MRE11 activates mitochondrial DNA–dependent cGAS immune signaling in cells with BRCA/FANC gene defects.
Abstract
Mitochondrial DNA (mtDNA) instability activates cGAS-dependent innate immune signaling by unknown mechanisms. Here, we find that Fanconi anemia suppressor genes are acting in the mitochondria to protect mtDNA replication forks from instability. Specifically, Fanconi anemia patient cells show a loss of nascent mtDNA through MRE11 nuclease degradation. In contrast to DNA replication fork stability, which requires pathway activation by FANCD2-FANCI monoubiquitination and upstream FANC core complex genes, mitochondrial replication fork protection does not, revealing a mechanistic and genetic separation between mitochondrial and nuclear genome stability pathways. The degraded mtDNA causes hyperactivation of cGAS-dependent immune signaling resembling the unphosphorylated ISG3 response. Chemical inhibition of MRE11 suppresses this innate immune signaling, identifying MRE11 as a nuclease responsible for activating the mtDNA-dependent cGAS/STING response. Collective results establish a previously unknown molecular pathway for mtDNA replication stability and reveal a molecular handle to control mtDNA-dependent cGAS activation by inhibiting MRE11 nuclease.
INTRODUCTION
Mitochondria, the organelle best known for functioning in cellular bioenergetics, are key signaling hubs for intracellular communication (1). A given cell contains 2 to 1000 mitochondria, with each harboring one or multiple circular ~16-kb genomes (2). Autonomously from nuclear DNA, mitochondrial DNA (mtDNA) is replicated by the DNA polymerase POLγ but involves many other factors including TFAM, a multifunctional high-mobility group protein that bends and compacts DNA, and assists transcription and RNA priming during replication (3). Mutations in mitochondrial replisome components cause mitochondrial disease, and mtDNA instability causes diverse physiological manifestations including premature aging, abnormal cellular energetics, and developmental abnormalities (3). More recently, mtDNA has emerged as a prominent inducer of inflammation as a damage-associated molecular pattern (DAMP), which can activate, so far, three known inflammatory signaling pathways (4): the endosomal Toll-like receptor–dependent response; the inflammasome, which is activated by aberrant mitophagy and increased newly synthesized mtDNA (5, 6); and the cyclic guanosine monophosphate–adenosine monophosphate synthase/stimulator of interferon response cGAMP interactor 1 (cGAS/STING) pathway, which activates type I interferon–stimulated gene (ISG) expression with (7) or without activating interferons themselves, such as seen with TFAM heterozygosity (8, 9).
Many of the mitochondrial disease–linked phenotypes are overlapping with those in Fanconi anemia (FA), a genetically diverse disorder that leads to bone marrow failure, strong cancer susceptibility, accelerated aging, congenital deformations, abnormal glucose metabolism, and chronically activated inflammation. The FA pathway consists of 23 FA suppressor genes (FANC) genes identified to date that promote nuclear genome stability by DNA interstrand cross-link repair involving homologous recombination (10). FANC genes including BRCA2/FANCD1 and RAD51C/FANCO also promote DNA replication fork protection, which is a more recently identified genome stability pathway that suppresses nascent DNA degradation by MRE11 (11–13). Pathway activation requires monoubiquitination of FANCD2-FANCI by FANC core complex proteins, which is a pivotal key step essential to both nuclear DNA repair and replication fork protection (10, 12).
Substantial evidence links FANC to unresolved oxidative stress response functions critical to the protection against bone marrow failure and cancer (14, 15). Because mitochondria are the site of aerobic respiration, mtDNA is prone to oxidative DNA damage, and the presence of FANC and other FANC pathway–associated proteins including MRE11 in the mitochondria and on mtDNA is established (6, 16–22). However, in contrast to the nucleus, DNA repair pathway employment aside from base excision repair in the mitochondria is controversial (23, 24). mtDNA stability is additionally regulated at the level of mtDNA replication with the exonuclease activity of POLγ polymerase and MGME1 nuclease degrading damaged mtDNA rather than by repair pathways mending it. Defects in mitochondrial genome maintenance exonuclease 1 (MGME1) promote recombination that causes deletion mutations (25). Thus, in mitochondria, canonical nuclear DNA repair pathways such as homologous recombination can also cause instability rather than suppressing it, rendering the presence and molecular role of FANC proteins in mitochondria enigmatic.
In investigating FANC gene functions during mtDNA replication, we here show that these genes are critical for the protection of nascent mtDNA replication forks upon oxidative damage, unveiling an unanticipated molecular mechanism for mitochondrial genome stability. We achieved this by developing single-cell replication assays for detecting mtDNA replication reactions that allow quantitative in situ molecular studies of mtDNA replication. Without FANC mtDNA fork protection, newly replicated mtDNA fragmentation caused by MRE11 nuclease activates cGAS-dependent innate immune signaling, thus uncovering MRE11 as a major nuclease responsible for mitochondria-dependent cGAS activation of ISGs.
RESULTS
Mitochondrial replication assay detects nascent mtDNA in situ
RAD51C is present within mitochondria, associates with replicating mtDNA, and controls mitochondrial genome copy number by unknown mechanisms (20, 26, 27). We therefore sought to better understand RAD51C functions during nascent mtDNA replication using FANCO/RAD51C patient–derived mutant skin fibroblasts (SH2038; which contain inactivating, biallelic RAD51C R258H mutations) and SH2038 cells stably complemented with wild-type RAD51C (RAD51C+) (28). mtDNA is replicated primarily by POLγ within ~1 to 2 hours (29, 30). Both 5′-bromo-2′-deoxyuridine (BrdU) and 5′-ethynyl-2′-deoxyuridine (EdU) are used to label mtDNA, traditionally using prolonged exposure. However, incorporation of these nucleoside analogs for 1 hour, which is a sufficiently short label time to monitor nascent mtDNA replication, does not produce signals suited for efficient and accurate quantitative analysis (fig. S1, A and B).
We therefore sought to develop an unambiguously quantifiable yet sensitive nascent in situ mtDNA replication assay (MIRA; Fig. 1A). By taking advantage of proximity ligation assay [PLA (31, 32)] between biotinylated EdU moieties for signal amplification (Fig. 1A), EdU signals with 1-hour labeling are visible outside the nucleus by conventional fluorescence microscopy (fig. S1C). Similar to conventional EdU and BrdU labeling, this method also visualizes nuclear DNA. However, only cytoplasmic signals are considered for the MIRA analysis, which are present irrespective of the cell cycle state and do not form without EdU (fig. S1, D and E).
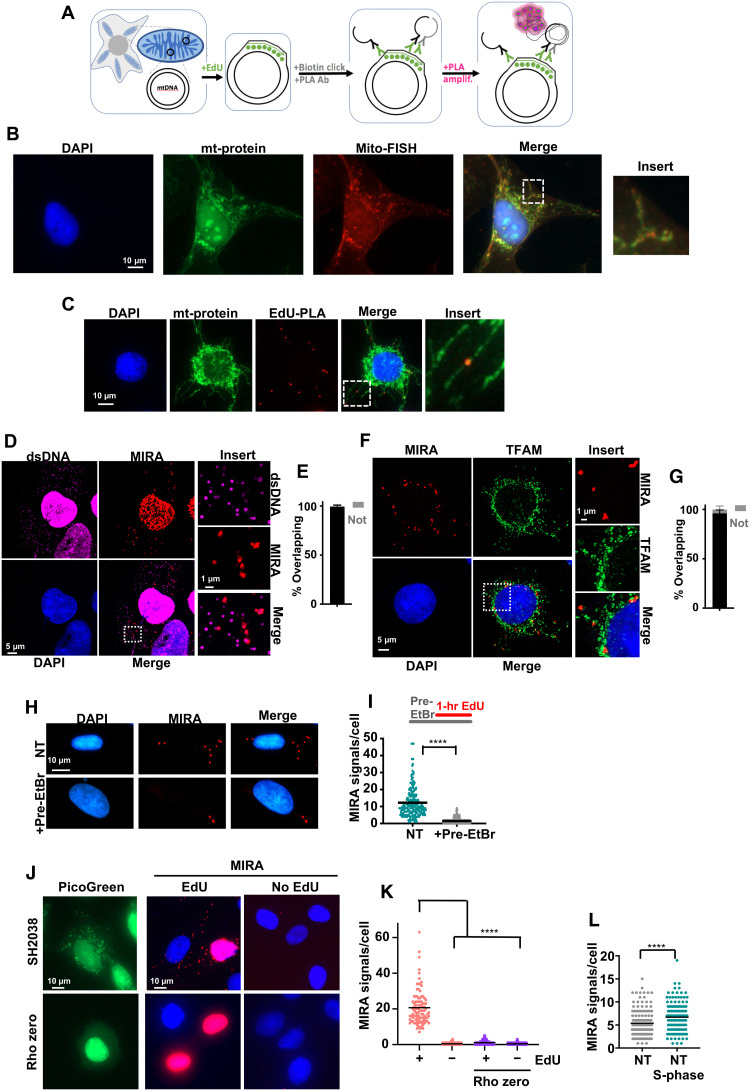
Fig. 1. MIRA detects nascent mtDNA.
(A) Schematic of MIRA. Incorporated EdU (green) is detected by PLA amplification (pink circles). Ab, antibody. (B) Representative image of wild-type RAD51C-complemented FA patient cells (RAD51C+) cells with antibody stain against mitochondria (mt-protein) and mito-FISH probes. (C) Representative images of MIRA signals in RAD51C+ cells with antibody stain against mitochondria (mt-protein). (D) Airyscan superresolution image of MIRA signals in RAD51C+ cells with antibody stain against double-stranded DNA (dsDNA). (E) Quantification of (D) MIRA signals overlapping (black bar) or not overlapping (gray bar) with dsDNA. (F) Airyscan superresolution image of MIRA signals in RAD51C+ cells with antibody stain against TFAM. (G) Quantification of (F) MIRA signals overlapping (black) or not (gray) with TFAM. (H) Representative images of MIRA in RAD51C+ cells with and without ethidium bromide (EtBr; 50 ng/ml) before and during EdU labeling. NT, no treatment. (I) Quantification of (H). Top: Experimental scheme. (J) Representative images of MIRA (red) in rho zero SH2038 cells with PicoGreen DNA stain (green). (K) Quantification of (J). (L) Quantification of MIRA signals in SH2038 S-phase cells (green) and non–S-phase cells (gray). Bars represent the mean of compiled data from biological repeats. P values are derived using the Mann-Whitney test. ****P < 0.0001.
mtDNA localizes near mitochondrial membrane proteins as seen by the membrane signals overlapping with those of fluorescence in situ hybridization (FISH) probes against mtDNA (Fig. 1B), similar to the cytoplasmic EdU-PLA signals (Fig. 1C and fig. S1F). Note that fluorescent signals of costains are fainter when overlapping with MIRA signals, consistent with the increased space occupancy of the PLA-DNA rolling circle amplifications. Using superresolution Airyscan imaging, MIRA signals further overlap with cytoplasmic double-stranded DNA (dsDNA) markers, which detect mitochondrial nucleoids, and with TFAM, the mitochondrial transcription factor (Fig. 1, D to G), consistent with the MIRA detecting nascent mtDNA.
Dideoxycytidine (ddC) or low concentrations of ethidium bromide (EtBr), either of which inhibit mtDNA replication but not nuclear DNA replication (33, 34), added 30 min before and during EdU incorporation markedly reduces the MIRA signals, suggesting that MIRA detects active EdU incorporation into mtDNA (Fig. 1, H and I, and fig. S1, G and H). No appreciable MIRA signals are produced in SH2038 cells fully depleted of mtDNA (rho zero) similar to omitting EdU (Fig. 1, J and K). Cytoplasmic MIRA signals are furthermore significantly increased in S-phase cells compared to non–S-phase SH2038 cells (Fig. 1L), consistent with increased mtDNA replication reported to occur during the S phase of the cell cycle (35). Collectively, these data confirm the suitability for MIRA to detect newly replicating mtDNA.
Nascent mtDNA is lost in FA cells with mtDNA replication stress
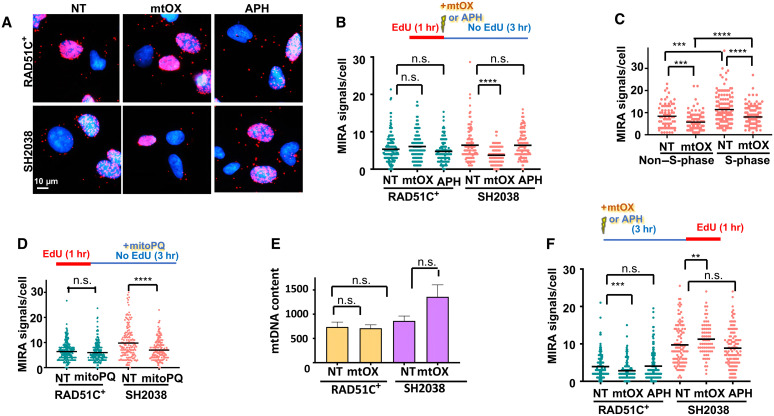
Mitochondria are the location for oxidative phosphorylation and therefore are exposed to high oxidative stress and damage, which potently stalls POLγ and mtDNA replication forks (36). To exogenously induce this lesion in mtDNA, we made use of mtOX, a small molecule targeted exclusively to the mitochondria that induces oxidative damage after light activation (fig. S2, A and B) (37). We labeled cells with EdU for 1 hour before activating mtOX and immediately replaced the medium after mtOX activation to remove the EdU (Fig. 2B, sketch). Thereby, only the nascent mtDNA that was replicated before stalling with mtOX is labeled and not the mtDNA synthesized during or after damage induction. In RAD51C+ cells, the number of mitochondrial MIRA signals remains unchanged 3 hours after mtOX activation comparable to undamaged cells (Fig. 2, A and B). In stark contrast, in FANCO/RAD51C-defective SH2038 cells, MIRA signals decrease significantly after mtOX damage activation, suggesting a loss of nascent mtDNA with oxidative damage in the patient cells. These results hold true for both S-phase and non–S-phase SH2038 cells (Fig. 2C), confirming that the MIRA signal changes are independent of nuclear DNA replication. In addition, EdU incorporation in the nucleus is not reduced with mtOX (fig. S2C) in both cell types, further supporting mitochondria-specific targeting of mtOX. Moreover, MIRA signals are unchanged with exposure to aphidicolin (APH), a chemical inhibitor of nuclear POLα/primase, which is absent in the mitochondria and causes nuclear but not mtDNA replication stalling (38), confirming that the observed replication phenotypes are independent of nuclear replication stress (Fig. 2, A and B, and fig. S2, D and E). As an alternative to mtOX, we used the reagent MitoPQ with similar results (Fig. 2D), another agent specifically targeted to mitochondria that interacts with mitochondrial complex I (39) and, so, causes oxidative damage without the need for light activation. EdU in these MIRA reactions solely marks the nascent DNA before mtDNA replication stalling, and it is not possible to measure replication restart or replication stalling using this labeling scheme. Thus, the data show that RAD51C-deficient FA patient cells exhibit a loss of nascent mtDNA that was replicated just before the exposure to oxidative stress in the mitochondria. To assess whether parental mtDNA is lost, we measured total mtDNA copy numbers. We found that, while statistically insignificant, SH2038 cells showed a consistently increased rather than decreased number of mtDNA copies after treatment with mtOX (Fig. 2E). We reasoned that whether mtDNA replicated before the damage is lost and mtDNA copy numbers increase, the nascent mtDNA loss may be compensated for by increased mtDNA synthesis after the damage. Consistently, labeling mtDNA replication with EdU after mtOX treatment resulted in increased MIRA signals in SH2038, but not with aphidicolin and/or in RAD51C+ cells (Fig. 2F).
Fig. 2. Replication stress causes loss of nascent mtDNA in FANCO/RAD51C patient cells.
(A) Representative images of MIRA with EdU label incorporated immediately before treatment with mtOX (4 μM), aphidicolin (APH; 7 μM), or no treatment. (B) Scatterplot quantification of (A). Top: Experimental sketch. (C) Scatterplot quantification of MIRA signals in S-phase cells and non–S phase in SH2038 cells following the experimental schemes in (B). (D) Scatterplot quantification of MIRA with or without treatment with MitoPQ (10 μM) for 3 hours after EdU incubation. Top: Experimental sketch. (E) mtDNA copy numbers measured by quantitative polymerase chain reaction (qPCR) without treatment, with mtOX, or with aphidicolin. (F) Scatter dot plot quantification MIRA with EdU labeled incorporated 3 hours after treatment with mtOX or aphidicolin, respectively. Top: Experimental sketch. Bars represent the mean of compiled data from biological repeats. P values for scatter dot blots are derived using the Mann-Whitney test and, for bar graph (mtDNA content), by Student’s t test. n.s., not significant; ****P < 0.0001, ***P < 0.001, and **P < 0.01.
MRE11 causes degradation of mtDNA
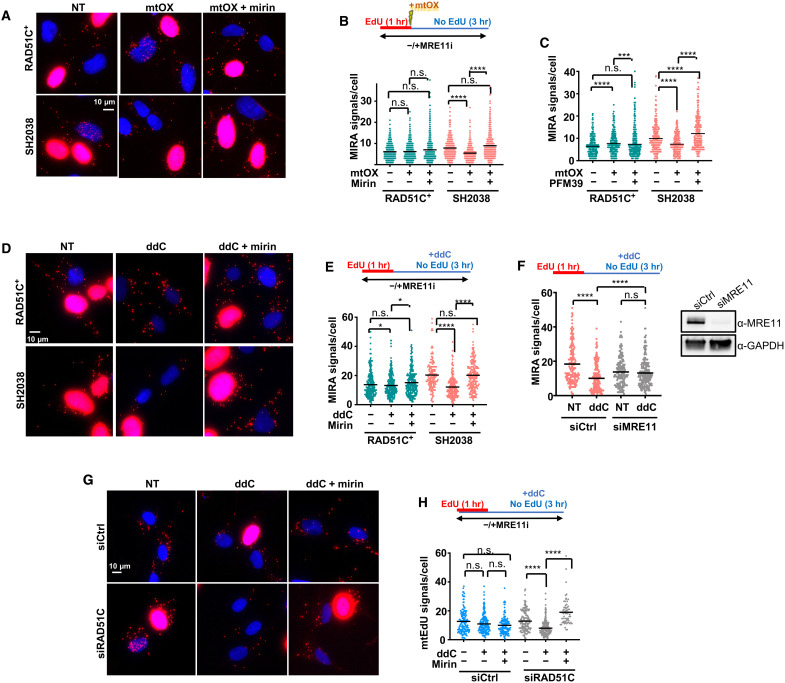
In the nucleus, RAD51C is canonically involved in DNA break repair. However, there is no measurable linearization of the mtDNA with mtOX, indicative of breaks that could cause the mtDNA loss (fig. S3, A to C). RAD51C, as do other FANC proteins, also stabilizes stalled replication forks to protect them from nucleolytic degradation by MRE11 (11, 12, 40, 41), a nuclease that localizes to mitochondria and mtDNA (17, 18). We therefore tested MRE11 activity in the MIRA (Fig. 3, A and B) and found that the addition of a chemical inhibitor of MRE11, mirin, restores the nascent MIRA signals in SH2038 cells. We confirmed these results with an additional, specific MRE11 inhibitor, PFM39 (Fig. 3C and fig. S4A) (42), suggesting that RAD51C protects nascent mitochondrial replication forks from MRE11-dependent degradation.
Fig. 3. MRE11 nuclease causes mtDNA degradation in FANCO/RAD51C patient cells.
(A) Representative images of MIRA with and without with mtOX (4 μM) and chemical inhibition of MRE11 nuclease with mirin (50 μM). Top: Experimental sketch. (B) Scatter dot plot quantification of (A), MIRA signals. (C) Scatter dot plot quantification of MIRA with and without mtOX and inhibition of MRE11 nuclease with PFM39 (100 μM), following the experimental schemes in (B). (D) Representative images of a MIRA with and without mitochondrial replication stalling with ddC (20 μM) in the presence or absence of mirin (50 μM). Top: Experimental sketch. (E) Scatter dot plot quantification of (D). (F) Scatter dot plot quantification of MIRA signals in SH2038 cells with MRE11 small interfering RNA (siRNA) knockdown (siMRE11) or control siRNA (siCtrl). Top: Experimental sketch. Inset: Western blot analysis of the MRE11 knockdown. GAPDH, glyceraldehyde phosphate dehydrogenase. (G) Representative images of a MIRA in BJ fibroblast with siRNA knockdown of RAD51C (siRAD51C) or siCtrl. Top: Experimental sketch. (H) Scatter dot plot quantification of (G). Inset: Western blot analysis of the RAD51C knockdown. Bars represent the mean of compiled data from biological repeats. P values are derived using the Mann-Whitney test. ****P < 0.0001, ***P < 0.001, and *P < 0.05.
Similar to mtOX exposure, the nascent mtDNA MIRA signals are reduced in the SH2038 patient cells but not RAD51C+ cells when ddC is added after the EdU labeling to stall replication (Fig. 3, D and E). ddC selectively stalls mtDNA replication (34), suggesting nascent mtDNA loss with mitochondrial replication stalling induced also by other stresses than oxidative damage. Note that, in this experimental setup, ddC is added solely after but not before or during EdU incorporation. Thus, changes in MIRA signals measure the potential loss of nascent mtDNA that was synthesized before stalling but not replication stalling or inhibition of EdU incorporation with or after addition of ddC. The observed loss in MIRA signals is rescued when MRE11 is chemically inhibited by mirin or by knockdown of MRE11 with small interfering RNA (siRNA) (Fig. 3, D to F). In contrast, the nascent DNA in the nucleus is not reduced with ddC (fig. S4B), confirming mitochondrial specificity of the agent and MRE11 nuclease activity as the cause for the reduction in nascent mtDNA. A similar trend in loss of mtDNA signals with ddC replication stalling is observed when labeling nascent mtDNA with BrdU (fig. S4, C and D) or EdU directly attached to Oregon Green azide by click chemistry and anti–Oregon Green antibodies (fig. S4, E and F) as an alternative signal amplification to PLA. While confirming the results observed by MIRA, these alternative methods show high signal background and are cumbersome to analyze in comparison to MIRA. Last, BJ fibroblast cells with RAD51C knockdown, but not with control siRNA, show a significant loss of mtDNA upon replication stalling with ddC that is rescued by inhibition of MRE11 (Fig. 3, G and H), confirming that the requirement of mtDNA fork protection is not restricted to FANC cells. Collectively, the data establish that RAD51C is necessary to protect mtDNA forks from excessive MRE11 processing.
mitoSIRF detects proteins bound to nascent mtDNA
We further sought to test whether these otherwise nuclear proteins directly bind to and act at the level of nascent mtDNA replication forks. We expanded from our recently described SIRF (in situ analysis of protein interactions at DNA replication forks) method for single-cell protein interactions with nascent replication forks (43) and adapted it for nascent mtDNA (mitoSIRF; Fig. 4). Briefly, protein interactions with nascent mtDNA are detected by PLA with primary antibodies against the biotinylated EdU and the protein of interest, so that a productive PLA signal is produced only when the protein is localized within 40 nm of the nascent mtDNA.
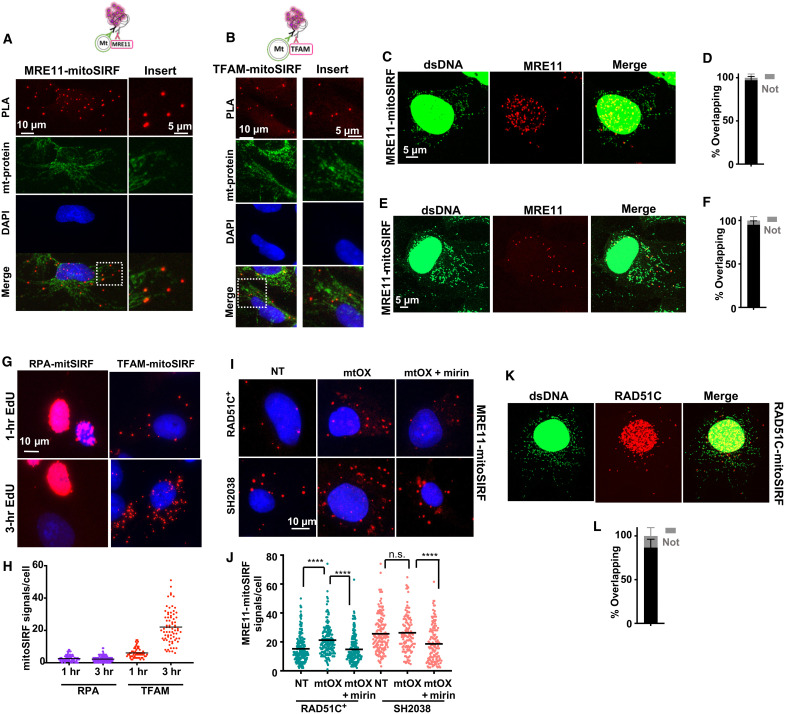
Fig. 4. mitoSIRF detects proteins bound to nascent mtDNA.
(A and B) Representative images of MRE11-mitoSIRF assay or TFAM-mitoSIRF assay (sketch to the left) in SH2038 cells with antibody stain against mitochondria (mt-protein). Only cytoplasmic signals are considered. Note that fluorescent signals of costains are fainter when overlapping with MIRA signals, consistent with spatial co-occupancy. (C) Airyscan superresolution image of SH2038 cells with immunofluorescent stain against dsDNA and MRE11-mitoSIRF signals. (D) Quantification of MRE11-mitoSIRF signals overlapping (black) or not overlapping (gray) with dsDNA signals. (E) Airyscan superresolution image of MRE11-mitoSIRF in BJ fibroblasts with antibody stain against dsDNA. (F) Quantification of (E) MRE11-mitoSIRF signals overlapping (black) or not (gray) with dsDNA. (G) Representative images of RPA-mitoSIRF and TFAM mitoSIRF with 1 or 3 hours of EdU. (H) Quantification of (G). (I) Representative images of a MRE11-mitoSIRF 3 hours after mtOX (4 μM) treatment. Mirin (50 μM) was present throughout the experiment where indicated. (J) Quantification of (I). (K) Airyscan superresolution image of RAD51C-mitoSIRF in BJ fibroblasts with antibody stain against dsDNA. (L) Quantification of (K) RAD51C-mitoSIRF signals overlapping (black) or not (gray) with dsDNA. Bars represent the mean of compiled data from biological repeats. P values are derived using the Mann-Whitney test. ****P < 0.0001.
MRE11 previously has been reported to localize to mitochondria and nucleoids (17, 18, 44). Using PLA antibodies against human MRE11 and EdU-biotin, we readily detected MRE11-mitoSIRF signals outside the nucleus that overlap with immunofluorescence signals of mitochondrial protein in SH2038 cells (Fig. 4A), similar to what is observed for the mitochondrial protein TFAM-mitoSIRF (Fig. 4B). In addition, superresolution images showed that the MRE11-mitoSIRF signals overlapped with staining against dsDNA, a commonly used marker for mitochondrial nucleoids, in both SH2038 cells and BJ fibroblasts as an independently tested cell line (Fig. 4, C to F), but not when EdU is omitted in the reactions with antibodies against MRE11 (fig. S5, A to D). Collectively, the data suggest that MRE11 binds to nascent mtDNA. To further validate the mitoSIRF method, we performed mitoSIRF assays against the nuclear protein replication protein A (RPA). While there were strong PLA signals in the nucleus of S-phase cells, RPA-mitoSIRF signals outside the nucleus were negligible (Fig. 4, G and H). In contrast, mitoSIRF against the mitochondrial TFAM protein resulted in strong signals outside the nucleus irrespective of cell cycle status and increased robustly with time of EdU incorporation (Fig. 4, G and H), validating that mitoSIRF detects protein interactions with mtDNA.
We observed significantly more MRE11-mitoSIRF signals with mtOX replication stalling in the RAD51C+ cells compared to without mtOX, suggesting increased MRE11 recruitment to damaged mtDNA (Fig. 4, I and J). Overall, there were more MRE11-mitoSIRF signals in the FANCO/RAD51C patient variant cells SH2038 with and without mtOX compared to RAD51C+ cells. The fact that the MIRA shows an MRE11-dependent decrease in nascent DNA in SH2038 cells with mtOX (compare to Fig. 2B for the amount of EdU present) suggests that the number of MRE11-mitoSIRF signals with mtOX in these cells could be an underestimation as the EdU necessary for signal formation may have been lost or, alternatively, MRE11 may have already dissociated. In both cell types, the signals are decreased with mirin, which chemically inhibits MRE11 binding to DNA (42, 45). Together, the data suggest that MRE11 functionally interacts with and is recruited to nascent mtDNA replication forks, whereby MRE11 is more abundantly associated with stalled mitochondrial replication forks or when RAD51C is defective, e.g., in the presence of patient FA mutant protein.
RAD51C has recently been reported to associate with stalled mtDNA forks (27). Consistently, we detect elevated RAD51C-mitoSIRF signals in RAD51C+ cells after mtOX treatment (fig. S5, E to G). To confirm nascent mtDNA localization in cells with endogenous RAD51C expression, we analyzed BJ cells and find RAD51C-mitoSIRF signals overlapping with dsDNA (Fig. 4, K and L). No RAD51C-mitoSIRF signals are observed without EdU (fig. S5H). Together, the data confirm the physical interaction of otherwise nuclear proteins with nascent mtDNA, including MRE11 and RAD51C, suggesting a direct protein action in the mitochondria.
Mitochondrial replication fork protection is genetically separable from nuclear DNA fork protection
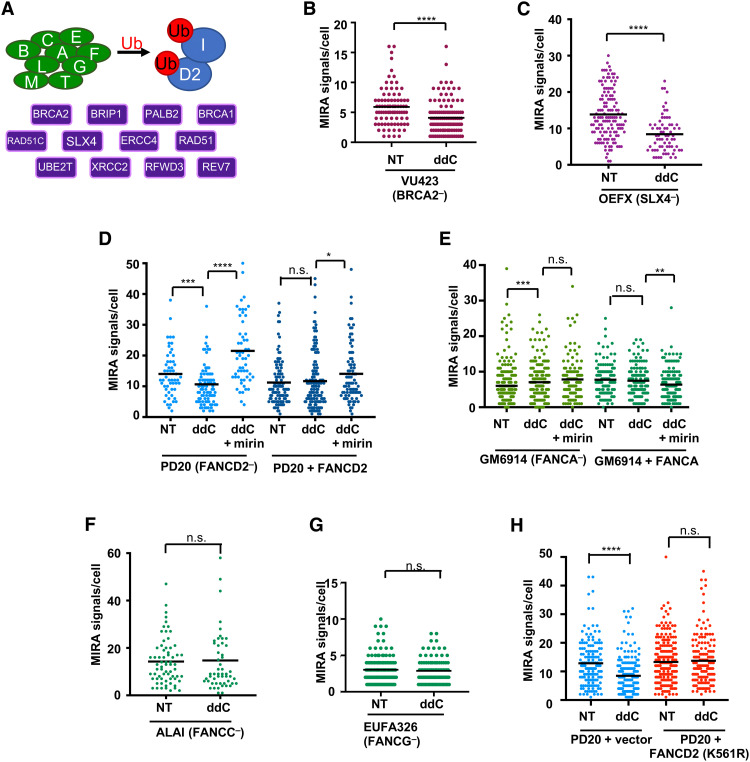
Aside from RAD51C, other FANC proteins have been reported to localize to mitochondria, including BRCA2/FANCD1, RAD51/FANCR, FANCD2, FANCM, FANCG, and FANCC (6, 16, 17, 20–22). We therefore tested additional FA patient cell lines using the MIRA to test whether mitochondrial fork protection requires the FANC pathway. Central to this pathway is its activation by FANCD2 monoubiquitination, which requires FANC core complex proteins and is essential for both nuclear fork protection and nuclear DNA repair (Fig. 5A). It further involves components downstream of FANCD2 including RAD51C/FANCO and BRCA2/FANCD1, as well as SLX4/FANCP. Similar to RAD51C/FANCO, BRCA2/FANCD1-defective FA patient cells (VU423) show a significant reduction in MIRA signals when mtDNA forks are stalled by ddC (Fig. 5B; treatment scheme as in Fig. 3F, sketch). Likewise, patient cells defective for the downstream FA pathway member SLX4/FANCP exhibit a significant mitochondrial fork protection defect tested independently in three separate patient-derived cell lines (OEFX, OERN, and OELA cells; Fig. 5C and fig. S6, A and B). FANCD2 is also required, whereby MRE11 inhibition with mirin can rescue the loss of MIRA signals in FANCD2-defective PD20 patient cells, while there is no loss in PD20 cells complemented with wild-type FANCD2 (Fig. 5D).
Fig. 5. FANCD2 monoubiquitination is dispensable for mtDNA fork protection.
(A) Schematic of FA pathway; core complex gene products (green), required for monoubiquitination (red, “Ub”) of FANCD2/FANCI (blue); and downstream gene products (purple). (B) Quantification of MIRA signals with and without ddC (20 μM) in BRCA2-defective VU423 patient fibroblasts. (C) Quantification of MIRA signals with and without ddC in SLX4-defective OEFX patient fibroblasts. (D) Quantification of MIRA signals with and without ddC in the presence or absence of mirin (50 μM) in FANCD2-defective PD20 patient fibroblasts and PD20 cells complemented with wild-type FANCD2 (PD20 + FANCD2). (E) Quantification of MIRA signals with and without ddC in the presence or absence of mirin (50 μM) in FANCA-defective GM6914 patient fibroblasts and GM6914 cells complemented with wild-type FANCA (GM6914 + FANCA). (F) Quantification of MIRA signals with and without ddC in FANCC-defective ALAI patient fibroblasts. (G) Quantification of MIRA signals with and without ddC in FANCG-defective EUFA326 patient fibroblasts. (H) Quantification of MIRA signals with and without ddC in FANCD2-defective PD20 patient fibroblasts and PD20 cells complemented with monoubiquitination defective mutant FANCD2 (PD20 + FANCD2 K561R). Bars represent the mean of compiled data. P values are derived using the Mann-Whitney test. ****P < 0.0001, ***P < 0.001, **P < 0.01, and *P < 0.05.
Unexpectedly and in stark contrast to nuclear fork protection (12), FANCA-defective patient cells do not exhibit nascent mtDNA degradation upon mitochondrial fork stalling, similar to wild-type FANCA-complemented cells (GM6914 cells; Fig. 5E). Moreover, cells derived from patients with defects in other FANC core complex components including FANCC (ALAI and ALDH cells; Fig. 5F and fig. S6C) and FANCG (EUFA326 cells; Fig. 5G) did not result in a mitochondrial fork protection defect, suggesting that FANC core complex proteins are dispensable in this process in the mitochondria.
To directly test whether FANCD2 monoubiquitination is required in mitochondrial fork protection, we tested FANCD2-deficient PD20 patient cells and PD20 cells expressing mutant FANCD2 K561R, which cannot be ubiquitinated (46). The data uncover that, while FANCD2-defective cells are incapable of protecting the nascent mtDNA from degradation upon mitochondrial replication stalling, expression of the ubiquitination mutant FANCD2 K561R rescues this phenotype (Fig. 5H), similar to the expression of wild-type FANCD2 (Fig. 5D). Together, the data show that, while FANCD2 and downstream FANC factors including BRCA2, RAD51C, and SLX4 are essential for mitochondrial fork protection, FANC core complex proteins and FANCD2 monoubiquitination are not. These results are in stark contrast to nuclear fork protection, which requires FANC core complex proteins and FANCD2 monoubiquitination for the protection of nuclear nascent DNA. Thus, the data reveal a functional separation of nuclear and mitochondrial fork protection.
mtDNA fork protection does not cause apparent physiological change but promotes cell proliferation
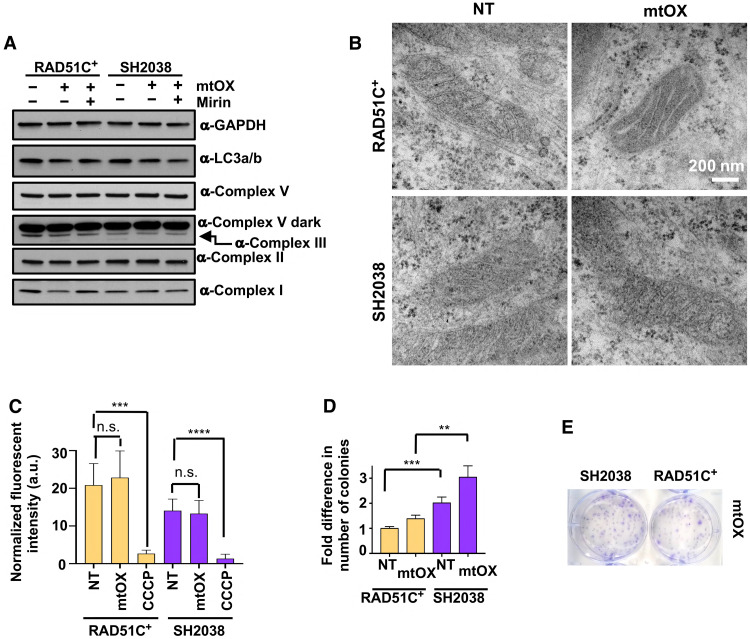
Nascent mtDNA previously has been shown to activate an inflammasome response in macrophages (5), and FANC core complex components suppress inflammasome reactions by promoting mitophagy (6). We therefore measured the expression of autophagy marker LC3, which remains unchanged in SH2038 and RAD51C+ cells with and without mtOX (Fig. 6A). There is furthermore no significant change in mitochondrial membrane structure lengths indicative of mitochondrial fragmentation (47) in either SH2038 or RAD51C+ cells with this low-dose mtOX replication stalling (fig. S7, A and B). Similarly, protein levels of members of the mitochondrial respiratory chain as another measure for mitochondrial membrane integrity remain unchanged (Fig. 6A), and electron microscope images of mitochondria with and without mtOX appeared normal in both SH2038 and RAD51C+ cells (Fig. 6B). Moreover, the mitochondrial membrane potential as an indicator of mitochondrial functional integrity was unchanged with mtOX (Fig. 6C). We detected a small but measurable increase in cellular survival with mtOX treatment (Fig. 6, D and E), consistent with an increased number of SH2038 cells in S phase after mtOX exposure (fig. S7C). Collectively, we find no evidence for mitochondrial abnormalities under the conditions measured.
Fig. 6. Mitochondria remain normal with replication fork degradation.
(A) Western blot of mitochondrial respiratory proteins and autophagy marker LC3 with or without mtOX for 3 or 24 hours and MRE11 inhibition by mirin. (B) Electron microscope images of SH2038 and RAD51C+ cells with and without mtOX treatment. (C) Bar graph quantification of mitochondrial membrane potential as measured by MitoTracker CMXRos 3 hours after treatment with mtOX (4 μM) or carbonyl cyanide m-chlorophenyl hydrazine (CCCP), used as a positive control. Error bars represent SD. a.u., arbitrary units. (D) Clonogenic survival of SH2038 FANCO patient cells and RAD51C+ cells with and without mtDNA replication stalling by mtOX, normalized to RAD51C+ without treatment. Error bars represent SEM. (E) Representative images of (C). Bars represent the mean of compiled data from biological repeats, and P values are derived using Student’s t test. ****P < 0.0001, ***P < 0.001, and **P < 0.01.
MRE11 causes cGAS-dependent ISG activation
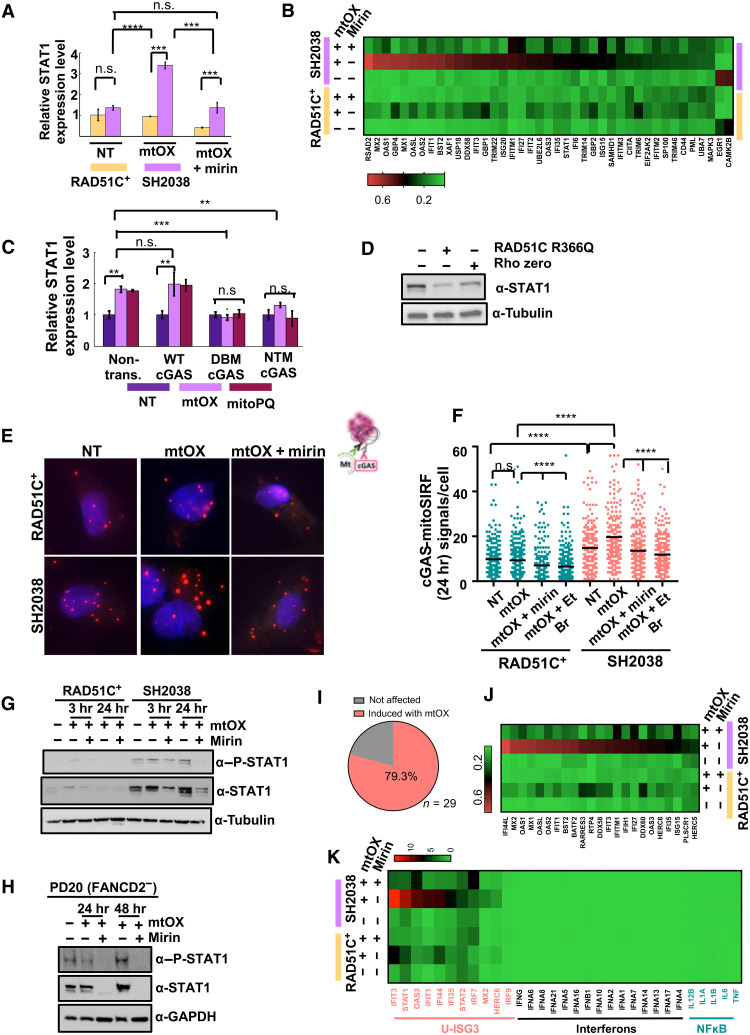
mtDNA stress can cause innate immune signaling via cytosolic mtDNA release that activates a cGAS/STING-dependent type I ISG expression via signal transducers and activators of transcription 1 (STAT1). Signaling by STAT1 is also known to promote cell proliferation (48), consistent with the increased cell survival and proliferation seen in SH2038 after mtOX (Fig. 6, D and E, and fig. S7C). We therefore measured STAT1 mRNA transcripts, which are significantly increased in SH2038 cells compared to RAD51C+ cells with replication stalling after mtOX treatment (Fig. 7A), correlating with the loss of nascent mtDNA in these cells. For more comprehensive testing, we used RNA sequencing (RNA-seq) analysis, which confirmed a strong increase in type I interferon pathway gene transcripts in FANCO/RAD51C-mutant SH2038 patient cells with mtOX exposure (Fig. 7B). Inhibition of MRE11 nuclease strongly suppresses both the induction of STAT1 and the induced ISG expression in the FA patient cells (Fig. 7, A and B), suggesting that the response greatly depends on nascent mtDNA degraded by MRE11 nuclease.
Fig. 7. MRE11 activates mtDNA-dependent cGAS immune signaling.
(A) qPCR of STAT1-mRNA in SH2038 and RAD51C+ with mtOX (4 μM) and mirin (50 μM). (B) Heatmap of type I interferon pathway gene expression in SH2038 and RAD51C+ with indicated treatments. (C) qPCR of STAT1-mRNA in SH2038 cells expressing wild-type, DNA-binding defective (C396A/C397A; “DBM”), or enzymatically inactive cGAS (G212A/S213A; “NTM”) with mtOX or MitoPQ (10 μM). (D) STAT1 Western blot in SH2038 with mtOX, expressing RAD51C R366Q protein, or depleted of mtDNA (rho zero). (E) Representative images of cGAS-mitoSIRF. (F) Quantification of (D). EtBr (50 ng/ml). (G) STAT1 and phosphorylated form of STAT1 (P-STAT1) Western blots in SH2038 and RAD51C+. (H) STAT1 and P-STAT1 Western blot in FANCD2-defective PD20. (I) Pie chart of unphosphorylated ISG3 (U-ISG3) pathway genes, statistically significantly up-regulated in SH2038 with mtOX (salmon). (J) RNA-seq data heatmap of (H). (K) RNA-seq data heatmap of ISG3 pathway (salmon), type I interferon (black), and NFκB pathway targets (turquoise) in SH2038 and RAD51C+ cells with or without mtOX and mirin. Error bars in (A) and (C) represent SEM, and P values are derived using the Student’s t test. Bars in (E) represent the mean, and P values are derived using the Mann-Whitney test. ****P < 0.0001, ***P < 0.001, and **P < 0.01.
To test an involvement of cGAS/STING in the observed STAT1 induction, we used a dominant-negative approach as previously reported (49) and expressed either wild-type, a DNA-binding cGAS mutant (DBM; C396A/C397A), or an enzymatically inactive cGAS mutant protein (NTM; G212A/S213A) to suppress cGAS activity. In SH2038 cells, RNA levels of STAT1 transcripts increase in the presence of wild-type cGAS upon mtOX treatment but not in the presence of mutant cGAS proteins (Fig. 7C). Similar results are obtained when using mtPQ as an alternative mitochondria-specific oxidative damaging agent. Moreover, STING is phosphorylated in SH2038 cells that can be suppressed by inhibition of MRE11 nuclease (fig. S8A), further correlating mtDNA fork protection defects with a cGAS-dependent innate immune signaling.
cGAS is activated by foreign DNA, nuclear DNA, and mtDNA. Western blot analysis of mtOX-treated SH2038 cells expressing RAD51C R366Q, a RAD51C separation of function mutant protein that is defective in nuclear but not mitochondrial localization (27), too, significantly lowered STAT1 protein levels (Fig. 7D and fig. S8B). This confirms that mitochondrial localization of RAD51C suppresses the observed STAT1 expression. Moreover, STAT1 protein expression levels are significantly lower in rho zero cells (Fig. 7D), suggesting that mtDNA is driving STAT1 protein expression in SH2038 cells. To directly test the involvement of nascent mtDNA in the observed cGAS activation, we measured the association of cGAS to nascently labeled mtDNA by a modified mitoSIRF assay (Fig. 7, E and F), assessing cGAS-mtDNA association 24 hours after EdU labeling and mtOX activation. We found a greater number of cGAS–nascent mtDNA signals in the SH2038 patient cells compared to RAD51C+ cells. cGAS–nascent mtDNA signals are decreased by rescuing mtDNA fork protection with inhibition of MRE11 by mirin (Fig. 7, E and F), suggesting that MRE11-excised nascent mtDNA is required for the observed interaction. The cGAS–nascent mtDNA signals are reduced when mtDNA replication is inhibited during the EdU labeling (low-dose EtBr; Fig. 7, E and F), confirming that the cGAS-DNA signals involve nascent mtDNA. As a second test for mtDNA dependence, we find the same cGAS-DNA signal pattern in both S-phase and non–S-phase cells (fig. S8C), suggesting an involvement of mtDNA because there is no nuclear EdU-labeled DNA in non–S-phase cells. Collectively, the data demonstrate that cGAS associates with mtDNA processed by MRE11 in mtDNA fork protection–defective FA patient cells.
Nuclear DNA instability via fork instability or via micronuclei formation also has been shown to activate cGAS (50–52). In contrast to nuclear replication stalling by hydroxyurea, neither mtOX nor ddC causes nuclear replication instability via degradation of DNA when measured by single-molecule DNA fiber spreading (fig. S8D). Moreover, the numbers of micronuclei remain unchanged in both FANCO/RAD51C-deficient and FANCO/RAD51C-complemented cells with the mitochondrially targeted treatments (fig. S8E). Together, the data render a significant contribution of nuclear DNA replication instability to the observed inflammation phenotypes reported here unlikely.
Canonical cGAS activation leads to the expression of type I interferons (4). Unlike the nuclear DNA-cGAS response, which leads to the phosphorylated form of STAT1 (P-STAT1) and results in induction of interferons (51, 52), mtDNA resulting from TFAM heterozygosity can stimulate an unphosphorylated ISGF3 (U-ISGF3) pathway that is controlled by total STAT1 protein amounts rather than its phosphorylated form and consequently does not induce the nuclear factor κB (NFκB) pathway or sustained interferons by the JAK/STAT (Janus kinase/STAT) pathways. This also distinguishes it from the mitochondrial MAV/RIG1 inflammation pathway that is activated by RNA, but not by mtDNA. Comparing the RNA-seq signatures, we find that the transcripts induced in SH2038 mutant cells closely follow the previously reported cGAS inflammation signature for mtDNA (fig. S8F) (8). Western blot analysis of SH2038 further reveals that total STAT1 protein levels notably increase with mtOX and are suppressed when mtDNA fork protection is restored by inhibition of the DNA nuclease MRE11. P-STAT1, on the other hand, solely trails with the total protein levels (Fig. 7G), suggesting low-level but unsustained INFβ-dependent processes, collectively consistent with the profile of an U-ISGF3 response in SH2038 cells. A similar pattern in STAT1 protein is observed in FANCD2-defective PD20 patient cells (Fig. 7H), suggesting that suppression of this type of immune signaling involves downstream FANC pathway members in addition to RAD51C. We further analyzed the expression of all U-ISGF3 pathway members in the FANCO/RAD51C-defective SH2038 cells and found that ~80% of them are significantly up-regulated in the mutant FA cells 24 hours after mtOX exposure, which are greatly reduced with mirin (Fig. 7, I and J). Notably, there is a lack of induction of interferons and NFκB target genes at this time point, characteristic of the U-ISGF3 pathway that is activated by mtDNA instability (Fig. 7K). Collectively, the data show that FANC deficiency results in activation of cGAS-dependent innate immune signaling that predominantly overlaps with the U-ISGF3 response. Importantly, this response is repressed by inhibition of MRE11 (Fig. 7, J and K). Together, the data uncover that MRE11 nuclease mediates mtDNA-induced cGAS activation.
DISCUSSION
mtDNA is being recognized as a prominent DAMP that stimulates innate immune signaling. Pioneering work established that mtDNA instability caused by TFAM heterozygosity activates the cGAS-mediated induction of ISGs (8, 9). The collective data presented here establish a previously unappreciated mtDNA stability pathway in mtDNA fork protection. DNA fork protection is evolutionarily conserved in Escherichia coli, where the BRCA functional homologues RecFOR protect stalled replication forks (53). The data presented here support the concept of endosymbiotic DNA replication fork protection of mtDNA, whereby nuclear fork protection genes function in the mitochondria to substitute for the lost ancestral bacterial genes in the protection of the mtDNA genome. Endosymbiotic DNA replication fork protection reconciles the previously reported mtDNA instability of FA cells and localization of DNA repair/replication proteins to mitochondria, although DNA break and cross-link repair are likely undesirable in human mitochondria (6, 16, 17, 20, 54).
mtDNA fork protection is mechanistically, genetically, and physiologically separable from nuclear fork protection and DNA cross-link repair, whereby FANC core complex components and FANCD2 monoubiquitination are essential in the nucleus but dispensable for mitochondrial mtDNA stability. Lower eukaryotes and invertebrates are devoid of FANC core complex proteins (55), suggesting that the necessity of the evolution of the full FANC pathway could, in part, relate to the size of the genome, with larger genomes requiring additional DNA stabilization that remained unnecessary for smaller genomes including that of the mitochondria. Alternatively, mtDNA genome stability may require distinct structural support by FANC proteins or use different posttranslational modification signals including phosphorylation. The distinct genetic requirements for fork protection in the mitochondria nevertheless can feasibly contribute to the pleiotropic phenotypes typifying patients with FA and may account for the more recently recognized genotypic differences associated with neuroinflammation in FA. Patients with mutations in FANCD2 and downstream pathway genes also often exhibit more severe congenital abnormalities compared to those with FANC core complex mutations (56), a phenotype that is also associated with diseases caused by mitochondrial dysfunction (57), suggesting that mitochondrial fork protection could play a role during development.
The data uncover a molecular mechanism for mitochondrial replication stress-dependent innate immune signaling identifying MRE11 as the nuclease responsible for mtDNA-mediated cGAS activation. MRE11 also plays an important role in viral recognition and defense of cytosolic viral DNA, which promotes cGAS pathway activation (58, 59). Over recent years, it has become evident that mitochondrial mtDNA extrusion is a major source for induction of the cGAS innate immune signaling (8, 9, 60, 61). As rare disease mechanisms including FA provide a magnifying view of fundamental biological processes that have gone awry, MRE11 feasibly may also contribute to mtDNA-mediated cGAS activation outside the context of FA. In support of this possibility, the expression profile of ISGs remarkably overlaps with the U-ISGF3 response, typifying the chronic cGAS-dependent innate immune signaling found with TFAM heterozygosity (8, 9). This response does not result in a sustained type I interferon response but result in STAT1 protein increase and an expression of a distinct subset of ISGs. While INFb can initiate this U-ISG3 response (62, 63) and low INFb levels difficult to detect by current methods may suffice to activate it (9), STAT1 is phosphorylated only during the initial phase (62) or at steady low levels (9). This is consistent with our data revealing low levels of P-STAT1 that, while detectable even in unchallenged cells, does not increase beyond total STAT1 protein levels otherwise expected from a canonical type I interferon response. In contrast, STAT1 protein substantially increases, which is entirely suppressed by inhibition of MRE11 nuclease. Given that no interferons are induced, it will be interesting to determine whether the U-ISG3 response results in inflammation to stimulate type I interferon–dependent immune reactions in future studies or perhaps even could suppress it. MRE11 also may not be the only nuclease capable of processing mtDNA for cGAS, and it may act in concert with other nucleases, as it does in the nucleus. In this context, DNA2, a nuclease downstream of MRE11 during replication stress, has also been reported to localize to the mitochondria, similar to MRE11 itself (64).
FANC genes, including FANC core complex genes, promote mitophagy for suppression of the inflammasome (6). mtDNA also activates the NLRP3-dependent inflammasome and interleukin-1β production in B cells and macrophages, which is dependent on newly synthesized DNA (5). While our data show that the mtDNA, which was synthesized just before exposure to the mtDNA damage, is degraded, mtDNA replication after the damage is increased, which, in principle, could stimulate inflammasome responses. Thus, it is possible that aberrant mtDNA fork protection reactions may also promote inflammasome responses in B cells and macrophages.
Our data here identifying mtDNA-dependent cGAS activation reveal that FANC genes control diverse and distinct mitochondrial innate immune signaling. The observed response also physiologically distinguishes mtDNA instability elicited by fork protection defects from nuclear genome instability, whereby the latter but not the former results in the induction of interferons. In contrast to nuclear cGAS-dependent reactions, mitochondrial fork protection defects do not elicit cell death nor elicit interferon induction as seen for acute responses to viral infection or nuclear DNA damage (51, 52). Instead, it follows a distinct U-ISGF3 response by cGAS that promotes proliferation and increases cellular viability, which may be protective. This type of cGAS response has previously been reported for chronic mtDNA instability seen with TFAM heterozygosity, which promotes resistance against chemotherapeutics (8, 9). It is well established that patients with FA exhibit chronically abnormally increased inflammation markers, yet their source is, so far, not fully understood (65–70). Because inflammation-induced proliferation rather than cell death causes hematopoietic stem cell exhaustion and anemia (71, 72), mtDNA-induced innate immune signaling reported here may ultimately contribute to bone marrow failure in patients with FA. Conversely, during tumorigenesis, promotion of cell proliferation aids tumor expansion, consistent with the role of chronic inflammation in tumorigenesis. The results presented here increase our knowledge on how nuclear genome instability suppressor genes cause diverse hallmarks of cancer including innate immune signaling (73) and identify MRE11 as a potential therapeutic target to offset mtDNA-specific reactions caused by cGAS activation and potentially chemotherapy resistance.
MATERIALS AND METHODS
Cell lines, siRNA, and reagents
RAD51C+ and SH2038 cells, previously described (28), were grown in Dulbecco’s modified Eagle’s medium (DMEM) (Life Technologies) supplemented with 10% fetal bovine serum (FBS) (Gemini Bio Products) and penicillin-streptomycin (100 U/ml) (Life Technologies). BJ cells were purchased from the American Type Culture Collection and grown in Eagle’s minimum essential medium with 15% FBS. OEFX, OERN, and OELA (mutant SLX4); GM6914 (mutant FANCA); PD20 (mutant FANCD2); EUFA326 (mutant FANCG); VU423 (mutant BRCA2); and ALAI and ALDH (mutant FANCC) were previously described (46, 74–80) and grown in DMEM with 15% FBS (74). All cell lines have been tested for mycoplasma contamination. All cells are grown at 37°C and 5% CO2. SH2038 cells were transfected with X-tremeGENE (Roche) and wild-type or mutant cGAS DNA constructs previously described (49) or with a RAD51C R366Q construct (pCMV2B backbone, provided by K. Bernstein laboratory) subsequently selected with neomycin (200 μg/ml) for 7 days. mtOX (37) and PFM39 (42) were synthesized at the MDACC Pharmaceutical Chemistry core facility. Mirin, hydroxyurea (HU), aphidicolin, BrdU, ddC, 5-iodo-2′-deoxyuridine (IdU), 5-chloro-2′-deoxyuridine thymidine (CldU), MitoTEMPO, and Duolink PLA reagents were purchased from Sigma-Aldrich. EdU and biotin azide were from Life Technologies; 4′,6-diamidino-2-phenylindole (DAPI) was from Thermo Fisher Scientific; ProLong Gold, PicoGreen, SYBR Green, and MitoSOX were from Invitrogen; MitoPQ was from Abcam; rotenone was from EMD Millipore; Puregene Core kit was from Qiagen; the Aurum total RNA mini kit and SYBR Green Master Mix were from Bio-Rad; and EtBr was from AMRESCO. siRNA against MRE11 and RAD51C were purchased from Dharmacon and transfected into cells using RNAiMAX (Invitrogen) according to the manufacturer’s protocol.
Antibodies
Antibodies used in MIRA, immunofluorescence, and Western blot assays are as follows: mouse anti-biotin (Sigma-Aldrich, BN-34), rabbit anti-biotin (1:200; Cell Signaling Technology, D5A7), rabbit anti-TFAM (Abcam, ab131607), mouse anti-MRE11 (Abcam, ab214), rabbit anti-cGAS (Novus, NBP1-86761), mouse anti-mitochondria (Abcam, ab3298), pY701 STAT1 (Cell Signaling Technology, 9167), STAT1 (Cell Signaling Technology, 9176, 14995), mouse anti-DNA (EMD Millipore, CBL186), rabbit anti-LC3A/B (Cell Signaling Technology, D3U4C), mouse anti-oxphos (Abcam, ab3601), mouse anti-BrdU (BD Pharmingen, 555627), rabbit anti-pS366 STING (Cell Signaling Technology, 50907), rabbit anti-TMEM173 (STING; Abcam, ab227704), and mouse anti-RAD51C (Abnova, H00005889-M01).
MIRA assay
Cells were seeded the day before treatment. FANC patient cells were plated with 20 μM MitoTEMPO. Cells were labeled with 20 μM EdU for 1 hour in the presence or absence of mtOX (4 μM), mirin (50 μM), or PFM39 (100 μM), followed by incubation in ambient light to allow for mtOX activation (37). Cells were washed in phosphate-buffered saline (PBS; pH 7.5) and allowed to recover in medium as indicated in the figures. Alternatively, after EdU labeling, cells were treated with ddC (20 μM) or MitoPQ (10 μM) for 3 hours. mtDNA replication was inhibited with EtBr (50 ng/ml), and nuclear replication was inhibited with aphidicolin (7 μM) where indicated. Cells were fixed in 2% paraformaldehyde, permeabilized with 0.25% Triton X-100/PBS, and blocked with 10% goat serum/PBS/0.1% Triton X-100, and a Click-iT reaction was performed using biotin azide (Life Technologies) following the manufacturer’s protocol. Where indicated to visualize colocalization of MIRA signal in the mitochondria, slides were incubated with anti-DNA (EMD Millipore), anti-mitochondria (Abcam), or anti-TFAM (Abcam) for 1 hour at 37°C in blocking solution, followed by the appropriate secondary antibody containing either Alexa Fluor 488 or Alexa Fluor 647. Slides were incubated overnight with mouse anti-biotin (Sigma-Aldrich) and rabbit anti-biotin (Cell Signaling Technology) in blocking solution, and a Duolink PLA was performed following the manufacturer’s protocol. Microscope slides were counterstained with DAPI (100 μg/ml) before mounting in ProLong Gold (Invitrogen).
mitoSIRF (mtDNA-protein PLA)
Assay conditions were adapted from SIRF (43) and followed the experimental conditions of the MIRA as indicated in the figures. Briefly, cells were labeled with 20 μM EdU for 1 to 3 hours and, where indicated, exposed to mtOX before fixation, Click-iT, incubation with primary antibodies against biotin, and MRE11, RAD51C, TFAM, RPA, or cGAS overnight as above. Duolink PLA was performed following the manufacturer’s protocol. Microscope slides were counterstained with DAPI (100 μg/ml) before mounting in ProLong Gold (Invitrogen).
Immunofluorescence
Cells were fixed with 2% paraformaldehyde/PBS for 15 min, permeabilized with 0.25% Triton X-100/PBS for 15 min, and blocked with 10% goat serum/0.1% Triton X-100 for 1 hour. For immunofluorescent visualization of mtDNA by BrdU or EdU, cells were labeled with EdU (20 μM) or BrdU (20 μM) for the indicated times. EdU was visualized by a Click-iT reaction with Alexa Fluor 488 azide following the manufacturer’s protocol. For BrdU reactions, before blocking, the cells were incubated for in 2 N HCl/0.5% Triton X-100/PBS and neutralized in 0.1 M Na borate (pH 8.5) before incubation with primary and secondary antibodies. Primary antibodies used include mouse anti-mitochondria (Abcam, ab3298) and mouse anti-BrdU (BD Biosciences, 555627). Cells were then counterstained with DAPI and mounted in ProLong Gold (Invitrogen). For mitochondrial fission immunofluorescence, cell lines were seeded in microscope eight-chamber slides and allowed to adhere overnight before incubation in Hank’s balanced salt solution (Life Technologies) for 24 hours before treatment. Cells were fixed as described above, 3 hours after treatment with mtOX (4 μM). Immunofluorescence was performed as described above using the anti-mitochondria (Abcam) primary antibody.
EdU and Oregon Green antibody amplification
Cells were cultured in eight-chamber slides overnight and then incubated with 20 μM EdU for 1 hour, washed with PBS, and treated with 20 μM ddC and 100 μM PFM39 where indicated. Three hours later, cells were fixed in 2% paraformaldehyde/PBS for 15 min at room temperature and permeabilized with 0.25% Triton X-100/PBS for 15 min at room temperature. EdU was detected using click chemistry reaction mix (2 mM CuSO4, 100 mM sodium ascorbate, and PBS) with 10 μM Oregon Green 488 azide (Thermo Fisher Scientific). The mixture was added to cells for 1 hour at room temperature, followed by three washes in PBS. Cells were then blocked with 10% goat serum/0.1% Triton X-100/PBS for 1 hour at 37°C and then incubated with rabbit Alexa Fluor 488–conjugated antibodies against Oregon Green (A-11090, Thermo Fisher Scientific) and mouse antibodies against mitochondria (Abcam) for 1 hour at 37°C. Cells were washed three times with PBS and incubated with goat Alexa Fluor 488 anti-rabbit and goat Alexa Fluor 555 anti-mouse. Microscope slides were counterstained with DAPI (100 μg/ml) before mounting in ProLong Gold (Invitrogen).
Mitochondrial FISH
Mitochondrial FISH (mito-FISH) was performed as previously described (81). Probes used were previously described and purchased from MYcroarray (82).
DNA fiber analysis
DNA fiber experiments were performed as previously described (11). Briefly, replication tracts of log-phase cells were pulse-labeled with 50 μM IdU and CldU before or after exposure to 4 mM HU, 4 μM mtOX, or 20 μM ddC, respectively, as indicated in the sketches. Cells were harvested, lysed, and spread to obtain single DNA molecules on microscope slides before standard immunofluorescence with antibodies against IdU and CldU (Novus Biologicals, BD Biosciences).
Imaging and statistical analyses
For MIRA, cGAS-mtDNA SIRF, DNA fibers, and immunofluorescence assays, slides were imaged using an Eclipse Ti-U inverted microscope with an Andor Zyla sCMOS camera or a Zeiss Airyscan LSM 800 microscope with a Hamamatsu C13440 camera. Images were analyzed using the Nikon NIS Elements or Zen2 software version 2.3. For ease of viewing, a black bar was placed over the scale bars of select images. Statistical analysis was performed using GraphPad Prism 7 software. Four to eight image fields were acquired for each condition, and the data from two to four biological replicates were compiled. For both MIRA and mitoSIRF assays, the cytoplasmic signals were counted irrespective of cell cycle status. P values were calculated using the Mann-Whitney test (GraphPad Prism 7 software).
Analysis of mtDNA topology and 7S DNA levels
The topological state of mtDNA was analyzed in total cellular nucleic acid extracts by one-dimensional agarose gels and Southern blotting essentially as described (83). For conformational studies, 1 μg of DNA was digested with Bgl II that does not cut mtDNA and separated over a 0.4% agarose gel in tris-borate EDTA without EtBr. Gels were blotted and hybridized with 32P-labeled DNA probes containing nucleotides 35 to 611. The signal was quantified using a phosphorimager (Molecular Imager FX, Bio-Rad).
Colony formation assay
RAD51C+ and SH2038 cells were plated in triplicate in six-well plates. Cells were left untreated or treated with 4 μM mtOX. Colonies were fixed with acetic acid/methanol, stained with 1.0% crystal violet in methanol, and hand counted. Data were combined from three biological repeats. P values were calculated using the Student’s t test (GraphPad Prism 7 software).
Mitochondrial membrane potential
Cells were cultured in a black-welled 96-well plate and incubated with either control medium or 4 μM mtOX for 1 hour before activation in ambient light for 5 min. The cells were allowed to recover for 3 hours. Medium was removed and replaced with 500 nM MitoTracker CMXRos diluted in warm culture medium with or without 80 μM CCCP (carbonyl cyanide m-chlorophenyl hydrazine) for 30 min. MitoTracker CMXRos fluorescent intensity was measured on a BioTek Gen5 microplate reader. Fluorescent intensity was normalized to protein content in each well measured by DC protein assay. The assay was performed with two biological repeats and five technical replicates each. P values were calculated using the Student’s t test (GraphPad Prism 7 software).
STAT1 quantitative polymerase chain reaction
Total RNA was isolated using an Aurum total RNA mini kit (Bio-Rad) according to the manufacturer’s protocols. cDNA was synthesized using random hexamers and oligo-dT primers (New England Biolabs), and quantitative polymerase chain reaction (qPCR) was performed using the SYBR Green Master Mix (Bio-Rad). The following primers were used: STAT1, CAGCTTGACTCAAAATTCCTGGA (forward) and TGAAGATTACGCTTGCTTTTCCT (reverse); TBP, CCCATGACTCCCATGACC (forward) and TTTACAACCAAGATTCACTGTGG (reverse). Expression values were normalized against TATA box–binding protein (TBP), and the relative expression level was calculated using standard methods (2 − ΔΔCt) and combined from three to five biological and multiple technical repeats. P values were calculated using the Student’s t test (GraphPad Prism 7 software).
mtDNA copy number assay by qPCR
Cells were seeded the day before the treatment. mtOX (20 μM) was added 1 hour before light activation. Total DNA was isolated using a PureGene Core kit (Qiagen) following the manufacturer’s instructions. mtDNA copy number was determined by real-time PCR as previously described (84) on an LightCycler 96 system (Roche) using the SYBR Green Master Mix (Bio-Rad). The following primers were used: tRNA-Leu, 5′-CACCCAAGAACAGGGTTTGT-3′ (forward) and 5′-TGGCCATGGGTATGTTGTTA-3′ (reverse); B2M, 5′-TGCTGTCTCCATGTTTGATGTATCT-3′ (forward) and 5′-TCTCTGCTCCCCACCTCTAAGT-3′ (reverse). Results were analyzed with LightCycler 96 software 1.1 (Roche). The fold change was calculated and combined from three biological with multiple technical repeats. P values were calculated using the Student’s t test (GraphPad Prism 7 software).
RNA-seq and analysis
Cells were treated for 24 hours as indicated and RNA-purified using the Aurum total RNA mini kit (Bio-Rad). RNA samples were sequenced and analyzed by Novogene. Raw data (fpkm values) are given in tables S1 to S4. Gene ontology enrichment analysis was done using ToppGene (85), and interferon and cytokine pathway proteins were identified according to the Reactome database (86) and according to (9). Heatmaps were generated using GraphPad Prism 7. Heatmaps for comparison between treatment conditions were generated by scaling by row, and heatmaps for comparison between interferon pathway groups (Fig. 7) were generated by scaling based on the population mean (87, 88).
Western blots
After indicated treatments, cells were lysed in 1% NP-40, 200 mM NaCl, and 50 mM tris-HCl (pH 7.4) containing protease (cOmplete Mini, Roche) and phosphatase (PhosSTOP, Roche) inhibitors. Lysates were cleared, and SDS–polyacrylamide gel electrophoresis was performed. Proteins were transferred to a nitrocellulose membrane, blocked in 5% milk/TBST (tris-buffered saline with Tween 20), incubated with the indicated primary antibodies for 1 hour, and visualized by enhanced chemiluminesence.
Electron microscopy
Cells were treated as indicated and fixed with a solution containing 3% glutaraldehyde and 2% paraformaldehyde in 0.1 M cacodylate buffer (pH 7.3), then washed in 0.1 M sodium cacodylate buffer and treated with 0.1% Millipore-filtered cacodylate-buffered tannic acid, postfixed with 1% buffered osmium, and stained en bloc with 1% Millipore-filtered uranylacetate. The samples were dehydrated in increasing concentrations of ethanol, infiltrated, and embedded in LX-112 medium. The samples were polymerized in a 60°C oven for approximately 3 days. Ultrathin sections were cut in a Leica Ultracut microtome (Leica, Deerfield, IL), stained with uranylacetate and lead citrate in a Leica EM stainer, and examined in a JEM 1010 transmission electron microscope (JEOL, USA Inc., Peabody, MA) at an accelerating voltage of 80 kV. Digital images were obtained using AMT Imaging System (Advanced Microscopy Techniques Corp., Danvers, MA) at the High Resolution Electron Microscopy Facility at the University of Texas MD Anderson Cancer Center.
Rho zero cells
Rho zero cells were created by growing SH2038 cells in medium containing EtBr (50 ng/ml) or 100 μM ddC with 1 mM sodium pyruvate and uridine (50 ng/ml) for 1 to 4 weeks. mtDNA depletion was confirmed by PicoGreen live cell imaging.
Acknowledgments
We thank S. Kelley, S. Wisnovsky, and T. Sack for advice on mtOX; K. Bernstein and G. Nagaraju for providing RAD51C R366Q plasmid constructs; and L.-J. C. Wong and D. Blackburn for helpful comments. mtOX and PFM39 were synthesized by the MD Anderson Cancer Center pharmaceutical chemistry core facility. Electron microscope images were captured at the MD Anderson Cancer Center EM facility.
Funding: This work was supported by a Rita Allen Foundation Fellowship and, in part, by NIEHS award 1R01ES029680, Cancer Prevention and Research Institute of Texas (CPRIT) RP180813, and R1312 to K.S. K.S. is a Rita Allen Foundation Fellow and a CPRIT scholar in Cancer Biology. EM facility is supported by CCSG grant NIH P30CA016672.
Author contributions: J.W.L. performed and analyzed MIRA, mitoSIRF assay, qPCR, RNA-seq, and Western blot experiments. R.A.B. developed and performed MIRA experiments. E.D. performed proliferation assays, membrane potential assays, and mitoSIRF assays. S.R. performed and analyzed MIRA. S.P. initiated qPCR experiments and performed clonogenic assays. S.K. performed DNA fiber spread assay. D.S. provided cell lines. S.G. performed mtDNA gel analysis. K.S. conceived the study; contributed to data interpretation, experimental design, and data analysis; and wrote the manuscript with input from J.W.L., D.S., and S.G.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Materials can be provided by K.S. pending scientific review and a completed material transfer agreement. Requests for materials should be submitted to kschlacher@mdanderson.org.
Supplementary Materials
This PDF file includes:
Figs. S1 to S8
Tables S1 to S4
REFERENCES AND NOTES
- 1.Chandel N. S., Evolution of mitochondria as signaling organelles. Cell Metab. 22, 204–206 (2015). [DOI] [PubMed] [Google Scholar]
- 2.D’Erchia A. M., Atlante A., Gadaleta G., Pavesi G., Chiara M., De Virgilio C., Manzari C., Mastropasqua F., Prazzoli G. M., Picardi E., Gissi C., Horner D., Reyes A., Sbisa E., Tullo A., Pesole G., Tissue-specific mtDNA abundance from exome data and its correlation with mitochondrial transcription, mass and respiratory activity. Mitochondrion 20, 13–21 (2015). [DOI] [PubMed] [Google Scholar]
- 3.DeBalsi K. L., Hoff K. E., Copeland W. C., Role of the mitochondrial DNA replication machinery in mitochondrial DNA mutagenesis, aging and age-related diseases. Ageing Res. Rev. 33, 89–104 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gong T., Liu L., Jiang W., Zhou R., DAMP-sensing receptors in sterile inflammation and inflammatory diseases. Nat. Rev. Immunol. 20, 95–112 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Zhong Z., Liang S., Sanchez-Lopez E., He F., Shalapour S., Lin X. J., Wong J., Ding S., Seki E., Schnabl B., Hevener A. L., Greenberg H. B., Kisseleva T., Karin M., New mitochondrial DNA synthesis enables NLRP3 inflammasome activation. Nature 560, 198–203 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sumpter R. Jr., Sirasanagandla S., Fernandez A. F., Wei Y., Dong X., Franco L., Zou Z., Marchal C., Lee M. Y., Clapp D. W., Hanenberg H., Levine B., Fanconi anemia proteins function in mitophagy and immunity. Cell 165, 867–881 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rongvaux A., Jackson R., Harman C. C., Li T., West A. P., de Zoete M. R., Wu Y., Yordy B., Lakhani S. A., Kuan C. Y., Taniguchi T., Shadel G. S., Chen Z. J., Iwasaki A., Flavell R. A., Apoptotic caspases prevent the induction of type I interferons by mitochondrial DNA. Cell 159, 1563–1577 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.West A. P., Khoury-Hanold W., Staron M., Tal M. C., Pineda C. M., Lang S. M., Bestwick M., Duguay B. A., Raimundo N., MacDuff D. A., Kaech S. M., Smiley J. R., Means R. E., Iwasaki A., Shadel G. S., Mitochondrial DNA stress primes the antiviral innate immune response. Nature 520, 553–557 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu Z., Oeck S., West P., Mangalhara K. C., Sainz A. G., Newman L. E., Zhang X., Wu L., Yan Q., Bosenberg M., Liu Y., Sulkowski P. L., Tripple V., Kaech S. M., Glazer P. M., Shadel G. S., Mitochondrial DNA stress signalling protects the nuclear genome. Nat. Metab. 1, 1209–1218 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niraj J., Farkkila A., D’Andrea A. D., The Fanconi anemia pathway in cancer. Annu. Rev. Cancer Biol. 3, 457–478 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schlacher K., Christ N., Siaud N., Egashira A., Wu H., Jasin M., Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell 145, 529–542 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schlacher K., Wu H., Jasin M., A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell 22, 106–116 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Somyajit K., Saxena S., Babu S., Mishra A., Nagaraju G., Mammalian RAD51 paralogs protect nascent DNA at stalled forks and mediate replication restart. Nucleic Acids Res. 43, 9835–9855 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y., Amarachintha S., Wilson A. F., Li X., Du W., Persistent response of Fanconi anemia haematopoietic stem and progenitor cells to oxidative stress. Cell Cycle 16, 1201–1209 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pagano G., Talamanca A. A., Castello G., Pallardo F. V., Zatterale A., Degan P., Oxidative stress in Fanconi anaemia: From cells and molecules towards prospects in clinical management. Biol. Chem. 393, 11–21 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Coene E. D., Hollinshead M. S., Waeytens A. A. T., Schelfhout V. R. J., Eechaute W. P., Shaw M. K., Van Oostveldt P. M. J., Vaux D. J., Phosphorylated BRCA1 is predominantly located in the nucleus and mitochondria. Mol. Biol. Cell 16, 997–1010 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dmitrieva N. I., Malide D., Burg M. B., Mre11 is expressed in mammalian mitochondria where it binds to mitochondrial DNA. Am. J. Physiol. Regul. Integr. Comp. Physiol. 301, R632–R640 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y., Shen Y., Jin K., Wen Z., Cao W., Wu B., Wen R., Tian L., Berry G. J., Goronzy J. J., Weyand C. M., The DNA repair nuclease MRE11A functions as a mitochondrial protector and prevents T cell pyroptosis and tissue inflammation. Cell Metab. 30, 477–492.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mukhopadhyay S. S., Leung K. S., Hicks M. J., Hastings P. J., Youssoufian H., Plon S. E., Defective mitochondrial peroxiredoxin-3 results in sensitivity to oxidative stress in Fanconi anemia. J. Cell Biol. 175, 225–235 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sage J. M., Gildemeister O. S., Knight K. L., Discovery of a novel function for human Rad51: Maintenance of the mitochondrial genome. J. Biol. Chem. 285, 18984–18990 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang T., Du W., Wilson A. F., Namekawa S. H., Andreassen P. R., Meetei A. R., Pang Q., Fancd2 in vivo interaction network reveals a non-canonical role in mitochondrial function. Sci. Rep. 7, 45626 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernal M., Yang X., Lisby M., Mazón G., The FANCM family Mph1 helicase localizes to the mitochondria and contributes to mtDNA stability. DNA Repair 82, 102684 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Gredilla R., DNA damage and base excision repair in mitochondria and their role in aging. J. Aging Res. 2011, 257093 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alexeyev M., Shokolenko I., Wilson G., LeDoux S., The maintenance of mitochondrial DNA integrity—Critical analysis and update. Cold Spring Harb. Perspect. Biol. 5, a012641 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Torregrosa-Munumer R., Hangas A., Goffart S., Blei D., Zsurka G., Griffith J., Kunz W. S., Pohjoismaki J. L. O., Replication fork rescue in mammalian mitochondria. Sci. Rep. 9, 8785 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thul P. J., Akesson L., Wiking M., Mahdessian D., Geladaki A., Ait Blal H., Alm T., Asplund A., Bjork L., Breckels L. M., Backstrom A., Danielsson F., Fagerberg L., Fall J., Gatto L., Gnann C., Hober S., Hjelmare M., Johansson F., Lee S., Lindskog C., Mulder J., Mulvey C. M., Nilsson P., Oksvold P., Rockberg J., Schutten R., Schwenk J. M., Sivertsson A., Sjostedt E., Skogs M., Stadler C., Sullivan D. P., Tegel H., Winsnes C., Zhang C., Zwahlen M., Mardinoglu A., Ponten F., von Feilitzen K., Lilley K. S., Uhlen M., Lundberg E., A subcellular map of the human proteome. Science 356, eaal3321 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Mishra A., Saxena S., Kaushal A., Nagaraju G., RAD51C/XRCC3 facilitates mitochondrial DNA replication and maintains integrity of the mitochondrial genome. Mol. Cell Biol. 38, e00489-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vaz F., Hanenberg H., Schuster B., Barker K., Wiek C., Erven V., Neveling K., Endt D., Kesterton I., Autore F., Fraternali F., Freund M., Hartmann L., Grimwade D., Roberts R. G., Schaal H., Mohammed S., Rahman N., Schindler D., Mathew C. G., Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat. Genet. 42, 406–409 (2010). [DOI] [PubMed] [Google Scholar]
- 29.Clayton D. A., Replication of animal mitochondrial DNA. Cell 28, 693–705 (1982). [DOI] [PubMed] [Google Scholar]
- 30.Korhonen J. A., Pham X. H., Pellegrini M., Falkenberg M., Reconstitution of a minimal mtDNA replisome in vitro. EMBO J. 23, 2423–2429 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fredriksson S., Gullberg M., Jarvius J., Olsson C., Pietras K., Gustafsdottir S. M., Ostman A., Landegren U., Protein detection using proximity-dependent DNA ligation assays. Nat. Biotechnol. 20, 473–477 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Soderberg O., Gullberg M., Jarvius M., Ridderstrale K., Leuchowius K. J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L. G., Landegren U., Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 3, 995–1000 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Leibowitz R. D., The effect of ethidium bromide on mitochondrial DNA synthesis and mitochondrial DNA structure in HeLa cells. J. Cell Biol. 51, 116–122 (1971). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feng J. Y., Johnson A. A., Johnson K. A., Anderson K. S., Insights into the molecular mechanism of mitochondrial toxicity by AIDS drugs. J. Biol. Chem. 276, 23832–23837 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Sasaki T., Sato Y., Higashiyama T., Sasaki N., Live imaging reveals the dynamics and regulation of mitochondrial nucleoids during the cell cycle in Fucci2-HeLa cells. Sci. Rep. 7, 11257 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stojkovic G., Makarova A. V., Wanrooij P. H., Forslund J., Burgers P. M., Wanrooij S., Oxidative DNA damage stalls the human mitochondrial replisome. Sci. Rep. 6, 28942 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wisnovsky S., Jean S. R., Kelley S. O., Mitochondrial DNA repair and replication proteins revealed by targeted chemical probes. Nat. Chem. Biol. 12, 567–573 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Lentz S. I., Edwards J. L., Backus C., McLean L. L., Haines K. M., Feldman E. L., Mitochondrial DNA (mtDNA) biogenesis: Visualization and duel incorporation of BrdU and EdU into newly synthesized mtDNA in vitro. J. Histochem. Cytochem. 58, 207–218 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robb E. L., Gawel J. M., Aksentijevic D., Cocheme H. M., Stewart T. S., Shchepinova M. M., Qiang H., Prime T. A., Bright T. P., James A. M., Shattock M. J., Senn H. M., Hartley R. C., Murphy M. P., Selective superoxide generation within mitochondria by the targeted redox cycler MitoParaquat. Free Radic. Biol. Med. 89, 883–894 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Lemaçon D., Jackson J., Quinet A., Brickner J. R., Li S., Yazinski S., You Z., Ira G., Zou L., Mosammaparast N., Vindigni A., MRE11 and EXO1 nucleases degrade reversed forks and lead to MUS81-dependent fork rescue in BRCA2-deficient cells. Nat. Commun. 8, 860 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kolinjivadi A. M., Sannino V., De Antoni A., Zadorozhny K., Kilkenny M., Techer H., Baldi G., Shen R., Ciccia A., Pellegrini L., Krejci L., Costanzo V., Smarcal1-mediated fork reversal triggers Mre11-dependent degradation of nascent DNA in the absence of Brca2 and stable Rad51 nucleofilaments. Mol. Cell 67, 867–881.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shibata A., Moiani D., Arvai A. S., Perry J., Harding S. M., Genois M. M., Maity R., van Rossum-Fikkert S., Kertokalio A., Romoli F., Ismail A., Ismalaj E., Petricci E., Neale M. J., Bristow R. G., Masson J. Y., Wyman C., Jeggo P. A., Tainer J. A., DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Mol. Cell 53, 7–18 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roy S., Luzwick J. W., Schlacher K., SIRF: Quantitative in situ analysis of protein interactions at DNA replication forks. J. Cell Biol. 217, 1521–1536 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hensen F., Potter A., van Esveld S. L., Tarres-Sole A., Chakraborty A., Sola M., Spelbrink J. N., Mitochondrial RNA granules are critically dependent on mtDNA replication factors Twinkle and mtSSB. Nucleic Acids Res. 47, 3680–3698 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dupre A., Boyer-Chatenet L., Sattler R. M., Modi A. P., Lee J. H., Nicolette M. L., Kopelovich L., Jasin M., Baer R., Paull T. T., Gautier J., A forward chemical genetic screen reveals an inhibitor of the Mre11-Rad50-Nbs1 complex. Nat. Chem. Biol. 4, 119–125 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garcia-Higuera I., Taniguchi T., Ganesan S., Meyn M. S., Timmers C., Hejna J., Grompe M., D’Andrea A. D., Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol. Cell 7, 249–262 (2001). [DOI] [PubMed] [Google Scholar]
- 47.Toyama E. Q., Herzig S., Courchet J., Lewis T. L. Jr., Loson O. C., Hellberg K., Young N. P., Chen H., Polleux F., Chan D. C., Shaw R. J., AMP-activated protein kinase mediates mitochondrial fission in response to energy stress. Science 351, 275–281 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meissl K., Macho-Maschler S., Muller M., Strobl B., The good and the bad faces of STAT1 in solid tumours. Cytokine 89, 12–20 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Aguirre S., Luthra P., Sanchez-Aparicio M. T., Maestre A. M., Patel J., Lamothe F., Fredericks A. C., Tripathi S., Zhu T., Pintado-Silva J., Webb L. G., Bernal-Rubio D., Solovyov A., Greenbaum B., Simon V., Basler C. F., Mulder L. C., Garcia-Sastre A., Fernandez-Sesma A., Dengue virus NS2B protein targets cGAS for degradation and prevents mitochondrial DNA sensing during infection. Nat. Microbiol. 2, 17037 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Coquel F., Silva M. J., Techer H., Zadorozhny K., Sharma S., Nieminuszczy J., Mettling C., Dardillac E., Barthe A., Schmitz A. L., Promonet A., Cribier A., Sarrazin A., Niedzwiedz W., Lopez B., Costanzo V., Krejci L., Chabes A., Benkirane M., Lin Y. L., Pasero P., SAMHD1 acts at stalled replication forks to prevent interferon induction. Nature 557, 57–61 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Harding S. M., Benci J. L., Irianto J., Discher D. E., Minn A. J., Greenberg R. A., Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature 548, 466–470 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mackenzie K. J., Carroll P., Martin C. A., Murina O., Fluteau A., Simpson D. J., Olova N., Sutcliffe H., Rainger J. K., Leitch A., Osborn R. T., Wheeler A. P., Nowotny M., Gilbert N., Chandra T., Reijns M. A. M., Jackson A. P., cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 548, 461–465 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chow K. H., Courcelle J., RecO acts with RecF and RecR to protect and maintain replication forks blocked by UV-induced DNA damage in Escherichia coli. J. Biol. Chem. 279, 3492–3496 (2004). [DOI] [PubMed] [Google Scholar]
- 54.Valentin-Vega Y. A., Kastan M. B., A new role for ATM: Regulating mitochondrial function and mitophagy. Autophagy 8, 840–841 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang A. T., Smogorzewska A., SnapShot: Fanconi anemia and associated proteins. Cell 160, 354–354.e1 (2015). [DOI] [PubMed] [Google Scholar]
- 56.Fiesco-Roa M. O., Giri N., McReynolds L. J., Best A. F., Alter B. P., Genotype-phenotype associations in Fanconi anemia: A literature review. Blood Rev. 37, 100589 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Siebel S., Solomon B. D., Mitochondrial factors and VACTERL association-related congenital malformations. Mol. Syndromol. 4, 63–73 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Syed A., Tainer J. A., The MRE11-RAD50-NBS1 complex conducts the orchestration of damage signaling and outcomes to stress in DNA replication and repair. Annu. Rev. Biochem. 87, 263–294 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kondo T., Kobayashi J., Saitoh T., Maruyama K., Ishii K. J., Barber G. N., Komatsu K., Akira S., Kawai T., DNA damage sensor MRE11 recognizes cytosolic double-stranded DNA and induces type I interferon by regulating STING trafficking. Proc. Natl. Acad. Sci. U.S.A. 110, 2969–2974 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Collins L. V., Hajizadeh S., Holme E., Jonsson I. M., Tarkowski A., Endogenously oxidized mitochondrial DNA induces in vivo and in vitro inflammatory responses. J. Leukoc. Biol. 75, 995–1000 (2004). [DOI] [PubMed] [Google Scholar]
- 61.McArthur K., Whitehead L. W., Heddleston J. M., Li L., Padman B. S., Oorschot V., Geoghegan N. D., Chappaz S., Davidson S., San Chin H., Lane R. M., Dramicanin M., Saunders T. L., Sugiana C., Lessene R., Osellame L. D., Chew T. L., Dewson G., Lazarou M., Ramm G., Lessene G., Ryan M. T., Rogers K. L., van Delft M. F., Kile B. T., BAK/BAX macropores facilitate mitochondrial herniation and mtDNA efflux during apoptosis. Science 359, eaao6047 (2018). [DOI] [PubMed] [Google Scholar]
- 62.Cheon H., Stark G. R., Unphosphorylated STAT1 prolongs the expression of interferon-induced immune regulatory genes. Proc. Natl. Acad. Sci. U.S.A. 106, 9373–9378 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cheon H., Holvey-Bates E. G., Schoggins J. W., Forster S., Hertzog P., Imanaka N., Rice C. M., Jackson M. W., Junk D. J., Stark G. R., IFNβ-dependent increases in STAT1, STAT2, and IRF9 mediate resistance to viruses and DNA damage. EMBO J. 32, 2751–2763 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zheng L., Zhou M., Guo Z., Lu H., Qian L., Dai H., Qiu J., Yakubovskaya E., Bogenhagen D. F., Demple B., Shen B., Human DNA2 is a mitochondrial nuclease/helicase for efficient processing of DNA replication and repair intermediates. Mol. Cell 32, 325–336 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ceccaldi R., Sarangi P., D’Andrea A. D., The Fanconi anaemia pathway: New players and new functions. Nat. Rev. Mol. Cell Biol. 17, 337–349 (2016). [DOI] [PubMed] [Google Scholar]
- 66.Morrell D., Chase C. L., Kupper L. L., Swift M., Diabetes mellitus in ataxia-telangiectasia, Fanconi anemia, xeroderma pigmentosum, common variable immune deficiency, and severe combined immune deficiency families. Diabetes 35, 143–147 (1986). [DOI] [PubMed] [Google Scholar]
- 67.Minton K., Inflammation: Inflammatory pathology of Fanconi anaemia. Nat. Rev. Immunol. 16, 336–337 (2016). [DOI] [PubMed] [Google Scholar]
- 68.Bregnard C., Guerra J., Dejardin S., Passalacqua F., Benkirane M., Laguette N., Upregulated LINE-1 activity in the Fanconi anemia cancer susceptibility syndrome leads to spontaneous pro-inflammatory cytokine production. EBioMedicine 8, 184–194 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Du W., Erden O., Pang Q., TNF-α signaling in Fanconi anemia. Blood Cells Mol. Dis. 52, 2–11 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dufour C., Corcione A., Svahn J., Haupt R., Poggi V., Beka’ssy A. N., Scime R., Pistorio A., Pistoia V., TNF-α and IFN-γ are overexpressed in the bone marrow of Fanconi anemia patients and TNF-α suppresses erythropoiesis in vitro. Blood 102, 2053–2059 (2003). [DOI] [PubMed] [Google Scholar]
- 71.Matatall K. A., Jeong M., Chen S., Sun D., Chen F., Mo Q., Kimmel M., King K. Y., Chronic infection depletes hematopoietic stem cells through stress-induced terminal differentiation. Cell Rep. 17, 2584–2595 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.King K. Y., Goodell M. A., Inflammatory modulation of HSCs: Viewing the HSC as a foundation for the immune response. Nat. Rev. Immunol. 11, 685–692 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hanahan D., Weinberg R. A., Hallmarks of cancer: The next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 74.Stoepker C., Hain K., Schuster B., Hilhorst-Hofstee Y., Rooimans M. A., Steltenpool J., Oostra A. B., Eirich K., Korthof E. T., Nieuwint A. W., Jaspers N. G., Bettecken T., Joenje H., Schindler D., Rouse J., de Winter J. P., SLX4, a coordinator of structure-specific endonucleases, is mutated in a new Fanconi anemia subtype. Nat. Genet. 43, 138–141 (2011). [DOI] [PubMed] [Google Scholar]
- 75.Freie B. W., Ciccone S. L. M., Li X., Plett P. A., Orschell C. M., Srour E. F., Hanenberg H., Schindler D., Lee S.-H., Clapp D. W., A role for the Fanconi anemia C protein in maintaining the DNA damage-induced G2 checkpoint. J. Biol. Chem. 279, 50986–50993 (2004). [DOI] [PubMed] [Google Scholar]
- 76.Hartmann L., Neveling K., Borkens S., Schneider H., Freund M., Grassman E., Theiss S., Wawer A., Burdach S., Auerbach A. D., Schindler D., Hanenberg H., Schaal H., Correct mRNA processing at a mutant TT splice donor in FANCC ameliorates the clinical phenotype in patients and is enhanced by delivery of suppressor U1 snRNAs. Am. J. Hum. Genet. 87, 480–493 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jakobs P. M., Sahaayaruban P., Saito H., Reifsteck C., Olson S., Joenje H., Moses R. E., Grompe M., Immortalization of four new Fanconi anemia fibroblast cell lines by an improved procedure. Somat. Cell Mol. Genet. 22, 151–157 (1996). [DOI] [PubMed] [Google Scholar]
- 78.Naf D., Kupfer G. M., Suliman A., Lambert K., D’Andrea A. D., Functional activity of the Fanconi anemia protein FAA requires FAC binding and nuclear localization. Mol. Cell. Biol. 18, 5952–5960 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Timmers C., Taniguchi T., Hejna J., Reifsteck C., Lucas L., Bruun D., Thayer M., Cox B., Olson S., D’Andrea A. D., Moses R., Grompe M., Positional cloning of a novel Fanconi anemia gene, FANCD2. Mol. Cell 7, 241–248 (2001). [DOI] [PubMed] [Google Scholar]
- 80.Howlett N. G., Taniguchi T., Olson S., Cox B., Waisfisz Q., De Die-Smulders C., Persky N., Grompe M., Joenje H., Pals G., Ikeda H., Fox E. A., D’Andrea A. D., Biallelic inactivation of BRCA2 in Fanconi anemia. Science 297, 606–609 (2002). [DOI] [PubMed] [Google Scholar]
- 81.Chatre L., Ricchetti M., Large heterogeneity of mitochondrial DNA transcription and initiation of replication exposed by single-cell imaging. J. Cell Sci. 126, 914–926 (2013). [DOI] [PubMed] [Google Scholar]
- 82.Phillips A. F., Millet A. R., Tigano M., Dubois S. M., Crimmins H., Babin L., Charpentier M., Piganeau M., Brunet E., Sfeir A., Single-molecule analysis of mtDNA replication uncovers the basis of the common deletion. Mol. Cell 65, 527–538.e6 (2017). [DOI] [PubMed] [Google Scholar]
- 83.Goffart S., Cooper H. M., Tyynismaa H., Wanrooij S., Suomalainen A., Spelbrink J. N., Twinkle mutations associated with autosomal dominant progressive external ophthalmoplegia lead to impaired helicase function and in vivo mtDNA replication stalling. Hum. Mol. Genet. 18, 328–340 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Venegas V., Wang J., Dimmock D., Wong L.-J., Real-time quantitative PCR analysis of mitochondrial DNA content. Curr. Protoc. Hum. Genet. Chapter 19, 19.7.1–19.7.12 (2011). [DOI] [PubMed] [Google Scholar]
- 85.Chen J., Bardes E. E., Aronow B. J., Jegga A. G., ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 37, W305–W311 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fabregat A., Jupe S., Matthews L., Sidiropoulos K., Gillespie M., Garapati P., Haw R., Jassal B., Korninger F., May B., Milacic M., Roca C. D., Rothfels K., Sevilla C., Shamovsky V., Shorser S., Varusai T., Viteri G., Weiser J., Wu G., Stein L., Hermjakob H., D’Eustachio P., The reactome pathway knowledgebase. Nucleic Acids Res. 46, D649–D655 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chi Y., Youn D. Y., Xiaoli A. M., Liu L., Pessin J. B., Yang F., Pessin J. E., Regulation of gene expression during the fasting-feeding cycle of the liver displays mouse strain specificity. J. Biol. Chem. 295, 4809–4821 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fall M. L., Xu D., Lemoyne P., Moussa I. E. B., Beaulieu C., Carisse O., A diverse virome of leafroll-infected grapevine unveiled by dsRNA sequencing. Viruses 12, 1142 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S8
Tables S1 to S4