Abstract
Objectives
This work aimed to analyse possible zoonotic spill-over of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). We report the spill-over of mink-adapted SARS-CoV-2 from farmed mink to humans after adaptation that lasted at least 3 months.
Methods
Next-generation sequencing and a bioinformatic approach were applied to analyse the data.
Results
In an isolate obtained from an asymptomatic patient testing positive for SARS-CoV-2, we found four distinguishing mutations in the S gene that gave rise to the mink-adapted variant (G75V, M177T, Y453F, and C1247F) and others.
Conclusions
Zoonotic spill-over of SARS-CoV-2 can occur from mink to human.
Keywords: Mink, SARS-CoV-2, Spill-over, Transmission, Zoonoses
Introduction
Coronaviruses are potential zoonotic pathogens, and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the third highly pathogenic member of this family to emerge in the 21st century [1]. Although mass vaccinations are currently under way, the fate of the virus remains unclear. Herd immunity and eradication are somewhat unrealistic given its prevalence, genetic diversity, and existing animal reservoirs. Thus far, SARS-CoV-2 infections have been reported in different mammalian species worldwide, including dogs, domestic cats, tigers, lions, ferrets, mink and deer, and it is maintained in rodents [[2], [3], [4], [5]]. SARS-CoV-2 infections in farmed mink have recently been confirmed in Europe [6], and transmission of the virus from infected mink to humans has been reported in Denmark and The Netherlands [[7], [8], [9]]. After Denmark, Poland is the second largest producer of mink pelts in Europe, with 354 active Polish mink farms and approximately 6.3 million mink [10]. Interspecies coronavirus transfer among mammals is not surprising. The transfer of highly pathogenic variants from bats usually occurs via intermediate hosts. Previous examples include infection of dromedary camels with Middle East respiratory syndrome (MERS) coronavirus and palm civets with SARS-associated coronavirus, followed by transfer of the viruses to humans [11]. A comprehensive analysis of existing strains suggests that these events are not exceptions. The human coronavirus OC43 is a betacoronavirus first described in the 1960s; it is associated with upper and lower respiratory tract disease in humans [12]. Of note, closely related virus species are found in cattle (bovine coronavirus) and dogs (canine respiratory coronavirus). The exact routes of transfer among different species remain puzzling, but one obvious possibility is that, as with SARS-CoV-2, these viruses spread between humans and companion and farmed animals [13]. Recently, we and others have detected SARS-CoV-2 infection in farmed mink in Northern Poland [14,15]. The first report identified SARS-CoV-2 in samples collected from mink in mid-November 2020. Even though the prevalence of the virus was low and cases were most likely isolated, we have sequenced the isolates and shown that the positive signal did not originate from contamination. The aim of this study was to detect possible zoonotic spill-over of SARS-CoV-2 from mink to farm workers.
Materials and methods
This study was approved by the Independent Bioethical Committee for Scientific Research at the Medical University of Gdansk, Gdansk, Poland (Statement no. KBBN/183/2020).
SARS-CoV-2 genome sequencing was performed at the University of Gdansk, Poland, using samples containing RNA isolated from positive swabs (amplification of two target genes in RT-PCR). ARTICv3 primer-based amplicon generation followed by an Oxford Nanopore Technology MinION run was performed with no live base-calling. Using Guppy v4 raw reads were base-called (high accuracy model), debarcoded, and trimmed to delete adapter, barcode and PCR primer sequences. ARTIC field bioinformatics v1.2.1 pipeline software with implemented Medaka final polishing were used to generate SARS-CoV-2 genomes. Phylogenetic analysis was performed using the procedure recommended by Nextstrain.org. In brief, Augur toolkit v10.03 use MAFFT v7 to align nucleotide genomic sequences and IQ-TREE v2 to infer phylogenetic trees by a maximum likelihood approach. Time of divergence was calculated by TreeTime v0.8.1. [16].
Results
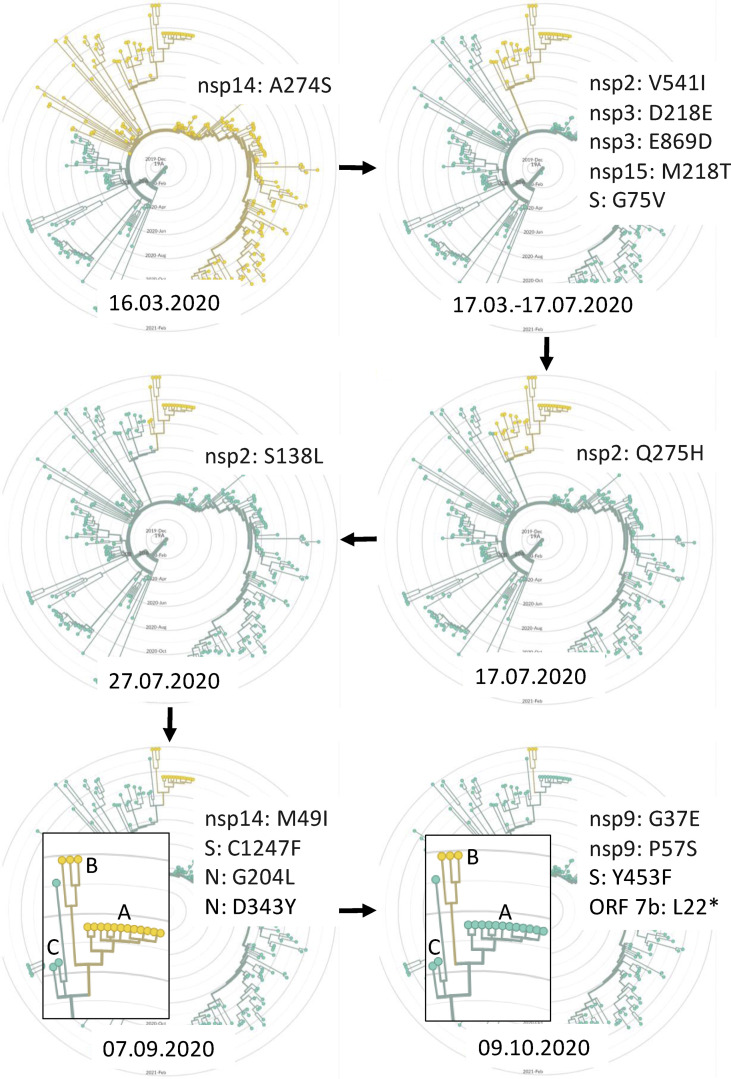
In this study we identified an index case of infection with a mink-adapted variant in humans. Following the identification of SARS-CoV-2 cases in farmed animals, exposed staff were tested for SARS-CoV-2 (nasopharyngeal swabs) using RT-qPCR. A single positive case involving an asymptomatic person was detected in a sample collected on 1st February 2021 and sequenced using the ARTIC Nanopore technology protocol. The resulting sequence was deposited in GISAID under the accession number EPI_ISL_1034274. Phylogenetic analysis (Fig. 1 ) shows that the virus clusters closely with those isolated from mink (group B, Fig. 1, bottom radial trees). Furthermore, mutations characteristic of the mink-adapted variant were present in the virus genomic sequence (Table 1 ), confirming that the person had contracted it from the mink.
Fig. 1.
Phylogenetic analysis of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) lineage B.1.1.279 combined with inferred time (bottom of each radial time tree) of fixing mutations (upper right of each radial time tree) in all isolates forming a group that leads to the generation of the mink variants. Yellow colour represents new variants. A: November 2020 mink isolates. B: January 2021 mink isolates and a single human isolate. C: nearest neighbours human isolates that share a common ancestor with A and B—Norway/4235/2020, Germany/NW-HHU-340/2020, and Iceland/4563/2021.
Table 1.
Representation of mutations in mink-originated severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) isolates. Group A: November 2020 mink isolates. Group B: January 2021 mink isolates and a single human isolate. Group C: nearest neighbours in lineage B.1.1.279 human isolates. The yellow colour represents mutations fixed in November 2020. The red colour represents mutations acquired during 3 months passages on the mink farm
Two SARS-CoV-2 isolates (GISAID: EPI_ISL_984305 and EPI_ISL_984307) from animals at the same farm were collected on 27th of January, sequenced, and deposited on 23rd of February by the laboratory of the National Veterinary Research Institute in Poland. Phylogenetic analysis of the data indicated that the virus belongs to the B.1.1.279 lineage (pangolin classification), which is not surprising considering the prevalence of this variant in Europe. Genome analysis for this isolate showed that they carry a combination of mutations noted in viruses already isolated from mink in November 2020 (Fig. 1, bottom left radial tree), and also four new changes in the genome (Fig. 1, bottom right radial tree).
Discussion
We report a SARS-CoV-2 zoonotic spill-over from farmed mink to a farm worker. In an isolate obtained from an asymptomatic farm employee who tested positive for SARS-CoV-2, we found four distinguishing mutations in the S gene that gave rise to the mink-adapted variant (G75V, M177T, Y453F, and C1247F) and others. These new changes include the Y453F mutation, which was previously reported to have emerged in mink during serial passages (e.g. in Denmark and recently Lithuania) [17], and a novel mutation that is not present in any global SARS-CoV-2 isolate which truncates ORF 7b at position L22. Considering all of the data, we speculate that the virus was already present in the mink population in November 2020, presumably after a single introduction during the late summer or fall season.
The emerging variant is clearly the result of adaptation of the virus to the mink host, with some point mutations fixed early during spread in the mink and additional changes accumulating over time (Fig. 1, bottom right). Although the exact role or roles of these mutations remain to be determined, one possible implication is improved fitness in the new host [18,19]. Any effects on disease course, transmissibility, or immunogenicity in humans are not known [20], but the variant should be tracked in the general population.
This study was limited by the access to the diagnostic material from citizens living close to the farm. Comprehensive screening of the population living in the nearby settlements would have enriched our study. We believe that such an approach should be conducted straight after the first SARS-CoV-2 infection in the mink and should last for an extended period to detect and analyse possible spill-over. Our results confirm the need for country-scale epizootiological monitoring and careful genomic analysis of SARS-CoV-2-positive human samples.
Data availability
The complete genome sequences of SARS-CoV-2 isolated from the patient was deposited in GISAID under the accession number: EPI_ISL_1034274.
Author contributions
The study was conceived and designed by LR and MG. Data handling: LR, MK, MG and KP. Material handling and laboratory work: LR, MK and NMP. Statistical and phylogenetic analysis: LR. Interpretation of data: LR, KP and MK. Data visualization: LR. Supervision: MG, KP and TS. The manuscript was written by LR, MG, KP, RK, TS, BS and KBS in consultation with all co-authors. MG, KP, LR, TS and RK revised the manuscript. All authors accepted the final version of the manuscript.
Transparency declaration
The authors declare that they have no conflicts of interest. We appreciate the support from the University of Gdańsk, the Medical University of Gdańsk, and the University of Helsinki. This research was funded through the 2018–2019 BiodivERsA joint call for research proposals, under the BiodivERsA3 ERA-Net COFUND program; the funding organizations ANR (France), DFG (Germany), EPA (Ireland), FWO (Belgium), and NCN (Poland). MG was supported by the National Science Centre, Poland, under the BiodivERsA3 programme (2019/31/Z/NZ8/04028) and funding for Young Scientists at Medical University of Gdańsk (664/772/61/71-1413) and The Polish National Agency for Academic Exchange Bekker Programme (PPN/BEK/2019/1/00337). LR was supported by the Ministry of Science and Higher Education Decision No. 54/WFSN/2020 “Co-infections with SARS-CoV-2, Database of COVID-19 Accompanying Infections”. KP was supported by the National Science Centre (UMO-2017/27/B/NZ6/02488). This study was supported by the VEO—European Union Horizon 2020 (grant number 874735), the Jane and Aatos Erkko Foundation, and by a subsidy from the Polish Ministry of Science and Higher Education for the research on SARS-CoV-2 for the Jagiellonian University.
Acknowledgements
We gratefully acknowledge the authors from the originating laboratories responsible for obtaining the specimens and the submitting laboratories where genetic sequence data were generated and shared via the GISAID Initiative, upon which this research is partially based. MG thanks Alicja Rost, Ewa Zieliniewicz, and Karolina Baranowicz for their assistance in the laboratory.
Editor: R. Chemaly
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cmi.2021.12.001.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Cucinotta D., Vanelli M. WHO declares COVID-19 a pandemic. Acta Biomed. 2020;91:157–160. doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palmer M.V., Martins M., Falkenberg S., Buckley A., Caserta L.C., Mitchell P.K., et al. Susceptibility of white-tailed deer (Odocoileus virginianus) to SARS-CoV-2. bioRxiv. 2021 doi: 10.1128/JVI.00083-21. 01.13.426628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abdel-Moneim A.S., Abdelwhab E.M. Evidence for SARS-CoV-2 infection of animal hosts. Pathogens. 2020;9:529. doi: 10.3390/pathogens9070529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Griffin B.D., Chan M., Tailor N., Mendoza E.J., Leung A., Warner B.M., et al. SARS-CoV-2 infection and transmission in the North American deer mouse. Nat Commun. 2021;12:3612. doi: 10.1038/s41467-021-23848-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martínez-Hernández F., Isaak-Delgado A.B., Alfonso-Toledo J.A., Muñoz-García C.I., Villalobos G., Aréchiga-Ceballos N., et al. Assessing the SARS-CoV-2 threat to wildlife: potential risk to a broad range of mammals. Perspect Ecol Conserv. 2020;18:223–234. doi: 10.1016/j.pecon.2020.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.World Organisation for Animal Health . 2021. World organisation for animal health. (cited 19th Aug 2021) [Google Scholar]
- 7.Hammer A.S., Quaade M.L., Rasmussen T.B., Fonager J., Rasmussen M., Mundbjerg K., et al. SARS-CoV-2 transmission between mink (Neovison vison) and humans, Denmark. Emerg Infect Dis. 2021;27 doi: 10.3201/eid2702.203794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koopmans M. SARS-CoV-2 and the human–animal interface: outbreaks on mink farms. Lancet Infect Dis. 2020;21:18–19. doi: 10.1016/S1473-3099(20)30912-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oude Munnink B.B., Sikkema R.S., Nieuwenhuijse D.F., Molenaar R.J., Munger E., Molenkamp R., et al. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science. 2021;371:172–177. doi: 10.1126/science.abe5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.EU Fur Association . 2020. EU Fur association. (cited 13th Dec 2020) [Google Scholar]
- 11.de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kahn J.S., McIntosh K. History and recent advances in coronavirus discovery. Pediatr Infect Dis J. 2005;24:S223–S227. doi: 10.1097/01.inf.0000188166.17324.60. [DOI] [PubMed] [Google Scholar]
- 13.Szczepanski A., Owczarek K., Bzowska M., Gula K., Drebot I., Ochman M., et al. Canine respiratory coronavirus, bovine coronavirus, and human coronavirus OC43: receptors and attachment factors. Viruses. 2019;11:328. doi: 10.3390/v11040328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rabalski L., Kosinski M., Smura T., Aaltonen K., Kant R., Sironen T., et al. Severe acute respiratory syndrome coronavirus 2 in farmed mink (Neovison vison), Poland. Emerg Infect Dis. 2021;27:2333–2339. doi: 10.3201/eid2709.210286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.GISAID 2021. https://www.epicov.org/epi3/EPI_ISL_984305 EPI_ISL_984307.
- 16.Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C., et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bayarri-Olmos R., Rosbjerg A., Johnsen L.B., Helgstrand C., Bak-Thomsen T., Garred P., et al. The SARS-CoV-2 Y453F mink variant displays a pronounced increase in ACE-2 affinity but does not challenge antibody neutralization. J Biol Chem. 2021;296:100536. doi: 10.1016/j.jbc.2021.100536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao J., Cui W., Tian B. The potential intermediate hosts for SARS-CoV-2. Front Microbiol. 2020;11:580137. doi: 10.3389/fmicb.2020.580137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Dorp L., Tan C.C.S., Lam S.D., Richard D., Owen C., Berchtold D., et al. Recurrent mutations in SARS-CoV-2 genomes isolated from mink point to rapid host-adaptation. bioRxiv. 2020;2020 11.16.384743. [Google Scholar]
- 20.Hayashi T., Yaegashi N., Konishi I. Effect of RBD mutation (Y453F) in spike glycoprotein of SARS-CoV-2 on neutralizing antibody affinity. bioRxiv. 2020 11.27.401893. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The complete genome sequences of SARS-CoV-2 isolated from the patient was deposited in GISAID under the accession number: EPI_ISL_1034274.