Abstract
Correlates of protection for COVID-19 vaccines are urgently needed to license additional vaccines. We measured immune responses to four COVID-19 vaccines of proven efficacy using a single serological platform. IgG anti-Spike antibodies were highly correlated with ID50 neutralization in a validated pseudoviral assay and correlated significantly with efficacies for protection against infection with wild-type, alpha and delta variant SARS-CoV-2 virus. The protective threshold for each vaccine was calculated for IgG anti-Spike antibody. The mean protective threshold for all vaccine studies for WT virus was 154 BAU/ml (95 %CI 42–559), and for studies with antibody distributions that enabled precise estimation of thresholds (i.e. leaving out 2-dose mRNA regimens) was 60 BAU/ml (95 %CI 35–102). We propose that the proportion of individuals with responses above the appropriate protective threshold together with the geometric mean concentration can be used in comparative non-inferiority studies with licensed vaccines to ensure that new vaccines will be efficacious.
Abbreviations: COP, Correlate of Protection; WHO, World Health Organisation
Keywords: COVID-19, SARS-CoV-2, COVID vaccines, Correlates of protection
1. Introduction
Severe acute respiratory syndrome-related coronavirus-2 (SARS-CoV-2) was first recognised in December 2019 and rapidly spread world-wide resulting in WHO declaring a COVID-19 pandemic on March 11th, 2020. Soon after the identification and genetic sequencing of the virus, numerous groups began developing vaccines with unprecedented speed and using a variety of approaches. Randomized controlled efficacy trials have shown a range of efficacies and have supported emergency use authorizations of more than eleven vaccines. Numerous additional vaccines are in development but confirming their efficacy in randomized placebo-controlled trials is becoming increasingly difficult. Given the urgent need for additional vaccines to meet the global demand, licensing new vaccines based on serologic correlates of protection is of critical importance.
Two recent studies have shown strong correlations between antibodies (both neutralizing and IgG binding) and protection in clinical efficacy trials although for comparison between studies relied on normalizing the antibody levels to those published alongside for human convalescent serum. Indeed, up to 90% of the variability in efficacy observed among different vaccines which used different technology platforms could be explained by their antibody levels, suggesting that post-immunization antibody levels can serve as a valid measure of short-term protection [1], [2].
An immunological correlate of protection (COP) has been established for many licensed vaccines based on a protective threshold or minimum protective level [3]. Two main methods have been used: individual-based correlates and population-based correlates [https://apps.who.int/iris/bitstream/handle/10665/84288/WHO_IVB_13.01_eng.pdf]. The individual-based correlate measures biomarkers prior to exposure in all vaccinated subjects and evaluates the relationship between these and the development of disease. The expectation is that a concentration of the relevant biomarker (most commonly a level of antibody) can be found above which individuals are reasonably likely to be protected. This method has been applied to a number of diseases such as measles [4], [5] and meningococcus [6], typically by evaluating outbreaks of disease in which, fortuitously, pre-outbreak sera were available. The method has rarely been used in large-scale vaccine trials because of the inconvenience and expense of collecting sera on all participants, but some COVID-19 vaccine trials are an exception. Indeed, the individual-based method has very recently been applied to the AstraZeneca and Moderna COVID-19 vaccine trials, both of which showed that spike-specific antibody binding is associated with lower risk of symptomatic disease, but a threshold above which subjects were reliably protected could not be identified [7], [8].
The population-based approach conceived by Chang and Kohberger requires the measurement of antibody in a representative sample of subjects after vaccination and calculates the protective threshold based on the observed efficacy by using the simplifying assumption that all subjects with antibody above the threshold are fully protected and all subjects below the threshold are fully at risk of disease [9]. This method has been applied to meningococcal C vaccine using post-licensure efficacy data in England [10] and pneumococcal vaccines based on multiple efficacy trials [11], [12] but not to viral vaccines. Protective thresholds identified by this method have been widely accepted by regulatory authorities and have proved useful for licensing multiple follow-on vaccines. This method does not rely on measurement of antibodies in individuals who have breakthrough infections but rather on defining the distribution of antibodies in a representative subset of the immunized population and hence is referred to as a population-based analysis.
A prerequisite for estimating a broadly applicable COP is an antibody assay, using similar or identical protocols, which has been shown to give equivalent results in different laboratories as urged by many including the CDC COVID-19 Response Team [13]. A WHO standard polyclonal antibody is now available, which was used here to normalize antibody levels in Binding Antibody Units (BAU/ml)[14]. To date, very few studies comparing immunogenicity between vaccines have been undertaken although support for licensure of new COVID-19 vaccines based on such comparisons with authorised/licensed vaccines has been announced by Valneva (https://valneva.com/press-release/valneva-initiates-phase-3-clinical-trial-for-its-inactivated-adjuvanted-covid-19-vaccine-candidate-vla2001) and the European Commission have approved and advance purchase agreement on this basis [https://valneva.com/press-release/valneva-announces-european-commission-approval-of-advance-purchase-agreement-for-up-to-60-million-doses-of-inactivated-covid-19-vaccine-vla2001/]. In such studies it will be important to compare immune responses based on the proportion of participants with antibody responses above the protective threshold and the geometric means, both of which have been used previously to compare vaccine immunogenicity in pivotal licensure studies.
In this study, we collected serum from vaccinees who received 1 or 2 doses of one of 4 vaccines: BNT162b2 (Pfizer), mRNA1273 (Moderna), ChadOx1/AZ 1222 (AstraZeneca), or Ad.26COV2.S (J&J), and compared IgG responses to spike and ACE2 Receptor blocking activity to the original wild-type strain (WT), alpha (B.1.117) and delta variant (B.1.617.2), RBD (WT and alpha) and pseudovirus neutralization (WT and Delta). We then applied population-based methods to assess the correlation of IgG binding with vaccine efficacy or effectiveness (Table S2) to estimate a protective threshold. Finally, we suggest a path toward formally establishing a correlate of protection for COVID-19 vaccines.
2. Results
2.1. Demographic characteristics of the four vaccine cohorts
Serum samples from 122 adults following a complete immunization schedule of the 4 vaccines were available for analysis. In addition, serum samples from 83 of these individuals were available following a first dose of BNT162b2 (Pfizer), mRNA1273 (Moderna), and ChadOx1/AZ 1222 (AstraZeneca). Individuals with positive anti-Nucleocapsid antibody at the time of evaluation or history/evidence of prior COVID 19 infection were excluded from the analysis. The demographic characteristic of the four cohorts of naïve subjects are provided in Table 1 . The median age of the different cohorts was between 35 and 60 years with ages ranging from 21 to 77 years. For each individual cohort neither age nor gender were significantly associated with vaccine responses, so antibody responses of cohorts were compared without adjustments. For the vaccines authorised for two dose use (Pfizer/Moderna), the median time between doses was 3 or 4 weeks. The interval for the AstraZeneca vaccine was 66 days reflecting the manner of its deployment in the United Kingdom where the vaccinees resided.
Table 1.
Demographics of Vaccine Cohorts. The demographics of the four vaccine cohorts and characteristics of their samples which are included in analysis are shown.
| Vaccine Name | Manufacturer | Number of Subjects One Dose/Two Dose | Gender Ratio Female/Male | Median Age Yrs (range) | Median Days Between Doses (range) | Median Days from First Dose to post dose 1 Bleed (range) | Median Days from Final Dose to Bleed (range) |
|---|---|---|---|---|---|---|---|
| mRNA 1273 | Moderna | 19/19 | 1.6 | 35 (20–55) | 27(26–28) | 27 (26–28) | 7 (7–8) |
| BNT 162b2 | Pfizer | 36/51 | 1.8 | 43 (21–77) | 21(20–60) | 23 (7–54) | 8 (7–29) |
| ChadOx1/AZ 1222 | AstraZeneca | 28/21 | 2.6 | 60 (23–70) | 66 (33–79) | 22 (19–31) | 8 (7–10) |
| Ad26.COV2.S | J&J | 25/NA | 1.2 | 48 (31–69) | N/A | N/A | 34 (20–31) |
2.2. Comparative IgG Spike binding antibody to wild type, alpha and delta variant for four vaccine cohorts
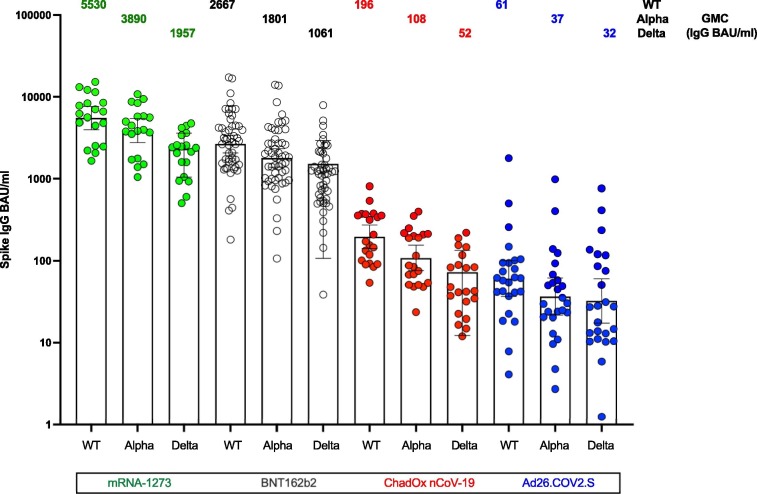
IgG binding antibody concentrations to Spike protein derived from WT virus (without D614G), B.1.1.7 (alpha variant) and B.1.617.2 (delta variant) were assessed by ELISA and expressed as binding antibody units/ml (BAU/ml) for each cohort utilizing the WHO standard as detailed in Methods. The comparative responses for Spike from WT, alpha and delta variants of the four vaccine cohorts are shown in Fig. 1 . The geometric mean concentrations (GMC) to Spike WT following immunization with Moderna and Pfizer vaccines were 5530 BAU/ml and 2653 BAU/ml respectively compared to 196 BAU /ml and 61 BAU/ml following AstraZeneca and J&J vaccines respectively (Table 2 ). A reduced GMC for alpha and delta Spike compared to WT was noted for each cohort with similar rank order among cohorts (Table 1). We performed rank sum pairwise tests to determine significance of the differences between cohorts for each response as specified in Table 2. All groups differed significantly except for AstraZeneca compared to J&J for delta responses. Binding antibody directed to RBD for original and alpha variant were also assessed with Pfizer and Moderna vaccine cohorts demonstrating between 13 and 115 times higher GMC compared to AstraZeneca and J&J cohorts for both RBD antigens with significant differences between cohorts as specified in Supplement Table S1. Of note, the assignment of units for the WHO standard for Spike and RBD antigens was the same at 1000 units/ml and thus values do not represent absolute amount of antibody directed to Spike compared to RBD.
Fig. 1.
Spike IgG antibody to original wild type (WT) virus, alpha and delta variant. IgG Concentrations, GMCs and 95% CI (BAU/ml) to Spike derived from WT as well as the B.1.1.7 (alpha variant) and B.617.2 (delta variant), following complete courses of four different SARS CoV 2 vaccines administered to naïve recipients. Concentrations are expressed in standardised binding antibody units (BAU)/ml calibrated against the WHO international standard, and GMC are displayed by text above the bars with 95% CI represented by whiskers.
Table 2.
Geometric mean concentrations (GMC) to spike derived from original wild-type virus, alpha or delta variants. IgG GMCs and 95% CI (BAU/ml) to Spike derived from original (without D614G), B.1.1.7 (alpha variant) and B.1.617.2 (delta variant), following complete courses of four different SARS CoV 2 vaccines administered to naïve recipients. GMCs are expressed in standardised binding antibody units (BAU) /ml calibrated against the WHO international standard. Fold reduction of GMCs for variants compared to original are displayed. Pairwise comparisons for groups using a rank based test demonstrated p < 0.0001 for all groups except for Moderna compared to Pfizer for WT (p < 0.006), alpha (p < 0.006), and delta (p < 0.02), and J&J compared to AstraZeneca for WT (p < 0.0008), alpha (p < 0.005) and not significant for delta.
| Vaccine | Manufacturer | GMC Original Spike BAU/ml (95% CI) | GMC Alpha Variant Spike BAU/ml (95% CI) | GMC Delta Variant Spike BAU/ml (95% CI) | Fold Reduction Alpha Compared to Original | Fold Reduction Delta Compared to Original |
|---|---|---|---|---|---|---|
| mRNA 1273 | Moderna | 5530 (4007–7633) |
3890 (2791–5421) |
1957 (1426–2686) |
1.2 | 2.3 |
| BNT162b2 | Pfizer | 2667 (2077–3425) |
1801 (1390–2332) |
1061 (811–1387) |
1.5 | 2.3 |
| ChadOx/AZ 1222 | AstraZeneca | 196 (141–273) |
108 (76–154) |
52 (35–77) |
1.8 | 3.8 |
| Ad.26COV2.S | J&J | 61 (37–101) |
37 (22–62) |
32 (17–60) |
1.7 | 1.9 |
2.3. Comparative neutralization assay results for wild type and delta variant for four vaccine cohorts
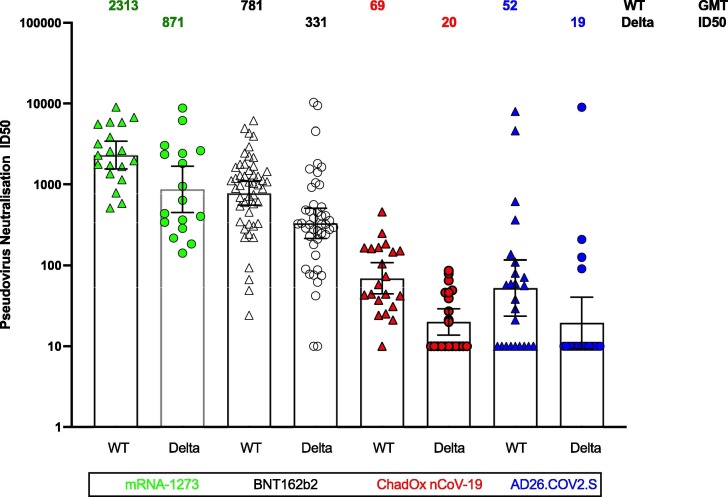
To further evaluate the differences among the vaccine cohorts observed for binding antibody, neutralization by a validated neutralization assay with lentivirus-based Spike-pseudotyped viruses (Montefiori laboratory) was performed for WT and delta variant. The comparative results for ID50 pseudovirus neutralization demonstrated similar rank order among the cohorts for both WT and delta variant with significant differences noted for all group comparisons except AstraZeneca and J&J delta responses (Fig. 2 ). Of note, many of the samples were below the limit of detection in the AstraZeneca and J&J vaccine cohorts for the delta variant. To further define the relationship of ELISA binding antibody and neutralization, we examined the correlation of IgG binding Spike antibody to ID50 neutralization for both WT and delta variant. Significant correlations were observed for both WT and delta (0.86 and 0.81 respectively, p < 0.0001) (Fig. S1a and 1c).
Fig. 2.
Pseudovirus neutralization of wild type and delta virus for the fully immunized cohorts. ID50 neutralisation geometric mean titres (GMT) for a lenti-pseudovirus Wuhan-1 spike containing D614G (WT) or B.1.617.2 (AY.3) (delta variant) in sera of vaccinees following complete courses of four different SARS CoV 2 vaccines administered to naïve recipients. Neutralization titers shown are the inhibitory dilution (ID) of serum samples at which relative luminescence units (RLU) were reduced by 50% (ID50) compared to virus control wells after subtraction of background RLUs (see methods for details). The cohorts were compared by pairwise rank sum test adjusted for multiple comparisons and significance determined as follows: p < 0.0001 for all except Moderna vs Pfizer for delta (p < 0.05), and AZ compared to J&J was not significant for WT or delta.
Finally, since functional assays for neutralization can be difficult to standardize among laboratories and are slow through-put, we also utilized a measurement of ACE-2 inhibition by ELISA format to compare the four cohorts. The relative amounts of ACE-2 inhibition to Spike derived from WT, alpha and delta for each cohort demonstrated the same hierarchy of responses seen for IgG Spike antibody assays and for pseudovirus neutralization ID50 assays (Fig. 2). Strong correlations were observed for ACE-2 inhibition to WT and delta spike with pseudovirus neutralization ID50 with correlation coefficients of 0.78 and 0.84 respectively (Fig. S1b and 1d) confirming the good agreement between ACE2 inhibition and neutralization reported previously [15], [16].
3. Correlation of IgG spike antibody GMC with vaccine efficacy across four vaccines
We and others have previously found a remarkably high correlation between ELISA binding antibody or neutralizing antibody and efficacy across different approved vaccines [1], [2]. To be able to compare GMC antibody levels measured by multiple assays and laboratories across the different trials, groups have calibrated antibody levels relative to human convalescent antibodies by the same assay reported in the same study. In contrast, in this study we utilized a standardized, commercially available ELISA binding assay performed in a single laboratory thereby circumventing the need for inter-laboratory assay standardization. We evaluated the correlation between GMC binding antibodies to Spike elicited by four approved vaccines with the efficacy determined in Phase 3 studies to WT virus or in large recent effectiveness studies of alpha and delta variants (see Supplementary Table S2 for studies utilized in this analysis).
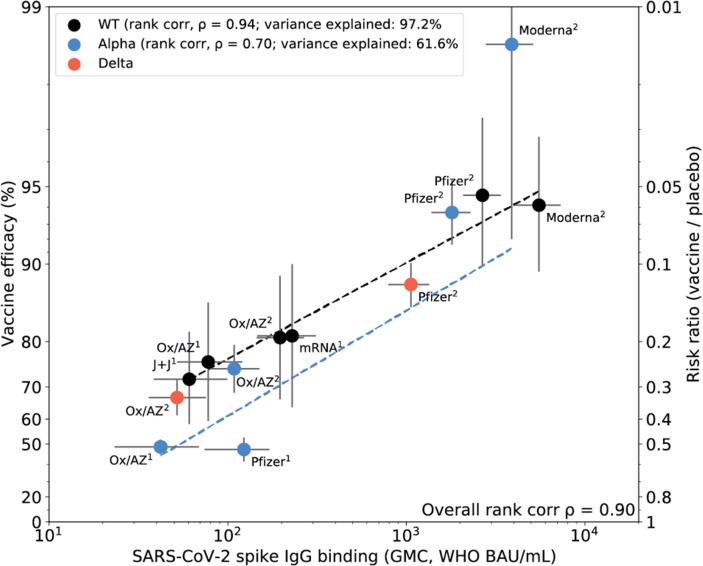
A strong correlation was observed between binding antibody to WT Spike antigen and efficacy with a rank correlation coefficient of 0.94 and with 97.4% of the variance explained by the antibody in a linear model (Fig. 3 A). Furthermore, all four of the vaccines in this study have reported clinical effectiveness data for the alpha variant and two of the vaccines have data available for effectiveness to the delta variant. We therefore examined the correlation of GMC antibody directed to each variant and effectiveness in prevention of PCR confirmed symptomatic disease to each of the variants (Fig. 3B). For alpha variant the rank correlation coefficient was 0.70 with 61.6% of the variance explained by the antibody. As only two points were available for delta variant, a specific rank correlation was not determined. However when delta was included in an overall rank analysis (wild type, alpha and delta), the rank correlation was 0.90.
Fig. 3.
Correlation of Spike IgG binding antibody with vaccine efficacy for wild type, alpha and delta variants. Vaccine efficacy/effectiveness (VE) and SARS-CoV-2 spike binding IgG GMC, against Original (WT), alpha and delta variants. Data included in correlation analyses are described in Table 2 and Table S2. Superscript 1 or 2 indicates the number of doses for the vaccine regimen. The y-axis is estimated log risk-ratio reported on the vaccine efficacy scale. The x-axis is the geometric mean concentration (GMC) of spike-specific IgG antibody binding measured by MSD and calibrated to the WHO standard (binding antibody units per mL). Error bars indicate 95% confidence intervals for either the GMC IgG level (x-axis) or VE (y-axis). Weighted least-squares linear regression fit using inverse variance weighting on VE estimates (dashed line black for WT, dashed line blue for alpha variant). Rank correlation coefficient, variance explained by the model, and mean squared error (MSE) are indicated for the WT, and alpha variant models. An overall model rank correlation includes Delta variant in Fig. 3B. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.1. Estimation of a protective threshold for each vaccine regimen using reverse cumulative distribution (RCD) curves
The high correlation between vaccine efficacy and the binding antibody to Spike (together with the high correlation of these binding antibodies with neutralization assays) provide a strong rationale to evaluate binding assays to estimate the protective threshold for COVID-19 vaccines. The purpose of the protective threshold, sometimes called the minimum protective level of antibody, is to define the cut-off between antibody levels deemed sufficient to provide protection in a population and those which are not deemed protective. Unlike the GMC of antibody, which has been shown to vary with vaccine efficacy, we expect the protective threshold to be similar for vaccines demonstrating different levels of efficacy, provided that factors affecting the amount of antibody needed for protection of the populations as a whole are similar among studies, including quality of antibody and other immune mechanisms, clinical endpoint, intensity of exposure and vaccine strain. The population of subjects with antibody levels above the threshold are expected to be protected and those with antibody levels below the threshold are expected to be at risk, regardless of which vaccine was given.
We estimated protective thresholds for each vaccine using the population-based model developed by Chang and Kohberger for pneumococcal vaccines [9]. The model makes one critical simplifying assumption which is that there is a sharp cut-off between protective and non-protective antibody levels: i.e. that all individuals, whether immunized or controls, with antibody above the threshold are protected and those with antibody below it are at risk. In the studies evaluated here control subjects had no pre-existing antibody so the threshold can simply be determined from the post-immunization RCD curves, such that the percent of subjects above the threshold is equal to the percent efficacy observed.
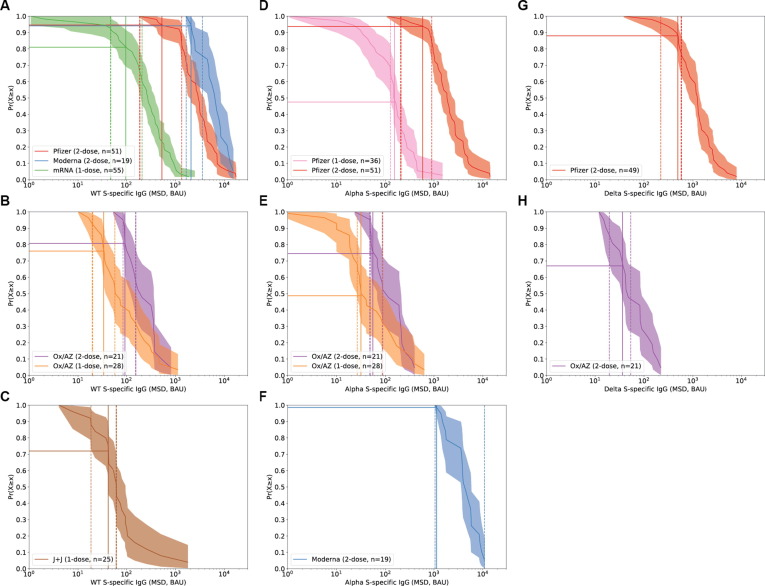
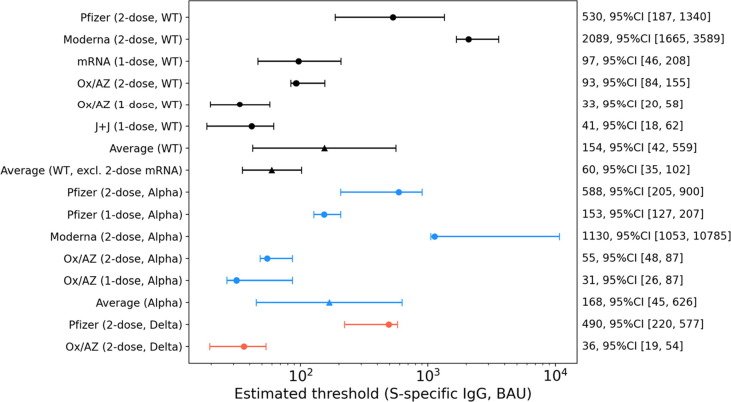
The RCD curves were estimated from immune responses to vaccine regimens for which efficacy estimates were available, including one or two doses of Pfizer, Moderna, and AstraZeneca vaccines, and one dose of J&J. (Fig. 4 ). For WT virus, when all vaccines and dose regimens are included, the overall protective threshold was estimated to be 154 BAU/ml (95 %CI 42–559) by taking the geometric mean of all thresholds across all six regimens using a random effects meta-analysis approach. A similar overall protective threshold was estimated for the alpha variant at 168 BAU/ml (95% CI; 45–626) (Fig. 5 ). With the limited data available for delta effectiveness, we estimate a protective threshold between 36 and 490 BAU/ml.
Fig. 4.
Distribution of spike-specific binding antibody vaccine responses and determination of a vaccine-specific protective threshold. Reverse cumulative distribution functions (RCDs) were estimated from spike-specific IgG vaccine responses measured in cohorts of individuals who received: one or two doses of the Pfizer or Moderna mRNA-based vaccines, one or two doses of the Oxford/AstraZeneca vaccine, and one dose of the J + J vaccine. Panel A, B, C for original virus, Panel D, E, F for alpha variant, and Panel G, H for delta variant. Each RCD along the y-axis represents the estimated proportion of participants who responded with at least as high a response as indicated along the x-axis. Shaded region indicates a point-wise 95% confidence interval (CI; see Methods for details). For each vaccine regimen, a published estimate of vaccine efficacy or effectiveness was used to compute a protective threshold by finding the VEth percentile of the RCD (solid horizontal line at VEth percentile). The protective threshold is shown (vertical sold line) with a 95% CI (dashed vertical line) that takes into account uncertainty in estimation of both the RCD and VE (see Methods for details).
Fig. 5.
Summary of protective antibody binding thresholds derived for each vaccine. A protective threshold (BAU/ml) for each vaccine regimen is displayed with 95% confidence interval (circle symbols). Thresholds were computed for original (WT, black), alpha (blue), and delta (orange) spike-specific responses paired with WT, alpha or delta variant specific estimates of vaccine efficacy. An average overall protective threshold was computed for WT and alpha independently (triangle symbols) using a random effects meta-analytic approach (see Methods for details). Separately an average was computed for WT excluding the two-dose mRNA vaccine regimens. The protective threshold and 95% CI are annotated along the right y-axis. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The thresholds calculated from the 2-dose mRNA vaccine regimens were outliers among the vaccine regimens as shown by the lack of overlap between 95% confidence intervals of the Moderna threshold and the average threshold (Fig. 5). It is also evident in Fig. 1 that neither 2-dose mRNA regimen induces antibody levels below the mean threshold of 154 BAU/ml and thus do not contribute information to estimating a mean threshold that would be useful in comparing efficacious vaccines with lower immunogenicity. Therefore, another analysis was performed on less immunogenic but clearly efficacious vaccines by omitting the 2-dose mRNA vaccine data but retaining the 1-dose mRNA vaccine data. The mean threshold excluding the two-dose mRNA vaccine data was 60 BAU/ml (95% CI 35–102). This lower value may be better suited for comparing vaccines which have lower but useful efficacy.
Finally, to verify that the threshold indeed discriminates between protected and non-protected populations in each study, we estimated the proportion of participants receiving each vaccine regimen that responded with antibody greater than the higher threshold of 154 BAU/mL (Fig. S3A) or the lower threshold of 60 BAU/ml (Fig. S3B). In the figures, the line of identity indicates that under a model of an absolute protective threshold, efficacy would be expected to be equal to the proportion of participants above the threshold; the mean squared error from the line of equivalence is a useful metric for evaluating this relationship. The data support that there is an association between vaccine efficacy and the proportion of individuals above either of the thresholds. In practice, a threshold could be used to directly compare two vaccines using a non-inferiority margin. It is noteworthy that for OX/AZ and J&J vaccines the proportion of subjects above the lower threshold of 60 BAU/ml is quite similar to their efficacy to wild type virus, whereas that is not the case with the higher threshold of 154 BAU/ml.
4. Discussion
Immunologic correlates of protection have played a critically important role in vaccine development by providing a framework for comparing newly developed vaccines to those which have already been approved based on demonstration of clinical efficacy. Such correlates are also needed for confirming consistent immunogenicity in initial lot-to-lot consistency trials, after major manufacturing changes of approved vaccines or in evaluating reduced doses of vaccines [17]. Multiple lines of evidence including pre-clinical studies and strong correlations between antibody levels and clinical efficacy after multiple vaccines show that antibodies are a major protective mechanism against primary infection with COVID-19 [1], [2]. Nevertheless, as of this writing, an antibody level associated with protection has not been defined for COVID-19 vaccines even though more than 10 large scale efficacy trials demonstrating a range of efficacies have been completed. Key reasons for this include the lack of well standardized antibody assays that facilitate direct comparisons between vaccines and lack of agreement on the methodology to be used to establish a protective correlate. Here we propose to overcome these obstacles by directly comparing immune responses of four vaccines in the same antibody assays in a single laboratory and by using an accepted population-based approach to estimate a threshold of protection based on observed vaccine efficacies in clinical trials.
The underlying assumptions of the population-based approach is that vaccines that induce antibodies above a protective threshold in a similar proportion of subjects can be expected to have similar clinical efficacy. A second assumption is that the protective threshold in a population can be modelled as a single cut-off concentration above which everyone is protected and below which everyone is at risk. Like all models, this is a gross simplification of the reality that individuals may require differing levels of antibody for protection for many reasons including host factors, pathogen virulence and intensity of exposure and thus cannot be used to determine the immune status of an individual. The population-based protective threshold can be estimated in each trial from the observed efficacy and the antibody distribution several weeks after immunization, without further assumptions. With other vaccines such as pneumococcal and meningococcal vaccines, the threshold of protection was estimated by this method and was subsequently used as the primary endpoint in numerous vaccine non-inferiority comparisons. The protective threshold provides the most sensitive ability to discriminate differences in immunogenicity [10], [11] and supports the use of a narrow non-inferiority margin for vaccines with very high efficacy. For example, the primary non inferiority outcome for pneumococcal and meningococcal vaccines is that the percent of subjects achieving the respective thresholds (IgG anti-polysaccharide of 0.35 μg/ml and BCA of 1/8, respectively) cannot be more than 10% lower than the licensed comparator [12]. GMCs serve as the secondary outcome which cannot be more than 1.5 fold lower than the licensed vaccine (i.e. < 0.67 GMC of comparator).
Estimates of the population-based threshold of protection are expected to be similar in each efficacy study, regardless of the efficacy that was observed, with the caveat that certain systematic differences among studies such as the quality of antibody induced, outcome definition, incidence of disease, strain causing disease or contribution of other immunological responses such as T cell responses may affect the amount of antibody needed for protection at a population level. For example, a range of estimates for thresholds for anti-polysaccharide antibodies were determined for the pneumococcal vaccine but a single consensus estimate of 0.35 μg/ml was agreed upon as a protective threshold. Post-introduction effectiveness studies of these vaccines have confirmed their efficacy and would similarly be required for COVID19 vaccines licensed in this way.
In this study the mean protective threshold for the original strain, based on data from all six vaccine regimens was 154 BAU/ml (95% CI 42–559) and a mean threshold omitting the 2-dose mRNA data was 60 BAU/ml with a narrower 95% CI of 35–102 BAU/ml. While the higher threshold would be useful for evaluating other vaccines with similarly high efficacy, using narrow 10% non-inferiority criteria, they would result in the failure to meet non-inferiority criteria for vaccines with lower but useful efficacy. For this reason, the use of the lower threshold may be more appropriate for comparing new COVID-19 vaccines in non-inferiority trials against approved vaccines other than Pfizer and Moderna mRNA vaccines.
Recently, concentrations of neutralizing antibody associated with protection were estimated after the AstraZeneca and Moderna vaccines using a different approach based on how well post-immunization antibody levels predicted risk of acquiring COVID-19 for the individual subjects in their efficacy studies [7], [18]. Both studies found broad overlap in antibody concentrations between individuals who developed COVID and those who did not. Consequently, it was not possible to identify a threshold antibody level above which subjects were reliably protected using analysis of breakthrough cases. Interestingly, however, Feng et al estimated a level of spike binding IgG antibody of 264 BAU/ml associated with 80% protection [18] by the AstraZeneca vaccine and Gilbert et al, estimated a level of 298 BAU/ml associated with 90% protection by the Moderna vaccine [7]. Although these estimates were not threshold values, they are in a similar range to our population-based method for determining a threshold value.
The goal of developing a threshold of protection is to serve as the primary endpoint for non-inferiority studies comparing new COVID-19 vaccines to vaccines already approved or authorized based on clinical efficacy. To accomplish this goal, the following key issues and questions should be resolved: First, which assays can be used as the primary basis for comparison? Binding assays have significant advantages over neutralization assays in reproducibility, lower variability, cost and convenience and have been much easier to standardize among laboratories. Given that for COVID-19 various binding and neutralization assays show a high degree of correlation with each other and both are highly correlated with clinical protection, it is reasonable to propose that binding assays can serve as the primary serologic outcome in vaccine comparisons. Interestingly though, virus neutralization is difficult to demonstrate after one dose of mRNA vaccines despite efficacy up to 81% measured from 14 d after the first dose [19]. In contrast, binding antibody responses assessed following one dose of mRNA vaccines are easily measured and even superior to some less immunogenic vaccines after two doses. A plausible explanation for this discordance between binding and neutralizing antibodies is that the first dose of mRNA vaccine activates a non-neutralizing recall response predominantly targeting epitopes in the S2 subunit which is highly conserved across human pathogenic coronaviruses [20]. We have recently completed a study of an experimental vaccine, SCB-2019 and adjuvanted S-trimer vaccine (Clover Biopharmaceuticals) and shown that the GMC of Delta variant specific IgG accurately predicted the recently published vaccine efficacy against the delta variant of 79% [https://www.cloverbiopharma.com/news/83.html] [21]. Second, a consensus should be obtained on the threshold level to be used for non-inferiority studies and the level of non-inferiority that must be demonstrated. Before such a threshold can be defined, regulatory and public health authorities will need to reach a consensus on what is the minimum acceptable vaccine efficacy for COVID-19 vaccine in the future. Based on our analyses in this study, a protective threshold of 60 BAU/ml would provide an appropriate basis for non-inferiority comparisons of vaccines when compared against existing approved vaccines although additional studies would be required to confirm this. Third, the ongoing evolution of variants of concern (VOC) poses a critical question about how protective thresholds can be estimated for VOCs. In this report, we used a population-based model to calculate threshold values for the alpha variant at 168 BAU/ml and a preliminary estimate for delta VOC in the range of 36–490 BAU/ml (a more accurate estimate will require additional delta specific VE data). The population-based method could readily be applied to other outcomes such as severe disease or asymptomatic infection but sufficient data to do these analyses are not yet available.
Limitations of our study include that a limited number of sera were assayed to construct the RCD curves, that our subjects were recruited from different population outside of clinical trials and thus may not be representative of those in the phase 3 efficacy studies and that only short term efficacy data are available. It would therefore be important to confirm the threshold estimates using larger numbers of sera from subjects enrolled in efficacy trials. It is noteworthy that the recent report by Gilbert et al estimated a Spike IgG threshold of 77 BAU/ml (95 %CI 60–94) using the population-based method on a large serum set drawn on day 29 after the 1st dose of the Moderna vaccine Phase 3 study [7]. When applied to sera after the 2nd dose, the threshold estimate was 1000 BAU/ml (95 %CI 860–1200) clearly demonstrating that the population-based method is not applicable with vaccine regimens inducing antibodies levels far above the threshold.
The population-based protective threshold for binding antibody proposed in this report could be useful not only to license new vaccines based on comparable immunogenicity but also to predict the need and timing for booster immunizations as concentrations of antibody wane. For example, the Pfizer vaccine has been shown to have reduced efficacy in Israel associated with both waning antibody and predominance of delta variant in Israel resulting in a decision to offer booster immunizations [22]. A recent study evaluating antibody concentrations associated with breakthrough cases in Israel, also showed that both neutralizing and IgG anti-Spike concentrations were lower in those cases compared to controls although a threshold value was not determined [23]. One could determine the threshold antibody concentrations associated with the reduced efficacy for the population at the time of reduced efficacy. Such a threshold could be applied to other populations to predict the proportion of the population at risk for that variant based on the percent of individuals waning below the determined threshold.
In conclusion, we propose an approach to defining a threshold of protection for IgG antibody to original spike protein that would serve as a basis for comparing new vaccines to existing vaccines in non-inferiority studies. This protective level should be calculated from larger data sets from efficacious vaccines than those available to us and a consensus would need to be achieved on defining a single protective threshold. Although other studies of COP have focused on neutralization assays, we believe that the data within this report and other studies support the use of binding antibody which has many practical advantages [24]. Finally, the COP could be useful for supporting boosting recommendations as serum antibody concentrations wane and new virus variants cause rapid increases in the number of CoVID-19 cases.
5. Methods
5.1. Sources of sera
This study was undertaken using a convenience sample obtained post vaccination from groups of individuals receiving vaccine as part of their government’s national rollout campaigns. Samples from vaccinees in Latvia and South Africa were obtained as part of a previous study of HCWs in paediatric facilities [25] originally initiated at Great Ormond Street Hospital (COSTARS, IRAS 282713, ClinicalTrials.gov Identifier: NCT04380896). Ethics approval was obtained locally by the lead investigators of each site. In the UK, volunteers who were part of the COSTARS Study as well as others who had received vaccines as part of the government rollout altruistically agreed to donate serum to help evaluate an assay for measuring post vaccine immunity being run the UCL laboratory. Vaccinees received one of four vaccines depending on local use.
In Latvia and South Africa, serum was aliquoted, given a unique identifier, and stored frozen until batch shipping to the WHO International Reference laboratory for Pneumococcal Serology at University College London, London, UK. Local UK samples had serum extracted and were stored frozen until batch tested.
5.2. Immunological assays
Samples were analysed for the presence IgG to SARS-CoV-2 nucleocapsid protein, receptor binding domain of S1 and trimeric spike antigen derived from the original wild type Wuhan strain as previously described [26] as well as spike and RBD responses to VOCs B.1.1.7 and B.1.617.2 (AY.3)(MSD® SARS-Coronavirus Plate 7, Rockville, MD). Responses to nucleocapsid protein and information about previous clinical symptoms compatible with COVID with or without tests confirming the diagnosis were used to stratify vaccinees into naïve or primed as this can influence antibody responses to the vaccine [27]. Only naïve vaccinees were include in this study. The MSD IgG assay was calibrated against the WHO international anti-SARS-Cov-2 antibody standard which assigns a value of 1000 Binding Antibody Units for the Spike, RBD and Nucleocapsid antigens. The lower limit of quantitation for wild type spike and RBD were 1.09 and 145 BAU/ml respectively. As no standards exist for the variants, the internal standard used was evaluated and adjusted for the variant antigens based on the binding signal obtained. The MSD assay was also used to evaluate ACE2 Receptor inhibition against the relevant antigens highlighted above as previously described [26].
Neutralization was measured in a validated assay that utilized lentiviral particles pseudotyped with SARS-CoV-2 spike and containing a firefly luciferase (Luc) reporter gene for quantitative measurements of infection by relative luminescence units (RLU). The assay was performed in 293 T/ACE2.MF provided by Drs. Michael Farzan and Huihui Mu. Pseudoviruses were prepared in HEK293T/17 cells and titrated for infectivity in 293 T/ACE2.MF cells as described [28]. For measurements of neutralization, a pre-titrated dose of pseudovirus (Wuhan-1 spike containing D614G) or B.1.617.2 (AY.3)
were incubated with 8 serial 5-fold dilutions of serum samples (1:20 starting dilution) in duplicate in a total volume of 150 µl for 1 hr at 37oC in 96-well flat-bottom poly-L-lysine-coated culture plates. 293 T/ACE2-MF cells were detached from T75 culture flasks using TrypLE Select Enzyme solution, suspended in growth medium (100,000 cells/ml) and immediately added to all wells (10,000 cells in 100 µl of growth medium per well). One set of 8 wells received cells + virus (virus control) and another set of 8 wells received cells only (background control). After 66–72 hrs of incubation, medium was removed by gentle aspiration and 30 µl of Promega 1X lysis buffer was added to all wells. After a 10-minute incubation at room temperature, 100 µl of Bright-Glo luciferase reagent was added to all wells. After 1–2 min, 110 µl of the cell lysate was transferred to a black/white plate. Luminescence was measured using a GloMax Navigator luminometer (Promega). Neutralization titers are the inhibitory dilution (ID) of serum samples at which RLUs were reduced by 50% (ID50) compared to virus control wells after subtraction of background RLUs. Serum samples were heat-inactivated for 30 min at 56 °C prior to assay. For calibration to the WHO International Standard 20/136, ID50 titers are multiplied by 0.242 and expressed as International Units per millilitre (IU/ml).
5.3. Statistical analysis
Geometric mean concentrations were calculated for all groups of vaccinees and groups were compared by pairwise using non-parametric rank-sum tests with correction of multiple comparisons using a family wise error adjustment. Empirical reverse cumulative distribution functions (RCDs) were estimated from the immune response datasets independently for each vaccine/regimen. The point-wise 95% confidence intervals (indicated by shaded regions in the figures) were constructed using the non-parametric “exact“ method of Hutson [29], based on fractional order statistics. To estimate a protective threshold for each regimen we imposed a model that assumes that all participants with an immune response above a specified level are protected [11]. Therefore, to estimate the protective threshold for a regimen we estimated the VE th quantile of the RCD, where VE is either the associated vaccine efficacy or effectiveness. To capture the uncertainty in both the RCD and the estimate of VE we used a double bootstrap approach; In each i th iteration of the bootstrap we sampled VEi from a normal distribution with mean and variance that reflected the published data (on a log-VE scale). Then the VEi th quantile was estimated from an RCD constructed from a resampling of the immune response data (fixed n, sampled with replacement). After 10,000 iterations the 2.5th and 97.5th percentiles were estimated from the distribution of computed thresholds providing a 95% confidence interval (indicated by vertical dashed lines in the RCD figures). For the 1-dose mRNA dataset we combined data from the Pfizer and Moderna vaccines. A weighted RCD was created such that the distribution of immune responses reflected that of the cohort of individuals studied in Pilishvili et al. [30] (23% 1-dose of Moderna and 77% 1-dose of Pfizer), since that was the associated vaccine effectiveness used in downstream analysis. To accommodate the weights, the RCD 95% CI was computed using a bias-corrected and accelerated (BCA) bootstrap as opposed to the exact method that was employed for the other regimens. The protective threshold was estimated using the double bootstrap approach described above. To derive an overall protective threshold, we used a random-effects meta-analysis of the estimated thresholds for each regimen. The model allows for each regimen to have its own true protective threshold, but provides an unbiased estimate of the geometric mean threshold given the uncertainty in estimating each individual threshold.
We conducted a population-level analysis to evaluate the association between vaccine efficacy/effectiveness (VE) and the distribution of immune responses. We call this a population-level analysis because we have estimates of VE and immune response and do not link immune responses to specific individuals in the VE studies, which is typical of individual-level analyses. The association of immune response with VE was assessed using a weighted least-squares linear regression. Both VE and immune responses were analyzed and fit after a log transformation. For the VE outcomes which were derived from relevant studies in the published literature for either wild type or variants of concern (Table S2) the weights were the inverse variances as published, which upweights studies with more precise estimates of VE. For immune response predictors we used a summary measure of the distribution of immune responses for each regimen; these summaries included estimates of: (i) geometric mean, or (ii) the proportion of participants above a fixed threshold (threshold estimated from the data using either all data or data excluding two-dose mRNA vaccines). Each of these was evaluated based on its ability to predict the observed VE, assessed using mean squared error (MSE). For the 1-dose mRNA responses each summary measure was computed by weighting the Pfizer and Moderna responses accordingly (see above). All analyses were conducted using Python3 with the numpy and statsmodels packages.
Author Contributions
All authors contributed to the design of the study, writing of the study and revision of the manuscript. Additionally, DG, JB, DZ, HZ, LW were responsible for sample collection, MJ, AH, CB, DM and XS were responsible for laboratory evaluation of samples and AFG for statistical analysis.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: [Dr. Plotkin consults for Janssen and Moderna; Dr. Siber reports personal fees and other from Clover Biopharmaceuticals, personal fees from AdVaccine, other from Vaxxinity personal fees from CanSino, personal fees from CureVac, personal fees from Valneva, personal fees from Vaxart, personal fees and other from Affinivax, outside the submitted work; Dr. Ambrosino reports personal fees from Vaxxinity, personal fees and other from Clover Biopharmaceuticals, outside the submitted work. Dr. Montefiori’s laboratory receives funding from Moderna for clinical sample testing].
Acknowledgments
Jim Wilbur of MesoScaleDiscovery for technical assay support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.vaccine.2021.12.006.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Khoury D.S., Cromer D., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A., et al. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat Med. 2021;27(7):1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- 2.Earle K.A., Ambrosino D.M., Fiore-Gartland A., Goldblatt D., Gilbert P.B., Siber G.R., et al. Evidence for antibody as a protective correlate for COVID-19 vaccines. Vaccine. 2021;39(32):4423–4428. doi: 10.1016/j.vaccine.2021.05.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Plotkin S.A. Correlates of protection induced by vaccination. Clin Vaccine Immunol. 2010;17(7):1055–1065. doi: 10.1128/CVI.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen R.T., Markowitz L.E., Albrecht P., Stewart J.A., Mofenson L.M., Preblud S.R., et al. Measles antibody: reevaluation of protective titers. J Infect Dis. 1990;162(5):1036–1042. doi: 10.1093/infdis/162.5.1036. [DOI] [PubMed] [Google Scholar]
- 5.Woudenberg T., van Binnendijk R., Veldhuijzen I., Woonink F., Ruijs H., van der Klis F., et al. Additional Evidence on Serological Correlates of Protection against Measles: An Observational Cohort Study among Once Vaccinated Children Exposed to Measles. Vaccines (Basel) 2019;7(4):158. doi: 10.3390/vaccines7040158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldschneider I., Gotschlich E.C., Artenstein M.S. Human immunity to the meningococcus. I. The role of humoral antibodies. J Exp Med. 1969;129:1307–1326. doi: 10.1084/jem.129.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gilbert P.B., Montefiori D.C., McDermott A., Fong Y., Benkeser D.C., Deng W., et al. Immune Correlates Analysis of the mRNA-1273 COVID-19 Vaccine Efficacy Trial. medRxiv. 2021 doi: 10.1101/2021.08.09.21261290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng S., Phillips D.J., White T., Sayal H., Aley P.K., Bibi S., et al. Correlates of protection against symptomatic and asymptomatic SARS-CoV-2 infection. Nat Med. 2021;27(11):2032–2040. doi: 10.1038/s41591-021-01540-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siber G.R. Methods for estimating serological correlates of protection. Dev Biol Stand. 1997;89:283–296. [PubMed] [Google Scholar]
- 10.Andrews N., Borrow R., Miller E. Validation of serological correlate of protection for meningococcal C conjugate vaccine by using efficacy estimates from postlicensure surveillance in England. ClinDiagnLab Immunol. 2003;10(5):780–786. doi: 10.1128/CDLI.10.5.780-786.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siber G.R., Chang I.h., Baker S., Fernsten P., O’Brien K.L., Santosham M., et al. Estimating the protective concentration of anti-pneumococcal capsular polysaccharide antibodies. Vaccine. 2007;25(19):3816–3826. doi: 10.1016/j.vaccine.2007.01.119. [DOI] [PubMed] [Google Scholar]
- 12.Jódar L., Butler J., Carlone G., Dagan R., Goldblatt D., Käyhty H., et al. Serological criteria for evaluation and licensure of new pneumococcal conjugate vaccine formulations for use in infants. Vaccine. 2003;21(23):3265–3272. doi: 10.1016/s0264-410x(03)00230-5. [DOI] [PubMed] [Google Scholar]
- 13.Gundlapalli A.V., Salerno R.M., Brooks J.T., Averhoff F., Petersen L.R., McDonald L.C., et al. SARS-CoV-2 Serologic Assay Needs for the Next Phase of the US COVID-19 Pandemic Response. Open Forum Infect Dis. 2021;8:ofaa555. doi: 10.1093/ofid/ofaa555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kristiansen P.A., Page M., Bernasconi V., Mattiuzzo G., Dull P., Makar K., et al. WHO International Standard for anti-SARS-CoV-2 immunoglobulin. Lancet. 2021;397(10282):1347–1348. doi: 10.1016/S0140-6736(21)00527-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Case J.B., Rothlauf P.W., Chen R.E., Liu Z., Zhao H., Kim A.S., et al. Neutralizing Antibody and Soluble ACE2 Inhibition of a Replication-Competent VSV-SARS-CoV-2 and a Clinical Isolate of SARS-CoV-2. Cell Host Microbe. 2020;28(3):475–485.e5. doi: 10.1016/j.chom.2020.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tan C.W., Chia W.N., Qin X., Liu P., Chen M.-C., Tiu C., et al. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2-spike protein-protein interaction. Nat Biotechnol. 2020;38(9):1073–1078. doi: 10.1038/s41587-020-0631-z. [DOI] [PubMed] [Google Scholar]
- 17.Jurgens G. Low Dose Regimens of BNT162b2 mRNA Vaccine Exceed SARS-Cov-2 Correlate of Protection Estimates for Symptomatic Infection, in those 19–55 Years of Age. medRxiv. 2021 doi: 10.1101/2021.03.06.21253058. [DOI] [Google Scholar]
- 18.Feng S., Phillips D.J., White T., Sayal H., Aley P.K., Bibi S., et al. Correlates of protection against symptomatic and asymptomatic SARS-CoV-2 infection. medRxiv. 2021 doi: 10.1101/2021.06.21.21258528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahase E. Covid-19: Longer interval between Pfizer doses results in higher antibody levels, research finds. BMJ. 2021;374:n1875. doi: 10.1136/bmj.n1875. [DOI] [PubMed] [Google Scholar]
- 20.Brewer R.C., Ramadoss N.S., Lahey L.J., Robinson W.H., Lanz T.V. BNT162b2 Vaccine Induces Divergent B cell responses to SARS-CoV-2 S1 and S2. medRxiv. 2021 doi: 10.1101/2021.07.20.21260822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ambrosino D., Han H.H., Hu B., Liang J., Clemens R., Johnson M., Siber G., Goldblatt D. Immunogenicity of SCB-2019 Coronavirus Disease 2019 Vaccine Compated With 4 Approved Vaccines. J Infect Dis. 2021:jiab574. doi: 10.1093/infdis/jiab574. PMID: 34888662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bar-On Y.M., Goldberg Y., Mandel M., Bodenheimer O., Freedman L., Kalkstein N., et al. Protection of BNT162b2 Vaccine Booster against Covid-19 in Israel. N Engl J Med. 2021 doi: 10.1056/NEJMoa2114255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bergwerk M., Gonen T., Lustig Y., Amit S., Lipsitch M., Cohen C., et al. Covid-19 Breakthrough Infections in Vaccinated Health Care Workers. N Engl J Med. 2021;385(16):1474–1484. doi: 10.1056/NEJMoa2109072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carreño J.M., Alshammary H., Singh G., Raskin A., Amanat F., Amoako A., et al. Reduced neutralizing activity of post-SARS-CoV-2 vaccination serum against variants B.1.617.2, B.1.351, B.1.1.7+E484K and a sub-variant of C.37. medRxiv. 2021 doi: 10.1101/2021.07.21.21260961. [DOI] [Google Scholar]
- 25.Goldblatt D., Johnson M., Falup-Pecurariu O., Ivaskeviciene I., Spoulou V., Tamm E., et al. Cross-sectional prevalence of SARS-CoV-2 antibodies in healthcare workers in paediatric facilities in eight countries. J Hosp Infect. 2021;110:60–66. doi: 10.1016/j.jhin.2020.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson M., Wagstaffe H.R., Gilmour K.C., Mai A.L., Lewis J., Hunt A., et al. Evaluation of a novel multiplexed assay for determining IgG levels and functional activity to SARS-CoV-2. J Clin Virol. 2020;130:104572. doi: 10.1016/j.jcv.2020.104572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krammer F., Srivastava K., Alshammary H., Amoako A.A., Awawda M.H., Beach K.F., et al. Antibody Responses in Seropositive Persons after a Single Dose of SARS-CoV-2 mRNA Vaccine. N Engl J Med. 2021;384(14):1372–1374. doi: 10.1056/NEJMc2101667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen X., Tang H., McDanal C., Wagh K., Fischer W., Theiler J., et al. SARS-CoV-2 variant B.1.1.7 is susceptible to neutralizing antibodies elicited by ancestral spike vaccines. Cell Host Microbe. 2021;29(4):529–539.e3. doi: 10.1016/j.chom.2021.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hutson A.D. Calculating nonparametric confidence intervals for quantiles using fractional order statistics. J Appl Stat. 1999;26(3):343–353. [Google Scholar]
- 30.Pilishvili T., Fleming-Dutra K.E., Farrar J.L., Gierke R., Mohr N.M., Talan D.A., et al. Interim Estimates of Vaccine Effectiveness of Pfizer-BioNTech and Moderna COVID-19 Vaccines Among Health Care Personnel — 33 U.S. Sites, January–March 2021. MMWR Morb Mortal Wkly Rep. 2021;70(20):753–758. doi: 10.15585/mmwr.mm7020e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.