Abstract
Cells isolated from individuals with Cockayne syndrome (CS) have a defect in transcription-coupled DNA repair, which rapidly corrects certain DNA lesions located on the transcribed strand of active genes. Despite this DNA repair defect, individuals with CS group A (CSA) or group B (CSB) do not exhibit an increased spontaneous or UV-induced cancer rate. In order to investigate the effect of CSB deficiency on spontaneous carcinogenesis, we crossed CSB−/− mice with cancer-prone mice lacking the p16Ink4a/p19ARF tumor suppressor locus. CSB−/− mice are sensitive to UV-induced skin cancer but show no increased rate of spontaneous cancer. CSB−/− Ink4a/ARF−/− mice developed 60% fewer tumors than Ink4a/ARF−/− animals and demonstrated a longer tumor-free latency time (260 versus 150 days). Moreover, CSB−/− Ink4a/ARF−/− mouse embryo fibroblasts (MEFs) exhibited a lower colony formation rate after low-density seeding, a lower rate of H-Ras-induced transformation, slower proliferation, and a lower mRNA synthesis rate than Ink4a/ARF−/− MEFs. CSB−/− Ink4a/ARF−/− MEFs were also more sensitive to UV-induced p53 induction and UV-induced apoptosis than were Ink4a/ARF−/− MEFs. In order to investigate whether the apparent antineoplastic effect of CSB gene disruption was caused by sensitization to genotoxin-induced (p53-mediated) apoptosis or by p53-independent sequelae, we also generated p53−/− and CSB−/− p53−/− MEFs. The CSB−/− p53−/− MEFs demonstrated lower colony formation efficiency, a lower proliferation rate, a lower mRNA synthesis rate, and a higher rate of UV-induced cell death than p53−/− MEFs. Collectively, these results indicate that the antineoplastic effect of CSB gene disruption is at least partially p53 independent; it may result from impaired transcription or from apoptosis secondary to environmental or endogenous DNA damage.
Genomic integrity is continuously threatened by endogenous and exogenous agents that damage DNA and have immediate and long-term effects on cellular functioning. Replication of damaged DNA can cause mutations that may ultimately lead to carcinogenic events or, when occurring in germ cells, to inborn defects. Immediate effects of DNA damage include a blockage to transcription which can result in cell death by apoptosis (22). To minimize the harmful effects of DNA damage, nature has equipped cells with a sophisticated network of DNA repair mechanisms. Nucleotide excision repair (NER) is a cut-and-paste repair mechanism for the removal of a variety of bulky DNA lesions, such as chemical adducts and UV-induced photolesions (reviewed in references 11 and 16). The NER apparatus utilizes approximately 30 gene products to (i) recognize DNA damage, (ii) introduce single-stranded nicks 5′ and 3′ to the lesion, resulting in the removal of a single-stranded oligonucleotide containing the lesion, and (iii) restore the integrity of the double helix by gap-filling DNA synthesis (using the undamaged DNA strand as a template) and strand ligation (16). NER can be divided into two subpathways that differ in the molecular mechanism of lesion recognition. The global genome repair machinery (ggNER), in which damage recognition is performed by the XPC-HR23B complex (54), can remove lesions located anywhere in the genome. However, some types of lesions are poorly recognized by the ggNER apparatus and therefore are inefficiently repaired. Such lesions, when present in the template strand of active genes, interfere with transcription and activate the transcription-coupled repair subpathway of NER (tcNER). In the tcNER reaction, stalling of RNA polymerase II at bulky lesions serves as the damage sensor and subsequently recruits the remainder of the NER apparatus (37, 38, 55, 56).
The importance of NER for genome preservation is illustrated by rare autosomal recessive disorders like xeroderma pigmentosum (XP), which is made up of seven complementation groups (XPA through XPG), and Cockayne syndrome (CS), which is made up of two complementation groups (CSA and CSB) (5). Both disorders are characterized by hypersensitivity to solar (UV) light. XP individuals demonstrate an increased rate of sunlight-induced skin cancer as well as carcinogen-induced internal tumors (5, 34). Except for XPC and XPE, where only the ggNER pathway is affected, XP is associated with a defect in both ggNER and tcNER. Individuals with CS exhibit small stature, intellectual impairment, cachexia, and sun sensitivity but no increased incidence of cancer (5, 15, 24). Cells from CS patients have been shown to be deficient in tcNER only (11, 16, 26, 57). An important question is why CS patients, despite their DNA repair deficiency, have not been reported to develop cancer of the skin or internal organs. A possible explanation is that CS cells can still perform ggNER, which, even though some types of lesions may be repaired at lower rates, could prevent accumulation of mutagenic lesions (15). Alternatively, since CSA and CSB cells cannot appropriately clear RNA polymerase II arrested at intragenic DNA lesions (39) and since stalled RNA polymerase II has been shown to promote intracellular accumulation of p53 and apoptosis (41, 53, 62), it has been suggested that in CS patients precancerous cells are more efficiently eliminated by apoptosis than they are in healthy persons (24).
Among the growing number of mouse mutants with engineered deletions of genes involved in cellular responses to DNA damage are mouse models for XPA (18, 42), CSA (21), and CSB (59). Totally NER-deficient XPA−/− mice resemble human XPA patients in their UV-induced skin cancer predisposition (18, 42) and also show elevated frequencies of skin and internal tumors after exposure to chemical carcinogens (17, 18). In contrast to human CS, CSB−/− mice demonstrate an elevated incidence of both UV- and chemically induced skin cancer (59). This difference between CSB patients and mice might be due to the lack of expression of the p53-inducible p48 gene, required for ggNER of UV-induced cyclobutane dimers (and presumably other carcinogen-induced DNA lesions) (27). Since removal of these types of lesions in rodents almost totally depends on tcNER, the CSB defect may have a more dramatic effect in mice than in humans. Alternatively, since UV-induced skin tumor formation in CSB mice requires a longer latency time than that required by totally NER-deficient XPA mice (3, 4), CSB patients may simply not get old enough (average life span, 12.5 years) to develop tumors (43).
Totally NER-deficient XPA mice, like human XP patients, develop spontaneous internal tumors upon aging (17). With the observation that, at least in mice, a specific tcNER defect predisposes to UV- and chemically induced skin cancer, the question of whether CS is also associated with increased sensitivity to spontaneous tumor formation arises. Mice with a homozygous disruption of the Ink4a/ARF locus have proven to be a particularly useful model system for studying factors (such as Ras and telomerase) contributing to neoplasia (9, 23, 48, 49). A great deal of evidence indicates that deletion of this region predisposes to cancer development (reviewed in references 8, 50, and 51). Humans with lesions in this genetic locus have an elevated incidence of melanoma, pancreatic carcinoma, and other neoplasms. Mice with homozygous disruption of the Ink4a/ARF locus are predisposed to developing spontaneous lymphomas and fibrosarcomas (49). The Ink4a gene product, p16Ink4a, helps regulate cellular proliferation by inhibiting cyclin-dependent kinases 4 and 6. The ARF gene product, p19ARF, is encoded by a gene overlapping that of the Ink4a gene but utilizing an alternative reading frame (ARF). p19ARF potentiates the function of p53 by antagonizing mdm2 function (52). If left unimpeded, mdm2 inhibits p53 function both by binding and by blocking p53's transcriptional activation domain as well as by catalyzing p53's ubiquitination, thus leading to its proteasomal degradation (32). Cancer-predisposed Ink4a/ARF−/− mice could therefore be used to investigate the role of the CSB gene product in spontaneous tumorigenesis.
In the present study we have generated CSB−/− Ink4a/ARF−/− mice to study the effect of a CSB deficiency on spontaneous tumor formation. Surprisingly, since DNA repair genes are usually considered tumor suppressor genes, we found that inactivation of the CSB gene reduced spontaneous tumor formation in Ink4a/ARF−/− mice. Comparison of CSB−/− Ink4a/ARF−/−, CSB−/−, Ink4a/ARF−/−, and wild-type (WT) mouse embryo fibroblasts (MEFs) for their colony formation rates after low density seeding, H-Ras oncogene-mediated transformation rates, proliferation rates, and mRNA transcription rates revealed that at the cellular level, the CSB gene deficiency diminished neoplastic potential. Moreover, we show that the CSB gene deficiency sensitizes Ink4a/ARF−/− MEFs to UV-induced accumulation of p53 and apoptosis. Similarly, the CSB defect is shown to reduce the neoplastic potential of p53−/− MEFs. The implications of a CSB deficiency for tumorigenesis are discussed.
MATERIALS AND METHODS
Generation of double knockout mice and cell lines.
CSB−/− Ink4a/ARF−/− mice and CSB−/− p53−/− mice were generated by crossing CSB−/− mice (knockout mice lacking the CSB gene) (59) with Ink4a/ARF−/− mice (knockout mice lacking the p16Ink4a gene as well as the overlapping p19ARF gene) (49) and p53−/− mice (knockout mice lacking the p53 gene) (30), respectively. p53−/− mice were obtained from Jackson Laboratory (Bar Harbor, Maine). All mice had a hybrid C57BL/6J-129/sv genetic background (28, 49, 59). WT, CSB−/−, Ink4a/ARF−/−, p53, CSB−/− Ink4a/ARF−/−, and CSB−/− p53−/− MEFs were isolated from embryos derived from crosses of (di)homozygous parents of the desired genotype according to published procedures (45). Briefly, mouse embryos at 13.5 days of gestation were isolated in utero and cells were dispersed by using a razor blade and a syringe with an 18-gauge needle attached. Resultant cells were cultured and frozen as soon as was practical. Cells were stored in liquid nitrogen and upon thawing were used within one to three passages. The presence or absence of p16Ink4a and p53 protein products was verified by immunoblot analysis (data not shown). Genotyping of mice and cell lines was performed by PCR, as described previously (3, 30, 49).
Spontaneous tumor formation.
When tumors were observed in CSB−/− Ink4a/ARF−/− or Ink4a/ARF−/− mice, they were fixed in 10% neutral buffered formalin, processed, and paraffin embedded and then sections were stained with hematoxylin and eosin according to standard procedures. Comparisons of tumor-free survival were charted by the method of Kaplan and Meier (percent survival versus time), and statistical significance of the curves generated for CSB−/− Ink4a/ARF−/− versus Ink4a/ARF−/− mice was established using the log rank test.
Growth rate and colony formation efficiency.
Three MEF lines were assayed for each genotype. For establishing the growth rate of each MEF line, 2 × 105 cells were plated in multiple 6-cm-diameter dishes and incubated in Dulbecco's modified Eagle's medium (DMEM) plus 15% fetal bovine serum (FBS). One dish of cells was counted every day. Colony formation efficiency was performed as described elsewhere (49). Briefly, for each MEF line analyzed, 3.5 × 103 cells were distributed on a total of 10 dishes of 6-cm diameter and incubated in DMEM plus 15% FBS. After 8 days, dishes were stained with crystal violet, and the number of visible colonies (>1.5 mm in diameter) was scored. Statistical significance (P value determined by Student's t test) was calculated using Microsoft Excel.
Ras transformation assay.
Transformed foci could be induced in Ink4a/ARF−/− MEFs by transfection of an H-Ras expression construct (49). The assay was performed as described previously (46). Early-passage MEFs derived from individual embryos (3 MEF lines per genotype) were seeded (8 × 105 cells) in plates of 10-cm diameter and grown in DMEM plus 10% FBS overnight. The medium was changed 4 h before transfections began. Transfections were done by standard calcium-phosphate procedures with DNA mixtures containing 10 μg of each of the relevant plasmids plus carrier DNA for a total of 30 μg of DNA. After 8 h of incubation with the precipitates, the incubation medium was changed and cultures were fed with fresh medium every 3 days. At day 14 posttransfection, foci were scored visually (49).
Flow cytometry analysis.
Assessment of a fraction of cells in S phase and analysis of apoptosis via a fraction of cells with a sub-G1 DNA content were performed by the fluorescence-activated cell sorter (FACS) facility of the Albert Einstein College of Medicine utilizing protocols described elsewhere (13). Briefly, early-passage MEFs from individual embryos (three MEF lines per genotype) were plated (106 cells) in 10-cm-diameter plates and incubated in DMEM plus 10% FBS for 24 h. For cell death (percentage of sub-G1) analysis, cells were subjected to UVC (UV radiation which is predominantly 254 nm in wavelength) irradiation at 5 or 10 J/m2, incubated another 12 or 24 h, harvested, and fixed with 70% ethanol. UVC irradiation was from a standard tissue culture germicidal hood; UVC dose was determined with a UVX radiometer (UVP, Upland, Calif.). The fixed cells are washed with phosphate-buffered saline and then stained with propidium iodide in a solution containing Triton X-100 as well as RNase A. Flow cytometry employed excitation with a blue light and detection of propidium iodide emission at red wavelengths. Statistical significance (P value determined by Student's t test) was calculated using Microsoft Excel.
In situ TUNEL assay.
Apoptosis of MEFs was also assayed via in situ DNA fragmentation using in situ terminal deoxyribonucleotide transferase-mediated dUTP nick-end labeling (TUNEL). TUNEL labeling was performed using the TACS 2 TdT (Fluor) kit (Trevigen, Gaithersburg, Md.). MEFs grown on glass slides in DMEM plus 15% FBS were UVC irradiated, incubated for 24 h, fixed, and treated using the manufacturer's recommended conditions. Percentages of apoptotic cells were determined by scoring TUNEL-positive (apoptotic) nuclei on a Leica (model Leitz DMRB) fluorescence microscope and dividing by the total cell number determined by using the microscope's phase-contrast optics.
mRNA synthesis.
The mRNA synthesis rate was measured by a modification of a method described elsewhere (1). Early-passage MEFs from individual embryos (three MEF lines per genotype) were plated (105 cells) onto 35-mm cell culture dishes and incubated in DMEM plus 10% FBS for 24 h before being prelabeled with [14C]thymidine for 48 h to provide a measure of total DNA content for cell normalization. The cells were then pulse labeled for 1 h with [3H]uridine. Cells were lysed and poly(A)+ RNA was isolated by using the Straight A's system following the manufacturer's recommendations (Novagen, Madison, Wis.). The relative transcription rate was defined as scintillations of 3H per scintillation of 14C. Statistical significance (P value determined by Student's t test) was calculated using Microsoft Excel.
Immunoblot analysis.
For p53 protein analysis, MEFs were subjected to UVC irradiation (5 J/m2) from a standard tissue culture germicidal hood. Whole-cell extracts were prepared and subjected to sodium dodecyl sulfate–8% polyacrylamide gel electrophoresis and immunoblot analysis as described previously (6) with p53 monoclonal antibody (PharMingen, San Diego, Calif.). Protein bands were visualized via chemiluminescence (Pierce, Rockford, Ill.) after being blotted with peroxidase-conjugated goat anti-mouse immunoglobulin G (diluted 1:5,000). Immunoblots for p16 utilized rabbit anti-p16 antibody (M-156) from Santa Cruz Biotechnology (Santa Cruz, Calif.); the antibody for beta tubulin (H-235) was also from Santa Cruz Biotechnology.
RESULTS
Effect of disrupting CSB gene product on tumor formation in Ink4a/ARF−/− mice.
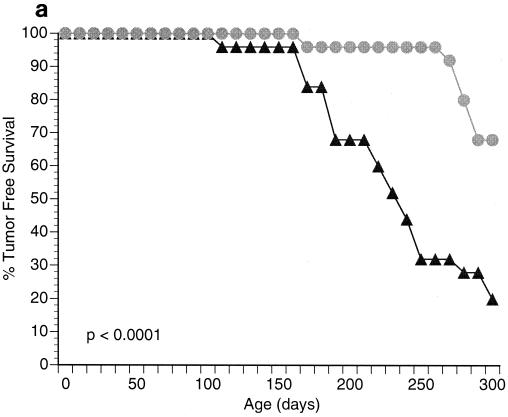
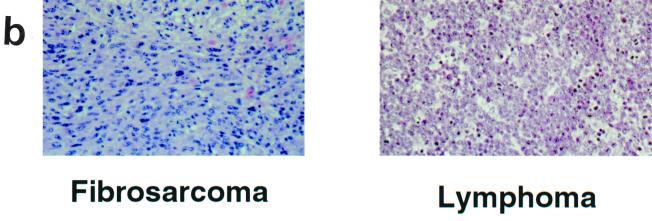
Both male and female CSB−/− and Ink4a/ARF−/− (null for both p16INK4a and p19ARF) mice are viable and fertile (49, 59). Intercrosses of these two strains generated colonies of CSB−/− Ink4a/ARF−/− mice in a hybrid C57BL/6J-129/sv genetic background. The size, behavior, fertility, and viability of CSB−/− Ink4a/ARF−/− animals were not significantly different from those of age-matched CSB−/−, Ink4a/ARF−/−, or WT controls. To assess the effect of CSB gene disruption on cancer susceptibility, a cohort of 4- to 6-week-old CSB−/− Ink4a/ARF−/− mice and age-matched Ink4a/ARF− controls (n = 25 for each group) were monitored for spontaneous tumor development over a 9-month period. Tumor-free survival was significantly greater in the CSB−/− Ink4a/ARF−/− mice than in Ink4a/ARF−/− animals (Fig. 1a; P < 0.0001). Although the tumor spectrum was similar, consisting primarily of fibrosarcomas and lymphomas (Fig. 1b), tumor incidence in the CSB−/− Ink4a/ARF−/− cohort was significantly lower than in the Ink4a/ARF−/− control (32 versus 80%, respectively). Moreover, latency to tumor development was increased from 150 days in the control to 260 days in the CSB−/− Ink4a/ARF−/− group (Fig. 1a).
FIG. 1.
Disruption of the CSB gene product reduces tumor incidence in Ink4a/ARF−/− mice. (a) Survival data for Ink4a/ARF−/− mice (triangles) versus CSB−/− Ink4a/ARF−/− mice (circles). Twenty-five Ink4a/ARF−/− mice (4 to 6 weeks old) and 25 CSB−/− Ink4a/ARF−/− mice of similar age were monitored for a total of 300 days. Points on the curve denote times when animals died or were sacrificed because of tumor formation. All deaths in this experiment were due to tumor formation. Of the 25 Ink4a/ARF−/− mice, 20 (80%) developed tumors (75% lymphomas and 25% fibrosarcomas). Of the 25 CSB−/− Ink4a/ARF−/− mice, 8 (32%) developed tumors (87% lymphomas and 13% fibrosarcomas). (b) Representative photomicrographs of the two tumor types (fibrosarcoma and lymphoma) observed in this experiment. Fibrosarcomas appeared as subcutaneous masses at various locations, including the shoulder, neck, flank, and legs. The fibrosarcomas were composed of elongated spindle cells forming whorls and bundles. Mice with lymphomas showed enlarged spleens, livers, lymph nodes, and sometimes thymuses. The lymphomas were composed of lymphoblastic cells, reticulum cells with abundant eosinophilic cytoplasm, lymphocytic mixed cells, and sometimes pleomorphic lymphoid cells.
Proliferation rate of Ink4a/ARF−/− MEFs is reduced by CSB gene disruption.
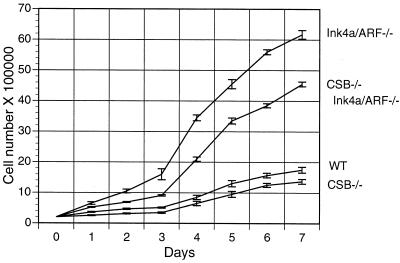
To understand how the CSB deficiency contributed to decreased tumor incidence in Ink4a/ARF−/− mice, we first examined the impact of CSB gene disruption on the cell proliferation rate of WT, CSB−/−, Ink4a/ARF−/−, and CSB−/− Ink4a/ARF−/− MEFs. Growth curve determination revealed that Ink4a/ARF−/− MEFs grew more rapidly than WT MEFs, in agreement with measurements obtained previously (49). The CSB deficiency significantly reduced the growth rate of Ink4a/ARF−/− MEFs (Fig. 2, compare Ink4a/ARF−/− with CSB−/− Ink4a/ARF−/−; P < 0.05). The proliferation rate of the CSB−/− MEFs was only slightly lower than that of the WT MEFs, suggesting that CSB gene disruption is more detrimental in rapidly growing (e.g., Ink4a/ARF−/−) cells (see Discussion).
FIG. 2.
Cell proliferation analysis of early-passage MEFs derived from individual embryos. For each genotype, 2 × 105 cells were plated on each of eight 6-cm-diameter dishes on day zero and incubated in DMEM plus 15% FBS. One dish of cells was counted per cell line for the next 7 days.
Another indicator of cellular proliferative capacity is the fraction of unsynchronized, logarithmically growing cells that are in S phase at any given time. MEFs of each of the four genotypes were assessed via flow cytometric analysis. The fraction of CSB−/− Ink4a/ARF−/− MEFs in S phase (20.7%) was significantly lower than the fraction of Ink4a/ARF−/− MEFs in S phase (27.1%, P < 0.05; data not shown). Consistent with the growth curves, CSB deficiency did not significantly reduce the fraction of WT MEFs in S phase (14.75% for CSB−/− MEFs as opposed to 15.3% for WT MEFs; data not shown).
CSB gene disruption reduces total mRNA synthesis rate.
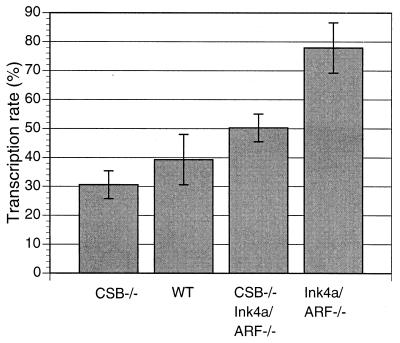
It has been shown that the rate of mRNA synthesis by RNA polymerase II is reduced in cells lacking CSB (19, 47). The basal transcription rate of RNA polymerase II is regulated by multiple cellular factors at the levels of initiation and elongation and is independent of the cellular proliferation rate (7). Under certain physiological conditions (e.g., cardiac hypertrophy), an enhanced basal transcription rate may be a cause of the increased cellular proliferation rate (1). When total mRNA synthesis rates were compared for WT, CSB−/−, Ink4a/ARF−/−, and CSB−/− Ink4a/ARF−/−MEFs, the level was significantly lower for CSB−/− Ink4a/ARF−/− cells than for Ink4a/ARF−/− cells (Fig. 3) (P < 0.05).
FIG. 3.
Effect of CSB disruption on mRNA synthesis rate. Cells were prelabeled with [14C]thymidine for 48 h and were then pulse labeled with [3H]uridine for 1 h. Cells were lysed and poly(A)+ RNA was isolated using Straight A's reagent (Novagen), as described in Materials and Methods. In addition, a fraction of each lysate was subjected to total DNA extraction. The relative transcription rate (polyadenylated mRNA) was calculated as scintillations of 3H per scintillation of 14C. P values were as follows: WT versus Ink4a/ARF−/−, P < 0.05; CSB−/− Ink4a/ARF−/− versus Ink4a/ARF−/−, P < 0.05; CSB−/− versus WT, P > 0.05.
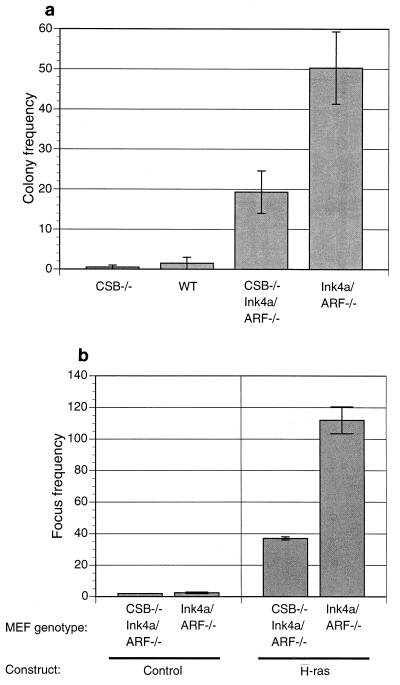
Disruption of CSB gene diminishes immortalization potential of Ink4a/ARF−/− MEFs.
In view of the impaired proliferation observed in CSB−/− Ink4a/ARF−/− MEFs, we sought to determine the colony formation rates of Ink4a/ARF−/− and CSB−/− Ink4a/ARF−/− MEFs after low-density seeding, a surrogate assay for immortalization potential (23). As shown in Fig. 4a, CSB deficiency dramatically reduced the colony formation rate of Ink4a/ARF−/− MEFs (P < 0.05). A similar trend held true for a CSB−/− Ink4a/ARF−/− MEF clone compared to a CSB+/+ Ink4a/ARF−/− MEF clone derived from a CSB+/− Ink4a/ARF−/− × CSB+/− Ink4a/ARF−/− heterozygous intercross (data not shown).
FIG. 4.
Disruption of the CSB gene product reduces the immortalization potential of Ink4a/ARF−/− MEFs. (a) Colony formation assay. For each genotype, 3,500 cells were plated onto 10 dishes (6-cm diameter) and colonies were visualized by crystal violet staining after 8 days. The colony frequencies on the histogram represent the average numbers of colonies (larger than 3 mm) per plate compiled from three colony formation assays. (b) H-Ras transformation assay. Early-passage MEFs derived from individual embryos of the indicated genotype were seeded (8 × 105 cells) on two plates of 10-cm diameter. Transfections were performed with the indicated H-Ras or control (vector) constructs the next day. At day 14 posttransfection, foci were scored visually. The histogram presents mean numbers of colonies per plate from two experiments.
Consistent with their reduced proliferative and immortalization potentials, CSB gene deficiency was found to render Ink4a/ARF−/− MEFs more resistant to neoplastic transformation in vitro, correlating with reduced tumor incidence in vivo. Specifically, Ink4a/ARF−/− MEFs were efficiently transformed by activated H-Ras alone, as previously shown (49). Focus formation was reduced by 65% in CSB−/− Ink4a/ARF−/− MEFs compared to Ink4a/ARF −/− MEFs (Fig. 4b, P < 0.05).
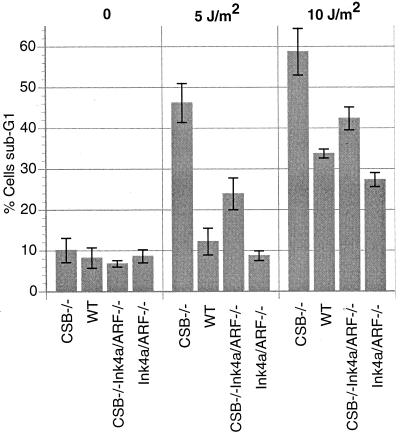
CSB gene disruption sensitizes cells to UV-induced apoptosis.
Previous studies have shown that CSB gene disruption renders cells more sensitive to apoptosis induced by UV radiation as well as ionizing radiation and oxidizing agents (36, 39, 40). To investigate the effect of CSB gene disruption upon UV-induced cell death in Ink4a/ARF−/− MEFs, we examined the sub-G1 fraction of MEFs of each of the four genotypes (WT, CSB−/−, Ink4a/ARF−/−, and CSB−/−Ink4a/ARF−/−) after various doses of UVC radiation (10, 5, or 0 J/m2). Consistent with previous observations, we found that the CSB deficiency sensitized both WT and Ink4a/ARF−/− MEFs to UVC-induced cell death (Fig. 5). Specifically, at a dose of 5 J/m2 we observed a 23.9% apoptosis rate in CSB−/− Ink4a/ARF−/− MEFs compared to 8.7% in Ink4a/ARF−/− cells (Fig. 5).
FIG. 5.
Effect of CSB gene disruption on UV-induced cell death. MEFs of the indicated genotypes were subjected to UVC radiation (0, 5, or 10 J/m2) and permitted to grow for another 24 h. The histogram shows data tabulated from average percentages of sub-G1 values for MEFs of each of the indicated genotypes (P value after 5 J/m2 for CSB−/− Ink4a/ARF−/− versus Ink4a/ARF−/− was <0.05).
In a separate experiment, we demonstrated that a UV dose as low as 2.5 J/m2 was sufficient to induce significant apoptosis in CSB−/− Ink4a/ARF−/− MEFs (but not in Ink4a/ARF−/− MEFs). By scoring for TUNEL-positive nuclei we observed 19% apoptotic nuclei in CSB−/− Ink4a/ARF−/− MEFs 24 h after 2.5 J/m2 versus 1% in Ink4a/ARF−/− MEFs (data not shown).
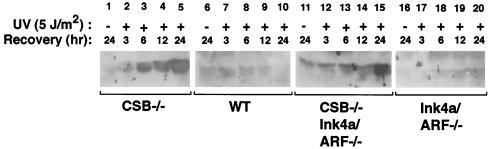
CSB gene disruption sensitizes cells to UV-mediated p53 induction.
An important mediator of genotoxin-induced apoptosis is p53 through its ability to induce the transcription of proapoptotic factors, such as bax (reviewed in reference 2). It has been shown that CSB gene disruption leads to p53 induction (increased steady-state level) at lower doses of UVC radiation than in WT cells, presumably because the CSB-deficient cells cannot clear stalled RNA polymerase II from UVC-induced lesions, which appear to stimulate p53 induction (35, 41). We therefore compared the sensitivity of WT, CSB−/−, Ink4a/ARF−/−, and CSB−/− Ink4a/ARF−/− MEFs for p53 induction after exposure to 5 J/m2 of UVC radiation (Fig. 6). As shown in Fig. 6, CSB deficiency enhanced p53 induction in WT and Ink4a/ARF−/− MEFs. Together these data suggest that UVC-induced apoptosis in this context is likely mediated by p53.
FIG. 6.
CSB gene disruption sensitizes MEFs to UV induction of p53. MEFs of each of the indicated genotypes were plated on 6-cm dishes and subjected to UVC radiation (5 J/m2) as indicated, and whole-cell extracts were prepared 3, 6, 12, or 24 h later. Cellular extracts from equivalent numbers of cells were subjected to immunoblot analysis with a monoclonal anti-p53 antibody (see Materials and Methods for details). An immunoblot with anti-tubulin antibody was performed to verify that equal amounts of protein were loaded (data not shown).
CSB gene disruption reduces proliferative and immortalization potential of p53−/− cells.
Since CSB−/− Ink4a/ARF−/− cells demonstrated sensitization to UV-induced p53 accumulation, we wished to determine to what extent the diminution of neoplastic properties caused by CSB gene disruption was p53 dependent. To address this question we crossed p53−/− mice (30) with CSB−/− mice to generate CSB−/− p53−/− mice. p53−/− and CSB−/− p53−/− MEF lines were generated from embryos obtained from p53−/− × p53−/− mice and CSB−/− p53−/− × CSB−/− p53−/− mice intercrosses, respectively.
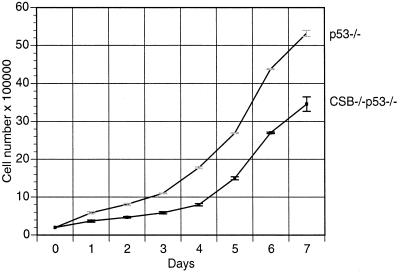
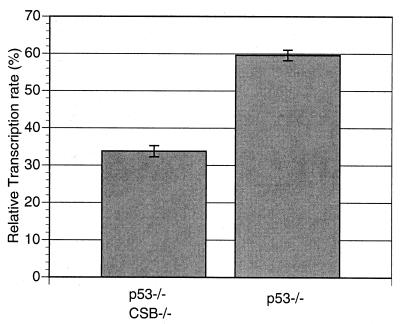
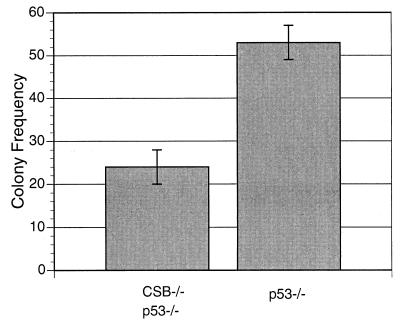
In line with previous reports (25), we observed that p53−/− MEFs proliferated more rapidly than WT MEFs (data not shown). In addition, CSB−/− p53−/− MEFs proliferated at a lower rate than p53−/− MEFs (Fig. 7). CSB−/− p53−/− MEFs also demonstrated a lower fraction of cells in S phase compared to p53−/− MEFs, as shown by FACS analysis of nonsynchronized, logarithmically growing samples of each genotype (23.0 versus 31.6%, P < 0.05; data not shown). CSB gene disruption reduced the basal mRNA transcription rate of p53−/− cells (Fig. 8) (P < 0.05), as it did for Ink4a/ARF−/− cells (Fig. 3). The transcription rate of p53−/− cells, like that of Ink4a/ARF−/− cells, was greater than that of WT cells. CSB−/− p53−/− MEFs also demonstrated a significantly reduced rate of colony formation after low-density seeding (Fig. 9) (P < 0.05). It had previously been reproducibly demonstrated that WT and CSB−/− MEFs form almost no colonies after low-density seeding (Fig. 4a).
FIG. 7.
CSB gene disruption leads to reduced proliferation in p53−/− MEFs. For each genotype, 2 × 105 aliquots of cells were plated in each of seven 6-cm-diameter dishes on day zero and incubated in DMEM plus 15% FBS. One dish of cells was counted per cell line every day thereafter. Each data point represents the mean cell number calculated from a MEF line of the indicated genotype on the indicated day (repeated three times) (P < 0.05).
FIG. 8.
CSB gene disruption reduces mRNA synthesis rate in p53−/− MEFs. The relative transcription rate (polyadenylated mRNA) was calculated as for Fig. 3. Each bar represents measurements obtained from two independent experiments (P < 0.05).
FIG. 9.
CSB gene disruption reduces the colony formation rate of MEFs lacking the p53 tumor suppressor gene. For each genotype, 3,500 cells were plated onto 6-cm dishes, and colonies were scored by crystal violet staining after 8 days as for Fig. 4. Each bar in the histogram represents the average number of colonies (larger than 3 mm) per plate from 10 plates from each of the indicated genotypes (repeated three times).
CSB gene disruption increases rate of UV-induced cell death.
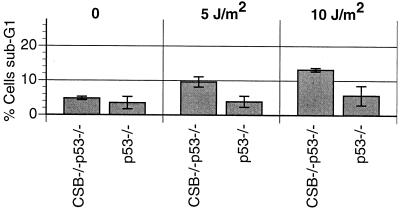
p53−/− and CSB−/− p53−/− MEFs were UVC irradiated (10, 5, or 0 J/m2), and the apoptotic rate was measured by FACS analysis, as for Fig. 5 (13). Interestingly, there is a small but statistically significant increase in the rate of apoptosis after irradiation with 5 or 10 J/m2 (P < 0.05) (Fig. 10). This is consistent with other studies that indicate that p53-independent mechanisms of UV-induced apoptosis exist (44, 58).
FIG. 10.
CSB gene disruption increases UV-induced apoptosis rate in p53−/− MEFs. MEFs of the indicated genotypes were subjected to UVC radiation (5 or 10 J/m2) and assayed for apoptosis 24 h later via flow cytometry (% sub-G1) as described in Materials and Methods. The histogram represents data obtained from two independent experiments (P < 0.05 for p53−/− versus CSB−/− p53−/− after irradiation with both 5 and 10 J/m2).
DISCUSSION
In the present study we have shown that inactivation of the CSB gene reduces the predisposition of Ink4a/ARF knockout mice to develop spontaneous tumors. Consistent with this observation, MEFs derived from CSB−/− Ink4a/ARF−/− animals were shown to possess a decreased neoplastic potential compared to animals with Ink4a/ARF−/− MEFs, as is evident from the reduced colony- and focus-forming abilities after low-density seeding or transfection with an H-Ras expression construct, respectively. Similarly, the neoplastic potential of p53−/− cells is reduced by inactivation of the CSB gene, suggesting that the antineoplastic effect of the CSB deficiency is generally applicable and is at least partially p53 independent. These findings appear to be in agreement with the absence of clear tumorigenic events in human CS, although the fact that the short life span of patients (10 to 12 years on average) masks a potential cancer predisposition at older ages cannot be excluded (15, 43). At the same time, however, the reduction in carcinogenic potential due to CSB inactivation seems inconsistent with the function of DNA repair genes as tumor suppressor genes as well as with the skin cancer predisposition of CSB mice following exposure to UV light and chemical carcinogens (3, 59). Several properties of the CSB gene product may help to explain the decreased neoplastic potential of CSB−/− mice and MEFs and the apparently opposing effects of CSB gene disruption on spontaneous and induced carcinogenesis.
First, the CSB protein is most closely associated with transcription-coupled repair of UV- and chemically induced lesions. Transcription-coupled repair may take place because the CSB gene product, along with the CSA protein and an RNA polymerase II molecule stalled at a DNA lesion, efficiently recruits the NER apparatus to the damage in the transcribed strand in a manner that remains to be clearly understood (3, 16, 56, 60). Evidence is accumulating that base excision repair (BER) (responsible for repair of damaged nucleotide bases), like NER, can also be subdivided into a global genome and a transcription-coupled subpathway and that the CSB protein is required for transcription-coupled BER (tcBER) (12, 36, 39). In addition (or perhaps, instead), the CSB gene product could play a role in displacing or removing the stalled RNA polymerase molecule from transcription-blocking lesions so that repair can take place and transcription can resume, a phenomenon that is distinct from rapid recruitment of the NER (and presumably the BER) apparatus (reviewed in reference 61). When subjected to exogenous mutagens (i.e., UV radiation), skin tumorigenesis is elevated in CSB−/− mice, indicating that tcNER acts as a tumor-suppressing mechanism by removing mutagenic lesions. However, since CSB−/− mice do not show increased frequencies of internal tumors and since inactivation of the CSB gene decreases rather than increases the spontaneous tumorigenesis rate in cancer-prone Ink4a/ARF−/− mice, loss of tcNER or tcBER does not appear to significantly promote spontaneous carcinogenesis (at least in mice). Instead, CSB−/− Ink4a/ARF−/− mice are less prone to develop spontaneous tumors than Ink4a/ARF−/−mice, which suggests that a functional CSB protein somehow assists (nonrepair) functions that promote rather than suppress cancer development.
One such function could relate to the essential process of transcription. The CSB defect has been shown to negatively affect the rate of mRNA synthesis, leading to the proposition that CSB is an auxiliary transcription elongation factor (19, 47). Further supporting this idea are studies showing a physical association of CSB with elongating RNA polymerase II (10, 56, 60). It has been suggested that the CS phenotype (including short stature and intellectual impairment) results from the inability of CS cells to transcribe their genome at rates sufficient to meet the metabolic and/or synthetic demands of highly active cells (e.g., myelin synthesis in the nervous system) (14, 15, 20, 24, 29, 61). Ink4a/ARF−/− and p53−/− cells are highly proliferative cell types and accordingly require a very active metabolism. Thus, it is conceivable that a reduction of transcription competence by the CSB defect as observed in CSB−/− Ink4a/ARF−/− and CSB−/− p53−/− cells (Fig. 3 and 8) contributes to the decrease in the proliferative capacity of these cells, and thus to the reduction of neoplastic potential.
Moreover, CSB-deficient cells are known to undergo apoptosis at lower doses of UV radiation, ionizing radiation, or the oxidizing agent hydrogen peroxide than WT cells (12, 39, 40). It has been demonstrated that RNA polymerase II persistently stalled at UV- or other genotoxin-induced DNA lesions is responsible for the elevated apoptosis rate (40). In accordance with these data, we have shown that inactivation of the CSB gene also increases the sensitivity of CSB−/− Ink4a/ARF−/− or CSB−/− p53−/− MEFs to UV-induced cell death (Fig. 5 and 10). Thus, it is possible that the reduced number of tumors in CSB−/− Ink4a/ARF−/− mice and the reduced neoplastic potential demonstrated by CSB−/− Ink4a/ARF−/− MEFs result from an increased rate of spontaneous apoptosis of precancerous cells. As these animals and cells have not been exposed to UV or other environmental agents, the latter process must be triggered by DNA damage produced by endogenous genotoxins. Recently, it was shown that DNA lesions (i.e., 8-oxo-guanine and thymine glycol) caused by reactive oxygen species continuously generated by cellular metabolic processes or upon exposure to environmental agents can also block transcription by RNA polymerase II (39). We hypothesize that the CSB defect in CSB−/− Ink4a/ARF−/− mice may make precancerous cells vulnerable to clearance by apoptosis for the following reasons. (i) Transformation of normal cells into highly proliferative (pre)cancerous cells is assumed to up-regulate cellular metabolism, which is expected to produce increased levels of oxidative DNA damage. (ii) CSB−/− cells cannot repair 8-oxo-guanine and thymine glycol in a transcription-coupled manner (12, 36). (iii) Stalled RNA polymerase II is a powerful signal for apoptosis (40).
In the present study we have provided strong evidence for the antineoplastic potential of the CSB defect in the background of p16Ink4a/p19ARF or p53 tumor suppressor deficiency. Yet, when CSB−/− mice are challenged with exogenous DNA-damaging agents, like UV light and the chemical carcinogen dimethyl benzanthracine (designated DMBA), a mild but obvious skin cancer predisposition is observed (3, 59). Although the CSB defect results in an enhanced sensitivity to apoptosis triggered by transcription-blocking lesions which is likely to reduce neoplastic susceptibility (40), apoptotic elimination of precancerous cells in carcinogen-exposed CSB−/− animals is apparently insufficient to counteract the increased mutagenesis originating from the tcNER defect. UV-induced tumor formation depends not only upon the initiation of neoplastic cells (determined by the rate of UV-induced mutagenesis, which is a function of UV-induced DNA lesion repair versus UV-induced apoptosis) but also on the ability of such cells to continue to proliferate and avoid apoptosis (tumor progression). It is noteworthy that although UV-exposed CSB−/− mice develop a fair number of early neoplastic lesions (benign papillomas), these lesions infrequently develop into invasive carcinomas (3). Thus, it seems that the decreased transcription rate and/or increased sensitivity to spontaneous apoptosis characteristic of the CSB−/− genotype is particularly detrimental to tumor progression.
Despite the role of mdm2 in maintaining low basal levels of p53 and the known function of p19ARF in antagonizing mdm2, ionizing radiation and actinomycin D can still induce elevated p53 levels in ARF−/− cells (31, 33). Hence, the genotoxin-induced stabilization of p53 has not been thought to involve up-regulation of ARF. Similarly, in the absence of functional p19ARF, UV radiation could still induce elevated levels of p53 in CSB−/− Ink4a/ARF−/− cells (Fig. 6, lane 15). It is likely that the increased UV-induced apoptotic rate in CSB−/− Ink4a/ARF−/− cells (compared to Ink4a−/− cells) directly relates to increased p53 induction. Nevertheless, CSB−/− p53−/− cells still demonstrated a slight but statistically significant elevation in UV-induced apoptosis compared to p53−/− cells, indicating that persistently stalled RNA polymerase II may activate the apoptotic machinery to at least some degree in a p53-independent manner (Fig. 10). However, the rate of UV-induced cell death was far lower in p53−/− than in p53+/+ cells (compare Fig. 5 and 10). On a CSB−/− background, the Ink4a/ARF deficiency appeared to partially, but significantly, decrease the rate of UV-induced apoptosis (Fig. 5, compare CSB−/− MEFs to CSB−/− Ink4a/ARF−/− MEFs). Moreover, p53 induction by 5 J of UVC radiation per m2 was found to be delayed and/or reduced in CSB−/− Ink4a/ARF−/− MEFs compared to CSB−/− MEFs (Fig. 6, lanes 3 through 5 versus lanes 13 through 15). A possible explanation is that the lack of p19ARF to antagonize mdm2 leads to decreased p53 expression.
In conclusion, we provide the first experimental evidence supporting an antineoplastic effect of CSB deficiency, which may explain the absence of cancer susceptibility in patients with CS. Moreover, our data suggest that the CSB gene could be considered an anticancer target. Inactivating this gene product may impede tumor cells and/or render them more susceptible to killing by chemo- or radiotherapeutic agents, many of which function by damaging DNA. On the other hand, such DNA lesions might potentially be mutagenic; thus, CSB inactivation could represent a very dangerous double-edged sword. However, human cells possess a potent back-up DNA repair pathway in the ggNER apparatus, which should remove many of the residual DNA lesions that might otherwise be converted into DNA mutations. Experiments under way support the idea that targeting the CSB gene product with antisense oligonucleotides can inhibit human cancer cells.
ACKNOWLEDGMENTS
We are grateful to Jianhua Zheng for managing the mouse colonies as well as to Diane Gaertner, Director of the Institute for Animal Studies of the Albert Einstein College of Medicine, and to David Gebhard of the FACS facility for advice and assistance. We acknowledge Ronald A. Depinho for providing Ink4a/ARF−/− mice.
This work was supported by grant 96-59 from the James S. McDonnell Foundation New Investigator Program, an intramural grant from the Albert Einstein College of Medicine Department of Pathology, and grant RO1 CA80171-01 from the NCI (to D.B.B.). Services provided by the Institute for Animal Studies and the FACS facility of the Albert Einstein College of Medicine were supported by the Cancer Center Core National Institutes of Health grant 5-P30-CA13330-26. Work performed at Erasmus University was supported by grants from the Dutch Cancer Society (EUR-1774, EUR-2004), NIH (grant AG17242-02), The Netherlands Organization for Scientific Research NWO (SPINOZA award), and the European Community (proposal no. QLRT-1999-02002).
REFERENCES
- 1.Abdellatif M, Packer S E, Michael L H, Zhang D, Charng M J, Schneider M D. A Ras-dependent pathway regulates RNA polymerase II phosphorylation in cardiac myocytes: implications for cardiac hypertrophy. Mol Cell Biol. 1998;18:6729–6736. doi: 10.1128/mcb.18.11.6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agarwal M L, Taylor W R, Chernov M V, Chernova O B, Stark G R. The p53 network. J Biol Chem. 1998;273:1–4. doi: 10.1074/jbc.273.1.1. [DOI] [PubMed] [Google Scholar]
- 3.Berg R J, Rebel H, van der Horst G T, van Kranen H J, Mullenders L H, van Vloten W A, de Gruijl F R. Impact of global genome repair versus transcription-coupled repair on ultraviolet carcinogenesis in hairless mice. Cancer Res. 2000;60:2858–2863. [PubMed] [Google Scholar]
- 4.Berg R J, Ruven H J, Sands A T, de Gruijl F R, Mullenders L H. Defective global genome repair in XPC mice is associated with skin cancer susceptibility but not with sensitivity to UVB induced erythema and edema. J Investig Dermatol. 1998;110:405–409. doi: 10.1111/j.1523-1747.1998.00173.x. [DOI] [PubMed] [Google Scholar]
- 5.Bootsma D, Kraemer K H, Cleaver J E, Hoeijmakers J H J. Nucleotide excision repair syndromes: xeroderma pigmentosum, Cockayne syndrome, and trichothiodystrophy. In: Vogelstein B, Kinzler K W, editors. The genetic basis of human cancer. New York, N.Y: McGraw-Hill; 1998. pp. 245–274. [Google Scholar]
- 6.Bregman D B, Halaban R, van Gool A J, Henning K A, Friedberg E C, Warren S L. UV-induced ubiquitination of RNA polymerase II: a novel modification deficient in Cockayne syndrome cells. Proc Natl Acad Sci USA. 1996;93:11586–11590. doi: 10.1073/pnas.93.21.11586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bregman D B, Pestell R G, Kidd V J. Cell cycle regulation and RNA polymerase II. Front Biosci. 2000;5:D244–D257. doi: 10.2741/bregman. [DOI] [PubMed] [Google Scholar]
- 8.Chin L, Pomerantz J, DePinho R A. The INK4a/ARF tumor suppressor: one gene—two products—two pathways. Trends Biochem Sci. 1998;23:291–296. doi: 10.1016/s0968-0004(98)01236-5. [DOI] [PubMed] [Google Scholar]
- 9.Chin L, Pomerantz J, Polsky D, Jacobson M, Cohen C, Cordon-Cardo C, Horner J W N, DePinho R A. Cooperative effects of INK4a and ras in melanoma susceptibility in vivo. Genes Dev. 1997;11:2822–2834. doi: 10.1101/gad.11.21.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Citterio E, Rademakers S, van der Horst G T, van Gool A J, Hoeijmakers J H, Vermeulen W. Biochemical and biological characterization of wild-type and ATPase-deficient Cockayne syndrome B repair protein. J Biol Chem. 1998;273:11844–11851. doi: 10.1074/jbc.273.19.11844. [DOI] [PubMed] [Google Scholar]
- 11.Citterio E, Vermeulen W, Hoeijmakers J H. Transcriptional healing. Cell. 2000;101:447–450. doi: 10.1016/s0092-8674(00)80854-5. [DOI] [PubMed] [Google Scholar]
- 12.Cooper P K, Nouspikel T, Clarkson S G, Leadon S A. Defective transcription-coupled repair of oxidative base damage in Cockayne syndrome patients from XP group G. Science. 1997;275:990–993. doi: 10.1126/science.275.5302.990. [DOI] [PubMed] [Google Scholar]
- 13.Darzynkiewicz Z, Juan G. Analysis of DNA content and DNA strand breaks for detection of apoptotic cells. In: Robinson J P, editor. Current protocols in cytometry. New York, N.Y: John Wiley & Sons, Inc.; 1997. pp. 7.4.1–7.4.8. [DOI] [PubMed] [Google Scholar]
- 14.de Boer J, Hoeijmakers J H. Cancer from the outside, aging from the inside: mouse models to study the consequences of defective nucleotide excision repair. Biochimie. 1999;81:127–137. doi: 10.1016/s0300-9084(99)80045-5. [DOI] [PubMed] [Google Scholar]
- 15.de Boer J, Hoeijmakers J H. Nucleotide excision repair and human syndromes. Carcinogenesis. 2000;21:453–460. doi: 10.1093/carcin/21.3.453. [DOI] [PubMed] [Google Scholar]
- 16.de Laat W L, Jaspers N G, Hoeijmakers J H. Molecular mechanism of nucleotide excision repair. Genes Dev. 1999;13:768–785. doi: 10.1101/gad.13.7.768. [DOI] [PubMed] [Google Scholar]
- 17.de Vries A, van Oostrom C T, Dortant P M, Beems R B, van Kreijl C F, Capel P J, van Steeg H. Spontaneous liver tumors and benzo[a]pyrene-induced lymphomas in XPA-deficient mice. Mol Carcinog. 1997;19:46–53. [PubMed] [Google Scholar]
- 18.de Vries A, van Oostrom C T, Hofhuis F M, Dortant P M, Berg R J, de Gruijl F R, Wester P W, van Kreijl C F, Capel P J, van Steeg H, et al. Increased susceptibility to ultraviolet-B and carcinogens of mice lacking the DNA excision repair gene XPA. Nature. 1995;377:169–173. doi: 10.1038/377169a0. [DOI] [PubMed] [Google Scholar]
- 19.Dianov G L, Houle J-F, Iyer N, Bohr V A, Friedberg E C. Reduced RNA polymerase II transcription in extracts of Cockayne syndrome and xeroderma pigmentosum/Cockayne syndrome cells. Nucleic Acids Res. 1997;25:3636–3642. doi: 10.1093/nar/25.18.3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friedberg E C. Cockayne syndrome—a primary defect in DNA repair, transcription, both or neither? Bioessays. 1996;18:731–738. doi: 10.1002/bies.950180908. [DOI] [PubMed] [Google Scholar]
- 21.Friedberg E C, Meira L B. Database of mouse strains carrying targeted mutations in genes affecting cellular responses to DNA damage. Version 4. Mutat Res. 2000;459:243–274. doi: 10.1016/s0921-8777(00)00006-9. [DOI] [PubMed] [Google Scholar]
- 22.Friedberg E C, Walker G C, Siede W. DNA repair and mutagenesis. Washington, D.C.: ASM Press; 1995. [Google Scholar]
- 23.Greenberg R A, Chin L, Femino A, Lee K H, Gottlieb G J, Singer R H, Greider C W, DePinho R A. Short dysfunctional telomeres impair tumorigenesis in the INK4a(delta2/3) cancer-prone mouse. Cell. 1999;97:515–525. doi: 10.1016/s0092-8674(00)80761-8. [DOI] [PubMed] [Google Scholar]
- 24.Hanawalt P C. DNA repair. The bases for Cockayne syndrome. Nature. 2000;405:415–416. doi: 10.1038/35013197. [DOI] [PubMed] [Google Scholar]
- 25.Harvey M, Sands A T, Weiss R S, Hegi M E, Wiseman R W, Pantazis P, Giovanella B C, Tainsky M A, Bradley A, Donehower L A. In vitro growth characteristics of embryo fibroblasts isolated from p53-deficient mice. Oncogene. 1993;8:2457–2467. [PubMed] [Google Scholar]
- 26.Henning K A, Li L, Narayan I, McDaniel L D, Reagan M S, Legerski R, Schultz R A, Stefanini M, Lehmann A R, Mayne L V, Friedberg E C. The Cockayne syndrome group A gene encodes a WD repeat protein that interacts with CSB protein and a subunit of RNA polymerase II TFIIH. Cell. 1995;82:555–564. doi: 10.1016/0092-8674(95)90028-4. [DOI] [PubMed] [Google Scholar]
- 27.Hwang B J, Toering S, Francke U, Chu G. p48 activates a UV-damaged-DNA binding factor and is defective in xeroderma pigmentosum group E cells that lack binding activity. Mol Cell Biol. 1998;18:4391–4399. doi: 10.1128/mcb.18.7.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ioffe E, Liu Y, Bhaumik M, Poirier F, Factor S M, Stanley P. WW6: an embryonic stem cell line with an inert genetic marker that can be traced in chimeras. Proc Natl Acad Sci USA. 1995;92:7357–7361. doi: 10.1073/pnas.92.16.7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iyer N, Reagan M S, Wu K J, Canagarajah B, Friedberg E C. Interactions involving the human RNA polymerase II transcription/nucleotide excision repair complex TFIIH, the nucleotide excision repair protein XPG, and Cockayne syndrome group B (CSB) protein. Biochemistry. 1996;35:2157–2167. doi: 10.1021/bi9524124. [DOI] [PubMed] [Google Scholar]
- 30.Jacks T, Remington L, Williams B O, Schmitt E M, Halachmi S, Bronson R T, Weinberg R A. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 31.Kamijo T, van de Kamp E, Chong M J, Zindy F, Diehl J A, Sherr C J, McKinnon P J. Loss of the ARF tumor suppressor reverses premature replicative arrest but not radiation hypersensitivity arising from disabled atm function. Cancer Res. 1999;59:2464–2469. [PubMed] [Google Scholar]
- 32.Kamijo T, Weber J D, Zambetti G, Zindy F, Roussel M F, Sherr C J. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc Natl Acad Sci USA. 1998;95:8292–8297. doi: 10.1073/pnas.95.14.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamijo T, Zindy F, Roussel M F, Quelle D E, Downing J R, Ashmun R A, Grosveld G, Sherr C J. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 34.Kraemer K H, Lee M M, Scotto J. DNA repair protects against cutaneous and internal neoplasia: evidence from xeroderma pigmentosum. Carcinogenesis. 1984;5:511–514. doi: 10.1093/carcin/5.4.511. [DOI] [PubMed] [Google Scholar]
- 35.Lakin N D, Jackson S P. Regulation of p53 in response to DNA damage. Oncogene. 1999;18:7644–7655. doi: 10.1038/sj.onc.1203015. [DOI] [PubMed] [Google Scholar]
- 36.Leadon S A, Cooper P K. Preferential repair of ionizing radiation-induced damage in the transcribed strand of an active human gene is defective in Cockayne syndrome. Proc Natl Acad Sci USA. 1993;90:10499–10503. doi: 10.1073/pnas.90.22.10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leadon S A, Lawrence D A. Preferential repair of DNA damage on the transcribed strand of the human metallothionein genes requires RNA polymerase II. Mutat Res. 1991;255:67–78. doi: 10.1016/0921-8777(91)90019-l. [DOI] [PubMed] [Google Scholar]
- 38.Leadon S A, Lawrence D A. Strand-selective repair of DNA damage in the yeast GAL7 gene requires RNA polymerase II. J Biol Chem. 1992;267:23175–23182. [PubMed] [Google Scholar]
- 39.Le Page F, Kwoh E E, Avrutskaya A, Gentil A, Leadon S A, Sarasin A, Cooper P K. Transcription-coupled repair of 8-oxoguanine: requirement for XPG, TFIIH, and CSB and implications for Cockayne syndrome. Cell. 2000;101:159–171. doi: 10.1016/s0092-8674(00)80827-2. [DOI] [PubMed] [Google Scholar]
- 40.Ljungman M, Zhang F. Blockage of RNA polymerase as a possible trigger for u. v. light-induced apoptosis. Oncogene. 1996;13:823–831. [PubMed] [Google Scholar]
- 41.Ljungman M, Zhang F, Chen F, Rainbow A J, McKay B C. Inhibition of RNA polymerase II as a trigger for the p53 response. Oncogene. 1999;18:583–592. doi: 10.1038/sj.onc.1202356. [DOI] [PubMed] [Google Scholar]
- 42.Nakane H, Takeuchi S, Yuba S, Saijo M, Nakatsu Y, Murai H, Nakatsuru Y, Ishikawa T, Hirota S, Kitamura Y, et al. High incidence of ultraviolet-B- or chemical-carcinogen-induced skin tumours in mice lacking the xeroderma pigmentosum group A gene. Nature. 1995;377:165–168. doi: 10.1038/377165a0. [DOI] [PubMed] [Google Scholar]
- 43.Nance M A, Berry S A. Cockayne syndrome: review of 140 cases. Am J Med Genet. 1992;42:68–84. doi: 10.1002/ajmg.1320420115. [DOI] [PubMed] [Google Scholar]
- 44.Nip J, Strom D K, Fee B E, Zambetti G, Cleveland J L, Hiebert S W. E2F-1 cooperates with topoisomerase II inhibition and DNA damage to selectively augment p53-independent apoptosis. Mol Cell Biol. 1997;17:1049–1056. doi: 10.1128/mcb.17.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pomerantz J, Schreiber-Agus N, Liegeois N J, Silverman A, Alland L, Chin L, Potes J, Chen K, Orlow I, Lee H-W, Cordon-Cardo C, DePinho R A. The Ink4a tumor suppressor gene product, p19ARF, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell. 1998;92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- 46.Schreiber-Agus N, Chin L, Chen K, Torres R, Rao G, Guida P, Skoultchi A I, DePinho R A. An amino-terminal domain of Mxil mediates anti-Myc oncogenic activity and interacts with a homolog of the yeast transcriptional repressor SIN3. Cell. 1995;80:777–786. doi: 10.1016/0092-8674(95)90356-9. [DOI] [PubMed] [Google Scholar]
- 47.Selby C P, Sancar A. Cockayne syndrome group B protein enhances elongation by RNA polymerase II. Proc Natl Acad Sci USA. 1997;94:11205–11209. doi: 10.1073/pnas.94.21.11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Serrano M. The INK4a/ARF locus in murine tumorigenesis. Carcinogenesis. 2000;21:865–869. doi: 10.1093/carcin/21.5.865. [DOI] [PubMed] [Google Scholar]
- 49.Serrano M, Lee H, Chin L, Cordon-Cardo C, Beach D, DePinho R A. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 50.Sharpless N E, DePinho R A. The INK4A/ARF locus and its two gene products. Curr Opin Genet Dev. 1999;9:22–30. doi: 10.1016/s0959-437x(99)80004-5. [DOI] [PubMed] [Google Scholar]
- 51.Sherr C J. Tumor surveillance via the ARF-p53 pathway. Genes Dev. 1998;12:2984–2991. doi: 10.1101/gad.12.19.2984. [DOI] [PubMed] [Google Scholar]
- 52.Sherr C J, Weber J D. The ARF/p53 pathway. Curr Opin Genet Dev. 2000;10:94–99. doi: 10.1016/s0959-437x(99)00038-6. [DOI] [PubMed] [Google Scholar]
- 53.Sugano T, Nitta M, Ohmori H, Yamaizumi M. Nuclear accumulation of p53 in normal human fibroblasts is induced by various cellular stresses which evoke the heat shock response, independently of the cell cycle. Jpn J Cancer Res. 1995;86:415–418. doi: 10.1111/j.1349-7006.1995.tb03072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sugasawa K, Ng J M, Masutani C, Iwai S, van der Spek P J, Eker A P, Hanaoka F, Bootsma D, Hoeijmakers J H. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol Cell. 1998;2:223–232. doi: 10.1016/s1097-2765(00)80132-x. [DOI] [PubMed] [Google Scholar]
- 55.Sweder K S, Hanawalt P C. Preferential repair of cyclobutane pyrimidine dimers in the transcribed strand of a gene in yeast chromosomes and plasmids is dependent on transcription. Proc Natl Acad Sci USA. 1992;89:10696–10700. doi: 10.1073/pnas.89.22.10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tantin D, Kansal A, Carey M. Recruitment of the putative transcription-repair coupling factor CSB/ERCC6 to RNA polymerase II elongation complexes. Mol Cell Biol. 1997;17:6803–6814. doi: 10.1128/mcb.17.12.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Troelstra C, van Gool A, de Wit J, Vermeulen W, Bootsma D, Hoeijmakers J H. ERCC6, a member of a subfamily of putative helicases, is involved in Cockayne's syndrome and preferential repair of active genes. Cell. 1992;71:939–953. doi: 10.1016/0092-8674(92)90390-x. [DOI] [PubMed] [Google Scholar]
- 58.Uberti D, Schwartz D, Almog N, Goldfinger N, Harmelin A, Memo M, Rotter V. Epithelial cells of different organs exhibit distinct patterns of p53-dependent and p53-independent apoptosis following DNA insult. Exp Cell Res. 1999;252:123–133. doi: 10.1006/excr.1999.4606. [DOI] [PubMed] [Google Scholar]
- 59.van der Horst G T, van Steeg H, Berg R J, van Gool A J, de Wit J, Weeda G, Morreau H, Beems R B, van Kreijl C F, de Gruijl F R, Bootsma D, Hoeijmakers J H. Defective transcription-coupled repair in Cockayne syndrome B mice is associated with skin cancer predisposition. Cell. 1997;89:425–435. doi: 10.1016/s0092-8674(00)80223-8. [DOI] [PubMed] [Google Scholar]
- 60.van Gool A J, Citterio E, Rademakers S, van Os R, Vermeulen W, Constantinous A, Egly J-M, Bootsma D, Hoeijmakers J H J. The Cockayne syndrome B protein, involved in transcription-coupled DNA repair, resides in an RNA polymerase II-containing complex. EMBO J. 1997;16:5955–5965. doi: 10.1093/emboj/16.19.5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Gool A J, van der Horst G T J, Citterio E, Hoeijmakers J H J. Cockayne syndrome: defective repair of transcription? EMBO J. 1997;16:4155–4162. doi: 10.1093/emboj/16.14.4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamaizumi M, Sugano T. U.V.-induced nuclear accumulation of p53 is evoked through DNA damage of actively transcribed genes independent of the cell cycle. Oncogene. 1994;9:2775–2784. [PubMed] [Google Scholar]