Abstract
Mycobacterium tuberculosis (Mtb) causes the human disease tuberculosis (TB) and remains the top global infectious pandemic after coronavirus disease 2019 (COVID-19). Furthermore, TB has killed many more humans than any other pathogen, after prolonged coevolution to optimise its pathogenic strategies. Full understanding of fundamental disease processes in humans is necessary to successfully combat this highly successful pathogen. While the importance of immunodeficiency has been long recognised, biologic therapies and unbiased approaches are providing unprecedented insights into the intricacy of the host–pathogen interaction. The nature of a protective response is more complex than previously hypothesised. Here, we integrate recent evidence from human studies and unbiased approaches to consider how Mtb causes human TB and highlight the recurring theme of extracellular matrix (ECM) turnover.
Keywords: tuberculosis, granuloma, bioinformatic analysis, extracellular matrix, metalloprotease
Human TB: the intricate and prolonged contest between host and pathogen
TB is a chronic and persistent human killer, causing more deaths in total over time than any other pathogen and, currently, is the most important infection after COVID-19. Furthermore, the TB pandemic is likely to worsen due to resources being diverted to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) control [1]. The causative organism, Mtb, has undergone long-term coevolution with humans and is an obligate human pathogen [2]. Although there have been significant steps forward, such as new antibiotics for drug-resistant disease, the GeneXpert for rapid diagnosis [3] and a promising new vaccine [4], standard treatment, diagnosis, and vaccination strategies in most high-incidence TB countries are unchanged. This partly reflects the fact that we still do not understand human TB sufficiently to design transformative strategies to achieve global TB control.
Accumulating evidence from biological therapeutics and genomic analyses have suggested that we need to refine our concepts of the spectrum of human disease [5,6]. Importantly, this includes confirmation in patients that an excessive immune response can be just as harmful as an insufficient response, as illustrated by increased TB incidence with PD-1 inhibition in cancer immunotherapy [6., 7., 8.]. These new data highlight the fine balance that exists between protection and disease, with either an insufficient or excessive immune response being harmful [9]. Furthermore, the concurrent progression and regression of lesions within the same individual highlights the intricacy of the host–pathogen interaction [10,11]. A recently emerging theme from unbiased analyses is that ECM turnover is a cardinal feature of human TB, which is well described clinically. Here, we consider human TB in light of these emerging phenomena and the accumulating omics data sets, interpreting these findings alongside clinical characteristics of disease.
The granuloma: the critical arena determining outcome
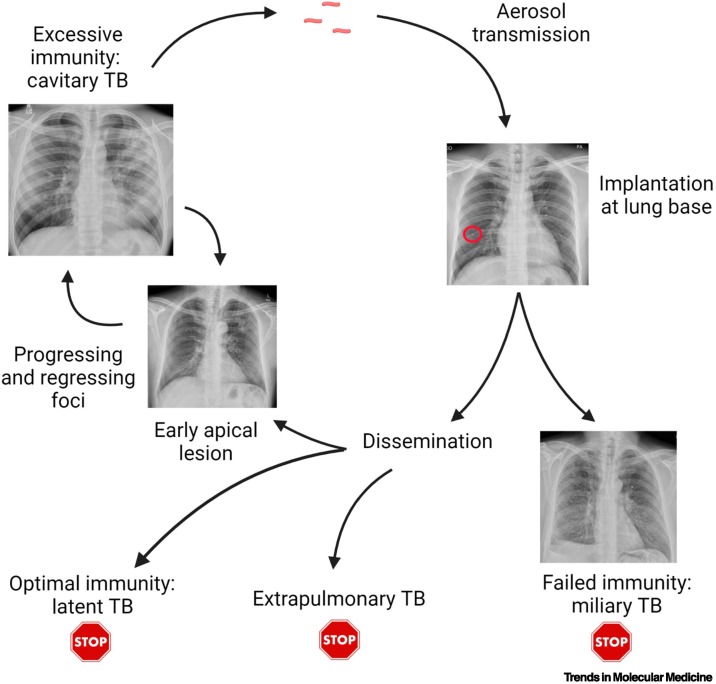
The Mtb human life cycle involves multiple stages and ironically for such a successful pathogen, Mtb usually reaches a dead end in most humans, failing to transmit to a new host (Figure 1 ) [12,13]. Infection is spread by aerosol from an individual with pulmonary TB, and those with lung cavities (see Glossary) are the most infectious and drive the epidemic [14]. Therefore, for efficient transmission, Mtb must cause immunopathology and lung matrix destruction at the apices of the lung to exit the host and spread onward [15]. In addition, recent PET-CT data suggest that propagation of TB within the lung starts with cavitation, followed by the seeding of new infection foci via bronchial spread [16]. Therefore, cavitation appears to be central for disease progression within the host as well as onward transmission in the population.
Figure 1.
The human tuberculosis (TB) life cycle.
A patient with pulmonary TB generates an aerosol by coughing, which is inhaled into the lower part of the lungs. Initial proliferation occurs, often leading to a Ghon focus visible on the chest X-ray (circle). In the absence of an efficacious immune response, disseminated miliary TB develops, with mycobacterial proliferation in many organs, but this is a dead end for the pathogen. Once the adaptive immune response activates, Mycobacterium tuberculosis (Mtb) proliferation is controlled, and a period of latency typically occurs. Infection can reactivate in other organs, such as lymph nodes, but again this does not typically transmit. In ~6% of individuals, typically those aged 20–25 with a robust immune response, Mtb drives extensive lung inflammation, leading to lung matrix destruction, cavitation, and transmission to new hosts. However, even extensive lesions can regress, with approximately one-third of ‘consumptives’ spontaneously healing during the pre-antibiotic era. Part of figure created with BioRender (www.BioRender.com').
In initial infection, Mtb aerosol droplets are typically inhaled to the well-ventilated lower lobes and phagocytosed by alveolar macrophages, although definitive proof in humans is difficult to obtain and not all early lesions are basal. Alveolar macrophages are poor at controlling Mtb [17], and an initial proliferation generates a large focus of infected cells, often over 5 mm in diameter, as demonstrated by the Ghon focus in the lung base [18]. During this period, Mtb proliferation is unrestricted by an adaptive host immune response, and it uses a variety of evasion capabilities to proliferate within a range of phagocytes, such as inhibiting phagolysosomal fusion [19]. Subsequently, at around 6 weeks, a T cell response develops, which is delayed relative to other respiratory pathogens [20] but ultimately leads to more efficacious control of Mtb. By this stage, Mtb needs to have spread to the lung apex, from where it will exit and restart the infectious cycle [13]. How Mtb travels from the lung base to apex is unknown [21], although it is likely that infected phagocytes act as Trojan horses carrying the mycobacteria [22,23]. In patients who never develop an adaptive response, Mtb disseminates throughout the body [24], with miliary TB nodules across the chest X-ray and in other organs, as described as early as 1700 by Manget [11]. This suggests that Mtb spreads extensively, with the goal of forming a niche in the upper lung, where factors favour persistence over immune eradication. Seminal postmortem studies by Opie confirmed Mtb survival in apical lung lesions in otherwise healthy individuals [25]. From this niche, Mtb must then cause inflammation, immunopathology, and cavitation to transmit and, although this can happen at any point, most cases reactivate in the first 2 years after infection [26]. With this time frame, disease evolution is typically a slow process, and changes in the peripheral transcriptome can be detected many months before presentation of active disease [27].
As the T cell response develops, Mtb needs to change strategy to reflect the more hostile environment of the host. The recent unpublished identification of changes in Mtb metabolism in response to IFN-γ gives some insight into these events. In sensing host IFN-γ, Mtb is able to change its metabolic rate and transcriptional programme, suggesting that it can respond to host immunological cues [28]. Once into this second phase of the host–pathogen interaction, Mtb must survive on a tightrope, ultimately needing to drive a host immune response that leads to cavitation while avoiding an effective immune response that causes its eradication. The critical structure during this ‘post-primary’ stage is the granuloma (Figure 2 ) [29]. This was historically thought to be restrictive to Mtb growth, but such concepts of granuloma function and structure have been questioned recently. For example, key studies in the Mycobacterium marinum/zebrafish model have shown that recruitment of monocytes to the granuloma can favour pathogen proliferation [22,30]. Indeed, in the same model system, limiting the formation of epithelioid macrophages, which help to wall off the granuloma, in fact helps to limit mycobacterial growth by allowing immune cells access to the granuloma [31]. In addition, the traditional ‘sphere-like’ structure of granulomas has been questioned by micro-CT approaches, which suggest a more complex root-like structure of interconnected areas [32], in which microenvironments may vary. In addition, whether cavities emerge from the middle of caseous necrotic granulomas or from confluent areas of lipoid pneumonia has also been disputed [33].
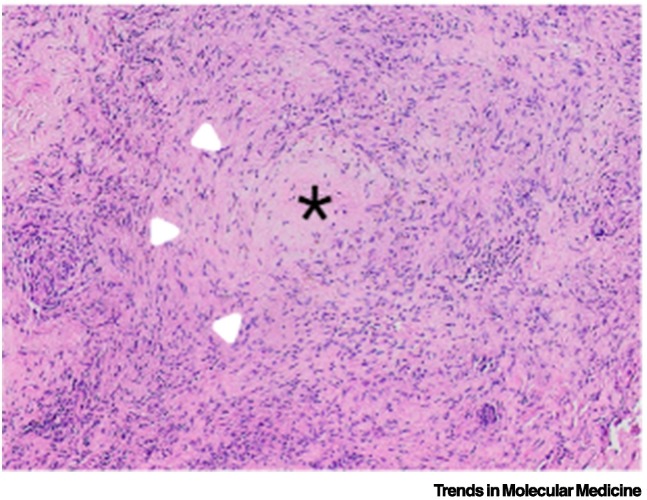
Figure 2.
Human tuberculosis (TB) granulomatous inflammation.
Haematoxylin and eosin staining of a lung nodule that cultured Mycobacterium tuberculosis (Mtb). The typical granuloma in the centre is an organised structure of activated macrophages, T cells, and fibroblasts. Star indicates central caseous necrosis; arrowheads indicate a ring of epithelioid macrophages. However, much of the surrounding inflammation is much more poorly organised than typically represented in schematics, and the concept of individual spherical granulomas being the source of TB cavities is being challenged.
Despite these uncertainties, it is clear that the immune response is both necessary to control infection and essential to drive the tissue destruction that leads to cavitation and spread [15]. Multiple types of immunodeficiency can lead to uncontrolled Mtb infection, such as advanced HIV infection, anti-TNF-α treatment, and mutations within the IFN-γ/IL-12/STAT signalling pathway [19]. This has led to research that primarily focuses on identifying what is missing from the immune response to Mtb that leads to disease. However, evidence that an absence of an immunological component(s) identified in individuals who progress to active TB disease does not mean that an excess will be beneficial [34], and, in fact, diverse evidence shows that inflammation, driven by excessive immunity, is damaging in TB. This debate is not new and dates back to bitter disputes between Koch and Virchow [35], over whether Koch’s tuberculin vaccine would cure infection or provoke an immune response that degraded the granuloma and enhanced disease. On the one hand, human studies and animal models provide clear evidence that immunological memory from TB exposure is protective [36,37]. On the other hand, in a seminal large-scale epidemiological study, Comstock demonstrated the surprising finding that, among tuberculin reactors, those with the greatest delayed-type hypersensitivity response had the highest risk of subsequent development of TB many years later [38]. One potential interpretation is that an excessive immune response to Mtb antigens is detrimental. With the onset of the HIV pandemic, the clinical features differentiating ‘standard’ TB from immunocompromised TB proved that the immune response contributes to lung immunopathology and spread, because cavities are rarely observed in individuals with advanced HIV-related immunocompromise, but occur on immune reconstitution with antiretroviral treatment [39]. The demonstration that T cell epitopes of Mtb are hyperconserved compared with nonepitope regions further suggests that the pathogen derives an evolutionary benefit from promoting the host T cell response [40,41].
Most recently, accumulating evidence that anti-PD-1 treatment for cancer can activate latent TB further highlights the danger of an excessive response, with enhanced T cell cytokine production implicated in driving immunopathology [7,8,42,43]. Taken together, these observations suggest a complex interplay between innate and adaptive responses, along with mycobacterial load, determining a range of outcomes from disseminated and noncavitary disease in the absence of an effective adaptive immune response, control/elimination with an optimal response, and matrix destruction, cavitation, and spread when excessive localised inflammation occurs [9]. Along similar lines, the concept that the optimal strategy for humans might be to sequester and tolerate Mtb has been proposed, and the breakdown of this tolerance leads to active disease [44,45]. The fraction of individuals defined as latently infected who in fact harbour viable bacteria is debated [46], but reactivation of Mtb can occur decades after initial infection [47], suggesting that this tolerant phenotype can be extremely durable.
Adding to the complexity of TB immunology is the fact that TB lesions can have diverse outcomes even in the same individual [11]. This was summed up neatly by Georges Canetti in 1955, based on examining thousands of tuberculous lungs before the advent of antimicrobial treatment: ‘Consider the bacillus in the lesion, experiencing such different fates in various foci of the same patient, and the same fate in widely different patients; destroyed in a certain histologic reaction and thriving in another nearby’ [48]. Likewise, Dubos wrote in 1952 ‘all these processes may occur in the same person either at different times or often simultaneously…which is still almost as much a puzzle today’. This concurrent progression and regression of lesions has been elegantly confirmed in modern imaging studies of infected non-human primates (NHPs) [10]. Consequently, it appears that the outcome of infectious foci is determined at a local granuloma level and not systemically, adding to the challenges of dissecting determinants of outcome. One proposed paradigm is that a balance within granulomas is necessary, with both proinflammatory and anti-inflammatory mediators leading to control of infection [49,50]. With their pivotal role in orchestrating the immune response, dendritic cells are also likely to have a central role in shaping the immune response and defining outcome [51]. However, because these events occur within tissue, they are challenging to investigate, and studying the host response in the periphery is unlikely to convey sufficient granularity about individual lesions [52].
Emerging insights from unbiased analyses
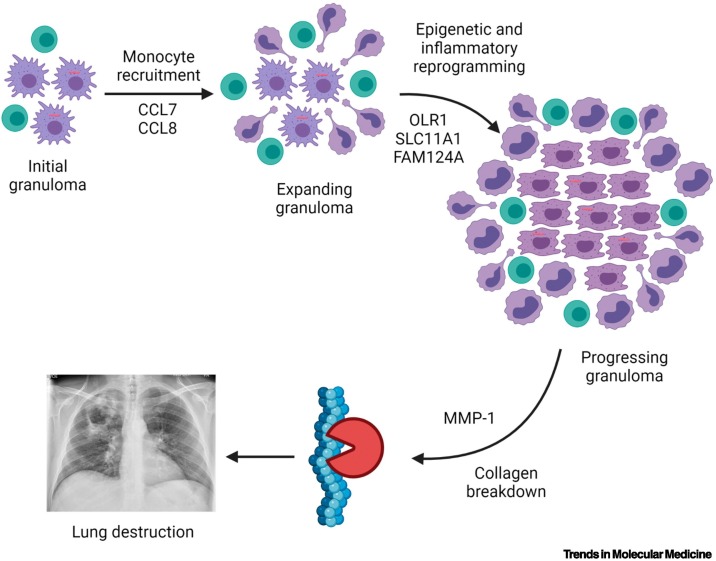
Therefore, events determining outcome within individual TB granulomas remain a highly pressing question, and omics analyses should provide a wealth of data to give mechanistic understanding. Recently, several studies reported unbiased analyses aiming to unpick the process. A strategy of comparing TB granulomas with sarcoidosis, a non-infectious granulomatous disease, was utilised to overcome the issue of cell-specific gene expression patterns [53]. Diverse analytical approaches demonstrated that the collagenase matrix metalloproteinase-1 (MMP1) was highly upregulated in TB and was the most significantly differentially expressed gene between TB and sarcoidosis. Analysis of gene correlations identified a seven-gene TB-specific cluster, comprising MMP1, the monocyte chemoattractants C-C motif chemokine ligand 7 and 8 ( CCL7 and CCL8 ), the divalent transition metal transporter solute carrier family 11 member 1 (SLC11A1; formerly known as NRAMP1), oxidised low-density lipoprotein receptor 1 (OLR1; formerly known as LOX1), family with sequence similarity 124 member A ( FAM124A ), and galectin 14 pseudogene ( LGALS17A ). Several of these genes have already been implicated in TB pathogenesis, and consideration of their known functions together informs a putative sequence of events that leads to progression of TB lesions (Figure 3 , Key figure). Thus, sequencing of clinical material followed by unbiased analysis generated a hypothesised cascade of disease evolution that can be experimentally investigated. Further bioinformatic analyses in combination with a 3D biomimetic model identified that sphingosine 1 kinase inhibition suppressed Mtb growth, thereby progressing from basic disease understanding to novel therapeutic targets in an unbiased manner [53].
Figure 3.
Key figure. Potential sequence of events in tuberculosis (TB) granuloma progression identified by gene co-expression analysis.
Unbiased analysis of RNA-sequencing data identified a seven-gene cluster unique to TB lymph nodes compared with the non-infectious granulomatous disease, sarcoidosis. Several of these genes have previously been implicated in TB pathogenesis. Considering their function together leads to a proposed sequence of events starting with excessive monocyte recruitment, which are then epigenetically reprogrammed to propagate inflammation, ultimately leading to excessive matrix metalloproteinase-1 (MMP1) expression, which causes matrix destruction. Part of figure created with BioRender (www.BioRender.com). Abbreviations: CCL, C-C motif chemokine ligand; FAM124A, family with sequence similarity 124 member A; OLR1, oxidised low-density lipoprotein receptor 1; SLC11A1, solute carrier family 11 member 1.
Using a similar transcriptomic approach, analysis of gene expression was compared in skin stimulated by tuberculin in patients with TB versus healthy controls [54]. Again, MMP1 emerged as a top divergently upregulated gene, and ingenuity pathway analysis suggested that an excessive IL-17 response was a key regulator. The IL-17/MMP1 profile resolved with treatment of infection, implying that Mtb actively primes an excessive, matrix-destructive immune response that can be replicated by a distal antigenic challenge. The authors highlighted the double-edged sword of IL-17 in TB, with data supporting a protective role [55,56] and a pathological role when present in excess [54,57]. These two recent studies have the limitation of analysing a distal compartment (mediastinal lymph node and skin), and an identical gene expression profile cannot be assumed in the lung. However, the emergence of MMP1 as a predominant mediator from these two RNA-sequencing (RNAseq) analyses is also consistent with several previous studies. In an early microarray analysis of restimulated macrophages, MMP1 was the most divergently regulated gene in patients with TB, although the authors then focused on a chemokine in validation stages [58]. Similarly, microarray analysis of lung tissue from patients failing treatment for multidrug-resistant TB found that MMP1 was very highly upregulated within lesions [59]. Comparison of modular signatures in lung cancer, TB, and sarcoidosis by RNAseq again showed over-representation in genes related to ECM organisation in TB [60]. A separate RNAseq study suggested that neuroendocrine signalling was downregulated at the air–caseum interface in drug-resistant TB, whereas the complement pathway was upregulated [61]. Although MMP regulation was not directly noted, the oncostatin M (OSM) pathway was upregulated, similar to observations in the skin tuberculin study [54], and OSM can induce MMP1 secretion [62].
Single-cell RNAseq (ssRNAseq) analysis is beginning to shed light on cellular subsets. One approach recently used in the NHP model of TB involved parallel ssRNAseq and quantification of viable Mtb bacilli from multiple individual granuloma [63]. This revealed a high degree of heterogeneity between different granulomas in the same individuals, and associations between T1/17 T cells and Mtb control, and mast cells, plasma B cells, and Mtb progression. Alternatively, by comparing ssRNAseq data from lung tissue isolated from NHPs with either progressive or latent Mtb infection, active TB was found to be associated with an influx of plasmacytoid dendritic cells (pDCs), activated macrophages, and T cells, and latency with enriched CD27+ natural killer (NK) cells [64]. In support of these data, NK cells emerged as a signature correlating with latency in a multi-omic study of human peripheral immune responses, suggesting they have a predominantly protective role [65]. In separate studies, ssRNAseq was combined with Mtb strains containing a bacterial stress reporter to identify macrophage subsets able to induce bacterial stress in vivo. This revealed distinct and epigenetically constrained macrophage subsets with differential degrees of permissiveness [66]. Finally, ssRNAseq of granuloma in zebrafish revealed an unexpected association between Th2 signalling and the generation of epithelioid macrophages, which help to ‘wall off’ Mtb within the granuloma [67]. Interestingly, as discussed in the preceding text, the same group previously showed that partial disruption of this epithelioid barrier improved Mtb control by enhancing immune cell access [31], whereas, in this most recent study, complete abrogation of the barrier led to increased Mtb growth. This neatly illustrates the fine balance between control and progression at the level of each individual granuloma.
In proteomic studies, a seminal laser capture study demonstrated the importance of spatial organisation within the TB granuloma [68]. A central proinflammatory environment was identified within the central caseous core, surrounded by a peripheral anti-inflammatory zone, with the arachidonic acid pathway having a key regulatory role. These findings parallel earlier reports of the importance of spatial organisation within the granuloma [69]. Similarly, multiplexed ion beam imaging by time-of-flight (MIBI-TOF) identified microenvironments within the TB granuloma, consistent with areas of immunosuppression [70]. A key question is whether this immunosuppression is part of the evasion strategy of the pathogen or, alternatively, the tolerance of the host to a persistent antigenic stimulus [44,45]. In plasma proteomic studies using SOMAscan methodology, MMP1 again emerged as one of the most divergently regulated proteins in teenagers who then progressed to TB [71], consistent with previous work identifying a critical role for MMP1 from a hypothesis-driven approach [72,73].
Taken together, three themes are emerging from these recent omics studies: (i) the necessary balance between pro- and anti-inflammatory pathways in controlling TB without causing immunopathology; (ii) the importance of cellular composition and crosstalk, 3D organisation, and microenvironments within TB granulomas; and (iii) the consistently observed role for MMP1 in TB immunopathology. A limitation to consider is that most cases studied represent failed control, because clinical disease occurred; thus, dissecting out determinants of protection versus pathology is challenging.
The recurring theme of the extracellular matrix
As outlined in the preceding text, unbiased studies from different groups and methodological approaches have recurrently identified MMP1 as one of the top few genes upregulated in TB. This raises the question of why MMP1 is so predominant. Within the granuloma, the goal of Mtb cannot purely be survival, because, ultimately, the host will die, and the pathogen will not transmit (Figure 1). Therefore, Mtb needs to cause cavities to transmit maximally [14]. However, the mechanism by which this happens remains poorly understood (Box 1 ). Strikingly, Mtb bacilli are frequently impossible to find by standard AFB staining techniques within human granulomas [74] and, yet, Mtb-driven inflammatory gene signatures are present through the granuloma. How Mtb causes widespread inflammation and reprogramming of granulomas in the apparent absence of high bacterial numbers is unknown. Several potential mechanisms could explain this; self-propagating intercellular proinflammatory cytokine networks, swarm behaviour by immune cells [75], microvesicles leading to transfer of Mtb antigens or mRNAs to uninfected cells [76], a progressive build-up of Mtb antigens [33], or Mtb that is not stained by standard approaches [77].
Box 1. Tuberculosis and the matrix.
The human lung is highly intricate, relying on the extracellular matrix to support a meshwork of alveoli to generate a total surface area the size of a tennis court [89]. Matrix destruction is fatal, because gas exchange then fails. Therefore, the basal environment of the lung is highly tolerogenic and skewed toward matrix protection. To effectively transmit, Mtb must overcome this matrix homeostasis to cause lung cavitation.
In individuals who progress to active disease, the immune equilibrium is lost, and excessive inflammation develops. Diverse unbiased approaches suggest that MMP1 is a final effector of collagen cleavage in this process. MMP1 is secreted as a pro-enzyme requiring proteolytic activation [90]. Numerous ex vivo and in vitro studies demonstrate that Mtb induces secretion of pro-MMP1 by host cells. In addition, Mtb secretes serine proteases [91,92], suggesting a potential proteolytic cascade whereby Mtb both directly induces and activates MMP1 within its microenvironment, propagating matrix breakdown. Intriguingly, one of the antigens in the novel M72/AS01E TB vaccine, the first candidate to improve on BCG in human trials, is an Mtb serine protease [4]. Therefore, it is possible that antiprotease antibodies generated by M72/AS01E vaccination help prevent TB reactivation by limiting MMP1 activation and initial matrix breakdown. This is pure speculation, but if proven, then matrix-protective vaccination strategies may be a novel way to prevent TB reactivation.
To date, investigating antiprotease strategies has been challenging, because standard TB mouse models do not develop the typical caseating lesions of human disease [93] and lack a functional orthologue of human MMP1 [94]. The C3HeB/FeJ or Kramnik mouse is a notable exception, developing cavitary lesions. However, these mice are immunodeficient and develop high bacterial loads [95]; thus, matrix destruction may occur via distinct mechanisms. Transgenic expression of human MMP1 in immunocompetent mice results in collagen destruction and caseation in granulomas without altering Mtb growth [96], supporting a central role for MMP1 in initiating the cavitary process.
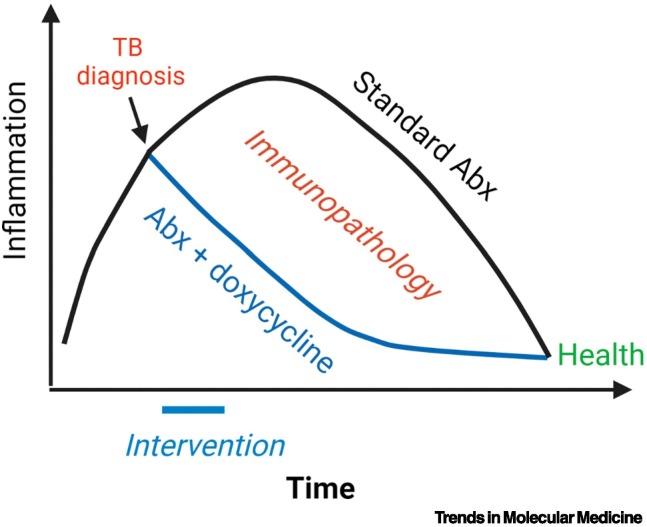
Multiple MMP inhibitors are available. However, MMP inhibitor therapy alone is harmful in preclinical models [97,98], whereas is beneficial when administered alongside antibiotics [99]. A Phase IIB trial of doxycycline as adjunctive therapy in patients with pulmonary TB showed that doxycycline suppressed MMP1 and reduced cavity size without affecting mycobacterial load [100]. Notably, 2 weeks of doxycycline caused changes that persisted at 8 weeks, suggesting that early events in TB treatment have long-lasting impact (Figure I), supporting the concept of host-directed therapies to improve outcome [101].
Figure I.
Persistent effects of early therapeutic intervention in tuberculosis (TB) .
Lung inflammation continues for the first few weeks after the start of efficacious TB antibiotic therapy, which results in ongoing damage. A short-term intervention of doxycycline for 2 weeks still had significant effects on gene expression and protein concentrations of tissue-damaging matrix metalloproteinases (MMPs) at 8 weeks, demonstrating that, if the immune response is diverted toward health early in treatment, this trajectory is maintained. This reduction in inflammation reduces the overall lung immunopathology during treatment. Figure created with BioRender (www.BioRender.com). Abbreviation: Abx, antibiotics.
Alt-text: Box 1
An additional factor that may contribute to excessive inflammation is that of trained innate immunity. Mtb evidently causes epigenetic modification [78,79] and innate immune training [80], and one of the mechanisms of protection through the BCG vaccine is thought to be via nonspecific protective training [81]. The recent demonstration that MMP1 is rapidly and highly upregulated upon purified protein derivative (PPD) stimulation in the skin of individuals with TB [54], remote to the site of lung infection, is consistent with circulating innate immune cells programmed to drive an excessive proinflammatory response. Interestingly, the seven-gene signature within TB granulomas includes a potential innate immune training component (Figure 3) [53], because OLR1 can regulate epigenetic modifications [82]. Of specific relevance to TB, OLR1 upregulation in atherosclerosis is associated with the formation of foamy macrophages [83], a cell type also induced in TB granulomas. However, the trained immune phenotype induced by TB infection that can lead to dysregulated inflammation has not been fully characterised. In addition, nonhaematopoietic cells, such as fibroblasts, may have significant roles in TB progression, because they are central players in matrix turnover. For example, OSM has emerged as one central hub from unbiased analysis [54,61] and can upregulate fibroblast MMP1 secretion [62].
Finally, it has previously been proposed that an autoimmune component contributes to TB progression, with immune cells responding to host stress antigens or matrix neoepitopes generated by matrix breakdown [84]. This phenomenon would explain many of the unusual clinical characteristics of human TB, such as uveitis and erythema nodosum, which overlap with autoimmune diseases. Genomic analyses support this concept, such as the similarities between TB and autoimmune disease signatures in peripheral blood [85]. Similarly, immunological network analysis in patients with HIV also supports an autoimmune process exacerbating pathology in TB via TH17 polarisation [57]. However, the possibility that host antigens contribute to pathogenesis in TB remains conjectural.
Tissue-dependent considerations in studying the immune response
Most research on host immunity to Mtb has focused on circulating immune cells [52]. However, it is becoming increasingly apparent that events within tissue may greatly differ from those in the periphery, just as they do between different TB lesions within the same lung [10,68,69,86]. For example, comparison of lung versus circulating T cells showed different immunological profiles at the site of disease relative to the periphery [55,87]. The fact that TB reactivates at the lung apices, not the base where it initially implants, suggests that, even within the same organ, there are broad immunological differences, which may be related to differential immune surveillance [13]. Alternatively, localisation could relate to differences in mechanotransduction across the lung, because there is an increased likelihood of collagen cleavage under tension [88]. Given that different lesions can progress and regress, one cannot assume that studying a single lesion is sufficient, presenting a significant experimental challenge. Once one adds the spatial immune organisation of the granuloma into this equation, comprehensive understanding of the host–pathogen interaction becomes complex. We propose that cross-correlation between human disease and model systems that incorporate the ECM, by studying the immune response in 3D within relevant tissue and accompanying biomimetic models in which outcomes differ stochastically, will be critical if this complexity is to be understood.
Concluding remarks
The lack of understanding of what determines protection versus pathology in TB is hindering progress (see Outstanding questions). The debate goes back to the previously mentioned hard-fought disputes between Koch and Virchow [35], and the greater granularity provided by the molecular era has further highlighted the complexity of this host–pathogen interaction. Both disputants could select recent data regarding IL-17 in human TB to support their argument that the host response is either protective or pathogenic [54,55] and, similarly, evidence from tissue microenvironments could support each position [63,68,70]. Ultimately, the historic concept of ‘good’ and ‘bad’ immune responses in TB is unlikely to be sufficient. New paradigms predicting determinants of outcome are needed, taking into consideration the multiplicity of inputs, spatial organisation, diverse outcomes, and even the potential for multiple routes to the same outcome. The wealth of data from omics technologies can only be successfully interpreted if analysis is framed within the clinical characteristics of human disease (Box 2 ). Emerging themes from recent unbiased analyses highlight the need for a balanced immune response for Mtb control and point to aberrant ECM turnover and excessive MMP1 activity as being critical effectors leading to disease progression (see Clinician’s corner). Ultimately, embracing the complexity of human TB is essential to understand the central unresolved question: what determines outcome in an individual TB lesion?
Outstanding questions.
What determines whether an individual TB granuloma will progress or regress?
Why are T cells necessary for not only protection from disease, but also cavitation and transmission?
Is it possible to determine a binary divide between protective and pathological pathways in TB or can multiple immunological profiles lead to the same outcome?
Why does the immune response to TB differ between the base and apex of lung lobes?
Why is excessive MMP1 expression emerging from multiple diverse approaches?
How do so few detectable Mtb bacilli cause such extensive inflammation and MMP1 expression within a granuloma?
What are the mycobacterial factors that contribute to immunopathology that results in excessive inflammation and matrix destruction? What regulates Mtb virulence factor activation and how does it engage the immune system to drive a specific host response that favours pathogen survival and concurrently cause matrix destruction?
What are the downstream effects of lung collagen destruction and the subsequent release of neoepitopes and matrikines?
What is the role of trained immunity in propagating inflammation?
Why do clinical features and gene expression profiles overlap between TB and autoimmune diseases?
How can the wealth of published and emerging data, including bulk granuloma, single-cell and spatial transcriptomics, proteomics, and metabolomics, be integrated to comprehensively understand the TB disease process and identify what determines outcome?
How can mechanistic insights from model systems, such as zebrafish, mouse, and NHPs, be most effectively bridged with human clinical investigation?
With many new therapeutic targets emerging, how can these be prioritised, especially given that combination therapy of multiple host-directed therapies may be most efficacious and clinical trials are lengthy and expensive? What is the best predictive model for target selection?
Alt-text: Outstanding questions
Box 2. Opportunities and challenges of omics data analysis for identification of novel therapies.
Advances in omics technologies over the past three decades have opened unprecedented opportunities for the investigation of complex biological events in human tissues. For the first time, analytical techniques can dissect not only expression levels of selected genes/proteins, but also, and often simultaneously, allow for the delineation of SNPs, the whole transcriptome, proteome, epigenome, and metabolome, of cell populations or single cells, and determine their spatial arrangement in tissues. While integrated analysis across these layers could provide a complete delineation of biological processes involved, current studies often focus these state-of-the-art approaches on a single information layer, such as transcriptomics or proteomics, and aim to demonstrate usefulness for identification of therapeutic targets.
Application of network analysis has proven extremely successful in this task. Based on mathematical graph theory, it allows interrogation of biological data in a hypothesis-free way and independently of the existing curated databases. Weighted and unweighted gene co-expression analysis [102,103] and mutual information and partial deconvolution of information analysis [104] allow delineation of underlying structure in experimental data and identification of candidate regulators for gene/protein modules. Importantly, by assigning eigenvector values to co-expressed modules of biological features, such as transcripts, genes, or proteins, it allows integration and co-analysis of clinical features and data from other high-throughput platforms.
For example, a recent study comparing TB and sarcoidosis, a non-infectious granulomatous disease, and modelling TB infection in a 3D biomimetic model, provides proof-of-concept of how gene co-expression analyses can be used for the identification of novel therapeutic targets in TB [53]. Similarly, by applying unbiased co-expression network analysis to clinical trial data, immunological processes were identified that were regulated by treatment with doxycycline, revealing selective modulation of innate immunity [100]. As an alternative approach, a module analysis approach was used to demonstrate a central role of the IL-17 response in exacerbating TB pathology [54]. In these analyses, extrapolation to events in the lung interstitium is now needed.
While significant progress is being made in the development of approaches to high-dimensional data analysis, including advanced mathematical modelling for single-cell and spatial data, and application of Bayes theory and machine learning/artificial intelligence methodologies, the key challenge yet to be overcome is bridging a mechanistic understanding of the biological process with data analysis. As advances are made in deriving causal network architecture from omics data, and in developing methods for analysis of dynamic signal flow through the network, this approach is likely to become the new frontier for predictive data analysis, allowing multilayer predictive modelling of therapeutic perturbations.
Alt-text: Box 2
Clinician’s corner.
The increase in TB incidence with both immunosuppressive anti-TNF-α and immunoactivating anti-PD-1 treatment highlights that an insufficient or excessive immune response to Mtb can be harmful to patients and further emphasises the complexity of the host–pathogen interaction. Clinicians need to be alert to reactivation of latent infections during cancer immunotherapy, which may mimic disease progression.
Several recent investigations using emerging unbiased methodologies have identified exaggerated inflammation and extracellular matrix destruction as critical pathological processes in human TB, including disproportionate upregulation of the collagenase MMP1.
New approaches to reduce immune-mediated tissue destruction in TB are being advanced using host-directed therapies, such as statins, metformin, imatinib, and doxycycline. Nonspecifically inhibiting lung matrix breakdown in patients with TB with doxycycline can suppress tissue-damaging collagenases and accelerate cavity resolution without affecting bacterial load.
Matrix-preserving strategies may not only reduce long-term lung damage, but also enhance immunological control of infection, just as cavity collapse through plombage, artificial pneumothorax, and thoracoplasty were successful during the pre-antibiotic era.
These recent studies demonstrate the power of combining network-based omics analysis of diseased human tissue and relevant control tissue with advanced 3D cell culture modelling to understand disease mechanisms and identify new therapeutic approaches. This tissue sequencing → bioinformatics → cellular modelling → clinical intervention pipeline can be used to investigate diverse human diseases and accelerate translation to new therapeutic interventions.
Alt-text: Clinician’s corner
Acknowledgments
Acknowledgments
P.E. was supported by MRC Global Challenges Research Fund MR/P023754/1; M.E.P. was supported by Wellcome Trust Sir Henry Dale Fellowship, 109377/Z/15/Z; M.T.R. was supported by Rosetrees Trust (M540); and A.L. was supported by Wellcome Trust (210662/Z/18/Z).
Declaration of interests
None declared by the authors.
Glossary
- C-C motif chemokine ligand 7 and 8 (CCL7, CCL8)
secreted chemokines that recruit monocytes to areas of inflammation.
- Cavities
air-filled holes within the lung that result from complete destruction of the extracellular matrix, as the end result of the process of cavitation. Mtb proliferates in the cavity walls exponentially, leading to patients who are highly infectious, chronic transmission, and an increased risk of treatment failure.
- Family with sequence similarity 124 member A (FAM124A)
although poorly characterised, may interact with NFκB activating protein, consistent with a role in propagating inflammation.
- Gene co-expression analysis
bioinformatic approach based on mathematical graph theory, in which clusters/modules of co-expressed genes are identified in an unbiased way based on the correlation in level of expression between each pair of genes/transcripts across study samples.
- Granuloma
organised collection of inflammatory cells, including activated macrophages, T cells, B cells, and fibroblasts, that forms in response to Mtb infection.
- Immunopathology
adverse outcome of the host immune response to persistent Mtb infection, involving cell death, matrix destruction, and impaired tissue function due to excessive cellular infiltration.
- Galectin 14 pseudogene (LGALS17A)
has an as-yet undefined function and, thus, its role in TB pathogenesis is uncertain.
- Matrix metalloproteinases (MMPs)
family of enzymes with the collective ability to degrade all fibrillar components of the extracellular matrix at neutral pH, in particular the triple helix of type I collagen, which provides the tensile strength of the lung.
- Miliary tuberculosis
disseminated infection, with appearance of millet seeds across the chest X-ray; frequently accompanied by central nervous system involvement.
- Oxidised low-density lipoprotein receptor 1 (OLR1)
formerly known as LOX1. Initially identified through its role in atherosclerosis, this receptor has a range of functions, including propagating inflammation and regulating foamy macrophage formation.
- Solute carrier family 11 member 1 (SLC11A1)
formerly known as NRAMP1. The first gene linked to TB susceptibility in population studies, when known as natural resistance-associated macrophage protein 1. Its functions include divalent cation transport and also regulation of macrophage activation.
- Trained innate immunity
modulation of innate immune responses over time by epigenetic reprogramming.
- Tuberculin
sterile protein extract from cultures of Mycobacterium tuberculosis, typically used to test for immunological memory by intradermal injection and measurement of resulting swelling 3 days later.
References
- 1.Cilloni L., et al. The potential impact of the COVID-19 pandemic on the tuberculosis epidemic a modelling analysis. EClinicalMedicine. 2020;28 doi: 10.1016/j.eclinm.2020.100603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brites D., Gagneux S. Co-evolution of Mycobacterium tuberculosis and Homo sapiens. Immunol. Rev. 2015;264:6–24. doi: 10.1111/imr.12264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walzl G., et al. Tuberculosis: advances and challenges in development of new diagnostics and biomarkers. Lancet Infect. Dis. 2018;18:e199–e210. doi: 10.1016/S1473-3099(18)30111-7. [DOI] [PubMed] [Google Scholar]
- 4.Tait D.R., et al. Final analysis of a trial of M72/AS01E vaccine to prevent tuberculosis. N. Engl. J. Med. 2019;381:2429–2439. doi: 10.1056/NEJMoa1909953. [DOI] [PubMed] [Google Scholar]
- 5.Boisson-Dupuis S., et al. Inherited and acquired immunodeficiencies underlying tuberculosis in childhood. Immunol. Rev. 2015;264:103–120. doi: 10.1111/imr.12272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morelli T., et al. Infections due to dysregulated immunity: an emerging complication of cancer immunotherapy. Thorax. 2021 doi: 10.1136/thoraxjnl-2021-217260. Published online October 4, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elkington P.T., et al. Implications of tuberculosis reactivation after immune checkpoint inhibition. Am. J. Respir. Crit. Care Med. 2018;198:1451–1453. doi: 10.1164/rccm.201807-1250LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barber D.L., et al. Tuberculosis following PD-1 blockade for cancer immunotherapy. Sci. Transl. Med. 2019;11:eaat2702. doi: 10.1126/scitranslmed.aat2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tezera L.B., et al. Reconsidering the optimal immune response to Mycobacterium tuberculosis. Am. J. Respir. Crit. Care Med. 2020;201:407–413. doi: 10.1164/rccm.201908-1506PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin P.L., et al. Sterilization of granulomas is common in active and latent tuberculosis despite within-host variability in bacterial killing. Nat. Med. 2014;20:75–79. doi: 10.1038/nm.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dubos R., Dubos J. The White Plague: Tuberculosis, Man, and Society. Rutgers University Press; 1987. [Google Scholar]
- 12.Ernst J.D. The immunological life cycle of tuberculosis. Nat. Rev. Immunol. 2012;12:581–591. doi: 10.1038/nri3259. [DOI] [PubMed] [Google Scholar]
- 13.Elkington P.T., Friedland J.S. Permutations of time and place in tuberculosis. Lancet Infect. Dis. 2015;15:1357–1360. doi: 10.1016/S1473-3099(15)00135-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Urbanowski M.E., et al. Cavitary tuberculosis: the gateway of disease transmission. Lancet Infect. Dis. 2020;20:e117–e128. doi: 10.1016/S1473-3099(20)30148-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elkington P.T., et al. Tuberculosis immunopathology: the neglected role of extracellular matrix destruction. Sci. Transl. Med. 2011;3:71ps6. doi: 10.1126/scitranslmed.3001847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen R.Y., et al. Radiological and functional evidence of the bronchial spread of tuberculosis: an observational analysis. Lancet Microbe. 2021;2:e518–e526. doi: 10.1016/S2666-5247(21)00058-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cohen S.B., et al. Alveolar macrophages provide an early Mycobacterium tuberculosis niche and initiate dissemination. Cell Host Microbe. 2018;24:439–446 e4. doi: 10.1016/j.chom.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghon A. The Primary Lung Focus of Tuberculosis in Children. Churchill; London: 1916. [Google Scholar]
- 19.O'Garra A., et al. The immune response in tuberculosis. Annu. Rev. Immunol. 2013;31:475–527. doi: 10.1146/annurev-immunol-032712-095939. [DOI] [PubMed] [Google Scholar]
- 20.Ravesloot-Chavez M.M., et al. The innate immune response to Mycobacterium tuberculosis infection. Annu. Rev. Immunol. 2021;39:611–637. doi: 10.1146/annurev-immunol-093019-010426. [DOI] [PubMed] [Google Scholar]
- 21.Murray J.F. Bill Dock and the location of pulmonary tuberculosis: how bed rest might have helped consumption. Am. J. Respir. Crit. Care Med. 2003;168:1029–1033. doi: 10.1164/rccm.200307-1016OE. [DOI] [PubMed] [Google Scholar]
- 22.Davis J.M., Ramakrishnan L. The role of the granuloma in expansion and dissemination of early tuberculous infection. Cell. 2009;136:37–49. doi: 10.1016/j.cell.2008.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schreiber H.A., et al. Inflammatory dendritic cells migrate in and out of transplanted chronic mycobacterial granulomas in mice. J. Clin. Invest. 2011;121:3902–3913. doi: 10.1172/JCI45113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dheda K., et al. Tuberculosis. Lancet. 2016;387:1211–1226. doi: 10.1016/S0140-6736(15)00151-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Opie E.L., Aronson J.D. Tubercle bacilli in latent tuberculous lesions and in lung tissue without tuberculous lesions. Arch. Pathol. Lab. Med. 1927;4:1–21. [Google Scholar]
- 26.Behr M.A., et al. Revisiting the timetable of tuberculosis. BMJ. 2018;362 doi: 10.1136/bmj.k2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scriba T.J., et al. Sequential inflammatory processes define human progression from M. tuberculosis infection to tuberculosis disease. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ahmed M., et al. Mycobacterium tuberculosis senses host interferon-γ via the membrane protein MmpL10. bioRxiv. 2021 doi: 10.1101/2021.11.12.468344. Published online November 12, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pagan A.J., Ramakrishnan L. The formation and function of granulomas. Annu. Rev. Immunol. 2018;36:639–665. doi: 10.1146/annurev-immunol-032712-100022. [DOI] [PubMed] [Google Scholar]
- 30.Cambier C.J., et al. Mycobacteria manipulate macrophage recruitment through coordinated use of membrane lipids. Nature. 2014;505:218–222. doi: 10.1038/nature12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cronan M.R., et al. Macrophage epithelial reprogramming underlies mycobacterial granuloma formation and promotes infection. Immunity. 2016;45:861–876. doi: 10.1016/j.immuni.2016.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wells G., et al. Micro-computed tomography analysis of the human tuberculous lung reveals remarkable heterogeneity in 3D granuloma morphology. Am. J. Respir. Crit. Care Med. 2021;204:583–595. doi: 10.1164/rccm.202101-0032OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hunter R.L. Tuberculosis as a three-act play: a new paradigm for the pathogenesis of pulmonary tuberculosis. Tuberculosis (Edinb) 2016;97:8–17. doi: 10.1016/j.tube.2015.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karp C.L., et al. Tuberculosis vaccines: barriers and prospects on the quest for a transformative tool. Immunol. Rev. 2015;264:363–381. doi: 10.1111/imr.12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaufmann S.H. A short history of Robert Koch's fight against tuberculosis: those who do not remember the past are condemned to repeat it. Tuberculosis (Edinb) 2003;83:86–90. doi: 10.1016/s1472-9792(02)00064-1. [DOI] [PubMed] [Google Scholar]
- 36.Andrews J.R., et al. Risk of progression to active tuberculosis following reinfection with Mycobacterium tuberculosis. Clin. Infect. Dis. 2012;54:784–791. doi: 10.1093/cid/cir951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cadena A.M., et al. Concurrent infection with Mycobacterium tuberculosis confers robust protection against secondary infection in macaques. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Comstock G.W., et al. The prognosis of a positive tuberculin reaction in childhood and adolescence. Am. J. Epidemiol. 1974;99:131–138. doi: 10.1093/oxfordjournals.aje.a121593. [DOI] [PubMed] [Google Scholar]
- 39.Kwan C.K., Ernst J.D. HIV and tuberculosis: a deadly human syndemic. Clin. Microbiol. Rev. 2011;24:351–376. doi: 10.1128/CMR.00042-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coscolla M., et al. M. tuberculosis T cell epitope analysis reveals paucity of antigenic variation and identifies rare variable TB antigens. Cell Host Microbe. 2015;18:538–548. doi: 10.1016/j.chom.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Comas I., et al. Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat. Genet. 2010;42:498–503. doi: 10.1038/ng.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kauffman K.D., et al. PD-1 blockade exacerbates Mycobacterium tuberculosis infection in rhesus macaques. Sci. Immunol. 2021;6:eabf3861. doi: 10.1126/sciimmunol.abf3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tezera L.B., et al. Anti-PD-1 immunotherapy leads to tuberculosis reactivation via dysregulation of TNF-alpha. eLife. 2020;9 doi: 10.7554/eLife.52668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olive A.J., Sassetti C.M. Tolerating the unwelcome guest; how the host withstands persistent Mycobacterium tuberculosis. Front. Immunol. 2018;9:2094. doi: 10.3389/fimmu.2018.02094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Divangahi M., et al. Beyond killing Mycobacterium tuberculosis: disease tolerance. Front. Immunol. 2018;9:2976. doi: 10.3389/fimmu.2018.02976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Behr M.A., et al. Latent tuberculosis: two centuries of confusion. Am. J. Respir. Crit. Care Med. 2021;204:142–148. doi: 10.1164/rccm.202011-4239PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lillebaek T., et al. Molecular evidence of endogenous reactivation of Mycobacterium tuberculosis after 33 years of latent infection. J. Infect. Dis. 2002;185:401–404. doi: 10.1086/338342. [DOI] [PubMed] [Google Scholar]
- 48.Canetti G. The Tubercle Bacillus in the Pulmonary Lesion of Man: Histobacteriology and Its Bearing on the Therapy of Pulmonary Tuberculosis. Springer; 1955. The Tubercle Bacillus in the Pulmonary Lesion of Man: Histobacteriology and its Bearing on the Therapy of Pulmonary Tuberculosis. [Google Scholar]
- 49.Cadena A.M., et al. Heterogeneity in tuberculosis. Nat. Rev. Immunol. 2017;17:691–702. doi: 10.1038/nri.2017.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gideon H.P., et al. Variability in tuberculosis granuloma T cell responses exists, but a balance of pro- and anti-inflammatory cytokines is associated with sterilization. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1004603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Polak M.E., Singh H. Tolerogenic and immunogenic states of Langerhans cells are orchestrated by epidermal signals acting on a core maturation gene module. Bioessays. 2021;43 doi: 10.1002/bies.202000182. [DOI] [PubMed] [Google Scholar]
- 52.Cliff J.M., et al. The human immune response to tuberculosis and its treatment: a view from the blood. Immunol. Rev. 2015;264:88–102. doi: 10.1111/imr.12269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reichmann M.T., et al. Integrated transcriptomic analysis of human tuberculosis granulomas and a biomimetic model identifies therapeutic targets. J. Clin. Invest. 2021;131 doi: 10.1172/JCI148136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pollara G., et al. Exaggerated IL-17A activity in human in vivo recall responses discriminates active tuberculosis from latent infection and cured disease. Sci. Transl. Med. 2021;13:eabg7673. doi: 10.1126/scitranslmed.abg7673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ogongo P., et al. Tissue-resident-like CD4+ T cells secreting IL-17 control Mycobacterium tuberculosis in the human lung. J. Clin. Invest. 2021;131 doi: 10.1172/JCI142014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dijkman K., et al. Prevention of tuberculosis infection and disease by local BCG in repeatedly exposed rhesus macaques. Nat. Med. 2019;25:255–262. doi: 10.1038/s41591-018-0319-9. [DOI] [PubMed] [Google Scholar]
- 57.Bruyn E.D., et al. Inflammatory profile of patients with tuberculosis with or without HIV-1 co-infection: a prospective cohort study and immunological network analysis. Lancet Microbe. 2021;2:e375–e385. doi: 10.1016/s2666-5247(21)00037-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thuong N.T., et al. Identification of tuberculosis susceptibility genes with human macrophage gene expression profiles. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim M.J., et al. Caseation of human tuberculosis granulomas correlates with elevated host lipid metabolism. EMBO Mol. Med. 2010;2:258–274. doi: 10.1002/emmm.201000079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chai Q., et al. Lung gene expression signatures suggest pathogenic links and molecular markers for pulmonary tuberculosis, adenocarcinoma and sarcoidosis. Commun. Biol. 2020;3:604. doi: 10.1038/s42003-020-01318-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dheda K., et al. Spatial network mapping of pulmonary multidrug-resistant tuberculosis cavities using RNA sequencing. Am. J. Respir. Crit. Care Med. 2019;200:370–380. doi: 10.1164/rccm.201807-1361OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.O'Kane C.M., et al. Monocyte-dependent oncostatin M and TNF-alpha synergize to stimulate unopposed matrix metalloproteinase-1/3 secretion from human lung fibroblasts in tuberculosis. Eur. J. Immunol. 2008;38:1321–1330. doi: 10.1002/eji.200737855. [DOI] [PubMed] [Google Scholar]
- 63.Gideon H.P., et al. Single-cell profiling of tuberculosis lung granulomas reveals functional lymphocyte signatures of bacterial control. bioRxiv. 2020 doi: 10.1101/2020.10.24.352492. Published online October 26, 2020. [DOI] [Google Scholar]
- 64.Esaulova E., et al. The immune landscape in tuberculosis reveals populations linked to disease and latency. Cell Host Microbe. 2021;29:165–178. doi: 10.1016/j.chom.2020.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chowdhury R.R., et al. A multi-cohort study of the immune factors associated with M. tuberculosis infection outcomes. Nature. 2018;560:644–648. doi: 10.1038/s41586-018-0439-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pisu D., et al. Single cell analysis of M. tuberculosis phenotype and macrophage lineages in the infected lung. J. Exp. Med. 2021;218 doi: 10.1084/jem.20210615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cronan M.R., et al. A non-canonical type 2 immune response coordinates tuberculous granuloma formation and epithelialization. Cell. 2021;184:1757–1774. doi: 10.1016/j.cell.2021.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marakalala M.J., et al. Inflammatory signaling in human tuberculosis granulomas is spatially organized. Nat. Med. 2016;22:531–538. doi: 10.1038/nm.4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mattila J.T., et al. Microenvironments in tuberculous granulomas are delineated by distinct populations of macrophage subsets and expression of nitric oxide synthase and arginase isoforms. J. Immunol. 2013;191:773–784. doi: 10.4049/jimmunol.1300113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McCaffrey E.F., et al. Multiplexed imaging of human tuberculosis granulomas uncovers immunoregulatory features conserved across tissue and blood. bioRxiv. 2020 doi: 10.1101/2020.06.08.140426. Published online June 9, 2020. [DOI] [Google Scholar]
- 71.Penn-Nicholson A., et al. Discovery and validation of a prognostic proteomic signature for tuberculosis progression: a prospective cohort study. PLoS Med. 2019;16 doi: 10.1371/journal.pmed.1002781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Elkington P.T., et al. Mycobacterium tuberculosis, but not vaccine BCG, specifically upregulates matrix metalloproteinase-1. Am. J. Respir. Crit. Care Med. 2005;172:1596–1604. doi: 10.1164/rccm.200505-753OC. [DOI] [PubMed] [Google Scholar]
- 73.Elkington P., et al. MMP-1 drives immunopathology in human tuberculosis and transgenic mice. J. Clin. Invest. 2011;121:1827–1833. doi: 10.1172/JCI45666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Park D.Y., et al. Comparison of polymerase chain reaction with histopathologic features for diagnosis of tuberculosis in formalin-fixed, paraffin-embedded histologic specimens. Arch. Pathol. Lab. Med. 2003;127:326–330. doi: 10.5858/2003-127-0326-COPCRW. [DOI] [PubMed] [Google Scholar]
- 75.Schrom E.C., 2nd, et al. Quorum sensing via dynamic cytokine signaling comprehensively explains divergent patterns of effector choice among helper T cells. PLoS Comput. Biol. 2020;16 doi: 10.1371/journal.pcbi.1008051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Biton M., et al. Tuberculosis's cargoman: bacteria load RNA into host extracellular vesicles. EMBO Rep. 2019;20 doi: 10.15252/embr.201947719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Seiler P., et al. Cell-wall alterations as an attribute of Mycobacterium tuberculosis in latent infection. J. Infect. Dis. 2003;188:1326–1331. doi: 10.1086/378563. [DOI] [PubMed] [Google Scholar]
- 78.DiNardo A.R., et al. DNA hypermethylation during tuberculosis dampens host immune responsiveness. J. Clin. Invest. 2020;130:3113–3123. doi: 10.1172/JCI134622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Budzik J.M., et al. Dynamic post-translational modification profiling of Mycobacterium tuberculosis-infected primary macrophages. eLife. 2020;9 doi: 10.7554/eLife.51461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Khan N., et al. M. tuberculosis reprograms hematopoietic stem cells to limit myelopoiesis and impair trained immunity. Cell. 2020;183:752–770. doi: 10.1016/j.cell.2020.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kaufmann E., et al. BCG educates hematopoietic stem cells to generate protective innate immunity against tuberculosis. Cell. 2018;172:176–190. doi: 10.1016/j.cell.2017.12.031. [DOI] [PubMed] [Google Scholar]
- 82.Mitra S., et al. Prior exposure to oxidized low-density lipoprotein limits apoptosis in subsequent generations of endothelial cells by altering promoter methylation. Am. J. Physiol. Heart Circ. Physiol. 2011;301:H506–H513. doi: 10.1152/ajpheart.00252.2011. [DOI] [PubMed] [Google Scholar]
- 83.Johnston J.M., et al. Myeloid Tribbles 1 induces early atherosclerosis via enhanced foam cell expansion. Sci. Adv. 2019;5:eaax9183. doi: 10.1126/sciadv.aax9183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Elkington P., et al. Tuberculosis: an infection-initiated autoimmune disease? Trends Immunol. 2016;37:815–818. doi: 10.1016/j.it.2016.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Clayton K., et al. Gene expression signatures in tuberculosis have greater overlap with autoimmune diseases than with infectious diseases. Am. J. Respir. Crit. Care Med. 2017;196:655–656. doi: 10.1164/rccm.201706-1248LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kauffman K.D., et al. Defective positioning in granulomas but not lung-homing limits CD4 T-cell interactions with Mycobacterium tuberculosis-infected macrophages in rhesus macaques. Mucosal Immunol. 2018;11:462–473. doi: 10.1038/mi.2017.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ogongo P., et al. Differential skewing of donor-unrestricted and gammadelta T cell repertoires in tuberculosis-infected human lungs. J. Clin. Invest. 2019;130:214–230. doi: 10.1172/JCI130711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ihms E.A., et al. Diverse cavity types and evidence that mechanical action on the necrotic granuloma drives tuberculous cavitation. Am. J. Pathol. 2018;188:1666–1675. doi: 10.1016/j.ajpath.2018.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Davidson J.M. Biochemistry and turnover of lung interstitium. Eur. Respir. J. 1990;3:1048–1063. [PubMed] [Google Scholar]
- 90.Brinckerhoff C.E., Matrisian L.M. Matrix metalloproteinases: a tail of a frog that became a prince. Nat. Rev. Mol. Cell Biol. 2002;3:207–214. doi: 10.1038/nrm763. [DOI] [PubMed] [Google Scholar]
- 91.Rowland S.S., et al. Identification of an elastolytic protease in stationary phase culture filtrates of M. tuberculosis. FEMS Microbiol. Lett. 1997;151:59–64. doi: 10.1016/s0378-1097(97)00138-9. [DOI] [PubMed] [Google Scholar]
- 92.Ribeiro-Guimaraes M.L., Pessolani M.C. Comparative genomics of mycobacterial proteases. Microb. Pathog. 2007;43:173–178. doi: 10.1016/j.micpath.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 93.Young D. Animal models of tuberculosis. Eur. J. Immunol. 2009;39:2011–2014. doi: 10.1002/eji.200939542. [DOI] [PubMed] [Google Scholar]
- 94.Balbin M., et al. Identification and enzymatic characterization of two diverging murine counterparts of human interstitial collagenase (MMP-1) expressed at sites of embryo implantation. J. Biol. Chem. 2001;276:10253–10262. doi: 10.1074/jbc.M009586200. [DOI] [PubMed] [Google Scholar]
- 95.Pan H., et al. Ipr1 gene mediates innate immunity to tuberculosis. Nature. 2005;434:767–772. doi: 10.1038/nature03419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Al Shammari B., et al. The extracellular matrix regulates granuloma necrosis in tuberculosis. J. Infect. Dis. 2015;212:463–473. doi: 10.1093/infdis/jiv076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ordonez A.A., et al. Matrix metalloproteinase inhibition in a murine model of cavitary tuberculosis paradoxically worsens pathology. J. Infect. Dis. 2019;219:633–636. doi: 10.1093/infdis/jiy373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Urbanowski M.E., et al. Repetitive aerosol exposure promotes cavitary tuberculosis and enables screening for targeted inhibitors of extensive lung destruction. J. Infect. Dis. 2018;218:53–63. doi: 10.1093/infdis/jiy127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Xu Y., et al. Matrix metalloproteinase inhibitors enhance the efficacy of frontline drugs against Mycobacterium tuberculosis. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1006974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Miow Q.H., et al. Doxycycline host-directed therapy in human pulmonary tuberculosis. J. Clin. Invest. 2021;131 doi: 10.1172/JCI141895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wallis R.S., et al. Adjunctive host-directed therapies for pulmonary tuberculosis: a prospective, open-label, phase 2, randomised controlled trial. Lancet Respir. Med. 2021;9:897–908. doi: 10.1016/S2213-2600(20)30448-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Theocharidis A., et al. Network visualization and analysis of gene expression data using BioLayout Express(3D) Nat. Protoc. 2009;4:1535–1550. doi: 10.1038/nprot.2009.177. [DOI] [PubMed] [Google Scholar]
- 104.Chan T.E., et al. Gene regulatory network inference from single-cell data using multivariate information measures. Cell Syst. 2017;5:251–267. doi: 10.1016/j.cels.2017.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]