The first genome-wide study of sulcal depth shows that it is highly genetically discoverable, associated with neurodevelopment.
Abstract
The folding of the human cerebral cortex is a highly genetically regulated process that allows for a much larger surface area to fit into the cranial vault and optimizes functional organization. Sulcal depth is a robust yet understudied measure of localized folding, previously associated with multiple neurodevelopmental disorders. Here, we report the first genome-wide association study of sulcal depth. Through the multivariate omnibus statistical test (MOSTest) applied to vertex-wise measures from 33,748 U.K. Biobank participants (mean age, 64.3 years; 52.0% female), we identified 856 genome-wide significant loci (P < 5 × 10−8). Comparisons with cortical thickness and surface area indicated that sulcal depth has higher locus yield, heritability, and effective sample size. There was a large amount of genetic overlap between these traits, with gene-based analyses indicating strong associations with neurodevelopmental processes. Our findings demonstrate sulcal depth is a promising neuroimaging phenotype that may enhance our understanding of cortical morphology.
INTRODUCTION
During early brain development, the cerebral cortical sheet folds into gyri and sulci in a highly regulated manner because of multiple intrinsic and extrinsic mechanical forces (1–3). This cortical folding not only allows for a much larger surface area to fit into the cranial vault but also reduces distance between neurons, leading to faster signal transmission (2). Accordingly, measures of sulcal morphology are associated with cognitive performance (4), and lack of cortical folding (lissencephaly) is accompanied by severe mental retardation (5). Atypical folding can result from defects in neuronal proliferation, migration, and differentiation, and has been associated with major neurodevelopmental (6–8) and neurodegenerative disorders (9).
Sulcal depth is a rather understudied measure of sulcal morphology, reflecting the convexity or concavity of any given point on the cortical surface. This measure is therefore highly suited for quantification of the primary folding pattern of the cortical sheet, and it has been shown to be insensitive to noise in the form of small wrinkles and to be relatively stable across individuals (10). Sulcal depth is also known to have high test-retest reliability across the brain (11, 12). The few studies using this measure have provided evidence of regional differences between men and women (13) and have shown that sulci become more shallow with aging (14, 15), yet this remains to be fully characterized.
Several studies have indicated that there is a strong genetic component to sulcal depth, which is mostly prenatally determined (11, 16, 17). Sulci are more similar in monozygotic than in dizygotic twins (18), and an estimated 56% of between-subject variance in average depth of the central sulcus is under genetic control (19). Furthermore, Williams syndrome, caused by deletion of a section of chromosome 7, is associated with widespread reductions in sulcal depth (20), which mediate its behavioral symptoms (21, 22). Yet, there has been no large-scale molecular genetics study of this measure.
Here, we provide the first genome-wide association study (GWAS) of sulcal depth, comparing its genetic architecture to the more commonly studied brain morphological measures of cortical thickness and surface area. Given that gene variants are likely to have distributed effects across magnetic resonance imaging (MRI) phenotypes, we targeted a multivariate analysis of a vertex-wise representation of the cortical surface, preventing the need for a multiple-comparison correction or data reduction strategies (23). We therefore applied the multivariate omnibus statistical test (MOSTest) (24) to data from 1153 vertices, using a common template (fsaverage3) with the medial wall vertices excluded. Our primary sample consisted of 33,748 unrelated White British participants of the UK Biobank (UKB), with a mean age of 64.3 years (SD, 7.5 years); 52.0% were female. See figs. S1 and S2 for vertex-wise brain maps showing the distribution of the sulcal depth metric in this sample as well as its Pearson’s correlation with cortical thickness and surface area. We further carried out analyses in two additional samples consisting of (i) 5199 UKB participants (mean age, 62.8 years) that were excluded from the main analyses as they were not of White British descent, and (ii) 8072 participants of the Adolescent Brain Cognitive Development (ABCD) study (mean age, 9.9 years). These samples differed substantially from the discovery sample in terms of ethnicity and age, providing a strong test of generalizability of the reported associations. Following surface reconstruction, we preresidualized all vertices for age, sex, scanner site, a proxy of image quality (25), and the first 20 genetic principal components to control for population stratification. After applying a rank-based inverse normal transformation, MOSTest was performed on the resulting residualized measures, yielding a multivariate association with each single-nucleotide polymorphism (SNP). We additionally repeated the main GWAS analyses while covarying for the mean across all vertices to remove global effects; the findings from these analyses were highly similar to the main analyses, as reported in the Supplementary Materials.
RESULTS
In the discovery sample, MOSTest revealed 856 independent loci, reaching the genome-wide significance threshold of α = 5 × 10−8 for sulcal depth (see Fig. 1A). In comparison, for surface area and thickness, we found 661 and 591 loci, respectively (see figs. S3 and S4). We found that the sets of discovered loci for sulcal depth had acceptable rates of generalization (see fig. S4 and table S2).
Fig. 1. Locus discovery.
(A) Manhattan plot of the multivariate GWAS on sulcal depth, with the observed −log10(P value) of each SNP shown on the y axis. The x axis shows the relative genomic location, grouped by chromosome, and the red dashed line indicates the whole-genome significance threshold of 5 × 10−8. The y axis is clipped at −log10(P value) = 150. (B) Lateral view of the cortex, depicting the color-coded vertex-wise z values for the top hit rs4924345 at chromosome 15, across the discovery sample (top) and the two replication samples (middle and bottom). The left column corresponds to the left hemisphere, and the right column to the right hemisphere. (C) Power plot showing the relation between variance explained by genome-wide significant hits (y axis) and sample size (x axis). The number of hits discovered per modality and the percent explained genetic variance are indicated between brackets in the legend. (D) Bar plot of the mean SNP-based heritability (with 95% confidence interval) across vertices (on the y axis) per modality (x axis). In (C) and (D), sulcal depth is represented in green, surface area in orange, and cortical thickness in purple. ****P ≤ 0.0001.
The effects of discovered top variants on sulcal depth followed gyral and sulcal patterns, and the spatial pattern of effects was highly similar in the two additional samples, as shown in Fig. 1B for the most significant SNP at chromosome 15. Data files S1 to S3 contain information on all discovered loci per trait, including mapped genes, and list the significance of each lead SNP in each sample.
Next, using the MiXeR tool (26, 27), we fitted a Gaussian mixture model of the null and non-null effects to the three GWAS summary statistics, estimating the polygenicity and effect size variance (“discoverability”). The results are summarized in Fig. 1C, depicting the estimated proportion of genetic variance explained by discovered SNPs for each trait as a function of sample size. The horizontal shift of the curve across the different traits indicates that the effective sample size is the highest for sulcal depth and lowest for cortical thickness. Furthermore, the mean heritability of sulcal depth, calculated through linkage disequilibrium score regression (LDSC) (28) applied to each vertex, was significantly higher than for the two other traits (see Fig. 1D), i.e., the higher genetic signal in sulcal depth is also captured by univariate measures.
We additionally performed gene-based analyses through multimarker analysis of genomic annotation (MAGMA) (29). We identified 2010 multiple comparison-corrected (α = 0.05/18,203) significant genes for sulcal depth (i.e., 11% of all 18,203 genes) versus 1486 for area and 1347 for thickness. The lists of all significant genes are provided in the Supplementary Materials.
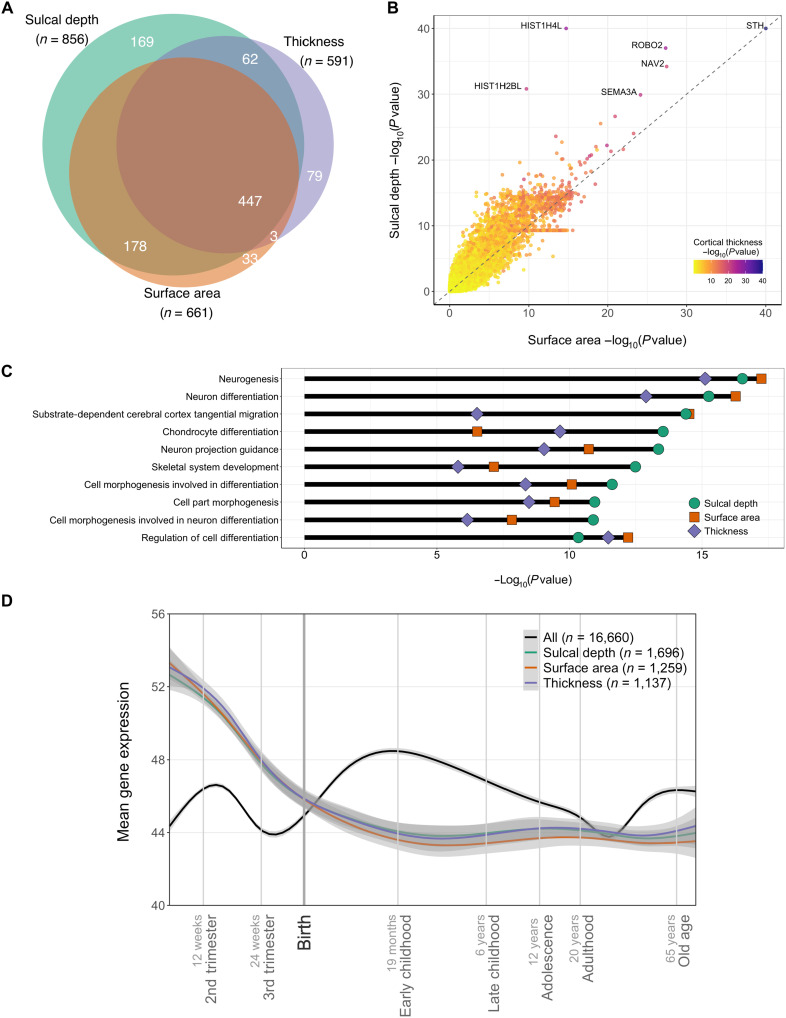
Next, we analyzed the genetic overlap between the three traits at the locus level, gene level, and pathway level. At the locus level, we found that sulcal depth and surface area had 625 loci with overlapping start and end genomic positions (Dice coefficient of 0.82), 509 loci overlapped between sulcal depth and cortical thickness (Dice = 0.70), and 450 loci overlapped between surface area and thickness (Dice = 0.72). A total of 447 loci overlapped across all three traits (see Fig. 2A). We further found that a substantial number of the loci discovered for these cortical traits overlap with those previously identified for a set of brain disorders and cognitive performance (see table S3).
Fig. 2. Genetic overlap.
(A) Venn diagram of the number of discovered loci overlapping between the three different traits. (B) Scatterplot of gene-based P values, with y axis indicating P values for sulcal depth and x axis for those for surface area, and the coloring indicating P values for cortical thickness. Note that −log10(P values) are clipped at 40. (C) Ten most significant gene pathways for sulcal depth, as listed on the y axis, with the −log10(P values) indicated on the x axis for each of the three traits. (D) Mean-normalized expression (y axis) of genes over time (x axis; log10 scale) per trait and over all available genes, as indicated by colors. Gray shading indicates 95% confidence bands.
The large genetic overlap between the traits was also evident at the gene level, as illustrated in Fig. 2B. The top gene STH, thought to play a role in phosphorylation of tau (30), was highly significantly associated with all three traits. ROBO2, NAV2, and SEMA3A, key players in neuronal outgrowth guidance (31–33), were also associated with all three traits. The two histone genes, HIST1H4L and HIST1H2BL, both located in the large histone gene cluster at chromosome 6, were relatively specifically associated with sulcal depth; among its many roles, histone activity is central in regulating gene expression patterns that determine neuronal proliferation and differentiation processes that shape the cerebral cortex (34).
Figure 2C shows results from gene set analyses through MAGMA (29), listing the top 10 most significant gene ontology pathways for sulcal depth together with its P values for the two other traits. We found strong associations with neurogenesis and neuron differentiation pathways, overlapping between all three traits. Associations with neuronal tangential migration were shared by sulcal depth and surface area but much less by cortical thickness, in line with the role of tangential migration of neurons in determining cortical folding (35). Notably, pathways related to chondrocyte differentiation and skeletal system development appeared more specific to sulcal depth, possibly pointing toward early life interactions between cortical folding and the shaping of the cranium (2). Full results from the pathway analyses are provided in data files S7 to S9.
We further coupled the findings of our gene-based analyses to cortical gene expression patterns, derived from postmortem brain tissue of clinically unremarkable donors across the life span (36). As shown in Fig. 2D, the probes tagging genes associated with the three traits showed a distinct profile over the life span, characterized by high prenatal expression and low postnatal expression.
DISCUSSION
Here, we reported the results from the first large-scale molecular genetics study of sulcal depth. With 856 loci discovered, explaining an estimated 32% of its genetic variance, this study has found the highest number of loci for any brain trait considered so far. We further provided evidence that our findings generalize to other populations and carry meaningful biological information.
The direct comparison with surface area and thickness indicated that sulcal depth is more heritable. This may reflect the evolutionary significance of cortical folding, the development of which enabled the advent of a larger brain and optimization of its functional organization (37). A synthesis of the literature suggests that human-specific folding follows from an interplay between mechanical forces and cellular mechanisms that have come about over the course of evolution through mutations of genes primarily coupled to cell cycling and neurogenesis (38). This is in accordance with the specific genes identified, known to play roles in the regulation of neuronal proliferation and migration (31–34). Our findings therefore suggest that the sulcal depth metric is closely aligned with these genetic processes that shape important brain morphological characteristics.
As indicated by the brain maps, genetic effects have opposing directions of effects on some neighboring points in the brain. This is in line with strong differences in the morphology and arrangement of neurons and fibers along cortical folds, varying widely from the gyral crown along the lateral wall down to the sulcal fundus (38, 39). Furthermore, the current findings indicate the presence of widespread genetic effects, illustrating this important characteristic of its genetic architecture; numerous variants are involved, each with a complex pattern of effects spread across the cortex. As shown by our estimates of generalization, these multivariate genetic effects on sulcal depth seem to be rather independent of ethnicity and age (40), emphasizing the fundamental neurobiological relevance for brain morphology.
We further found large genetic overlap between all three morphological brain traits, extending our previous findings that surface area and thickness share the majority of their genetic determinants (24, 41). We found that the genetics of sulcal depth overlaps more with surface area than with cortical thickness, indicating a closer relation between the neurobiological mechanisms determining the degree of cortical folding and surface area. Patterns of tangential migration of neurons are thought to be important drivers of both folding of the cortical sheet and determination of its surface area (3), while its thickness is influenced more by radial migration (42). The results from the pathway analyses appear to confirm this distinction. However, the estimates of overlap, together with the observed phenotypic correlations, also indicate that sulcal depth and surface area do partly capture distinct genetic processes. Mammalian species also vary in their degree of folding independent of surface area (2). Sulcal depth is therefore likely to provide additional information on the molecular genetic influences on brain morphology, capturing variation in the mechanical processes that determine the folding of the cortical sheet, to complement what is found through studies of surface area and cortical thickness. The identified genetic overlap with brain disorders and cognitive ability additionally indicates that the genetics of these brain measures carry clinically relevant information.
In addition to the reported locus overlap, the specific identified genetic variants, genes, and pathways further inform our understanding of cortical morphology and associated disorders. The most significant pathways were particularly relevant for early brain development, with neurogenesis and differentiation ranking highest. This fits very well with a large body of literature on the genetic regulation of the mechanical forces that drive cortical folding (38). It is also in accordance with our findings that the sets of identified genes showed highest expression in fetal cortical tissue. Furthermore, cortical folding has been shown to take place almost entirely prenatally (17), with sulcal patterns at birth being predictive of neurobehavioral outcomes (16). Follow-ups on our work with neuroimaging data across the life span, including infants, are needed to replicate these findings and to further determine spatiotemporal patterns of genetic effects on sulcal depth. Given the reported associations of sulcal morphology with a range of neurodevelopmental and neurodegenerative disorders (6–9) as well as our exploratory findings of genetic overlap with brain disorders, it will also be of interest to investigate more thoroughly how sulcal depth genetics relates to the development of brain disorders over the life span.
We note as a limitation that we are currently lacking a strong mechanistic understanding of what the sulcal depth metric captures. Still, the evolutionary and ontogenetic importance of cortical folding appears to be reflected in the higher heritability compared to cortical thickness and surface area. Our exploration of multivariate genetic associations with sulcal depth attests to this, providing novel insight into the complexity of the human cortex, warranting further investigation into this measure to enhance our understanding of the brain and associated disorders.
MATERIALS AND METHODS
Participants
For the primary analyses, we made use of data from participants of the UKB population cohort, obtained from the data repository under accession number 27412. The composition, setup, and data gathering protocols of the UKB have been extensively described elsewhere (43). UKB has received ethics approval from the National Health Service National Research Ethics Service (reference: 11/NW/0382) and obtained informed consent from its participants. For this study, we selected White British individuals, as determined by self-report and confirmed by genetic principal components analysis (UKB Data-Field 22006), which had undergone the neuroimaging protocol. We made use of T1 MRI scan data released up to March 2020, excluding 771 individuals with bad structural scan quality as indicated by an age- and sex-adjusted Euler number (44) more than 3 SDs lower than the scanner site mean. We further excluded one of each pair of related individuals, as determined through genome-wide complex trait analysis (GCTA), using a threshold of 0.0625 (n = 1138). Our sample size for this analysis was n = 33,748, with a mean age of 64.3 years (SD, 7.5). A total of 52.0% of the sample were female.
For the analyses of generalizability, we made use of the same UKB data and preprocessing steps but restricted our sample to those individuals who were not classified as White British. This left us with n = 5199 individuals, with a mean age of 62.8 years (SD, 7.7), of which 54.1% were female.
We additionally included data from children participating in the ABCD neurodevelopmental study, with complete genetic data and baseline T1 MRI scans from data release 3.0 [NIMH Data Archive (NDA) DOI:10.151.54/1519007] that passed the ABCD quality control procedures (n = 8072). These children had a mean age of 9.9 years (SD, 0.6), and 46.9% were female. All procedures were approved by a central Institutional Review Board (IRB) at the University of California, San Diego, and, in some cases, by individual site IRBs. Parents or guardians provided written informed consent, and children assented before participation.
Data preprocessing
UKB T1-weighted scans were collected from three scanning sites throughout the United Kingdom, all on identically configured Siemens Skyra 3T scanners, with a 32-channel receive head coils. The UKB core neuroimaging team has published extensive information on the applied scanning protocols and procedures, which we refer to for more details (45). ABCD data were collected from 21 acquisition sites using Siemens Prisma, GE 750, and Philips 3T scanners to collect the T1-weighted scans. Scanning protocols were harmonized across sites. Full details of all imaging acquisition protocols used in ABCD are outlined by Casey et al. (46).
All scans were stored locally at the secure computing cluster of the University of Oslo. We applied the standard “recon-all -all” processing pipeline of FreeSurfer v5.3, followed by extracting vertex-wise data for sulcal depth, surface area, and thickness, at ico3 (1284 vertices) and ico4 (5124) resolutions, without applying smoothing. We included both the left and right hemisphere measures and excluded noncortical vertices belonging to the medial wall.
Note that we have chosen sulcal depth as a metric of cortical folding, as it captures vertex-wise localized folding, providing the signed distance from the inflated surface.
We subsequently regressed out age, sex, scanner site, Euler number, and the first 20 genetic principal components from each vertex measure. Following this, we applied rank-based inverse normal transformation (47) to the residuals of each measure, leading to normally distributed measures as input for the GWAS.
We reran the MOSTest analyses as described above on the primary UKB sample, additionally regressing out the mean across all vertices for each of the three traits. The resulting number of loci is shown in table S1.
MOSTest procedure
The MOSTest software is freely available at https://github.com/precimed/mostest, and details about the procedure and its extensive validation have been described previously (24). In brief, consider N variants and M (preresidualized) phenotypes. Let zij be a z score from the univariate association test between ith variant and jth (residualized) phenotype, and zi = (zi1, …, ziM) be the vector of z scores of the ith variant across M phenotypes. Let Z = {zij} be the matrix of z scores, with variants in rows and phenotypes in columns. For each variant, consider a random permutation of its genotypes and let be the matrix of z scores from the univariate association testing between variants with permuted genotypes and phenotypes. A random permutation of genotypes is done once for each variant, and the resulting permuted genotype is tested for association with all phenotypes, therefore preserving correlation structure between phenotypes.
Let be the correlation matrix of , and is its singular valued decomposition (U and V are the orthogonal matrixes, and S is the diagonal matrix, with singular values of on the diagonal). Consider the regularized version of the correlation matrix , where Sr is obtained from S by keeping r largest singular values and replacing the remaining with rth largest. The MOSTest statistic for the ith variant (scalar) is then estimated as , where regularization parameter r is selected separately for cortical area and thickness to maximize the yield of genome-wide significant loci. In this study, we observed the largest yield for cortical surface area with r = 10; the optimal choice for cortical thickness was r = 20 and for sulcal depth r = 30. The distribution of the test statistics under null () is approximated from the observed distribution of the test statistics with permuted genotypes, using the empirical distribution in the 99.99th percentile and gamma distribution in the upper tail, where shape and scale parameters of gamma distribution are fitted to the observed data. The P value of the MOSTest test statistic for the ith variant is then obtained as .
Univariate GWAS procedure
We made use of the UKB v3 imputed data, which have undergone extensive quality control procedures as described by the UKB genetics team (48). After converting the BGEN format to PLINK binary format, we additionally carried out standard quality check procedures, including filtering out individuals with more than 10% missingness, SNPs with more than 5% missingness, and SNPs failing the Hardy-Weinberg equilibrium test at P = 1 × 10−9. We further set a minor allele frequency threshold of 0.005, leaving 9,061,022 SNPs.
For ABCD, we took the genetic data that were part of the third data release, imputed through Trans-Omics for Precision Medicine (TOPMED), and mapped this back from genome build hg38 to hg19. We subsequently applied identical postimputation quality check procedures and filters for the UKB genetic data, leaving 13,131,314 SNPs.
We have previously calculated that the number of features provided by fsaverage3, 1153 vertices following exclusion of the medial wall, leads to the maximum number of loci identified through MOSTest, compared to other resolutions (49). We therefore used the fsaverage3 resolution data for the input to MOSTest and for calculating heritability of the univariate vertex-wise data (described below). For visualization of the regionalization of the results, in the form of brain maps, we additionally carried out univariate GWAS on the 5124 vertices that make up fsaverage4, i.e., one level of resolution above fsaverage3. This was done only to improve the resolution of the visualizations. The univariate GWAS on each of the preresidualized and normalized measures were carried out using the standard additive model of linear association between genotype vector, gj, and phenotype vector, y.
Heritability
We calculated the SNP-based heritability for each vertex at fsaverage3 resolution by applying LDSC to the univariate GWAS summary statistics with default settings (28).
Locus definitions
Independent significant SNPs and genomic loci were identified from the MOSTest summary statistics in accordance with the psychiatric genomics consortium (PGC) locus definition, as also used in Functional Mapping and Annotation of GWAS (FUMA) SNP2GENE (50). First, we select a subset of SNPs that pass the genome-wide significance threshold of 5 × 10−8 and use PLINK to perform a clumping procedure at linkage disequilibrium (LD) r2 = 0.6 to identify the list of independent significant SNPs. Second, we clump the list of independent significant SNPs at LD r2 = 0.1 threshold to identify lead SNPs. Third, we query the reference panel for all candidate SNPs in LD r2 of 0.1 or higher with any lead SNPs. Furthermore, for each lead SNP, its corresponding genomic loci is defined as a contiguous region of the lead SNPs’ chromosome, containing all candidate SNPs in r2 = 0.1 or higher LD with the lead SNP. Last, adjacent genomic loci are merged together if they are separated by less than 250 kb. Allele LD correlations are computed from the European (EUR) population of the 1000 Genomes phase 3 data. We additionally performed clumping according to the definition used by the Enhancing Neuroimaging Genetics through Meta-analysis Consortium to allow for comparison with previous imaging GWAS studies. According to this definition, loci were formed through PLINK using a P value threshold of 5 × 10−8 (--clump-p1) and LD cutoffs of 1 Mb (--clump-kb) and r2 < 0.2 (--clump-r2). Please see table S1 for the number of lead SNPs and loci according to both definitions. We made use of the FUMA online platform (https://fuma.ctglab.nl/) to map significant SNPs from the MOSTest analyses to genes.
Genetic overlap
Genetic overlap between the different MOSTest feature sets was operationalized as the number of significant loci that were physically overlapping between each pair of summary statistics. Loci from the pair are considered physically overlapping if their boundaries, their start and end genomic positions as determined through clumping, overlap. Contrary to metrics of global genetic correlation, this approach can be applied to MOSTest summary statistics and is insensitive to mixed directions of effects that would lower estimates of genetic correlation.
The Dice coefficient for each pair of traits was calculated as the number of overlapping loci divided by the sum of the total number of discovered loci for both traits.
MiXeR analysis
We applied a causal mixture model (26, 27) to estimate the percentage of variance explained by genome-wide significant SNPs as a function of sample size. For each SNP, i, MiXeR models its additive genetic effect of allele substitution, βi, as a point normal mixture, , where π1 represents the proportion of non-null SNPs (“polygenicity”) and represents variance of effect sizes of non-null SNPs (discoverability). Then, for each SNP, j, MiXeR incorporates LD information and allele frequencies for 9,997,231 SNPs extracted from 1000 Genomes phase 3 data to estimate the expected probability distribution of the signed test statistic; , where N is sample size, Hi indicates heterozygosity of ith SNP, rij indicates allelic correlation between ith and jth SNPs, and is the residual variance. Furthermore, the three parameters, , are fitted by direct maximization of the likelihood function. Fitting the univariate MiXeR model does not depend on the sign of zj, allowing us to calculate ∣zj∣ from MOSTest P values. Last, given the estimated parameters of the model, the power curve S(N) is then calculated from the posterior distribution p(δj∣zj, N).
Gene set analyses
We carried out gene-based analyses using MAGMA v1.08 with default settings, which entail the application of an SNP-wide mean model and use of the 1000 Genomes phase 3 EUR reference panel to the three MOSTest summary statistics. Gene set analyses were carried out by applying MAGMA to the gene-level output, restricting the sets under investigation to those that are part of the Gene Ontology biological processes subset (n = 7522), as listed in the Molecular Signatures Database (c5.bp.v7.1).
Regarding the results from the gene-based analyses, in Fig. 2, we note that there is a horizontal line visible at –P = 5 × 10−10, caused by many genes having this exact P value. This is due to MAGMA switching to permutation when its numerical integration approach fails. MAGMA uses 1 × 10−9 permutations, so when the observed is more extreme than this, this is the resulting P value.
Gene expression analyses
We made use of gene expression data derived from brain tissue from 56 clinically unremarkable donors, ranging in age from 5 weeks post conception to 82 years (36). We took the data as preprocessed in (36), selecting for each gene the probe with the highest differential stability, n = 16,660. Given the relatively high homogeneity of expression patterns across cortical brain samples (51), we subsequently averaged over 13 cortical regions, within donor, and normalized the expression values, within probe, across donors, to a range between 0 (lowest observed value) and 100 (highest observed value). As a check, we reran these analyses while restricting to samples from the individual cortical lobes. For each of the lobes, we observed expression patterns that were highly similar to those reported in the main analyses, with the gene sets showing high prenatal expression that dropped off at birth. Plotting of the mean expression over time per gene set was done with ggplot2 in R v4.0.3., with geom_smooth(method=”gam”) using default settings.
Acknowledgments
This work was partly performed on the TSD (Tjenester for Sensitive Data) facilities, owned by the University of Oslo, operated and developed by the TSD service group at the University of Oslo, IT Department (USIT) (tsd-drift@usit.uio.no). Computations were also performed on resources provided by UNINETT Sigma2, the National Infrastructure for High-Performance Computing and Data Storage in Norway. Data used in the preparation of this article were obtained from the ABCD Study (https://abcdstudy.org), held in the NIMH Data Archive (NDA). This is a multisite, longitudinal study designed to recruit more than 10,000 children aged 9 to 10 and follow them over 10 years into early adulthood. The ABCD Study is supported by the National Institutes of Health and additional federal partners under award numbers U01DA041048, U01DA050989, U01DA051016, U01DA041022, U01DA051018, U01DA051037, U01DA050987, U01DA041174, U01DA041106, U01DA041117, U01DA041028, U01DA041134, U01DA050988, U01DA051039, U01DA041156, U01DA041025, U01DA041120, U01DA051038, U01DA041148, U01DA041093, U01DA041089, U24DA041123, and U24DA041147. A full list of supporters is available at https://abcdstudy.org/federal-partners.html. A listing of participating sites and a complete listing of the study investigators can be found at https://abcdstudy.org/consortium_members/. ABCD consortium investigators designed and implemented the study and/or provided data but did not necessarily participate in the analysis or writing of this report. This manuscript reflects the views of the authors and may not reflect the opinions or views of the NIH or ABCD consortium investigators. The ABCD data repository grows and changes over time. The ABCD data used in this report came from http://dx.doi.org/10.15154/1519007.
Funding: The authors were funded by the Research Council of Norway (276082, 213837, 223273, 204966/F20, 229129, 249795/F20, 225989, 248778, 249795, 298646, and 300767), the South-Eastern Norway Regional Health Authority (2013-123, 2014-097, 2015-073, 2016-064, 2017-004, and 2019-101), Stiftelsen Kristian Gerhard Jebsen (SKGJ-Med-008), the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (ERC Starting Grant, grant agreement no. 802998), ERA-Net Cofund through the ERA PerMed project “Implement,” and the National Institutes of Health (R01MH100351, R01GM104400, U24DA055330, R01MH111359, and U24DA041123; principal investigator: A.M.D.).
Author contributions: Conceptualizations: D.v.d.M., O.A.A., and A.M.D. Data preprocessing: D.v.d.M., A.A.S., and T.K. Analyses: D.v.d.M. and A.A.S., with conceptual input from A.M.D., D.R., J.R., C.d.L., O.A.A., L.T.W., and T.K. Results interpretation: All authors. Writing: D.v.d.M. Review and editing: All authors.
Competing interests: C.d.L. is funded by Hoffmann-La Roche. O.A.A. has received speaker’s honorarium from Lundbeck and is a consultant to HealthLytix. A.M.D. is the founder of and holds equity in CorTechs Labs Inc. and serves on its Scientific Advisory Board. A.M.D. is also a member of the Scientific Advisory Board of Human Longevity Inc. and receives funding through research agreements with General Electric Healthcare and Medtronic Inc. The terms of these arrangements have been reviewed and approved by UCSD in accordance with its conflict of interest policies. A.M.D. is an inventor on a patent related to this work, filed by CorTechs Labs Inc. (9 US-7324842B2, filed 22 January 2002, published 29-01-2008). The other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The data incorporated in this work were gathered from the UKB under accession number 27412. The code used for the analyses is available via https://github.com/precimed/mostest (GPLv3 license) and at 10.5281/zenodo.5266328.
Supplementary Materials
This PDF file includes:
Supplementary Text
Figs. S1 to S4
Tables S1 to S3
Legends for data S1 to S9
References
Other Supplementary Material for this manuscript includes the following:
Data S1 to S9
REFERENCES AND NOTES
- 1.Van Essen D. C., A tension-based theory of morphogenesis and compact wiring in the central nervous system. Nature 385, 313–318 (1997). [DOI] [PubMed] [Google Scholar]
- 2.Striedter G. F., Srinivasan S., Monuki E. S., Cortical folding: When, where, how, and why? Annu. Rev. Neurosci. 38, 291–307 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Ronan L., Voets N., Rua C., Alexander-Bloch A., Hough M., Mackay C., Crow T. J., James A., Giedd J. N., Fletcher P. C., Differential tangential expansion as a mechanism for cortical gyrification. Cereb. Cortex 24, 2219–2228 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gautam P., Anstey K. J., Wen W., Sachdev P. S., Cherbuin N., Cortical gyrification and its relationships with cortical volume, cortical thickness, and cognitive performance in healthy mid-life adults. Behav. Brain Res. 287, 331–339 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Ross M. E., Walsh C. A., Human brain malformations and their lessons for neuronal migration. Annu. Rev. Neurosci. 24, 1041–1070 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Penttilä J., Paillère-Martinot M.-L., Martinot J.-L., Ringuenet D., Wessa M., Houenou J., Gallarda T., Bellivier F., Galinowski A., Bruguière P., Pinabel F., Leboyer M., Olié J.-P., Duchesnay E., Artiges E., Mangin J.-F., Cachia A., Cortical folding in patients with bipolar disorder or unipolar depression. J. Psychiat. Neurosci. JPN 34, 127–135 (2009). [PMC free article] [PubMed] [Google Scholar]
- 7.Cachia A., Paillère-Martinot M.-L., Galinowski A., Januel D., de Beaurepaire R., Bellivier F., Artiges E., Andoh J., Bartrés-Faz D., Duchesnay E., Rivière D., Plaze M., Mangin J.-F., Martinot J.-L., Cortical folding abnormalities in schizophrenia patients with resistant auditory hallucinations. NeuroImage 39, 927–935 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Nordahl C. W., Dierker D., Mostafavi I., Schumann C. M., Rivera S. M., Amaral D. G., Van Essen D. C., Cortical folding abnormalities in autism revealed by surface-based morphometry. J. Neurosci. 27, 11725–11735 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pereira J. B., Ibarretxe-Bilbao N., Marti M., Compta Y., Junqué C., Bargallo N., Tolosa E., Assessment of cortical degeneration in patients with Parkinson’s disease by voxel-based morphometry, cortical folding, and cortical thickness. Hum. Brain Mapp. 33, 2521–2534 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischl B., Sereno M. I., Dale A. M., Cortical surface-based analysis: II: Inflation, flattening, and a surface-based coordinate system. NeuroImage 9, 195–207 (1999). [DOI] [PubMed] [Google Scholar]
- 11.Pizzagalli F., Auzias G., Yang Q., Mathias S. R., Faskowitz J., Boyd J. D., Amini A., Rivière D., McMahon K. L., de Zubicaray G. I., Martin N. G., Mangin J.-F., Glahn D. C., Blangero J., Wright M. J., Thompson P. M., Kochunov P., Jahanshad N., The reliability and heritability of cortical folds and their genetic correlations across hemispheres. Commun. Biol. 3, 510 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Madan C. R., Robust estimation of sulcal morphology. Brain Informatics 6, 5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang G., Bozek J., Han M., Gao J., Constructing and evaluating a cortical surface atlas and analyzing cortical sex differences in young Chinese adults. Hum. Brain Mapp. 41, 2495–2513 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin K., Zhang T., Shaw M., Sachdev P., Cherbuin N., Relationship between sulcal characteristics and brain aging. Front. Aging Neurosci. 10, 339 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Díaz-Caneja C. M., Alloza C., Gordaliza P. M., Fernández-Pena A., de Hoyos L., Santonja J., Buimer E. E. L., van Haren N. E. M., Cahn W., Arango C., Kahn R. S., Hulshoff Pol H. E., Schnack H. G., Janssen J., Sex differences in lifespan trajectories and variability of human sulcal and gyral morphology. Cereb. Cortex 31, 5107–5120 (2021). [DOI] [PubMed] [Google Scholar]
- 16.Dubois J., Benders M., Borradori-Tolsa C., Cachia A., Lazeyras F., Ha-Vinh Leuchter R., Sizonenko S. V., Warfield S. K., Mangin J. F., Hüppi P. S., Primary cortical folding in the human newborn: An early marker of later functional development. Brain 131, 2028–2041 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garel C., Chantrel E., Brisse H., Elmaleh M., Luton D., Oury J.-F., Sebag G., Hassan M., Fetal cerebral cortex: Normal gestational landmarks identified using prenatal MR imaging. Am. J. Neuroradiol. 22, 184–189 (2001). [PMC free article] [PubMed] [Google Scholar]
- 18.Lohmann G., Von Cramon D. Y., Steinmetz H., Sulcal variability of twins. Cereb. Cortex 9, 754–763 (1999). [DOI] [PubMed] [Google Scholar]
- 19.McKay D. R., Kochunov P., Cykowski M. D., Kent J. W., Laird A. R., Lancaster J. L., Blangero J., Glahn D. C., Fox P. T., Sulcal depth-position profile is a genetically mediated neuroscientific trait: Description and characterization in the central sulcus. J. Neurosci. 33, 15618–15625 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kippenhan J. S., Olsen R. K., Mervis C. B., Morris C. A., Kohn P., Meyer-Lindenberg A., Berman K. F., Genetic contributions to human gyrification: Sulcal morphometry in williams syndrome. J. Neurosci. 25, 840–7846 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan C. C., Schork A. J., Brown T. T., Spencer B. E., Akshoomoff N., Chen C.-H., Kuperman J. M., Hagler D. J. J., Steen V. M., Le Hellard S., Håberg A. K., Espeseth T., Andreassen O. A., Dale A. M., Jernigan T. L., Halgren E.; Pediatric Imaging, Neurocognition and Genetics Study, Alzheimer’s Disease Neuroimaging Initiative , Williams syndrome neuroanatomical score associates with GTF2IRD1 in large-scale magnetic resonance imaging cohorts: A proof of concept for multivariate endophenotypes. Transl. Psychiatry 8, 114 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fan C. C., Brown T. T., Bartsch H., Kuperman J. M., Hagler D. J. J., Schork A., Searcy Y., Bellugi U., Halgren E., Dale A. M., Williams syndrome-specific neuroanatomical profile and its associations with behavioral features. NeuroImage Clin. 15, 343–347 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Der Sluis S., Verhage M., Posthuma D., Dolan C. V., Phenotypic complexity, measurement bias, and poor phenotypic resolution contribute to the missing heritability problem in genetic association studies. PLOS ONE 5, e13929 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van der Meer D., Frei O., Kaufmann T., Shadrin A. A., Devor A., Smeland O. B., Thompson W. K., Fan C. C., Holland D., Westlye L. T., Andreassen O. A., Dale A. M., Understanding the genetic determinants of the brain with MOSTest. Nat. Commun. 11, 3512 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sánchez J. M., de Jong J. J. A., Drenthen G. S., Beran M., Backes W. H., Stehouwer C. D. A., Schram M. T., Linden D. E. J., Jansen J. F. A., Monereo-Sánchez J., de Jong J. J. A., Drenthen G. S., Beran M., Backes W. H., Stehouwer C. D. A., Schram M. T., Linden D. E. J., Jansen J. F. A., Quality control strategies for brain MRI segmentation and parcellation: Practical approaches and recommendations-insights from The Maastricht Study. NeuroImage 237, 118174 (2021). [DOI] [PubMed] [Google Scholar]
- 26.Holland D., Frei O., Desikan R., Fan C.-C., Shadrin A. A., Smeland O. B., Sundar V. S., Thompson P., Andreassen O. A., Dale A. M., Beyond SNP heritability: Polygenicity and discoverability of phenotypes estimated with a univariate Gaussian mixture model. PLOS Genet. 16, e1008612 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frei O., Holland D., Smeland O. B., Shadrin A. A., Fan C. C., Maeland S., O’Connell K. S., Wang Y., Djurovic S., Thompson W. K., Andreassen O. A., Dale A. M., Bivariate causal mixture model quantifies polygenic overlap between complex traits beyond genetic correlation. Nat. Commun. 10, 2417 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bulik-Sullivan B. K., Loh P.-R., Finucane H. K., Ripke S., Yang J., Patterson N., Daly M. J., Price A. L., Neale B. M., LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Leeuw C. A., Mooij J. M., Heskes T., Posthuma D., MAGMA: Generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 11, e1004219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Conrad C., Vianna C., Freeman M., Davies P., A polymorphic gene nested within an intron of the tau gene: Implications for Alzheimer’s disease. Proc. Natl. Acad. Sci. 99, 7751–7756 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.López-Bendito G., Flames N., Ma L., Fouquet C., Di Meglio T., Chedotal A., Tessier-Lavigne M., Marín O., Robo1 and Robo2 cooperate to control the guidance of major axonal tracts in the mammalian forebrain. J. Neurosci. 27, 3395–3407 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maes T., Barceló A., Buesa C., Neuron navigator: A human gene family with homology to unc-53, a cell guidance gene from Caenorhabditis elegans. Genomics 80, 21–30 (2002). [DOI] [PubMed] [Google Scholar]
- 33.Hanchate N. K., Giacobini P., Lhuillier P., Parkash J., Espy C., Fouveaut C., Leroy C., Baron S., Campagne C., Vanacker C., Collier F., Cruaud C., Meyer V., García-Piñero A., Dewailly D., Cortet-Rudelli C., Gersak K., Metz C., Chabrier G., Pugeat M., Young J., Hardelin J.-P., Prevot V., Dodé C., SEMA3A, a gene involved in axonal pathfinding, is mutated in patients with Kallmann syndrome. PLOS Genet. 8, e1002896 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lilja T., Heldring N., Hermanson O., Like a rolling histone: Epigenetic regulation of neural stem cells and brain development by factors controlling histone acetylation and methylation. Biochim. Biophys. Acta, Gen. Subj. 1830, 2354–2360 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Reillo I., de Juan Romero C., García-Cabezas M. Á., Borrell V., A role for intermediate radial glia in the tangential expansion of the mammalian cerebral cortex. Cereb. Cortex 21, 1674–1694 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Kang H. J., Kawasawa Y. I., Cheng F., Zhu Y., Xu X., Li M., Sousa A. M. M., Pletikos M., Meyer K. A., Sedmak G., Guennel T., Shin Y., Johnson M. B., Krsnik Ž., Mayer S., Fertuzinhos S., Umlauf S., Lisgo S. N., Vortmeyer A., Weinberger D. R., Mane S., Hyde T. M., Huttner A., Reimers M., Kleinman J. E., Šestan N., Spatio-temporal transcriptome of the human brain. Nature 478, 483–489 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fernández V., Llinares-Benadero C., Borrell V., Cerebral cortex expansion and folding: What have we learned? EMBO J. 35, 1021–1044 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Llinares-Benadero C., Borrell V., Deconstructing cortical folding: Genetic, cellular and mechanical determinants. Nat. Rev. Neurosci. 20, 161–176 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Smart I. H., McSherry G. M., Gyrus formation in the cerebral cortex of the ferret. II. Description of the internal histological changes. J. Anat. 147, 27 (1986). [PMC free article] [PubMed] [Google Scholar]
- 40.Loughnan R. J., Shadrin A. A., Frei O., van der Meer D., Zhao W., Palmer C. E., Thompson W. E., Makowski C., Jernigan T., Andreassen O., Fan C. C., Dale A., Generalization of cortical MOSTest genome-wide associations within and across samples. bioRxiv, 2021.04.23.441215 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van der Meer D., Frei O., Kaufmann T., Chen C.-H., Thompson W. K., O’Connell K. S., Monereo Sánchez J., Linden D. E. J., Westlye L. T., Dale A. M., Andreassen O. A., Quantifying the polygenic architecture of the human cerebral cortex: Extensive genetic overlap between cortical thickness and surface area. Cereb. Cortex 30, 5597–5603 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lui J. H., Hansen D. V., Kriegstein A. R., Development and evolution of the human neocortex. Cell 146, 18–36 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sudlow C., Gallacher J., Allen N., Beral V., Burton P., Danesh J., Downey P., Elliott P., Green J., Landray M., UK Biobank: An open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 12, e1001779 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosen A. F. G., Roalf D. R., Ruparel K., Blake J., Seelaus K., Villa L. P., Ciric R., Cook P. A., Davatzikos C., Elliott M. A., de La Garza A. G., Gennatas E. D., Quarmley M., Schmitt J. E., Shinohara R. T., Tisdall M. D., Craddock R. C., Gur R. E., Gur R. C., Satterthwaite T. D., Quantitative assessment of structural image quality. NeuroImage 169, 407–418 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller K. L., Alfaro-Almagro F., Bangerter N. K., Thomas D. L., Yacoub E., Xu J., Bartsch A. J., Jbabdi S., Sotiropoulos S. N., Andersson J. L. R., Griffanti L., Douaud G., Okell T. W., Weale P., Dragonu I., Garratt S., Hudson S., Collins R., Jenkinson M., Matthews P. M., Smith S. M., Multimodal population brain imaging in the UK Biobank prospective epidemiological study. Nat. Neurosci. 19, 1523–1536 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Casey B. J., Cannonier T., Conley M. I., Cohen A. O., Barch D. M., Heitzeg M. M., Soules M. E., Teslovich T., Dellarco D. V., Garavan H., Orr C. A., Wager T. D., Banich M. T., Speer N. K., Sutherland M. T., Riedel M. C., Dick A. S., Bjork J. M., Thomas K. M., Chaarani B., Mejia M. H., Hagler D. J. J., Cornejo M. D., Sicat C. S., Harms M. P., Dosenbach N. U. F., Rosenberg M., Earl E., Bartsch H., Watts R., Polimeni J. R., Kuperman J. M., Fair D. A., Dale A. M.; ABCD Imaging Acquisition Workgroup , The Adolescent Brain Cognitive Development (ABCD) study: Imaging acquisition across 21 sites. Dev. Cogn. Neurosci. 32, 43–54 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beasley T. M., Erickson S., Allison D. B., Rank-based inverse normal transformations are increasingly used, but are they merited? Behav. Genet. 39, 580–595 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bycroft C., Freeman C., Petkova D., Band G., Elliott L. T., Sharp K., Motyer A., Vukcevic D., Delaneau O., O’Connell J., Cortes A., Welsh S., Young A., Effingham M., McVean G., Leslie S., Allen N., Donnelly P., Marchini J., The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shadrin A. A., Kaufmann T., van der Meer D., Palmer C. E., Makowski C., Loughnan R., Jernigan T. L., Seibert T. M., Hagler D. J., Smeland O. B., Chu Y., Lin A., Cheng W., Hindley G., Thompson W. K., Fan C. C., Holland D., Westlye L. T., Frei O., Andreassen O. A., Dale A. M., Multivariate genome-wide association study identifies 780 unique genetic loci associated with cortical morphology. bioRxiv, 2020.10.22.350298 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Watanabe K., Taskesen E., Bochoven A., Posthuma D., Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 8, 1826 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hawrylycz M., Miller J. A., Menon V., Feng D., Dolbeare T., Guillozet-Bongaarts A. L., Jegga A. G., Aronow B. J., Lee C.-K., Bernard A., Glasser M. F., Dierker D. L., Menche J., Szafer A., Collman F., Grange P., Berman K. A., Mihalas S., Yao Z., Stewart L., Barabási A.-L., Schulkin J., Phillips J., Ng L., Dang C., Haynor D. R., Jones A., Van Essen D. C., Koch C., Lein E., Canonical genetic signatures of the adult human brain. Nat. Neurosci. 18, 1832–1844 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alemán-Gómez Y., Janssen J., Schnack H., Balaban E., Pina-Camacho L., Alfaro-Almagro F., Castro-Fornieles J., Otero S., Baeza I., Moreno D., Bargalló N., Parellada M., Arango C., Desco M., The human cerebral cortex flattens during adolescence. J. Neurosci. 33, 15004–15010 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O’Reilly P. F., Hoggart C. J., Pomyen Y., Calboli F. C. F., Elliott P., Jarvelin M.-R., Coin L. J. M., MultiPhen: Joint model of multiple phenotypes can increase discovery in GWAS. PLOS ONE 7, e34861 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van der Sluis S., Posthuma D., Dolan C. V., TATES: Efficient multivariate genotype-phenotype analysis for genome-wide association studies. PLOS Genet. 9, e1003235 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Willer C. J., Li Y., Abecasis G. R., METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wightman D. P., Jansen I. E., Savage J. E., Shadrin A. A., Bahrami S., Rongve A., Børte S., Winsvold B. S., Drange O. K., Martinsen A. E., Skogholt A. H., Willer C., Bråthen G., Bosnes I., Nielsen J. B., Fritsche L., Thomas L. F., Pedersen L. M., Gabrielsen M. E., Johnsen M. B., Meisingset T. W., Zhou W., Proitsi P., Hodges A., Dobson R., Velayudhan L.; 23andMe Research Team, Sealock J. M., Davis L. K., Pedersen N. L., Reynolds C. A., Karlsson I. K., Magnusson S., Stefansson H., Thordardottir S., Jonsson P. V., Snaedal J., Zettergren A., Skoog I., Kern S., Waern M., Zetterberg H., Blennow K., Stordal E., Hveem K., Zwart J.-A., Athanasiu L., Saltvedt I., Sando S. B., Ulstein I., Djurovic S., Fladby T., Aarsland D., Selbæk G., Ripke S., Stefansson K., Andreassen O. A., Posthuma D., Largest GWAS (N=1,126,563) of Alzheimer’s disease implicates microglia and immune cells. medRxiv, 2020.11.20.20235275 (2020). [Google Scholar]
- 57.Mullins N., Forstner A. J., O’Connell K. S., Coombes B., Coleman J. R. I., Qiao Z., Als T. D., Bigdeli T. B., Børte S., Bryois J., Charney A. W., Drange O. K., Gandal M. J., Hagenaars S. P., Ikeda M., Kamitaki N., Kim M., Krebs K., Panagiotaropoulou G., Schilder B. M., Sloofman L. G., Steinberg S., Trubetskoy V., Winsvold B. S., Won H.-H., Abramova L., Adorjan K., Agerbo E., Eissa M. A., Albani D., Alliey-Rodriguez N., Anjorin A., Antilla V., Antoniou A., Awasthi S., Baek J. H., Bækvad-Hansen M., Bass N., Bauer M., Beins E. C., Bergen S. E., Birner A., Pedersen C. B., Bøen E., Boks M. P., Bosch R., Brum M., Brumpton B. M., Brunkhorst-Kanaan N., Budde M., Bybjerg-Grauholm J., Byerley W., Cairns M., Casas M., Cervantes P., Clarke T.-K., Cruceanu C., Cuellar-Barboza A., Cunningham J., Curtis D., Czerski P. M., Dale A. M., Dalkner N., David F. S., Degenhardt F., Djurovic S., Dobbyn A. L., Douzenis A., Elvsåshagen T., Escott-Price V., Ferrier I. N., Fiorentino A., Foroud T. M., Forty L., Frank J., Frei O., Freimer N. B., Frisén L., Gade K., Garnham J., Gelernter J., Pedersen M. G., Gizer I. R., Gordon S. D., Gordon-Smith K., Greenwood T. A., Grove J., Guzman-Parra J., Ha K., Haraldsson M., Hautzinger M., Heilbronner U., Hellgren D., Herms S., Hoffmann P., Holmans P. A., Huckins L., Jamain S., Johnson J. S., Kalman J. L., Kamatani Y., Kennedy J. L., Kittel-Schneider S., Knowles J. A., Kogevinas M., Koromina M., Kranz T. M., Kranzler H. R., Kubo M., Kupka R., Kushner S. A., Lavebratt C., Lawrence J., Leber M., Lee H.-J., Lee P. H., Levy S. E., Lewis C., Liao C., Lucae S., Lundberg M., Mac Intyre D. J., Magnusson S. H., Maier W., Maihofer A., Malaspina D., Maratou E., Martinsson L., Mattheisen M., McCarroll S. A., McGregor N. W., Guffin P. M., McKay J. D., Medeiros H., Medland S. E., Millischer V., Montgomery G. W., Moran J. L., Morris D. W., Mühleisen T. W., O’Brien N., O’Donovan C., Olde Loohuis L. M., Oruc L., Papiol S., Pardiñas A. F., Perry A., Pfennig A., Porichi E., Potash J. B., Quested D., Raj T., Rapaport M. H., De Paulo J. R., Regeer E. J., Rice J. P., Rivas F., Rivera M., Roth J., Roussos P., Ruderfer D. M., Sánchez-Mora C., Schulte E. C., Senner F., Sharp S., Shilling P. D., Sigurdsson E., Sirignano L., Slaney C., Smeland O. B., Smith D. J., Sobell J. L., Hansen C. S., Artigas M. S., Spijker A. T., Stein D. J., Strauss J. S., Świątkowska B., Terao C., Thorgeirsson T. E., Toma C., Tooney P., Tsermpini E.-E., Vawter M. P., Vedder H., Walters J. T. R., Witt S. H., Xi S., Xu W., Yang J. M. K., Young A. H., Young H., Zandi P. P., Zhou H., Zillich L.; HUNT All-In Psychiatry, Adolfsson R., Agartz I., Alda M., Alfredsson L., Babadjanova G., Backlund L., Baune B. T., Bellivier F., Bengesser S., Berrettini W. H., Blackwood D. H. R., Boehnke M., Børglum A. D., Breen G., Carr V. J., Catts S., Corvin A., Craddock N., Dannlowski U., Dikeos D., Esko T., Etain B., Ferentinos P., Frye M., Fullerton J. M., Gawlik M., Gershon E. S., Goes F. S., Green M. J., Grigoroiu-Serbanescu M., Hauser J., Henskens F., Hillert J., Hong K. S., Hougaard D. M., Hultman C. M., Hveem K., Iwata N., Jablensky A. V., Jones I., Jones L. A., Kahn R. S., Kelsoe J. R., Kirov G., Landén M., Leboyer M., Lewis C. M., Li Q. S., Lissowska J., Lochner C., Loughland C., Martin N. G., Mathews C. A., Mayoral F., McElroy S. L., McIntosh A. M., McMahon F. J., Melle I., Michie P., Milani L., Mitchell P. B., Morken G., Mors O., Mortensen P. B., Mowry B., Müller-Myhsok B., Myers R. M., Neale B. M., Nievergelt C. M., Nordentoft M., Nöthen M. M., O’Donovan M. C., Oedegaard K. J., Olsson T., Owen M. J., Paciga S. A., Pantelis C., Pato C., Pato M. T., Patrinos G. P., Perlis R. H., Posthuma D., Ramos-Quiroga J. A., Reif A., Reininghaus E. Z., Ribasés M., Rietschel M., Ripke S., Rouleau G. A., Saito T., Schall U., Schalling M., Schofield P. R., Schulze T. G., Scott L. J., Scott R. J., Serretti A., Weickert C. S., Smoller J. W., Stefansson H., Stefansson K., Stordal E., Streit F., Sullivan P. F., Turecki G., Vaaler A. E., Vieta E., Vincent J. B., Waldman I. D., Weickert T. W., Werge T., Wray N. R., Zwart J.-A., Biernacka J. M., Nurnberger J. I., Cichon S., Edenberg H. J., Stahl E. A., Quillin A. M., Florio A. D., Ophoff R. A., Andreassen O. A., Genome-wide association study of more than 40,000 bipolar disorder cases provides new insights into the underlying biology. Nat. Genet. 53, 817–829 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Howard D. M., Adams M. J., Shirali M., Clarke T.-K., Marioni R. E., Davies G., Coleman J. R. I., Alloza C., Shen X., Barbu M. C., Wigmore E. M., Gibson J.; 23andMe Research Team, Hagenaars S. P., Lewis C. M., Ward J., Smith D. J., Sullivan P. F., Haley C. S., Breen G., Deary I. J., McIntosh A. M., Genome-wide association study of depression phenotypes in UK Biobank identifies variants in excitatory synaptic pathways. Nat. Commun. 9, 1–10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schizophrenia Working Group of the Psychiatric Genomics Consortium , Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Savage J. E., Jansen P. R., Stringer S., Watanabe K., Bryois J., de Leeuw C. A., Nagel M., Awasthi S., Barr P. B., Coleman J. R. I., Grasby K. L., Hammerschlag A. R., Kaminski J. A., Karlsson R., Krapohl E., Lam M., Nygaard M., Reynolds C. A., Trampush J. W., Young H., Zabaneh D., Hägg S., Hansell N. K., Karlsson I. K., Linnarsson S., Montgomery G. W., Muñoz-Manchado A. B., Quinlan E. B., Schumann G., Skene N. G., Webb B. T., White T., Arking D. E., Avramopoulos D., Bilder R. M., Bitsios P., Burdick K. E., Cannon T. D., Chiba-Falek O., Christoforou A., Cirulli E. T., Congdon E., Corvin A., Davies G., Deary I. J., DeRosse P., Dickinson D., Djurovic S., Donohoe G., Conley E. D., Eriksson J. G., Espeseth T., Freimer N. A., Giakoumaki S., Giegling I., Gill M., Glahn D. C., Hariri A. R., Hatzimanolis A., Keller M. C., Knowles E., Koltai D., Konte B., Lahti J., Le Hellard S., Lencz T., Liewald D. C., London E., Lundervold A. J., Malhotra A. K., Melle I., Morris D., Need A. C., Ollier W., Palotie A., Payton A., Pendleton N., Poldrack R. A., Räikkönen K., Reinvang I., Roussos P., Rujescu D., Sabb F. W., Scult M. A., Smeland O. B., Smyrnis N., Starr J. M., Steen V. M., Stefanis N. C., Straub R. E., Sundet K., Tiemeier H., Voineskos A. N., Weinberger D. R., Widen E., Yu J., Abecasis G., Andreassen O. A., Breen G., Christiansen L., Debrabant B., Dick D. M., Heinz A., Hjerling-Leffler J., Ikram M. A., Kendler K. S., Martin N. G., Medland S. E., Pedersen N. L., Plomin R., Polderman T. J. C., Ripke S., van der Sluis S., Sullivan P. F., Vrieze S. I., Wright M. J., Posthuma D., Genome-wide association meta-analysis in 269,867 individuals identifies new genetic and functional links to intelligence. Nat. Genet. 50, 912–919 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grasby K. L., Jahanshad N., Painter J. N., Colodro-Conde L., Bralten J., Hibar D. P., Lind P. A., Pizzagalli F., Ching C. R. K., McMahon M. A. B., Shatokhina N., Zsembik L. C. P., Thomopoulos S. I., Zhu A. H., Strike L. T., Agartz I., Alhusaini S., Almeida M. A. A., Alnæs D., Amlien I. K., Andersson M., Ard T., Armstrong N. J., Ashley-Koch A., Atkins J. R., Bernard M., Brouwer R. M., Buimer E. E. L., Bülow R., Bürger C., Cannon D. M., Chakravarty M., Chen Q., Cheung J. W., Couvy-Duchesne B., Dale A. M., Dalvie S., de Araujo T. K., de Zubicaray G. I., de Zwarte S. M. C., den Braber A., Doan N. T., Dohm K., Ehrlich S., Engelbrecht H.-R., Erk S., Fan C. C., Fedko I. O., Foley S. F., Ford J. M., Fukunaga M., Garrett M. E., Ge T., Giddaluru S., Goldman A. L., Green M. J., Groenewold N. A., Grotegerd D., Gurholt T. P., Gutman B. A., Hansell N. K., Harris M. A., Harrison M. B., Haswell C. C., Hauser M., Herms S., Heslenfeld D. J., Ho N. F., Hoehn D., Hoffmann P., Holleran L., Hoogman M., Hottenga J.-J., Ikeda M., Janowitz D., Jansen I. E., Jia T., Jockwitz C., Kanai R., Karama S., Kasperaviciute D., Kaufmann T., Kelly S., Kikuchi M., Klein M., Knapp M., Knodt A. R., Krämer B., Lam M., Lancaster T. M., Lee P. H., Lett T. A., Lewis L. B., Lopes-Cendes I., Luciano M., Macciardi F., Marquand A. F., Mathias S. R., Melzer T. R., Milaneschi Y., Mirza-Schreiber N., Moreira J. C. V., Mühleisen T. W., Müller-Myhsok B., Najt P., Nakahara S., Nho K., Loohuis L. M. O., Orfanos D. P., Pearson J. F., Pitcher T. L., Pütz B., Quidé Y., Ragothaman A., Rashid F. M., Reay W. R., Redlich R., Reinbold C. S., Repple J., Richard G., Riedel B. C., Risacher S. L., Rocha C. S., Mota N. R., Salminen L., Saremi A., Saykin A. J., Schlag F., Schmaal L., Schofield P. R., Secolin R., Shapland C. Y., Shen L., Shin J., Shumskaya E., Sønderby I. E., Sprooten E., Tansey K. E., Teumer A., Thalamuthu A., Tordesillas-Gutiérrez D., Turner J. A., Uhlmann A., Vallerga C. L., van der Meer D., van Donkelaar M. M. J., van Eijk L., van Erp T. G. M., van Haren N. E. M., van Rooij D., van Tol M.-J., Veldink J. H., Verhoef E., Walton E., Wang M., Wang Y., Wardlaw J. M., Wen W., Westlye L. T., Whelan C. D., Witt S. H., Wittfeld K., Wolf C., Wolfers T., Wu J. Q., Yasuda C. L., Zaremba D., Zhang Z., Zwiers M. P., Artiges E., Assareh A. A., Ayesa-Arriola R., Belger A., Brandt C. L., Brown G. G., Cichon S., Curran J. E., Davies G. E., Degenhardt F., Dennis M. F., Dietsche B., Djurovic S., Doherty C. P., Espiritu R., Garijo D., Gil Y., Gowland P. A., Green R. C., Häusler A. N., Heindel W., Ho B.-C., Hoffmann W. U., Holsboer F., Homuth G., Hosten N., Jack C. R., Jang M., Jansen A., Kimbrel N. A., Kolskår K., Koops S., Krug A., Lim K. O., Luykx J. J., Mathalon D. H., Mather K. A., Mattay V. S., Matthews S., Van Son J. M., McEwen S. C., Melle I., Morris D. W., Mueller B. A., Nauck M., Nordvik J. E., Nöthen M. M., O’Leary D. S., Opel N., Martinot M.-L. P., Pike G. B., Preda A., Quinlan E. B., Rasser P. E., Ratnakar V., Reppermund S., Steen V. M., Tooney P. A., Torres F. R., Veltman D. J., Voyvodic J. T., Whelan R., White T., Yamamori H., Adams H. H. H., Bis J. C., Debette S., Decarli C., Fornage M., Gudnason V., Hofer E., Ikram M. A., Launer L., Longstreth W. T., Lopez O. L., Mazoyer B., Mosley T. H., Roshchupkin G. V., Satizabal C. L., Schmidt R., Seshadri S., Yang Q., Alvim M. K. M., Ames D., Anderson T. J., Andreassen O. A., Arias-Vasquez A., Bastin M. E., Baune B. T., Beckham J. C., Blangero J., Boomsma D. I., Brodaty H., Brunner H. G., Buckner R. L., Buitelaar J. K., Bustillo J. R., Cahn W., Cairns M. J., Calhoun V., Carr V. J., Caseras X., Caspers S., Cavalleri G. L., Cendes F., Corvin A., Crespo-Facorro B., Dalrymple-Alford J. C., Dannlowski U., de Geus E. J. C., Deary I. J., Delanty N., Depondt C., Desrivières S., Donohoe G., Espeseth T., Fernández G., Fisher S. E., Flor H., Forstner A. J., Francks C., Franke B., Glahn D. C., Gollub R. L., Grabe H. J., Gruber O., Håberg A. K., Hariri A. R., Hartman C. A., Hashimoto R., Heinz A., Henskens F. A., Hillegers M. H. J., Hoekstra P. J., Holmes A. J., Hong L. E., Hopkins W. D., Pol H. E. H., Jernigan T. L., Jönsson E. G., Kahn R. S., Kennedy M. A., Kircher T. T. J., Kochunov P., Kwok J. B. J., Le Hellard S., Loughland C. M., Martin N. G., Martinot J.-L., McDonald C., McMahon K. L., Meyer-Lindenberg A., Michie P. T., Morey R. A., Mowry B., Nyberg L., Oosterlaan J., Ophoff R. A., Pantelis C., Paus T., Pausova Z., Penninx B. W. J. H., Polderman T. J. C., Posthuma D., Rietschel M., Roffman J. L., Rowland L. M., Sachdev P. S., Sämann P. G., Schall U., Schumann G., Scott R. J., Sim K., Sisodiya S. M., Smoller J. W., Sommer I. E., Pourcain B. S., Stein D. J., Toga A. W., Trollor J. N., Van der Wee N. J. A., van’t Ent D., Völzke H., Walter H., Weber B., Weinberger D. R., Wright M. J., Zhou J., Stein J. L., Thompson P. M., Medland S. E.; Enhancing NeuroImaging Genetics through Meta-Analysis Consortium (ENIGMA)—Genetics working group , The genetic architecture of the human cerebral cortex. Science 367, eaay6690 (2020).32193296 [Google Scholar]
- 62.Cheng W., Frei O., van der Meer D., Wang Y., O’Connell K. S., Chu Y., Bahrami S., Shadrin A. A., Alnæs D., Hindley G. F. L., Lin A., Karadag N., Fan C.-C., Westlye L. T., Kaufmann T., Molden E., Dale A. M., Djurovic S., Smeland O. B., Andreassen O. A., Genetic association between schizophrenia and cortical brain surface area and thickness. JAMA Psychiat. 78, 1020–1030 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van der Meer D., Shadrin A. A., O’Connell K., Bettella F., Djurovic S., Wolfers T., Alnæs D., Agartz I., Smeland O. B., Melle I., Sánchez J. M., Linden D. E. J., Dale A. M., Westlye L. T., Andreassen O. A., Frei O., Kaufmann T., Improved prediction of schizophrenia by leveraging genetic overlap with brain morphology. medRxiv, 2020.08.03.20167510 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Figs. S1 to S4
Tables S1 to S3
Legends for data S1 to S9
References
Data S1 to S9