Abstract
In February 1996, a Hospital Infection Control Practices Advisory Committee-style screening program was commenced to isolate and subsequently characterize glycopeptide-resistant enterococci (GRE) from patients at a hospital trust in Glasgow, Scotland. Over the next 30 months, GRE were isolated from 154 patients. GRE were isolated from patients in traditionally high-risk areas such as the renal unit and intensive care unit and also in areas considered to be lower risk, including medical wards and associated long-stay geriatric hospitals. The majority (90%) of isolates were Enterococcus faecium vanB. The remaining isolates consisted of seven E. faecalis (vanA), three E. gallinarum (vanC), and a further six E. faecium (five vanA, one both vanA and vanB) isolates. Analysis of SmaI-digested DNA by pulsed-field gel electrophoresis revealed that 34 of 40 (85%) VanB E. faecium isolates were identical or closely related, while 11 of 13 (85%) VanA GRE were distinct. High-level aminoglycoside resistance was seen in less than 8% of isolates. VanB E. faecium isolates were almost uniformly resistant to ampicillin and tetracycline. In this study, GRE have been isolated over a prolonged period from a broad range of patients. Glycopeptide resistance within the study hospital trust appeared to be mainly due to the clonal dissemination of a single strain of E. faecium VanB.
Following their initial discovery in the United Kingdom and France in 1986 (15, 26), glycopeptide-resistant enterococci (GRE) have now spread worldwide, with the percentage of enterococcal infections in the United States resistant to vancomycin rising from 0.3% in 1989 to 7.9% in 1993 (4). A number of outbreaks of colonization and infection with GRE have been described. VanA-type Enterococcus faecium strains typically predominate in clinical areas such as transplantation and oncology units and the intensive care unit (ICU) (3, 7, 9, 19, 21).
The Hospital Infection Control Practices Advisory Committee (HICPAC) of the Centers for Disease Control and Prevention has published recommendations for prevention of the spread of glycopeptide resistance (12) which, although published in the United States, are believed to be relevant in the United Kingdom also (10). The recommendations include screening of enterococcal isolates for vancomycin resistance and establishment of a fecal screening program to detect patients with intestinal colonization with GRE. It is also recommended that control efforts be intensified once GRE are known to be present within a hospital, particularly in high-risk areas such as transplantation and oncology units and the ICU.
An isolate of GRE was obtained from the routine culture of a femoral line tip from a renal unit patient in the ward of a Scottish hospital trust in October 1995. In response to this, all enterococcal isolates from clinical specimens were examined for glycopeptide resistance. A fecal screening program was also established and involved thrice-weekly screening of all ICU patients and routine screening of all fecal specimens or rectal swabs submitted to the laboratory.
This paper presents the findings of this program over a 30-month period.
MATERIALS AND METHODS
Hospital setting.
The study hospital trust is a tertiary referral center that comprises two major hospitals with a combined total of 1,100 beds, together with three smaller, long-stay geriatric hospitals. One of the major hospitals has a 33-bed renal unit located in two adjacent wards on a single floor that comprises a mixture of single- and four-bed rooms. An eight-bed ICU is located two floors below.
Fecal screening program.
Routine screening of all fecal specimens submitted to the laboratory was instituted in February 1996. Additionally, rectal swabs were taken thrice weekly from all patients on the ICU. Fecal suspensions or rectal swabs were inoculated directly onto esculin azide agar base (Oxoid, Basingstoke, United Kingdom) supplemented with 6 μg of vancomycin (Sigma, Poole, United Kingdom) per ml and with 10 μg of colistin sulfate per ml and 15 μg of nalidixic acid per ml (both from Oxoid). The plates were incubated for 18 h in air at 37°C. Isolates that were esculin positive, catalase negative, gram-positive cocci, and vancomycin resistant by the E-test were tentatively identified as GRE.
Identification.
Clinical enterococci and those from the fecal screens were identified to the species level in the laboratory with the API 20 STREP system (Biomerieux, Marcy l'Etoile, France). The identities of all isolates were confirmed by PCR and biochemical tests (see Table 1).
TABLE 1.
Tests used for differentiating between E. faecalis, E. faecium, E. gallinarum, and E. casseliflavus species groups
| Test | Test result
|
|||
|---|---|---|---|---|
| E. faecalis | E. faecium | E. gallinarum | E. casseliflavus | |
| ITS-PCR | ||||
| Pattern 1 | − | + | − | − |
| Pattern 2 | + | − | + | + |
| Arabinose fermentation | − | + | + | + |
| Van PCR | ||||
| vanC1 | − | − | + | − |
| vanC2 | − | − | − | + |
PCR amplification of intergenic rRNA spacer regions (ITS-PCR) was based on the methods of Jensen et al. (13) and Tyrrell et al. (25) with primers L1 (5′-CAAGGCATCCACCGT) and G1 (5′-GAAGTCGTAACAAGG). Amplification was performed with a Hybaid Omn-E thermal cycler with 20 pmol of each primer, 100 μM deoxynucleoside triphosphates, 1.5 mM MgCl2, 10 mM Tris-HCl (pH 8.8), 50 mM KCl, and 0.1% Triton X-100. Template DNA was prepared from whole cells by mixing 200 μl of a culture grown overnight in brain heart infusion broth (Oxoid) with 800 μl of water and boiling for 10 min. Following centrifugation, 5 μl of supernatant was added to each PCR mixture, and the final volume was adjusted to 50 μl. The PCR mixture was heated to 95°C for 5 min, and 1 U of BioTaq DNA polymerase (Bioline, London, United Kingdom) was added. Amplification conditions were 94°C for 1 min, 55°C for 7 min, and 72°C for 2 min for a total of 25 cycles, followed by an additional 7 min at 72°C.
Arabinose fermentation was detected in a 1% (wt/vol) sugar solution in Andrade's peptone water. The identities of E. gallinarum and E. casseliflavus strains were confirmed by PCR amplification of the vanC ligase gene specific for each organism (6).
Antimicrobial susceptibility.
All isolates were tested for susceptibility to vancomycin, ampicillin, gentamicin, and streptomycin (all from Sigma), teicoplanin (Marion Merrell, Uxbridge, United Kingdom), and ciprofloxacin (Bayer, Newbury, United Kingdom) by an agar incorporation method in accordance with the British Society for Antimicrobial Chemotherapy guidelines (28).
Glycopeptide-resistant isolates were characterized phenotypically on the basis of susceptibility levels to vancomycin and teicoplanin. Isolates were defined as having a VanA phenotype if the vancomycin MIC was ≥64 μg/ml and the teicoplanin MIC was ≥2 μg/ml. For isolates with a VanB phenotype the vancomycin MIC was ≥8 μg/ml and the teicoplanin MIC was <2 μg/ml.
The MIC breakpoints for susceptibility to an antibiotic were as follows: ampicillin, ≤8 μg/ml; tetracycline, ≤1 μg/ml; and ciprofloxacin, ≤4 μg/ml. The numbers of isolates with high-level gentamicin resistance (MICs, >500 μg/ml) and high-level streptomycin resistance (MICs, >2,000 μg/ml) were determined. Cell suspensions were tested for β-lactamase production in a solution of 20 μg of nitrocefin per ml.
PCR amplification of glycopeptide resistance elements.
Glycopeptide resistance determinants vanA, vanB, vanC1, and vanC2-vanC3 were detected by multiplex PCR with the primers described by Dutka-Malen et al. (6). Amplification reactions were performed in 50-μl volumes containing 20 μl of template (prepared from boiled whole cells as for ITS-PCR), 20 pmol of each primer, 200 μM deoxynucleoside triphosphates, 3 mM MgCl2, 16 mM (NH4)2SO4, 67 mM Tris-HCl (pH 8.8), 0.01% Tween-20, and 1 U of BioTaq DNA polymerase. The reaction mixtures were denatured at 94°C for 2 min, followed by 30 cycles of 94°C for 1 min, 54°C for 1 min, and 72°C for 2 min, with a final extension at 72°C for 10 min.
PFGE typing.
Genomic DNA was prepared in agarose plugs as described by Murray et al. (20). DNA was digested with 20 U of SmaI (Promega, Madison, Wis.) for 6 h at 25°C and was electrophoresed with the contour-clamped homogenous electric fields device (CHEF-DRII; Bio-Rad, Hercules, Calif.). The switch interval was ramped from 5 to 35 s over a 24-h period at 6 V/cm2. A pulsed-field gel electrophoresis (PFGE) type was assigned to each strain in accordance with the criteria outlined by Tenover et al. (23).
Control strains.
E. faecium NCTC 12202 (vanA), E. faecium ATCC 19434 (glycopeptide susceptible), E. faecalis ATCC 51299 (vanB), E. faecalis ATCC 19433 (glycopeptide susceptible), E. gallinarum ATCC 49573 (vanC1), and E. casseliflavus ATCC 25788 (vanC2) were used as control strains.
RESULTS
Isolates.
Between February 1996 and July 1998, GRE were obtained from 154 patients. Only the initial isolate from each patient was used for further study. The majority of isolates (134 of 154) were obtained from the two major hospitals.
Thirty-six isolates were from clinical specimens, including 21 from urine, 10 from wounds, 3 from continuous ambulatory peritoneal dialysis effluents, and 2 from blood cultures. Twenty nine (80.5%) of the 36 clinical GRE were from patients in the primary hospital unit, of which 17 were renal unit patients. A single clinical GRE isolate was recovered from an ICU patient during the study period. Two isolates were from patients in the long-stay geriatric hospitals. These patients represented 0.87% of the 4,130 patients from whom clinically significant enterococci were isolated by the laboratory during the study period.
The remaining 118 GRE were from 8,549 patients screened for fecal carriage (carriage rate, 1.4%). Of these, 23 were recovered from 1,117 ICU patients (carriage rate, 2.1%), 36 were recovered from 316 renal unit patients (11.5%), and 6 were recovered from 153 oncology and hematology unit patients (1.2%). A further 45 were isolated from 6,843 patients (0.7%) in the two major hospitals, including patients from surgical and medical wards, patients from the obstetrics unit, and outpatients. The final eight fecal isolates were recovered from 124 patients (6.5%) within the long-stay geriatric hospitals. There were no apparent associations between colonization with GRE and the clinical indication for the submission of fecal specimens.
Identification.
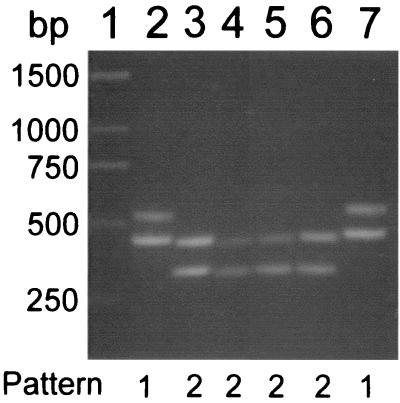
The use of ITS-PCR with primers L1 and G1 to produce unique patterns of bands for different enterococcal species has been described previously (25). In our hands, these primers amplified only two distinct patterns of bands from isolates collected for this study and control strains (Fig. 1). Isolates of E. faecium gave two bands of approximately 430 and 520 bp (pattern 1). A single distinct pattern (pattern 2), with bands of approximately 295 and 420 bp, was produced by E. faecalis, E. gallinarum, and E. casseliflavus isolates. Thus, species identity was determined by a combination of ITS-PCR, arabinose fermentation, and PCR amplification of van genes, as outlined in Table 1. By using these criteria, 144 (93.5%) isolates were identified as E. faecium, 7 (4.5%) were identified as E. faecalis, and 3 (2%) were identified as E. gallinarum. No E. casseliflavus isolates were obtained.
FIG. 1.
ITS-PCR profiles of enterococcal species. Lanes: 1, Generuler 1-kb DNA ladder (MBI Fermentas); 2, E. faecium ATCC 19434; 3, E. faecalis ATCC 19433; 4, E. casseliflavus ATCC 25788; 5, E. gallinarum ATCC 49573; 6, strain G-089 (E. faecalis); 7, strain G-090 (E. faecium). The image was generated with Adobe Photoshop, version 4.0.
The identities of the isolates determined by these tests did not always correlate with the identifies determined with the API 20 STREP system (Table 2). The API 20 STREP kit failed to identify any of the three E. gallinarum strains. Furthermore, 29 strains of E. faecium were incorrectly identified as E. casseliflavus by the API 20 STREP system.
TABLE 2.
Comparison of species identification methods for enterococcal isolates
| Speciesa | No. of strains identified with API 20 STREP systemb
|
||||
|---|---|---|---|---|---|
| E. faecalis | E. faecium | E. gallinarum | E. casseliflavus | Total | |
| E. faecalis | 6 | 1 | 0 | 0 | 7 |
| E. faecium | 1 | 114 | 0 | 29 | 144 |
| E. gallinarum | 0 | 1 | 0 | 2 | 3 |
| E. casseliflavus | 0 | 0 | 0 | 0 | 0 |
| Total | 7 | 116 | 0 | 31 | 154 |
Species determined by using the criteria described in Table 1.
API 20 STREP profiles analyzed by using APILAB, version 5.1, software.
Characterization of glycopeptide resistance elements.
The Van phenotype of each isolate was determined on the basis of its levels of susceptibility to vancomycin and teicoplanin (Table 3). All seven E. faecalis isolates had a VanA phenotype. The majority, 138 of 142 (97%), of E. faecium strains were VanB, while the remaining 6 isolates had susceptibility levels that suggested a VanA phenotype. The PCR-determined genotypes were consistent with the phenotypes for all but one isolate. One strain of E. faecium with a VanA phenotype produced both vanA and vanB PCR products. A vanC1 gene was amplified from the three E. gallinarum isolates.
TABLE 3.
In vitro susceptibilities of E. faecalis and E. faecium isolates to various antimicrobial agents
| Organism (Van phenotype) and antimicrobial agent | MIC (μg/ml)a
|
No. (%) resistant | ||
|---|---|---|---|---|
| 50% | 90% | Range | ||
| E. faecalis (VanA) (n = 7) | ||||
| Vancomycin | 128 | 512 | 64–512 | 7 (100) |
| Teicoplanin | 8 | 64 | 4–64 | 7 (100) |
| Ampicillin | 2 | 4 | 1–4 | 0 (0) |
| Tetracycline | 128 | >128 | 64–>128 | 7 (100) |
| Streptomycin | 512 | >2,048 | 512–>2,048 | 2 (28.6) |
| Gentamicin | 64 | >1,024 | 32–>1,024 | 1 (14.3) |
| Ciprofloxacin | 1 | 1 | 0.5–1 | 0 (0) |
| E. faecium (VanA) (n = 6b) | ||||
| Vancomycin | 256 | >256 | 128–>256 | 6 (100) |
| Teicoplanin | 32 | >128 | 8–>128 | 6 (100) |
| Ampicillin | 32 | >256 | 0.5–>256 | 5 (83.3) |
| Tetracycline | 0.25 | 128 | <0.25–128 | 1 (16.7) |
| Streptomycin | 256 | >2,048 | 128–>2,048 | 1 (16.7) |
| Gentamicin | 32 | >1,024 | 16–>1,024 | 1 (16.7) |
| Ciprofloxacin | 2 | >128 | 0.5–>128 | 2 (33.3) |
| E. faecium (VanB) (n = 138) | ||||
| Vancomycin | 16 | 16 | 8–128 | 138 (100) |
| Teicoplanin | 0.5 | 0.5 | <0.25–1 | 0 (0) |
| Ampicillin | 128 | 128 | 1–256 | 137 (99.3) |
| Tetracycline | 32 | 64 | 0.25–64 | 134 (97.1) |
| Streptomycin | 256 | 256 | 64–>2,048 | 5 (3.6) |
| Gentamicin | 64 | 64 | 32–>1,024 | 1 (0.7) |
| Ciprofloxacin | 2 | 4 | 0.5–32 | 4 (2.9) |
50% and 90%, MICs at which 50 and 90% of isolates are inhibited, respectively.
Includes an E. faecium strain with both vanA and vanB ligase genes.
Interestingly, the VanB E. faecium strains showed low-level resistance to vancomycin, and the vancomycin MIC was greater than 32 μg/ml for only one isolate.
Antimicrobial susceptibility.
Strains were tested for susceptibility to ampicillin, tetracycline, streptomycin, gentamicin, and ciprofloxacin (Table 3). E. faecium isolates were almost uniformly resistant to ampicillin, while E. faecalis isolates remained susceptible. Twenty isolates for which ampicillin MICs covered a wide range were tested for β-lactamase activity. None produced a detectable β-lactamase enzyme. Of the VanB E. faecium isolates, 97% were resistant to both ampicillin and tetracycline. Overall, 4% of isolates were resistant to ciprofloxacin. High-level aminoglycoside resistance was seen in less than 8% of isolates, and no isolate was resistant to both streptomycin and gentamicin. The three E. gallinarum strains were susceptible to all antibiotics tested.
Typing.
The 13 VanA GRE were typed by PFGE. All E. faecalis isolates had distinct PFGE patterns. Of the six E. faecium isolates, three were distinct types and three were identical. The three identical isolates were recovered from fecal screens for patients located in a single four-bed room within the renal unit over a 2-week period in June and July 1997, suggesting that person-to-person spread had occurred.
Forty VanB E. faecium isolates were selected for typing, including both fecal and nonfecal isolates from the 30-month period. Thirty-five isolates were from the primary hospital unit (from which 70% of GRE were isolated) and comprised 9 nonfecal isolates from the renal unit and ICU and 26 fecal isolates (23 from the renal unit and ICU, 3 from other wards). Four GRE were isolated from patients in the second hospital unit, and one was from a patient in a geriatric hospital. Twenty-eight of 40 (70%) VanB E. faecium isolates had identical PFGE patterns, including both fecal and nonfecal isolates from the two major hospitals. A further six isolates were closely related (two or three band differences), including the isolate from the patient in the geriatric hospital. Five isolates were possibly related (four to six band differences), and one (a fecal isolate from the renal unit) was unrelated.
DISCUSSION
GRE have become an increasing problem in hospitals from the standpoint of nosocomial infection and infection control. HICPAC has published guidelines on the control of GRE within hospitals, and these recommend the routine screening of enterococcal isolates for resistance to vancomycin and intensified fecal screening to detect patients with gastrointestinal colonization. In this study the findings from such a screening program are presented. These differ from many other studies of colonization with GRE in that they are not restricted solely to patients in high-risk areas over short time periods.
The vancomycin-containing indicator medium used in this study proved useful for the isolation of GRE from fecal specimens. Identification to the species level proved more problematic. Other investigators have reported discrepancies between the identities obtained with the API 20 STREP system and by genotypic methods (16, 27). In this study, the API 20 STREP kit was particularly unreliable at distinguishing E. faecium and E. casseliflavus isolates. The epidemiological and infection control implications of enterococci with intrinsic (vanC) and acquired (vanA, vanB) resistance to vancomycin are very different, with intrinsically resistant species failing to demonstrate person-to-person spread (24). Failure to correctly identify patients colonized with intrinsically resistant strains entails subjecting them to unnecessary infection control measures. Reanalysis of the API 20 STREP profiles on uprated software correctly identified all strains of E. faecium but was still unable to identify two of the three strains of E. gallinarum. It would appear that accurate differentiation of intrinsically glycopeptide-resistant species from those with acquired resistance during screening requires a molecular biological methodology such as PCR.
One such method uses amplification of species-specific ddl ligase genes to identify E. faecalis and E. faecium (6). This method appears to be particularly useful as it can be performed in combination with amplification of glycopeptide resistance elements. In this study, we assessed the ITS-PCR method, which has been reported to give distinct bands for a number of enterococcal species, in addition to E. faecalis and E. faecium (25), for species identification. In our hands, this method could be used to identify E. faecium isolates, but further tests were required to distinguish E. faecalis from the intrinsically resistant species.
Although only 12% of isolates of GRE in the United Kingdom have the vanB genotype (18), the predominance of the vanB genotype within a hospital setting has been reported previously (2, 17, 22). However, the low level of vancomycin resistance present within the vanB strains (MIC at which 90% of isolates are inhibited, 16 μg/ml) in this study is unusual and may be partially explained by the lower concentration of vancomycin in the selective medium than that used in earlier studies. The incidence of high-level gentamicin resistance was also low, being found only in a single strain. The rate of high-level gentamicin resistance found in this study (0.7%) is much lower than rates of 6.7 to 13.4% found among all species of enterococci, both vancomycin resistant and sensitive, in the United Kingdom (11). The retention of sensitivity to teicoplanin is fortuitous in that it provides a synergistic therapeutic option in combination with an aminoglycoside should the need arise (14). All E. faecalis isolates retained sensitivity to ampicillin, giving an alternative therapeutic option.
The resistance genotype determined by PCR was consistent with the phenotype for all but one isolate, despite reports of mismatching in previous studies (1, 5, 8, 16). There does, however, exist an area of overlap between MICs at the bottom of the range for VanB strains (an area into which most of the VanB study isolates fell) and those at the top of the range for VanC strains, making distinction by phenotypic methods between intrinsic and acquired resistance impossible for certain strains. The failure of phenotypic methods of identification and antimicrobial sensitivity testing to distinguish between acquired and intrinsic resistance consequently necessitates the use of a molecular biological method for accurate differentiation of all strains.
The carriage of GRE described in this study gives a much fuller picture than that often obtained, given the prolonged study period and broad area screened. GRE colonization occurred throughout the study period among patients admitted to the renal unit and ICU, areas traditionally regarded as high risk (12). However, this may reflect the regular nature of the screening protocol in these areas. Among patients in the hematology and oncology units colonization was infrequent in this study, despite frequent use of glycopeptide agents in both units. Colonization was detected in patients in traditionally lower-risk areas, such as medicine and surgery, as well as patients within the long-stay geriatric hospitals over the 30-month period.
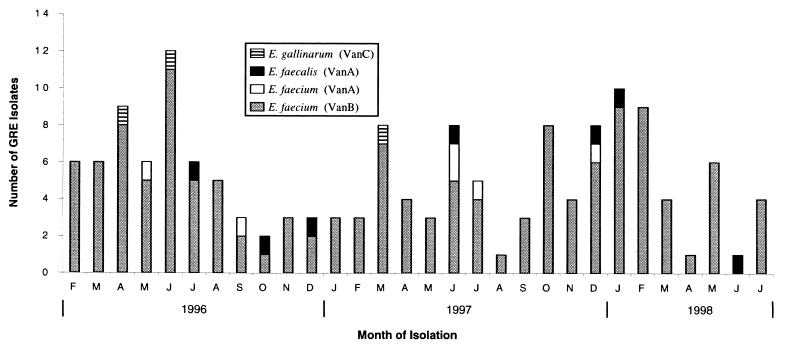
Carriage and infection with VanB E. faecium occurred at a relatively constant level throughout the 30-month period (Fig. 2). Although only a selection of VanB E. faecium isolates were typed in this study, it appears that a dominant clone is present in a number of hospital units and patient groups in Glasgow. In contrast, isolation of VanA and VanC GRE was sporadic. PFGE analysis suggests these isolates were distinct (except for one cluster of VanA E. faecium isolates for which nosocomial spread was indicated).
FIG. 2.
Isolation of GRE from a Scottish hospital trust over a 30-month period.
The finding of a single vanB E. faecium clone that disseminated within and between hospitals parallels that seen in other studies (17, 22). The free movement of patients between the two major hospital units in this study may well have contributed to the dissemination of the clone in this instance. Furthermore, medical staff rotated between both sites, and although nursing staff usually worked on only a single ward, many worked extra shifts at other sites. Patients within the long-stay geriatric hospitals had often been previously admitted to either of the major hospitals.
The introduction of a fecal screening program greatly increased the rate of detection of GRE among inpatients. Thirty-six of 154 isolates originated from nonfecal sources and would have been detected by routine methods, as all were ampicillin resistant, necessitating testing for glycopeptide resistance. The isolates from the remaining 118 colonized patients, however, would not otherwise have manifested themselves, as during the study period none of the patients yielded GRE from nonfecal routine specimens. Detection of colonized patients allowed their isolation and reduced their risk as a source of infection. However, steady acquisition continued to occur despite these precautions. As detection of GRE outside the ICU relied on clinical staff deciding to send fecal specimens for other reasons, detection rates are probably far from complete and many patients colonized with GRE but in whom GRE were not detected are likely to have remained on the open ward.
The HICPAC-style fecal screening program did demonstrate a far greater extent of colonization than that which would have been revealed by more selective programs. However, colonization continued to occur, despite the knowledge obtained, and resources may have been better directed toward improving overall infection control and hand washing, as any patient, even in traditionally low-risk areas, may be potentially colonized with GRE. A point prevalence study in which every patient in the hospital on a given day is screened for the presence of GRE would give useful information on colonization and potential empirical treatment with considerable savings in resources.
Data collected from this program have given us an indication of the background level of GRE at the study hospital trust, and this information will be of benefit should an outbreak of clinically significant GRE occur.
ACKNOWLEDGMENTS
We are grateful to A. Speekenbrink, Department of Clinical Microbiology, Western Infirmary, Glasgow, Scotland, for retrieving computerized patient data.
This study was supported by the Scottish Office Department of Health (grant reference no. K/MRS/50/C2607).
REFERENCES
- 1.Bell J M, Paton J C, Turnidge J. Emergence of vancomycin-resistant enterococci in Australia: phenotypic and genotypic characteristics of isolates. J Clin Microbiol. 1998;36:2187–2190. doi: 10.1128/jcm.36.8.2187-2190.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyce J M, Opal S M, Chow J W, Zervos M J, Potterbynoe G, Sherman C B, Romulo R L C, Fortna S, Medeiros A A. Outbreak of multidrug-resistant Enterococcus faecium with transferable vanB class vancomycin resistance. J Clin Microbiol. 1994;32:1148–1153. doi: 10.1128/jcm.32.5.1148-1153.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown A R, Amyes S G B, Paton R, Plant W D, Stevenson G M, Winney R J, Miles R S. Epidemiology and control of vancomycin-resistant enterococci (VRE) in a renal unit. J Hosp Infect. 1998;40:115–124. doi: 10.1016/s0195-6701(98)90090-1. [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. Nosocomial enterococci resistant to vancomycin—United States, 1989–1993. Morbid Mortal Weekly Rep. 1993;42:597–600. [PubMed] [Google Scholar]
- 5.Clark N C, Cooksey R C, Hill B C, Swenson J M, Tenover F C. Characterization of glycopeptide-resistant enterococci from U.S. hospitals. Antimicrob Agents Chemother. 1993;37:2311–2317. doi: 10.1128/aac.37.11.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dutka-Malen S, Evers S, Courvalin P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol. 1995;33:24–27. doi: 10.1128/jcm.33.1.24-27.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edmond M B, Ober J F, Weinbaum D L, Pfaller M A, Hwang T, Sanford M D, Wenzel R P. Vancomycin-resistant Enterococcus faecium bacteremia: risk-factors for infection. Clin Infect Dis. 1995;20:1126–1133. doi: 10.1093/clinids/20.5.1126. [DOI] [PubMed] [Google Scholar]
- 8.Eliopoulos G M, Wennersten C B, Gold H S, Schulin T, Souli M, Farris M G, Cerwinka S, Nadler H L, Dowzicky M, Talbot G H, Moellering R C. Characterization of vancomycin-resistant Enterococcus faecium isolates from the United States and their susceptibility in vitro to dalfopristin-quinupristin. Antimicrob Agents Chemother. 1998;42:1088–1092. doi: 10.1128/aac.42.5.1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Endtz H P, van den Braak N, van Belkum A, Kluytmans J A J W, Koeleman J G M, Spanjaard L, Voss A, Weersink A J L, Vandenbroucke-Grauls C M J E, Buiting A G M, van Duin A, Verbrugh H A. Fecal carriage of vancomycin-resistant enterococci in hospitalized patients and those living in the community in The Netherlands. J Clin Microbiol. 1997;35:3026–3031. doi: 10.1128/jcm.35.12.3026-3031.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fraise A P. The treatment and control of vancomycin-resistant enterococci. J Antimicrob Chemother. 1996;38:753–756. doi: 10.1093/jac/38.5.753. [DOI] [PubMed] [Google Scholar]
- 11.Gray J W, Pedler S J. Antibiotic resistant enterococci. J Hosp Infect. 1992;21:1–14. doi: 10.1016/0195-6701(92)90149-g. [DOI] [PubMed] [Google Scholar]
- 12.Hospital Infection Control Practices Advisory Committee. Recommendation of preventing the spread of vancomycin resistance. Infect Control Hosp Epidemiol. 1995;16:105–113. doi: 10.1086/647066. [DOI] [PubMed] [Google Scholar]
- 13.Jensen M A, Webster J A, Straus N. Rapid identification of bacteria on the basis of polymerase chain reaction-amplified ribosomal DNA spacer polymorphisms. Appl Environ Microbiol. 1993;59:945–952. doi: 10.1128/aem.59.4.945-952.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Landman D, Quale J M. Management of infection due to resistant enterococci: a review of the therapeutic options. J Antimicrob Chemother. 1997;40:161–170. doi: 10.1093/jac/40.2.161. [DOI] [PubMed] [Google Scholar]
- 15.LeClercq R, Derlot E, Duval J, Courvalin P. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N Engl J Med. 1988;319:157–161. doi: 10.1056/NEJM198807213190307. [DOI] [PubMed] [Google Scholar]
- 16.Liassine N, Frei R, Jan I, Auckenthaler R. Characterization of glycopeptide-resistant enterococci from a Swiss hospital. J Clin Microbiol. 1998;36:1853–1858. doi: 10.1128/jcm.36.7.1853-1858.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moreno F, Grota P, Crisp C, Magnon K, Melcher G P, Jorgensen J H, Patterson J E. Clinical and molecular epidemiology of vancomycin-resistant Enterococcus faecium during its emergence in a city in southern Texas. Clin Infect Dis. 1995;21:1234–1237. doi: 10.1093/clinids/21.5.1234. [DOI] [PubMed] [Google Scholar]
- 18.Morrison D, Woodford N, Cookson B. Enterococci as emerging pathogens of humans. J Appl Microbiol. 1997;83:89S–99S. doi: 10.1046/j.1365-2672.83.s1.10.x. [DOI] [PubMed] [Google Scholar]
- 19.Morrison D, Woodford N, Cookson B D. Epidemic vancomycin-resistant Enterococcus faecium in the UK. J Clin Microbiol Infect. 1999;1:146–147. doi: 10.1111/j.1469-0691.1995.tb00460.x. [DOI] [PubMed] [Google Scholar]
- 20.Murray B E, Singh K V, Heath J D, Sharma B R, Weinstock G M. Comparison of genomic DNAs of different enterococcal isolates using restriction endonucleases with infrequent recognition sites. J Clin Microbiol. 1990;28:2059–2063. doi: 10.1128/jcm.28.9.2059-2063.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pegues D A, Pegues C F, Hibberd P L, Ford D S, Hooper D C. Emergence and dissemination of a highly vancomycin-resistant vanA strain of Enterococcus faecium at a large teaching hospital. J Clin Microbiol. 1997;35:1565–1570. doi: 10.1128/jcm.35.6.1565-1570.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perlada D E, Smulian A G, Cushion M T. Molecular epidemiology and antibiotic susceptibility of enterococci in Cincinnati, Ohio: a prospective citywide survey. J Clin Microbiol. 1997;35:2342–2347. doi: 10.1128/jcm.35.9.2342-2347.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toye B, Shymanski J, Bobrowska M, Woods W, Ramotar K. Clinical and epidemiologic significance of enterococci intrinsically resistant to vancomycin (possessing the vanC genotype) J Clin Microbiol. 1997;35:3166–3170. doi: 10.1128/jcm.35.12.3166-3170.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tyrrell G J, Bethune R N, Willey B, Low D E. Species identification of enterococci via intergenic ribosomal PCR. J Clin Microbiol. 1997;35:1054–1060. doi: 10.1128/jcm.35.5.1054-1060.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uttley A H C, Collins C H, Naidoo J, George R C. Vancomycin-resistant enterococci. Lancet. 1988;i:57–58. doi: 10.1016/s0140-6736(88)91037-9. [DOI] [PubMed] [Google Scholar]
- 27.VanDamme P, Vercauteren E, Lammens C, Pensart N, Ieven M, Pot B, LeClercq R, Goossens H. Survey of enterococcal susceptibility patterns in Belgium. J Clin Microbiol. 1996;34:2572–2576. doi: 10.1128/jcm.34.10.2572-2576.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Working Party of Antibiotic Sensitivity Testing of the British Society for Antimicrobial Chemotherapy. A guide to sensitivity testing. J Antimicrob Chemother. 1991;27:13–35. [PubMed] [Google Scholar]