using serum-free neural stem cell conditions (Supplementary Table 1).19 Further, we included two IDH-mutant (IDHmut) cell lines, NCH551b (glioblastoma) and NCH1681 (anaplastic astrocytoma).20 EVs were isolated from conditioned culture supernatants and their typical cup-shaped morphology23 was proven by electron microscopy (Figure 1A and 1B). EVs fell within the expected size range of exosomes and microvesicles (Supplementary Figure 1A) and expressed tetraspanin markers CD9, CD63, and CD81 (Figure 1C, Supplementary Figure 1B–E).
Fig. 1.
Extracellular vesicle (EV) isolation and characterization. A, Glioma cells release small EVs from multivesicular bodies (MVB), coordinated by the endosomal sorting complex required for transport (ESCRT), while larger EVs are generated by membrane budding. Glioma tissue was cultured and EVs secreted by tumor cells were analyzed. B, Electron microscopy demonstrates the cup-shaped morphology of EVs (arrows). C, Detection of CD9, CD63, and CD81 by imaging flow cytometry. D, Direct stochastic optical reconstruction microscopy of EVs identified by CD63 or CD81 and stained with nucleic acid dye. DNase treatment removed extra-vesicular DNA, and permeabilization accessed intra-vesicular DNA. E, Quantification of the number of DNA fragment localizations per EV, based on co-localization with CD63 and/or CD81. Box plots with 10-90 percentile (whiskers), median (line) and 25-75 percentile (box); ****P < .0001, Kruskal-Wallis analysis.
To precisely assess the localization and quantify EV-DNA, we performed super-resolution imaging of single EVs using dSTORM. To distinguish DNA inside EVs from DNA associated with the outer membrane, intact EVs were treated with DNase to digest external DNA and subsequently permeabilized to facilitate dye access to internal DNA. EVs were identified by their expression of CD63 and/or CD81, while DNA was detected by nucleic acid dye staining (Figure 1D). More than 4000 single-EV images were acquired and analyzed. Without DNase treatment, 95.3% of tetraspanin-positive EVs exhibited co-localization with DNA, indicating that nearly all EVs carried DNA. After DNase removal of external DNA and permeabilization, 76.4% of the EVs contained membrane-protected DNA inside.
The comparison of DNA fragments per EV between DNase-treated and permeabilized samples (D+/P+, internal DNA) vs untreated and non-permeabilized samples (D−/P−, external DNA) showed that the amount of DNA inside EVs was 3.3-fold lower than the amount of DNA on the outer membrane (D+/P+, mean 8.2 ± 33.6 localizations per EV; D−/P−, mean 26.8 ± 87.3 localizations per EV; P < .0001) (Figure 1E). There was no correlation with any particular subtype of EVs, as defined by staining for CD63 and/or CD81. These findings indicate that the vast majority of EVs carry DNA, and that more DNA localizes to the EV surface than the inside.
DNA Methylome Analysis of Glioma EVs
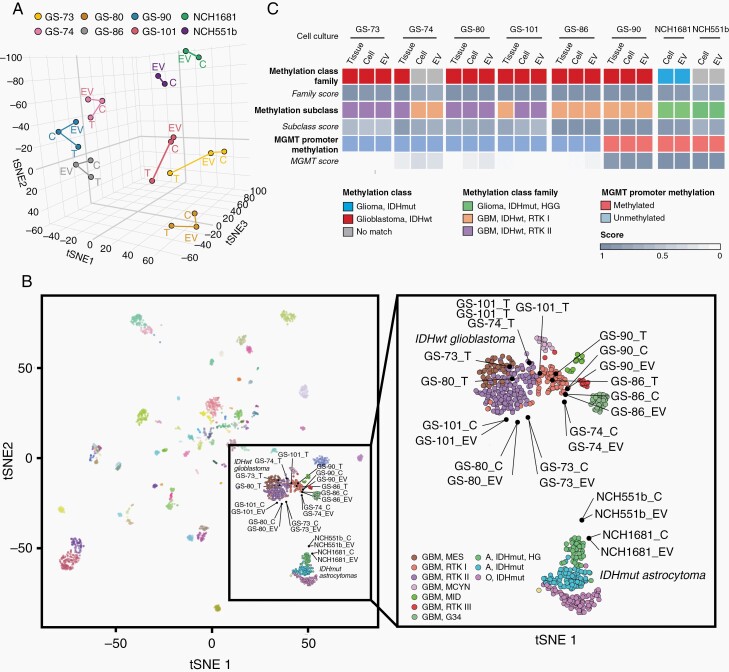
Next, we determined whether EV-associated DNA reflects the global DNA methylation pattern of parental glioma cells and original tumors. DNA was purified from EVs secreted by cultured glioma cells as well as from corresponding cells and tumors, and was subjected to Infinium MethylationEPIC BeadChip profiling. Using t-distributed stochastic neighbor embedding dimensionality reduction (t-SNE) of normalized methylation intensities in EVs (n = 8), cells (n = 8), and tissues (n = 6), we found that EVs mapped in close proximity to their corresponding parental cells and tumor tissue (Figure 2A). We then aligned our samples to the CNS tumor reference database, including over 2800 reference samples, representative of more than 80 tumor methylation classes.18 All samples derived from IDHwt tumors clustered in close proximity to the IDHwt glioblastoma RTKI and RTKII clusters (Figure 2B). EVs and cells were slightly further outlying than tumors, which could be either due to the reduced complexity of their methylome signature without the microenvironmental component present in tissue samples, or due to sampling variation between tissue used for direct analysis vs culturing, or due to in vitro cell selection and adaptation. The methylation patterns of EVs and cells derived from two IDHmut tumors were most closely related to the cluster of IDH-mutant high-grade gliomas (Figure 2B).
Fig. 2.
DNA methylation profiling of EV-DNA. A, t-SNE analysis of genome-wide methylation profiles. B, Comparison of samples to the CNS tumor reference cohort18 (left). All samples from IDHwt tumors EVs, cells (C), and tumors (T), cluster in close spatial proximity to RTKI and RTKII reference clusters, while cells and EVs from IDHmut tumors cluster with IDH-mutant high-grade astrocytomas (right, magnified glioma clusters). C, Summarized results of methylation profiling reports. Abbreviations: EV, extracellular vesicles; IDHwt, isocitrate dehydrogenase wild-type; t-SNE, t-distributed stochastic neighbor embedding dimensionality reduction.
The Heidelberg brain tumor methylation classifier assigns unknown CNS tumors to different methylation classes and subclasses based on prediction scores.24 A score threshold of >0.5 is required to predict the methylation subclass and a score >0.9 is required for methylation class family assignment. The family score represents the sum of all subclass scores generated for a tumor, so that in heterogeneous tumors the scores for different subclasses are added up.24 Applying these thresholds, EVs from five of six cases in which original tissue was available were correctly classified as glioblastoma IDHwt, and methylation subclasses were correctly identified as RTKI or RTKII in four cases (Figure 2C). GS-74 cells and EVs were also most closely related to IDHwt glioblastoma according to Figure 2B and to the methylation analysis report, however, the methylation class family score did not quite reach the threshold of 0.9 (cells 0.81, EVs 0.75). In addition, the subclass RTKI did not match the tissue subclass RTKII (RTKI score: cells 0.77, EVs 0.71). Notably, the GS-74 culture was difficult to establish because most cells died before reaching the first passage. Therefore, in vitro selection and/or intratumoral heterogeneity likely account for the epigenetic divergence in culture. Similarly, in case of T101, the tumor was heterogeneous, mapping between RTKI and RTKII (Figure 2B) and displaying a subclass score of 0.81 for RTK1 and 0.17 for RTK2. In vitro, this ratio shifted toward RTK2 (cells: RTKII 0.68, RTKI 0.27; EVs: RTKII 0.62, RTKI 0.33). For IDHmut cell lines no original tissue was available, however, NCH1681 EVs, cells, and tumor were correctly classified as IDH-mutant glioma, subclass high-grade astrocytoma, and NCH551b was also most similar to the methylation class IDH-mutant glioma, matching the subclass IDH-mutant high-grade astrocytoma, although the family score was slightly below the threshold (cells 0.73, EVs 0.82). EVs and cells from both IDHmut cell lines displayed methylation of the MGMT promoter, as observed in the majority of IDH-mutant tumors25 (Figure 2C, Supplementary Figure 2). In all other cases, the MGMT promoter methylation status was also identical between EVs, corresponding cells, and tissues. These findings show that the methylation pattern of malignant gliomas is maintained in the EV-DNA, indicating that EV profiling can predict the methylation class and MGMT status of the original tumor.
CNV Analysis of Glioma EVs
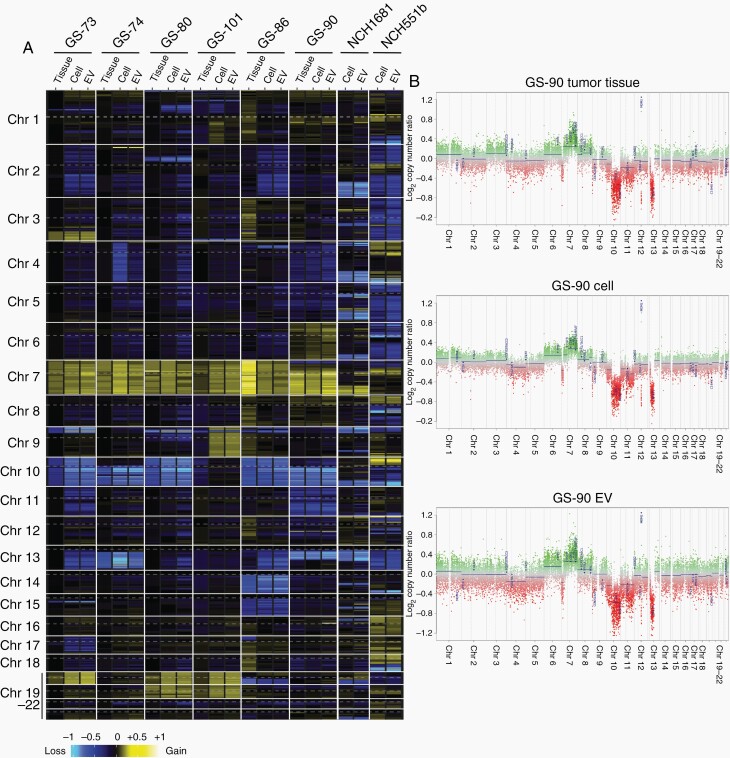
Methylome analysis can also be used to detect copy number alterations,24 and we compared the CNV profiles of all analyzed samples. Heatmap representation of gains and losses highlights the CNV similarities between EV-DNA, cells, and matching tissues (Figure 3A). Complete or partial gains of chromosome 7 were present in all specimens (Figure 3A and 3B, Supplementary Figure 3). Focal amplification of the EGFR gene was present in two tumors (T80, T101) but was lost in corresponding cells and EVs, consistent with the common rapid loss of this alteration in vitro.26 Hemizygous loss of chromosome 10 was detected in three tumors (T73, T80, T86) and loss of 10q occurred in two tumors (T74, T90). All chromosome 10 or 10q losses were maintained and often even more pronounced in corresponding cells and EVs. Chromosome 10 losses in the two IDHmut cell lines were also reflected in EVs. Gains of chromosome 20 were present in two tumors (T80, T101) and were also found in corresponding cells and EVs. Gains of chromosome 19 were absent in original tumors but became apparent in cells and EVs derived from three tumors (T73, T80, T101), indicating that they were selected for by in vitro culturing.
Fig. 3.
CNV analysis of EV-DNA. A, Heatmap representation of genome-wide copy number gains and losses inferred from the DNA methylation analysis. B, Example of CNV profiles for tumor T90 with corresponding cells and EVs. Abbreviations: CNV, copy number variations; EV, extracellular vesicles.
In addition to larger chromosomal losses, hemizygous gene deletions and focal high-level amplifications were also maintained in EV-DNA. Homozygous deletion of CDKN2A/B was present in four tumors as well as in cells and corresponding EVs (T73, T80, T86, T101) (Supplementary Figure 3). Other amplifications detected in EVs affected MDM2 (T90 tumor, cells, and EVs), MYCN (T74 cells and EVs only), and PDGFRA, MYC, CCND2, and CDK4 (NCH551b cells and EVs). Collectively, tumors, cells, and EVs displayed highly similar CNV profiles. Any divergences usually occurred between tumors and cells, rather than between cells and EVs and are therefore most likely due to sampling heterogeneity and/or in vitro selection.
Mutation Analysis of Glioma EVs
All samples were subjected to gene panel NGS investigating 47 genes recurrently altered in gliomas and to ddPCR for detecting TERT promoter mutations. The somatic origin of DNA sequence alterations was determined through public databases (Supplementary Table 2). Heatmap representation of the variant allele frequencies (VAF) highlights the similarities between EV-DNA, cells, and matching tissues (Figure 4). All IDHwt tumors carried TERT promoter mutations, and these were readily detectable in EVs (Figure 4, Supplementary Table 2). PTEN mutations occurred in three tumors, and the frequency of the mutated allele was even higher in cells and EVs in vitro, similar to the VAF of mutated TP53, RB1, or CDKN2A in four other cases (Figure 4). The IDH mutation in two cell lines was also present in EVs, as well as several other mutations in these lines, including TP53, ATRX, CIC, and PDGFRA. In only one case a tumor mutation (NF1) was not detected in EVs or cells, however, the VAF in the tumor was very low (6.9%) so that sampling bias and subclonal heterogeneity likely explain the disappearance in culture.
Fig. 4.
Mutation profiles of EV-DNA. The variant allele frequency (VAF) was determined by NGS. *TERT promoter mutations were analyzed by ddPCR since NGS reads are usually low in this region. Abbreviations: ddPCR, digital droplet PCR; NGS, next-generation sequencing.
NGS further confirmed relevant copy number alterations in many samples, such as homozygous deletion of CDKN2A/B or PTEN, or amplification of MDM2 or EGFR (Supplementary Table 2), however, methylation arrays were generally more informative and sensitive in detecting CNV. In summary, these findings demonstrate that comprehensive mutational profiling of EVs is feasible and that mutations in original tumors are retained in EV-DNA.
Proteomics of Glioma EVs
Previous studies suggested that specific proteins are enriched in plasma EVs from glioblastoma patients compared to healthy individuals and could provide a glioblastoma-specific signature.2,27 To determine whether glioblastoma signatures are reflected in EVs, we investigated the protein content of tissues, cells, and EVs from four glioblastoma patients by differential quantitative proteomics.
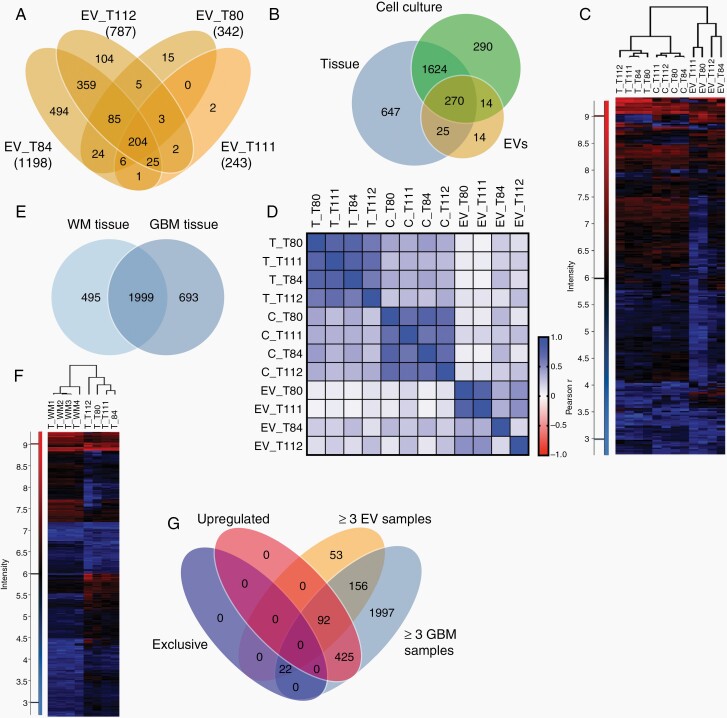
The total number of detected proteins in the EV samples varied considerably, ranging from 1198 proteins in EVs from tumor T84 to only 243 proteins in EVs from T111 (Figure 5A, Supplementary Table 3). Of 323 proteins present in at least 3 of 4 EV specimens, 270 were also detected in tumor tissue and cells (Figure 5B, Supplementary Figure 4A). Unsupervised clustering of these proteins showed that tumor cells and EVs each clustered together and that tumors and cells have more in common than they have with EVs (Figure 5C). Correlation analysis confirmed the similarity within the three sample groups but did not show any consistent correlations between EVs and corresponding cells or tumors (Figure 5D, Supplementary Figure 4B and C). Although EVs from T84 and T112 were most closely related to their respective parental tumors, EVs from T84 also exhibited closest similarity with T80 and T111, followed by EVs from T112. Similar results were obtained when comparing EVs and cells, indicating that the higher number of total proteins in two EV preparations and their quantitative overlap with tissue and cellular proteins is responsible for the correlations rather than the specific composition of the protein profiles (Figure 5D, Supplementary Figure 4B–D).
Fig. 5.
Proteomic profiles of glioma extracellular vesicles (EVs). A, Numbers of proteins detected in EVs from cultured cells of four glioblastomas by differential quantitative proteomics. B, Proteins detected in at least three of four samples each of EVs, cells, and original tumors. C, Unsupervised clustering based on 270 proteins present in all three sample types. D, Pearson correlation analysis of the proteomes of EVs, cells, (C) and tumor tissue (T). E, Overlap between proteins presents in at least three of four white matter (WM) or glioblastoma (GBM) samples. F, Unsupervised hierarchical clustering based on proteins (2172) detected in GBMs and WM. G, Proteins either exclusively detected in GBM tissue, upregulated in GBMs vs WM (≥2-fold), present in ≥3EV samples and in ≥3 GBMs.
In order to identify proteins that could be useful to specifically enrich glioblastoma-derived EVs, we compared the proteins contained in glioblastoma samples to four samples of non-tumorous white matter tissue and assessed their overlap with EV proteins. In total, 1999 proteins were detected in ≥3 white matter samples as well as in ≥3 glioblastoma samples (Figure 5E and 5F). Of the proteins that were either exclusively present in glioblastomas or upregulated ≥2-fold in tumors vs white matter, 114 were also detected in ≥3 EV samples (Figure 5G). Gene ontology (GO) analysis showed that many of these proteins were related to vesicles or extracellular matrix (ECM) interactions (Supplementary Figure 5A and B). The ECM proteins included tenascin, fibronectin, vimentin, collagen, SPARC, and ECM-binding molecules, such as CD44 or integrin-β1 (Supplementary Table 4). Other proteins included nestin, pleiotrophin, midkine, profilin-1, tetraspanin-14, cathepsin B, moesin, filamin A, transgelin-2, myosin-9, S100-A6, and others. These findings demonstrate that while proteomic EV analysis does not allow tumor specification, it can identify proteins potentially useful for enriching glioma EVs.
EV Methylome Analysis Is Not Affected by Different Isolation Techniques
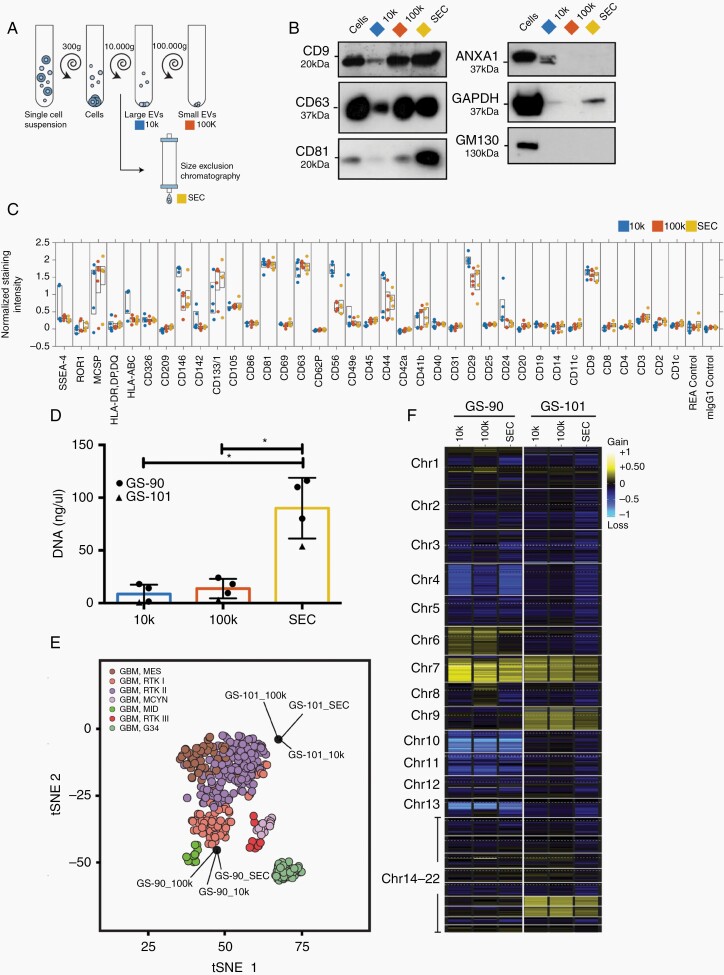
An important point to consider is that the EV protein and DNA methylation profiles may be affected by EV isolation techniques, and it is a much-debated controversy in the International Society for Extracellular Vesicles (ISEV) how to optimize and standardize isolation methods across different laboratories to obtain comparable results. To investigate the effects of different isolation techniques, we divided culture supernatants into three aliquots. Large vesicles were pelleted through centrifugation at 10,000 × g (10k EVs), while small EVs were either isolated by SEC EVs or by ultracentrifugation at 100,000 × g (100k EVs) (Figure 6A). Particle yields were highest when using SEC, and the morphology of SEC EVs was indistinguishable from 100k EVs, while 10k EVs tended to be larger and had the lowest yield (Supplementary Figure 6A–C). EVs prepared by all techniques were positive for CD9, CD63, and CD81, but levels were lowest in 10k EVs (Figure 6B). EV purity was substantiated by the absence of the Golgi marker GM130, while the microvesicle marker ANXA1 was present in 10k EVs. GAPDH, which is involved in EV biogenesis, was mainly present in SEC EVs. Characterization of EV surface epitopes using a multiplex flow cytometry assay showed that 10k EVs clustered separately, whereas SEC EVs and 100k EVs were virtually indistinguishable (Figure 7C, Supplementary Figure 6D). Multiplex profiling confirmed that CD44 and integrin β-1, which were identified as upregulated in glioblastomas vs white matter and present in EVs by proteomic analysis, were detectable on the EV surface by specific antibodies, suggesting that they could potentially be useful for enriching glioma EVs (Figure 6C).
Fig. 6.
EV-DNA methylation profiling is independent of isolation techniques. A, EVs were isolated by centrifugation at 10,000 × g (10k), ultracentrifugation (100k), or size exclusion chromatography (SEC). B, Immunoblot analysis of GS-90 cells and EVs. C, Characterization of EV surface proteins by multiplex bead-based flow cytometry analysis (GS-57, GS-73, GS-74, GS-90, GS-101). D, DNA amount in EVs measured by qBit (GS-90, n = 3, GS-101, n = 1), ANOVA analysis. Values are means ± SD, *P < .05. E, Methylation profiling of EV-DNA analyzed in comparison to the CNS tumor reference cohort.18 F, Heatmap representation of genome-wide CNV inferred from the DNA methylation analysis. Abbreviations: CNV, copy number variations; EV, extracellular vesicles.
Identical volumes of starting material were used for DNA extraction and the yield was highest for SEC EVs (Figure 6D). Methylation analysis of EV-DNA from GS-90 and GS-101 showed that all three EV samples derived from the same cell line clustered virtually on the same spot, indicating high similarity between their methylation profiles (Figure 6E). All EV preparations from GS-101 were classified as IDHwt glioblastoma, subclass RTKII. 100k EVs from GS-90 were correctly classified as IDHwt glioblastoma RTKI, although classification scores for SEC EVS and 10k EVs were borderline (RTKI score: 100k EVs 0.97, SEC EVs 0.65, 10k EVs 0.46). CNV profiles of differently isolated EVs were also very similar and neither isolation technique appeared to be superior (Supplementary Figure 7).
Discussion
Elevated circulating levels of EVs have been observed in patients with glioblastoma and other types of cancer, and circulating tumor-derived EVs could be a valuable biomarker source to monitor treatment response and aid tumor diagnosis.1,2,28 While previous studies on glioma EVs largely focused on their proteome and RNA content, DNA analyses were so far limited to the detection of IDH1 mutations.9 Our study presents the following major findings: (1) genome-wide DNA methylation analysis of EV-DNA allows correct tumor identification and glioma subtype classification as well as the determination of the MGMT promoter methylation status; (2) DNA copy number alterations can be extracted from EV-DNA methylome analysis; (3) targeted gene panel NGS allows the detection of glioma-associated mutation profiles, including potentially targetable driver mutations; (4) different EV isolation techniques yield comparable methylation and CNV results; (5) DNA is located on the surface as well as inside of EVs; (6) proteomic EV analysis does not permit specific tumor identification but recognizes tumor-associated proteins, potentially useful for enriching glioma EVs.
Our study provides proof of principle that EV-DNA reflects genome-wide methylation profiles as well as the mutational and CNV status of original glioblastomas. All mutations detected by NGS in original tumors were also found in EVs, except for a single NF1 mutation which was limited to a minor subpopulation in the original tumor. Methylation profiles of all EV samples were highly similar to their corresponding parental cells and tumors. Methylation classes and subclasses of tumors and cells were correctly identified in EVs in almost all cases. In case of T101, the methylation class of EVs and cells did not match the tissue, however, the original tumor was heterogeneous, while in T74, massive cell death occurred in early culture, suggesting that in both cases, subpopulation selection in vitro was responsible for the divergence. EVs, cells, and tumors displayed very similar CNV profiles, and any minor divergences occurred at the transition from tumors to cells rather than between cells and EVs. These findings demonstrate that EVs reflect the genomic and epigenetic alterations present in glioblastomas with high accuracy.
Since EVs of different sizes have been described to contain different amounts of DNA,17,29 we compared smaller EVs isolated by ultracentrifugation or SEC with larger EVs.17,30 The EV and DNA yield was highest when using SEC. DNA from all three EV preparations displayed very similar methylation profiles, and all samples were correctly identified as IDHwt glioblastoma, subclass RTKI or RTKII, indicating that the quality of methylation analysis is independent of EV isolation techniques and that epigenetic marks on EV-DNA are consistent.
Single-EV analysis using dSTORM showed that the amount of DNA on the surface of EVs was ~3-fold higher than the internal amount. This topography is consistent with studies in other cell types (using different techniques), which also localized the majority of EV-DNA to the surface.29,31 Notably, Thakur et al. found the majority of external EV-DNA from cancer cells to be larger than 2.5 kb, while internal DNA was mainly in the range of 100 bp to 2.5 kb.16 Although multiple groups provided strong evidence of DNA in EVs,13–17,29,31–33 a recent report questioned these findings and suggested that DNA is mainly associated with non-vesicular components.3 Our observation that even after robust digestion of surface-associated DNA and any possibly contaminating free-floating DNA, we still detected DNA in 76.4% of the CD63/CD81-positive vesicles strongly supports the notion that EVs contain DNA inside. Further work is necessary to clarify the precise nature of different DNA-carrying vesicles and particles (eg, exosomes, exomeres, microvesicles, oncosomes, apoptotic bodies, amphisomes, virtosomes).
To our knowledge, only a single study exists that detected mutated DNA in EVs from glioma patient blood, and the IDH1 mutations were often inconsistent between EVs and tumors.9 Other studies investigated free circulating tumor DNA (ctDNA) as a source for tumor mutation detection in CSF or blood. While tumor-specific alterations were detectable in the majority of CSF samples from high-grade glioma patients,34,35 the detection rate of ctDNA in plasma was <10%,36 unless a preselected approach was used in which mutations were pre-identified in tumor tissue to create tumor-guided sequencing panels, resulting in a detection rate of 83%.37 Another recent study showed that cell-free methylated DNA immunoprecipitation and high-throughput sequencing (cfMeDIP-seq) of plasma cfDNA fragments could separate different types of brain tumors, including IDH-mutant and IDHwt gliomas, meningiomas, hemangiopericytomas, and metastases.38 Our results extend these findings by showing that 850k methylation array analysis of EV-DNA allows methylation-based classification of gliomas using the Heidelberg classifier and that targeted NGS of EV-DNA provides comprehensive information on genomic tumor alterations.
A major challenge in obtaining tumor-specific epigenetic and genetic profiles from circulating tumor EV-DNA as well as ctDNA is that both are rare in patient plasma, compromising detection sensitivity. Glioma EVs constitute <10% of the total EV pool in patient plasma.39 However, tumor-derived EVs may potentially be isolated or enriched via capture of tumor-associated surface proteins. Our proteomic analysis showed that protein signatures in EVs could not identify the tumors of origin and EV samples with high total numbers of proteins correlated better with tissue and cells, regardless of their correspondence. However, we identified 104 proteins as upregulated in glioblastomas and present in EVs. Among these were CD44 and integrin-β1, confirming previous findings by others,40,41 and these were also detected on the EV surface by multiplex flow cytometry. Other promising candidates included nestin, tenascin, profilin-1, tetraspanin-14, and annexin A2. These data show that while proteomic EV analysis provides only limited insight into the specific biology of corresponding tumors, it may facilitate surface marker identification for the development of enrichment techniques, which are greatly needed for blood-based EV analysis.
In conclusion, our study demonstrates that DNA extracted from glioma EVs allows comprehensive methylation profiling and glioma subtype assignment as well as detection of tumor-specific genome-wide mutations and CNVs. Limitations of this proof of principle study include the small cohort size and the in vitro approach. In some of the analyzed cases, sampling variation between tissue used for tumor analysis vs cell culturing, as well as in vitro selection, apparently accounts for partial genetic and epigenetic divergences between tumors and cells/EVs. Presumably, tumor-derived EVs circulating in patient blood or CSF better reflect the full spectrum of heterogeneity present in original tumors. Further studies in larger numbers of patients are necessary to investigate how EVs in patient biofluids can be used for tumor classification and disease monitoring, and whether EV enrichment via capture of glioma-associated proteins may aid in this task.
Funding
This study was supported by grants from the Deutsche Forschungsgemeinschaft to F.L.R. and K.L. (RI2616/3-1), the Anni Hofmann Stiftung (K.L.), Wilhelm Sander-Stiftung (F.L.R., U.S.), Deutsche Krebshilfe (K.P.) and Fördergemeinschaft Kinderkrebszentrum Hamburg (U.S.).
Supplementary Material
Acknowledgments
We thank Mareike Holz, Katharina Kolbe, Svenja Zapf, and Anne Reichstein for technical assistance and the FACS core facility at the University Medical Center Hamburg-Eppendorf for help with flow cytometry. We are grateful to Dr. Christel Herold-Mende (Heidelberg University Hospital, Heidelberg, Germany) for providing NCH1681 and NCH 551b cells.
Conflict of interest statement. A.G. is a consultant for and has equity interest in Evox Therapeutics Ltd., Oxford, UK.
Authorship statement. F.L.R., K.L., and C.L.M. conceived the study, designed and analyzed experiments, and prepared the manuscript. C.L.M., F.L.R., I.S., and S.A.J. performed experiments. M.M.F. and H.S. performed differential quantitative proteomics. K.K. and G.R. performed gene panel sequencing. K.D.F., D.H.H., J.A.W., J.C.J., A.G., and T.R. performed bioinformatic analyses. L.D. and T.S. provided human samples. K.P., M.G., and M.W. provided scientific input. A.G.M., J.H.F., M.C., and V.P. provided the dSTORM instrument and analyzed the data. R.R. performed electron microscopy. U.S. performed methylation analyses.
References
- 1. Ricklefs FL, Maire CL, Reimer R, et al. . Imaging flow cytometry facilitates multiparametric characterization of extracellular vesicles in malignant brain tumours. J Extracell Vesicles. 2019;8(1):1588555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Osti D, Del Bene M, Rappa G, et al. . Clinical significance of extracellular vesicles in plasma from glioblastoma patients. Clin Cancer Res. 2019;25(1):266–276. [DOI] [PubMed] [Google Scholar]
- 3. Jeppesen DK, Fenix AM, Franklin JL, et al. . Reassessment of exosome composition. Cell. 2019;177(2):428–445.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Akers JC, Ramakrishnan V, Kim R, et al. . miRNA contents of cerebrospinal fluid extracellular vesicles in glioblastoma patients. J Neurooncol. 2015;123(2):205–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ebrahimkhani S, Vafaee F, Hallal S, et al. . Deep sequencing of circulating exosomal microRNA allows non-invasive glioblastoma diagnosis. NPJ Precis Oncol. 2018;2:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Manterola L, Guruceaga E, Gállego Pérez-Larraya J, et al. . A small noncoding RNA signature found in exosomes of GBM patient serum as a diagnostic tool. Neuro Oncol. 2014;16(4):520–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen WW, Balaj L, Liau LM, et al. . BEAMing and droplet digital PCR analysis of mutant IDH1 mRNA in glioma patient serum and cerebrospinal fluid extracellular vesicles. Mol Ther Nucleic Acids. 2013;2:e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Figueroa JM, Skog J, Akers J, et al. . Detection of wild-type EGFR amplification and EGFRvIII mutation in CSF-derived extracellular vesicles of glioblastoma patients. Neuro Oncol. 2017;19(11):1494–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. García-Romero N, Carrión-Navarro J, Esteban-Rubio S, et al. . DNA sequences within glioma-derived extracellular vesicles can cross the intact blood-brain barrier and be detected in peripheral blood of patients. Oncotarget. 2017;8(1):1416–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Graner MW, Alzate O, Dechkovskaia AM, et al. . Proteomic and immunologic analyses of brain tumor exosomes. FASEB J. 2009;23(5):1541–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee K, Fraser K, Ghaddar B, et al. . Multiplexed profiling of single extracellular vesicles. ACS Nano. 2018;12(1):494–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Skog J, Würdinger T, van Rijn S, et al. . Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10(12):1470–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Balaj L, Lessard R, Dai L, et al. . Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun. 2011;2:180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kahlert C, Melo SA, Protopopov A, et al. . Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J Biol Chem. 2014;289(7):3869–3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Takahashi A, Okada R, Nagao K, et al. . Exosomes maintain cellular homeostasis by excreting harmful DNA from cells. Nat Commun. 2017;8:15287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Thakur BK, Zhang H, Becker A, et al. . Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res. 2014;24(6):766–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vagner T, Spinelli C, Minciacchi VR, et al. . Large extracellular vesicles carry most of the tumour DNA circulating in prostate cancer patient plasma. J Extracell Vesicles. 2018;7(1):1505403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Capper D, Jones DTW, Sill M, et al. . DNA methylation-based classification of central nervous system tumours. Nature. 2018;555(7697):469–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Günther HS, Schmidt NO, Phillips HS, et al. . Glioblastoma-derived stem cell-enriched cultures form distinct subgroups according to molecular and phenotypic criteria. Oncogene. 2008;27(20):2897–2909. [DOI] [PubMed] [Google Scholar]
- 20. Dettling S, Stamova S, Warta R, et al. . Identification of CRKII, CFL1, CNTN1, NME2, and TKT as novel and frequent T-cell targets in human IDH-mutant glioma. Clin Cancer Res. 2018;24(12):2951–2962. [DOI] [PubMed] [Google Scholar]
- 21. Ricklefs FL, Alayo Q, Krenzlin H, et al. . Immune evasion mediated by PD-L1 on glioblastoma-derived extracellular vesicles. Sci Adv. 2018;4(3):eaar2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ricklefs FL, Maire CL, Matschke J, et al. . FASN is a biomarker enriched in malignant glioma-derived extracellular vesicles. Int J Mol Sci. 2020;21(6):1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rikkert LG, Nieuwland R, Terstappen LWMM, Coumans FAW. Quality of extracellular vesicle images by transmission electron microscopy is operator and protocol dependent. J Extracell Vesicles. 2019;8(1):1555419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Capper D, Stichel D, Sahm F, et al. . Practical implementation of DNA methylation and copy-number-based CNS tumor diagnostics: the Heidelberg experience. Acta Neuropathol. 2018;136(2):181–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bady P, Delorenzi M, Hegi ME. Sensitivity analysis of the MGMT-STP27 model and impact of genetic and epigenetic context to predict the MGMT methylation status in gliomas and other tumors. J Mol Diagn. 2016;18(3):350–361. [DOI] [PubMed] [Google Scholar]
- 26. Schulte A, Günther HS, Martens T, et al. . Glioblastoma stem-like cell lines with either maintenance or loss of high-level EGFR amplification, generated via modulation of ligand concentration. Clin Cancer Res. 2012;18(7):1901–1913. [DOI] [PubMed] [Google Scholar]
- 27. Indira Chandran V, Welinder C, Månsson AS, et al. . Ultrasensitive immunoprofiling of plasma extracellular vesicles identifies syndecan-1 as a potential tool for minimally invasive diagnosis of glioma. Clin Cancer Res. 2019;25(10):3115–3127. [DOI] [PubMed] [Google Scholar]
- 28. Becker A, Thakur BK, Weiss JM, Kim HS, Peinado H, Lyden D. Extracellular vesicles in cancer: cell-to-cell mediators of metastasis. Cancer Cell. 2016;30(6):836–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lázaro-Ibáñez E, Lässer C, Shelke GV, et al. . DNA analysis of low- and high-density fractions defines heterogeneous subpopulations of small extracellular vesicles based on their DNA cargo and topology. J Extracell Vesicles. 2019;8(1):1656993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yekula A, Minciacchi VR, Morello M, et al. . Large and small extracellular vesicles released by glioma cells in vitro and in vivo. J Extracell Vesicles. 2020;9(1):1689784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fischer S, Cornils K, Speiseder T, et al. . Indication of horizontal DNA gene transfer by extracellular vesicles. PLoS One. 2016;11(9):e0163665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee TH, Chennakrishnaiah S, Audemard E, Montermini L, Meehan B, Rak J. Oncogenic ras-driven cancer cell vesiculation leads to emission of double-stranded DNA capable of interacting with target cells. Biochem Biophys Res Commun. 2014;451(2):295–301. [DOI] [PubMed] [Google Scholar]
- 33. Yokoi A, Villar-Prados A, Oliphint PA, et al. . Mechanisms of nuclear content loading to exosomes. Sci Adv. 2019;5(11):eaax8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Miller AM, Shah RH, Pentsova EI, et al. . Tracking tumour evolution in glioma through liquid biopsies of cerebrospinal fluid. Nature. 2019;565(7741):654–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang Y, Springer S, Zhang M, et al. . Detection of tumor-derived DNA in cerebrospinal fluid of patients with primary tumors of the brain and spinal cord. Proc Natl Acad Sci U S A. 2015;112(31):9704–9709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bettegowda C, Sausen M, Leary RJ, et al. . Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6(224):224ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wan JCM, Heider K, Gale D, et al. . ctDNA monitoring using patient-specific sequencing and integration of variant reads. Sci Transl Med. 2020;12(548):eaaz8084. [DOI] [PubMed] [Google Scholar]
- 38. Nassiri F, Chakravarthy A, Feng S, et al. . Detection and discrimination of intracranial tumors using plasma cell-free DNA methylomes. Nat Med. 2020;26(7):1044–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fraser K, Jo A, Giedt J, et al. . Characterization of single microvesicles in plasma from glioblastoma patients. Neuro Oncol. 2019;21(5):606–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lane R, Simon T, Vintu M, et al. . Cell-derived extracellular vesicles can be used as a biomarker reservoir for glioblastoma tumor subtyping. Commun Biol. 2019;2:315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mallawaaratchy DM, Hallal S, Russell B, et al. . Comprehensive proteome profiling of glioblastoma-derived extracellular vesicles identifies markers for more aggressive disease. J Neurooncol. 2017;131(2):233–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.