Abstract
Inflammatory breast cancer (IBC) is an aggressive BC subtype with poor outcomes. A targetable somatic PIK3CA mutation is reported in 30% of IBC, allowing for treatment by PI3Kα-specific inhibitors, such as alpelisib. The aim of this study was to evaluate the detection rate of circulating PIK3CA mutation in locally-advanced IBC (LAIBC) patients harbouring a PIK3CA mutation on initial biopsy. This monocentric retrospective study was based on available stored plasma samples and tumour biopsies at diagnosis from all LAIBC patients treated with neo-adjuvant chemotherapy (NCT) between 2008 and 2018 at the Centre Henri Becquerel. PIK3CA mutations (E542K, E545K, H1047R/L) were assessed by droplet digital PCR (ddPCR) in plasma samples and tumoral tissue at diagnosis. A total of 55 patients were included. Overall, 14/55 patients (25%) had a PIK3CA mutation identified on baseline biopsy (H1047R = 8; H1047L = 3; E545K = 2; E542K = 1). Among them, 11 (79%) patients had enough DNA for circulating DNA analyses, and corresponding circulating PIK3CA mutations were found in 6/11 (55%). Among the 41 patients without PIK3CA mutations on biopsy, 32 (78%) had enough DNA for circulating DNA analysis, and no circulating PIK3CA mutation was identified. Our results revealed no prognostic or predictive value of PIK3CA mutations at the diagnosis of non-metastatic IBC but highlighted the prognostic value of the cfDNA rate at diagnosis. Our study showed that a corresponding circulating PIK3CA mutation was identified in 55% of LAIBC patients with PIK3CA-mutated tumours, while no circulating mutation was found among patients with PI3KCA wild-type tumours.
Subject terms: Cancer, Molecular biology, Oncology
Introduction
Inflammatory breast cancer (IBC) is a rare form of breast cancer that accounts for approximately only 2% to 4% of all cases1–3 and contributes to 10% of breast cancer-caused mortality4. IBC is characterized by an early age at diagnosis, aggressiveness and poor survival5,6. Data on IBC risk factors are limited, but there is a higher incidence in young African-American women, and a high body mass index (BMI) is more frequently associated with IBC than with non-inflammatory breast cancer7. Originally described by Sir Charles Bell in 18148, the diagnosis of IBC is commonly based on clinical criteria, described by the American Joint Committee of Cancer (AJCC) as rapid onset of breast skin erythema with oedema (known as “peau d’orange”) and considered T4d stage according to TNM classification. IBC patients also have more frequent lymph node involvement, and 30% are metastatic at diagnosis9,10.
Treatment is multimodal, including neoadjuvant chemotherapy (NCT) followed by mammectomy with axillary dissection if a tumour-free resection margin is expected and locoregional radiotherapy11. Until now, the median overall survival (OS) of IBC patients has remained poor, with a median OS of 43 months in the entire population12. Although the presence of a pathological complete response (pCR) after NCT is considered a significant prognostic factor in all biological subtypes of IBC13, there is no consensus predictive marker of pCR. For the past 20 years, several studies have tried to provide a molecular description of IBC14 but were relatively limited by the rarity of this entity and the small sample size. Compared to non-inflammatory BC, IBC is generally characterized by important genomic instability and a lower frequency of luminal A subtypes15. However, IBCs do not share a specific pattern of molecular alteration16. The most frequent somatic mutations are those located on the TP53 and PIK3CA genes, which are observed in approximately 75% and 40% of cases and have a higher prevalence than within non-inflammatory breast cancer17.
PIK3CA activating mutation induces hyperactivation of the alpha isoform (p110alpha) of phosphatidylinositol-3-kinase (PI3K) and activates the PI3K/AKT/mTOR pathway, which is the most frequently activated pathway in breast cancer and one of the most important mechanisms in endocrine therapy resistance18. PIK3CA mutations are found in 22 to 30% of breast cancers19 and in 40% of hormone receptor-positive (HR +) HER2− tumours20,21. More than 90% of these mutations are restricted to two hotspots: E542K or E545K in exon 9 and H1047R or H1047L in exon 2022, which are easily identified by sensitive methods such as digital PCR. In the era of liquid biopsy, a high concordance between tumour tissue and circulating tumoral DNA (ctDNA) mutation status23,24 has been reported. Moreover, while the prognostic value of PIK3CA mutations remains controversial21,25, their predictive value as a marker of response to PI3K pathway inhibitors is now established26,27. In particular, the PI3Kα-specific inhibitor alpelisib has recently shown manageable toxicity and good clinical activity in PIK3CA-mutated BC. Moreover, patients with circulating PIK3CA mutations rather than biopsy-based PIK3CA mutations have a better predictive value for response to PI3K inhibitors28,29. In this context, the aim of this study was to investigate the association and the clinical impact of PI3KCA mutational status in paired tumour and plasma samples at diagnosis in patients with locally advanced IBC (LAIBC) undergoing NCT.
Methods
Patients
We retrospectively screened all patients with LAIBC undergoing NCT at the Centre Henri Becquerel from 2008 to 2018. IBC was defined by clinical stage T4d, and pathological evidence of tumour emboli in the dermal lymphatics was not mandatory. Only patients with available tumour tissue from diagnostic biopsies and corresponding blood sample collection were included in the analysis dataset. Tumour biopsies and corresponding plasma samples at diagnosis were analysed for PIK3CA mutations using ddPCR. PIK3CA mutation status was also analysed in surgical resections and plasma samples after neoadjuvant treatment when available in patients with PIK3CA-mutated BC at diagnosis. The last update for survival follow-up was July 2020.
This study was conducted in accordance with French laws regarding retrospective studies. All patients received a non-opposition form, and the study was authorized by our local institutional review board (IRB) (Centre Henri Becquerel, No. 1913B).
DNA extraction in formalin-fixed, paraffin-embedded (FFPE) and plasma samples
DNA of FFPE breast tumour biopsies was extracted with the Maxwell 16FFPE Plus LEV DNA Purification Kit (Promega, Madison, Wisconsin, USA) using two cuts of 2 µM.
Blood samples were remnants of blood analyses performed during IBC patient treatment. Blood samples were collected in heparinized or EDTA tubes and processed within two hours after collection with one centrifugation at 2000 g 10min at 4 °C before storage at −20 °C. cDNA was isolated using the QIAamp Circulating Nucleic Acid Kit (Qiagen, Hilden, Germany). Double-stranded DNA quantification was performed by the fluorometric method using a Qubit ds DNA HS Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA). DNA was pre-amplified as previously described30,31.
ddPCR
Analyses for PIK3CA mutation detection were performed blind to the clinical data. ddPCR from the Stilla system (Stilla Technologies, Villejuif, France) was used for PIK3CA mutation detection in the plasma and FFPE samples. We used a Bio-Rad (Hercules, CA, USA) ddPCR assay for the four mutations, E542K (dHsaMDV2010073), E545K (dHsaMDV2010123), H1047R (dHsaMDV2010077), and H1047L (dHsaMDV2010123).The results were analysed using CrystalMiner software (Stilla Technologies, Villejuif, France) which enables a visualization of each chamber (visualization of the droplets appearing as empty, wild-type positive or mutant positive) and provides a count of the generated droplets, the positive droplets for wild-type signal and the positive droplets for mutant signal. The variant allele fraction (VAF) was defined as the proportion of mutant DNA copies compared with wild-type (WT) DNA copies obtained by ddPCR. To validate the run, we verified 2 criteria: a minimum of 15,000 total droplets generated, and a minimum of 200copies/µL (wild type copies + mutant copies) obtained. In contributive runs according to these 2 previous criteria, we confronted the number of mutant positive droplets to the limit of detection (LOD) value: the sample was considered as positive if the number of positive droplets was larger than the LOD. Each sample was tested in duplicated. For every duplicate, the same qualitative conclusion (mutated or not mutated) was obtained. The VAF mentioned in Table 2 represents the mean of the 2 duplicates.
Table 2.
Clinical outcomes and survival in FFPE PIK3CA mutated patients.
| Patients | FFPE PIK3CA mutation at diagnosis (VAF%) | Circulating PIK3CA mutation at diagnosis (VAF%) | Outcomes post neoadjuvant chemotherapy | PIK3CA mutation on mastectomy post neoadjuvant chemotherapy (VAF%) | Circulating PIK3CA mutation post neoadjuvant chemotherapy (VAF%) | Relapse | DFS or follow-up (months) |
|---|---|---|---|---|---|---|---|
| No. 1 | H1047L 40.3% | H1047L 11% | Pathological partial response | NA | NA | Yes | 30.2 |
| No. 2 | E545K 47.80% | E545K 3.99% | Refractory | 0 | 0 | Yes | 8.5 |
| No. 3 | H1047R 6.55% | NC | Refractory | NA | 0 | Yes | 5.7 |
| No. 4 |
E542K 23% |
NC | pCR | NA | No | 83.9 | |
| No. 5 | H1047R 58.9% | H1047R 0.32% | Pathological partial response | NA | NA | No | 37.6 |
| No. 6 | E545K 16.31% | E545K 6.26% | Pathological partial response | E545K 0.23% | 0 | Yes | 8.7 |
| No. 7 | H1047R 25.3% | 0 | Pathological partial response | 0 | 0 | No | 69 |
| No. 8 | H1047L 39.6% | H1047L 11% | Pathological partial response | H1047L 27% | H1047L 0.98% | No | 20.7 |
| No. 9 | H1047R 40.7% | 0 | Pathological partial response | H1047R 14% | NA | No | 60.4 |
| No. 10 | H1047R 41.7% | H1047R 8.13% | Pathological partial response | H1047R 41% | NA | No | 9 |
| No. 11 | H1047R 3.31% | 0 | pCR | NA | No | 71.6 | |
| No. 12 | H1047R 25.2% | 0 | Pathological partial response | H1047R 18% | NA | No | 61.9 |
| No. 13 | H1047L 0.60% | 0 | Pathological partial response | NA | NA | No | 97.2 |
| No. 14 | H1047R 2.02% | NC | pCR | NA | Yes | 47.6 |
NC non contributive, NA non available, pCR pathological complete response, VAF variant allele fraction.
Statistics
The primary endpoint was the association between PIK3CA mutation status at diagnosis between tumour tissue and corresponding plasma. The key secondary endpoints were to evaluate the association between PI3KCA mutational status and IBC molecular subtype, pathologic response and disease-free survival (DFS) and overall survival (OS). The impact of the total circulating DNA level at diagnosis on the pCR rate, DFS and OS was also analysed as well as the association between pCR and survival. pCR was considered in our study as the absence of invasive disease after mastectomy and lymphadenectomy (ypT0/is, N0), according to the Residual Cancer Burden calculator of the MD Anderson Center. DFS and OS were defined as the time from diagnosis to relapse, death, or death only, respectively. Patients were defined as refractory in the absence of a response to neoadjuvant chemotherapy, including clinically progressive disease and stable disease.
The chi-square test was used for the comparison of patient characteristics according to their mutational status at diagnosis. The Kaplan–Meier method was used to estimate the DFS and OS endpoints. The log-rank test was used to compare survival curves according to the observed determinants. P-values < 0.05 were considered significant. All reported P-values are two-sided, and confidence intervals (CIs) are at the 95% level. Statistical analyses were performed using R statistical software (version 4.0.2).
Ethics approval and consent to participate
Informed patient consent was obtained by sending a non-objection form. The study was approved by the Institutional Review Board of the Henri Becquerel Center (register order 1913B).
Results
Patient characteristics
A total of 55 LAIBC patients were considered for this study, and 43/55 (78%) had sufficient quality samples for tumour and circulating DNA analyses, as illustrated in the CONSORT diagram of Fig. 1. The main characteristics of the population are summarized in Table 1. The median age was 55 years (range 33–87), with 43.6% premenopausal patients. Most LAIBC patients were obese with a median BMI of 30.6 kg/m2 and had an aggressive profile with high tumour grade, lymph node invasion and a higher rate of HR-negative tumours.
Figure 1.
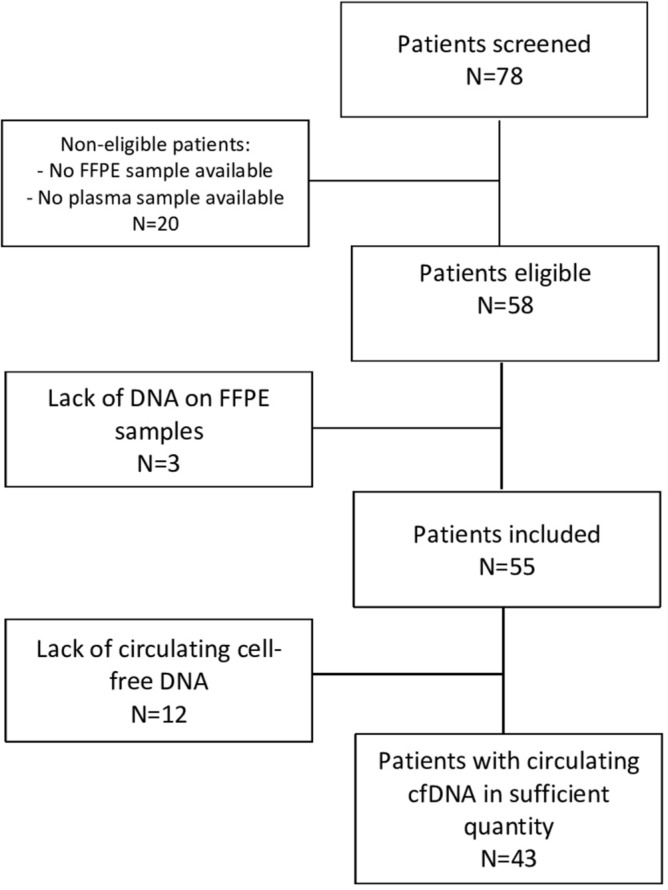
CONSORT diagram. Among the 78 patients screened, 20 were non-eligible because of non-available FFPE samples or plasma samples. Among them, 3 had a lack of DNA on FFPE samples and 55 patients were included. Finally, there was a lack of circulating cell-free DNA for 12 patients with 43 patients with circulating cfDNA in sufficient quantity.
Table 1.
Characteristics.
| Total N = 55 |
FFPE PIK3CA mutated patients N = 14 |
FFPE PIK3CA non-mutated patients N = 41 |
p | ||
|---|---|---|---|---|---|
| Median age at diagnosis, years [min–max] | 54.8 [33–87] | 55.9 [43–78] | 54.11 [33–87] | 0.27 | |
| Histological subtype IDC | 55 (100%) | 14 (100%) | 41 (100%) | 1 | |
| Lymph node status | Positive | 53 (96.4%) | 13 (92.9%) | 40 (97.6%) | 1 |
| Negative | 2 (3.6%) | 1 (7.1%) | 1 (2.4%) | ||
| Molecular subtype | HER2 + and HR + /− | 16 (29%) | 2 (14%) | 14 (34%) | 0.26 |
| HER2− and HR− | 16 (29%) | 6 (43%) | 10 (24%) | ||
| HER2− and HR + | 23 (42%) | 6 (43%) | 17 (42%) | ||
| Tumor grade | 1–2 | 20 (36.4%) | 6 (42.9%) | 14 (34.1%) | 0.79 |
| 3 | 34 (61.8%) | 7 (50%) | 27 (65.9%) | ||
| NA | 1 (1.8%) | 1 (7.1%) | 0 (0%) | ||
| Median BMI at diagnosis kg/m2 [min–max] | 30.6 [19–44.2] | 28.6 [22.7–37.5] | 31.1 [19–44.2] | 0.37 | |
| Menopausal status | Premenopausal | 24 (43.6%) | 5 (35.7%) | 19 (46.3%) | 0.7 |
| Postmenopausal | 31 (56.4%) | 9 (64.3%) | 22 (53.7%) | ||
| Neoadjuvant chemotherapy | 55 (100%) | 14 (100%) | 41 (100%) | 1 |
HER2 + HER2 positive, defined as 3 + overexpression by immunohistochemical testing or 2 + with HER2 amplification by fluorescent in-situ hybridization, HR hormone receptor, BMI body mass index, NA non available, IDC invasive ductal carcinoma.
PIK3CA mutational status in tumour and corresponding plasma samples at diagnosis
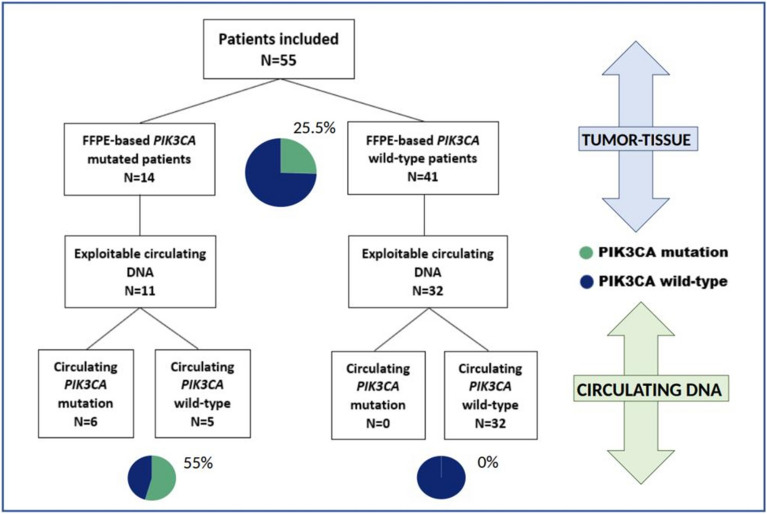
A total of 14/55 patients (25.5%) had a PIK3CA mutation identified on baseline biopsy (H1047R = 8; H1047L = 3; E545K = 2; E542K = 1), with no significant difference in baseline characteristics between the patients with and without mutations. All mutations were single, and the prevalence of FFPE-based PIK3CA mutations at diagnosis was 12.5% (2/16), 26% (6/23) and 37.5% (6/16) in the HER2-positive, HR-positive/HER2-negative and HR-negative/HER2-negative subtypes, respectively. Among the 43 patients with analysable plasma samples, 6 (14%) had detectable circulating PIK3CA mutations, corresponding to 6/11 patients (55%) with PIK3CA-mutated tumours and with detectable cfDNA and 0/32 non-mutated tumours. All mutations were single. Thus, there was no additional PIK3CA mutation identified in ctDNA compared to FFPE-based mutational status. Those results are summarized in Fig. 2.
Figure 2.
Distribution of FFPE-based and circulating PIK3CA mutation at diagnosis. Among the 55 patients of this cohort, FFPE-based PIK3CA mutation were detected in 14 patients (25.5%); among them, 11 had exploitable circulating DNA, and 6 patients (55%) harboured a corresponding circulating PIK3CA mutation. No other circulating mutation was identified among the 43 patients with fully interpretable circulating and biopsy mutational analyses.
Association between PI3KCA mutational status and pCR
A total of 52/55 patients underwent surgery; the 3 remaining patients were refractory to neoadjuvant chemotherapy. Pathological assessment in the operated patients showed 13 pCRs, 38 partial responses and 1 non-responder.
There was no difference in the pathological response rate according to PI3KCA tumour mutation status, with pCRs of 21.4% and 24.4% in the patients with and without mutations, respectively. Among the 14 patients with PIK3CA tumour mutations at diagnosis, 3/14 (21.4%) achieved a pCR after neoadjuvant chemotherapy, and 2/14 (14.3%) were non-responders (Table 2). Seven tumours had enough tissue to analyse PIK3CA mutation status after neoadjuvant chemotherapy. We found the same PIK3CA mutations as those described at diagnosis in 5 patients (71%), with a lower rate of VAF. Of note, plasma samples were available after neoadjuvant treatment in 5/14 patients with mutations (36%), with only one circulating mutation found (20%). These results are detailed in Table 2.
Among the 14 patients harbouring somatic PIK3CA mutations, compared to 3/8 (37.5%) without circulating mutations, 0/6 patients with circulating PIK3CA mutations at diagnosis had pCR (p = 0.09). Thus, there is no predictive value of circulating PIK3CA mutations for pCR.
Association between cfDNA and pCR
The median cfDNA level at diagnosis was 1.22 ng/µL. The rate of pCR was not different among patients above or below the median cfDNA level at diagnosis (22.2% and 25%, respectively). In contrast, 3 out of the 4 patients refractory to neoadjuvant chemotherapy had a cfDNA above 1.22 ng/µL at diagnosis. Overall, when using the median cfDNA value as the cut-off, the baseline cfDNA level was not associated with the response to neoadjuvant chemotherapy (p = 0.68). These results are detailed in Table 3.
Table 3.
Response to neoadjuvant chemotherapy according to PIK3CA mutation status and cfDNA rate.
| Total N = 55 (%) |
Patients without PIK3CA mutation N = 41 (%) |
Patients with biopsy-based PIK3CA mutation but no corresponding circulating mutation N = 8 (%) |
Patients with biopsy-based and corresponding circulating PIK3CA mutation N = 6 (%) |
p | Patients with cfDNA ≤ 1.22 ng/µl N = 28 |
Patients with cfDNA > 1.22 ng/µl N = 27 |
p | ||
|---|---|---|---|---|---|---|---|---|---|
| Outcomes post neoadjuvant treatment | Pathological complete response | 13 (23.6%) | 10 (24.4%) | 3 (37.5%) | 0 (0%) | 0.26 | 7 (25%) | 6 (22.2%) | 0.68 |
| Refractory | 4 (7.3%) | 2 (4.9%) | 1 (12.5%) | 1 (16.7%) | 1 (3.6%) | 3 (11.1%) | |||
| Pathological partial response | 38 (69%) | 29 (70.7%) | 4 (50%) | 5 (83.3%) | 20 (71.4%) | 18 (66.7%) | |||
cfDNA cell-free DNA.
Association between PIK3CA mutation status and cfDNA with OS and DFS
After a median follow-up of 52.1 months [7.7–140.6], the median OS was not reached in our retrospective cohort, with 20 deaths among our 55 included patients. No significant difference was found in OS according to FFPE-based PIK3CA mutation status at diagnosis (HR = 0.95 CI[0.35–2.63], p = 0.93), according to circulating PIK3CA mutation status at diagnosis (HR = 2.27 CI[0.66–7.81], p = 0.18) and according to circulating and biopsy-based PIK3CA mutation status at diagnosis (p = 0.29).
The median DFS was 104.8 months, and 22 relapses were observed. No significant difference was found in DFS according to FFPE-based PIK3CA mutation status at diagnosis (HR = 0.92 CI[0.34–2.52], p = 0.88), according to circulating PIK3CA mutation status at diagnosis (HR = 2.24 CI[0.64–7.81], p = 0.19) and according to circulating and biopsy-based PIK3CA mutation status at diagnosis (p = 0.29).
Using the median baseline cfDNA level as the threshold, the patients with low cfDNA had a significantly better OS outcome (HR = 0.36 CI[0.14–0.93], p = 0.028) Fig. 3. Regarding DFS, a non-significant trend also identified a low baseline cfDNA level at diagnosis as a marker of better outcome (HR = 0.45 CI[0.19–1.07], p = 0.063), Fig. 4.
Figure 3.
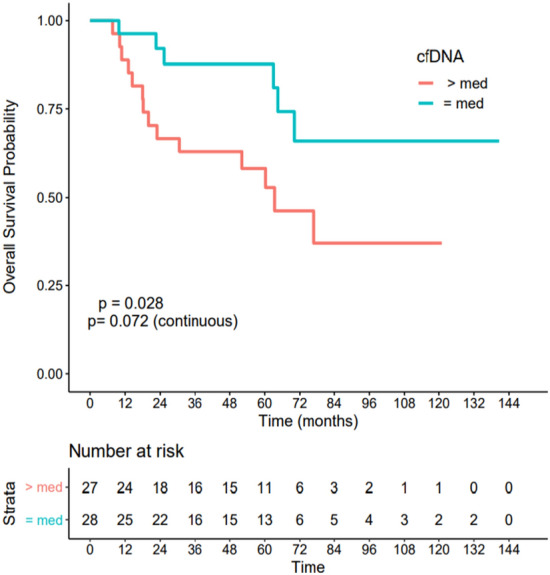
Association between cell-free DNA level at diagnosis and overall survival. Patients with cfDNA below the median had a significantly better OS outcome (HR = 0.36 CI[0.14–0.93], p = 0.028).
Figure 4.
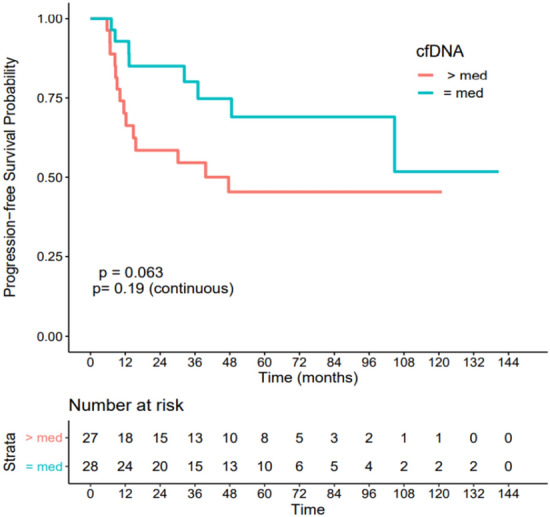
Association between cell-free DNA level at diagnosis and disease-free survival. Patients with cfDNA below the median had a non-significant better DFS outcome (HR = 0.45 CI[0.19–1.07], p = 0.063).
Indeed, the 3-year DFS rate was 54.6% [38.5–77.4] for patients with cfDNA greater than the median value of 1.22 ng/µL and 80.1% [65.7–97.7] for patients with cfDNA equal to or lower than 1.22 ng/µL. Similarly, the 3-year OS rates were 63% [47.1–84.1%] and 87.7% [75.6–100], respectively.
Association between pCR and survival data
Among the 13 patients with pCR after neoadjuvant treatment, only 2 (15%) experienced tumoral relapse during follow-up, compared to 20 relapses among the 42 patients with partial or refractory histological response (47.6%). A significant difference was found in OS according to the response to neoadjuvant treatment (HR = 0.25 IC[0.06–1.08], p = 0.044) Fig. 5, and in DFS (HR = 0.23 IC[0.05–1], p = 0.032), Fig. 6.
Figure 5.
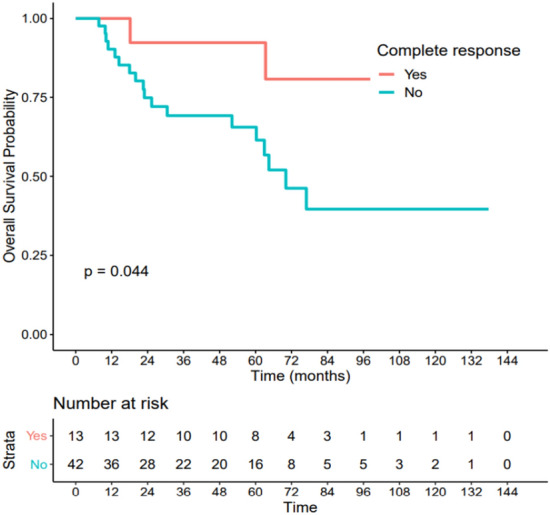
Association between pCR and overall survival. Patients with pCR had a significantly better overall survival (HR = 0.25 IC[0.06–1.08], p = 0.044).
Figure 6.
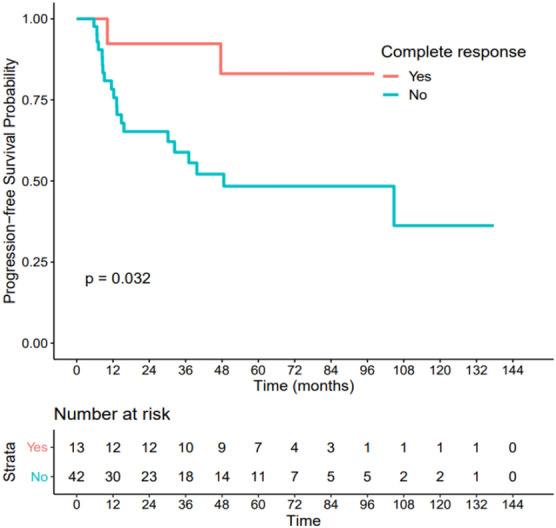
Association between pCR and disease-free survival. Patients with pCR had a significantly better disease-free survival (HR = 0.23 IC[0.05–1], p = 0.032).
Discussion
This retrospective study included 55 LAIBC patients, among which 25.5% had PIK3CA-mutated tumours. Corresponding circulating PIK3CA mutations were identified in 55% of patients with mutations, while no circulating mutations were found among patients with PI3KCA WT tumours. There was no predictive value for pCR of PIK3CA mutations or baseline cfDNA level and no prognostic value of PIK3CA mutation status. In contrast, patients with baseline cfDNA below the median or those with pCR after NCT had a better prognosis.
To our knowledge, this study is the first to address circulating PIK3CA mutations in patients treated for LAIBC.
Indeed, IBC is a sub-type excluded from most studies, including SOLAR-129. We found one study dealing with cell-free DNA in 19 patients with IBC, but only one PIK3CA mutation was found in tumour samples without circulating corresponding mutations32.
Despite the limited number of patients included, our cohort seems representative of the non-metastatic IBC population. Indeed, this cohort was characterized by a majority of obese patients, with an aggressive tumoral profile, as already described in the IBC population33,34. As expected, a higher rate of HER2-positive and triple-negative tumours in comparison to non-inflammatory breast cancer was observed, as well as a pCR rate of 23.6%, comparable to the pCR rate of 23.2% reported in the study of Van Uden et al.11. In our study, a PIK3CA mutation was found at a rate of 25.5% on initial biopsy, corroborating recently published data about IBC, with a rate of 29.5% among 156 patients and a rate of 28% among 53 patients, reported by Liang et al. and Ross et al., respectively35,36. Similar results were described in non-inflammatory early-stage breast cancer, with a rate of 32% among 10 319 patients and a rate of 23% among 1008 patients, reported by Zardavas et al. and Papaxoinis et al.37,38. Only single-hotspot mutations were detected in this study, whereas multiple PIK3CA mutations were described in 12 to 15% of PIK3CA-mutated breast cancers39–41. As expected, a majority of PIK3CA mutations were localized in the H1047R hotspot in exon 2042,43. Our results highlight that a corresponding circulating PIK3CA mutation was identified in 55% of non-metastatic IBC patients with a baseline somatic PIK3CA mutation in tumour tissue and with detectable cfDNA, while no circulating mutation was found among patients with no PIK3CA mutations. Despite its aggressiveness, PIK3CA-mutated LAIBC appears to have a PIK3CA circulating detection rate comparable to localized (47%) rather than metastatic breast cancer (approximately 80%)23,44,45. Thus, those results do not encourage the use of cfDNA testing to find actionable findings earlier during patient management. Based on the favourable results of the SOLAR-1 study, therapeutic trials are expected in PIK3CA-mutated positive hormone receptor LAIBC with the use of alpelisib in neoadjuvant treatment or in therapeutic intensification after surgery with residual invasive cancer. In our study, PIK3CA mutation status does not appear to have prognostic value, as in non-inflammatory early breast cancer, or predictive value, but no definite conclusion can be formulated given the small number of patients with mutations.
Interestingly, our results highlight the prognostic value of baseline cfDNA, showing worse survival outcome for LAIBC patients with cfDNA above the median, suggesting that baseline cfDNA could reflect tumour burden in LAIBC. The predictive and prognostic value of cfDNA has been demonstrated in several studies, mostly in lung cancer46, rectal cancer during neoadjuvant chemotherapy47, and metastatic breast cancer30. In the study of Park et al., among 72 early-stage triple-negative breast cancer patients who underwent NCT, patients with baseline cfDNA levels > 264 ng/mL demonstrated a higher risk of relapse than those with baseline cfDNA levels ≤ 264 ng/mL (HR, 2.84; 95% CI, 1.11–7.24; P = 0.029)48. Otherwise, as expected, pathological complete response (pCR) after neoadjuvant treatment in LAIBC is a predictor of favourable long-term outcome, corroborating literature data. Indeed, among 1061 early breast cancer patients of all subtypes, improved survival was previously reported for patients who achieved pCR, especially for HER2 + /HR− tumour subtypes with a 5-year overall survival rate of 83% with pCR versus 50% without pCR49. Similarly, Pierga et al. demonstrated the prognostic value of pCR and circulating tumour cells rate at baseline in inflammatory breast cancer in a pooled analysis of BEVERLY-1 and -250.
Our study has some limitations. First, given the limited number of patients, our results cannot be considered definitive. Nevertheless, it must be taken into consideration that IBC is a rare disease, explaining the limited literature data available. Moreover, the confirmation of the pCR status and cfDNA level as prognostic factors highlights the internal validity of our results. Second, due to its retrospective design, some FFPE or plasma samples could not be used, with a lack of quality DNA mostly due to storage constraints and long storage times. Moreover, taking into account a limited quantity of material and a majority of heparinized plasma samples, we could not study genomic alterations by targeted next-generation sequencing. Taken together, these technical limitations prevented us from studying genomic tumoral heterogeneity which could have provided precious new information within the mutational landscape of IBC.
Finally, since we focused our analysis on the four main PIK3CA mutations by ddPCR, we cannot exclude the presence of rare mutations, and we could not analyse AKT mutations or PTEN deletion that result in the same oncogenic activation pathway, which could participate in the resistance mechanisms of PIK3CA therapies.
Conclusion
In conclusion, this study showed that a corresponding circulating PIK3CA mutation was identified in 55% of non-metastatic IBC patients with baseline somatic PIK3CA mutations in tumour tissue and with detectable cfDNA, while no circulating mutation was found among patients with no PIK3CA mutations. Despite its aggressiveness, LAIBC surprisingly appears to have quite a low circulating ctDNA release. These results suggest that future therapeutic trials based on PIK3CA mutation status within LAIBC should focus mostly on primary material. Nevertheless, the cfDNA rate seems to be a discriminatory predictor of survival, allowing us to better stratify patients according to their level of risk (Suppl. Information).
Supplementary Information
Abbreviations
- AJCC
American Joint Committee of Cancer
- BC
Breast cancer
- BMI
Body mass index
- CI
Confidence interval
- ctDNA
Circulating tumoral DNA
- cfDNA
Cell-free DNA
- ddPCR
Digital droplet polymerase chain reaction
- DFS
Disease-free survival
- FFPE
Formalin-fixed, paraffin-embedded
- HR
Hormone receptor
- IBC
Inflammatory breast cancer
- IRB
Institutional Review Board
- LAIBC
Locally-advanced inflammatory breast cancer
- LOD
Limit of detection
- NCT
Neoadjuvant chemotherapy
- OS
Overall survival
- pCR
Pathological complete response
- VAF
Variant allele fraction
- WT
Wild-type
Author contributions
Conception and Design of the study: F.C., A.P., V.A. Supervision: A.P., F.D.F., F.C. Collection of clinical data: V.A. Collection and preparation of biological samples: A.P., C.C., A.B. Experiments: N.S.V., A.B., A.P., P.E. Data analysis: V.A., J.L., F.D.F., F.C. Preparation of the manuscript, table and figures (all originals): V.A., F.C., J.L., F.D.F. All authors read and approved the final manuscript.
Funding
This work was supported by Centre Henri Becquerel and La Ligue contre le Cancer.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-02643-y.
References
- 1.Anderson WF, Chu KC, Chang S. Inflammatory breast carcinoma and noninflammatory locally advanced breast carcinoma: Distinct clinicopathologic entities? J. Clin. Oncol. 2003;21(12):2254–2259. doi: 10.1200/JCO.2003.07.082. [DOI] [PubMed] [Google Scholar]
- 2.Wingo PA, Jamison PM, Young JL, Gargiullo P. Population-based statistics for women diagnosed with inflammatory breast cancer (United States) Cancer Causes Control. 2004;15(3):321–328. doi: 10.1023/B:CACO.0000024222.61114.18. [DOI] [PubMed] [Google Scholar]
- 3.Hance KW, Anderson WF, Devesa SS, Young HA, Levine PH. Trends in inflammatory breast carcinoma incidence and survival: The surveillance, epidemiology, and end results program at the National Cancer Institute. JNCI J. Natl. Cancer Inst. 2005;97(13):966–975. doi: 10.1093/jnci/dji172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Menta A, Fouad TM, Lucci A, Le-Petross H, Stauder MC, Woodward WA, et al. Inflammatory breast cancer. Surg. Clin. N. Am. 2018;98(4):787–800. doi: 10.1016/j.suc.2018.03.009. [DOI] [PubMed] [Google Scholar]
- 5.Dawood S, Ueno NT, Valero V, Woodward WA, Buchholz TA, Hortobagyi GN, et al. Differences in survival among women with stage III inflammatory and noninflammatory locally advanced breast cancer appear early. Cancer. 2011;117(9):1819–1826. doi: 10.1002/cncr.25682. [DOI] [PubMed] [Google Scholar]
- 6.Iuliis FD, Iuliis FD, D’Aniello D, Cefali K, Corvino R, Ferraro E, et al. Inflammatory breast cancer management: a single centre experience. Ann. Oncol. 2015;26:24. [Google Scholar]
- 7.Anderson WF, Schairer C, Chen BE, Hance KW, Levine PH. Epidemiology of inflammatory breast cancer (IBC)1. Breast Dis. 2006;22(1):9–23. doi: 10.3233/bd-2006-22103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bell SC. A System of Operative Surgery: Founded on the Basis of Anatomy. Hale & Hosmer; 1812. [Google Scholar]
- 9.Biswas T, Efird JT, Prasad S, James SE, Walker PR, Zagar TM. Inflammatory TNBC breast cancer: Demography and clinical outcome in a large cohort of patients with TNBC. Clin. Breast Cancer. 2016;16(3):212–216. doi: 10.1016/j.clbc.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 10.Levine PH, Zolfaghari L, Young H, Hafi M, Cannon T, Ganesan C, et al. What is inflammatory breast cancer? Revisiting the case definition. Cancers. 2010;2(1):143–152. doi: 10.3390/cancers2010143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Uden DJP, van Laarhoven HWM, Westenberg AH, de Wilt JHW, Blanken-Peeters CFJM. Inflammatory breast cancer: An overview. Crit. Rev. Oncol. Hematol. 2015;93(2):116–126. doi: 10.1016/j.critrevonc.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 12.Pan X, Yang W, Chen Y, Tong L, Li C, Li H. Nomogram for predicting the overall survival of patients with inflammatory breast cancer: SEER-based study. Breast. 2019;47:56–61. doi: 10.1016/j.breast.2019.05.015. [DOI] [PubMed] [Google Scholar]
- 13.Biswas T, Jindal C, Fitzgerald TL, Efird JT. Pathologic complete response (pCR) and survival of women with inflammatory breast cancer (IBC): An analysis based on biologic subtypes and demographic characteristics. Int. J. Environ. Res. Public Health. 2019;16(1):124. doi: 10.3390/ijerph16010124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costa R, Santa-Maria CA, Rossi G, Carneiro BA, Chae YK, Gradishar WJ, et al. Developmental therapeutics for inflammatory breast cancer: Biology and translational directions. Oncotarget. 2016;8(7):12417–12432. doi: 10.18632/oncotarget.13778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Laere SJ, Van den Eynden GG, Van der Auwera I, Vandenberghe M, van Dam P, Van Marck EA, et al. Identification of cell-of-origin breast tumor subtypes in inflammatory breast cancer by gene expression profiling. Breast Cancer Res. Treat. 2006;95(3):243–255. doi: 10.1007/s10549-005-9015-9. [DOI] [PubMed] [Google Scholar]
- 16.Bertucci F, Finetti P, Vermeulen P, Van Dam P, Dirix L, Birnbaum D, et al. Genomic profiling of inflammatory breast cancer: A review. Breast. 2014;23(5):538–545. doi: 10.1016/j.breast.2014.06.008. [DOI] [PubMed] [Google Scholar]
- 17.Matsuda N, Lim B, Wang Y, Krishnamurthy S, Woodward W, Alvarez RH, et al. Identification of frequent somatic mutations in inflammatory breast cancer. Breast Cancer Res. Treat. 2017;163(2):263–272. doi: 10.1007/s10549-017-4165-0. [DOI] [PubMed] [Google Scholar]
- 18.Goncalves MD, Hopkins BD, Cantley LC. Phosphatidylinositol 3-kinase, growth disorders, and cancer. N. Engl. J. Med. 2018;379(21):2052–2062. doi: 10.1056/NEJMra1704560. [DOI] [PubMed] [Google Scholar]
- 19.Fan H, Li C, Xiang Q, Xu L, Zhang Z, Liu Q, et al. PIK3CA mutations and their response to neoadjuvant treatment in early breast cancer: A systematic review and meta-analysis. Thorac. Cancer. 2018;9(5):571–579. doi: 10.1111/1759-7714.12618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.André F, Bachelot T, Commo F, Campone M, Arnedos M, Dieras V, et al. Comparative genomic hybridisation array and DNA sequencing to direct treatment of metastatic breast cancer: A multicentre, prospective trial (SAFIR01/UNICANCER) Lancet Oncol. 2014;15(3):267–274. doi: 10.1016/S1470-2045(13)70611-9. [DOI] [PubMed] [Google Scholar]
- 21.Mukohara T. PI3K mutations in breast cancer: Prognostic and therapeutic implications. Breast Cancer Targets Ther. 2015;7:111–123. doi: 10.2147/BCTT.S60696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Samuels Y, Velculescu VE. Oncogenic mutations of PIK3CA in human cancers. Cell Cycle. 2004;3(10):1221–1224. doi: 10.4161/cc.3.10.1164. [DOI] [PubMed] [Google Scholar]
- 23.Kodahl AR, Ehmsen S, Pallisgaard N, Jylling AMB, Jensen JD, Lænkholm A-V, et al. Correlation between circulating cell-free PIK3CA tumor DNA levels and treatment response in patients with PIK3CA-mutated metastatic breast cancer. Mol. Oncol. 2018;12(6):925–935. doi: 10.1002/1878-0261.12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buono G, Gerratana L, Bulfoni M, Provinciali N, Basile D, Giuliano M, et al. Circulating tumor DNA analysis in breast cancer: Is it ready for prime-time? Cancer Treat. Rev. 2019;73:73–83. doi: 10.1016/j.ctrv.2019.01.004. [DOI] [PubMed] [Google Scholar]
- 25.Aleskandarany MA, Rakha EA, Ahmed MAH, Powe DG, Paish EC, Macmillan RD, et al. PIK3CA expression in invasive breast cancer: A biomarker of poor prognosis. Breast Cancer Res. Treat. 2010;122(1):45–53. doi: 10.1007/s10549-009-0508-9. [DOI] [PubMed] [Google Scholar]
- 26.Baselga, J., Cortes Castan, J., De Laurentiis, M., Dieras, V., Harbeck, N., Hsu, J., et al. SANDPIPER: Phase III study of the PI3-kinase (PI3K) inhibitor taselisib (GDC-0032) plus fulvestrant in patients (pts) with oestrogen receptor (ER)-positive, HER2-negative locally advanced or metastatic breast cancer (BC) enriched for pts with PIK3CA-mutant tumours. Ann. Oncol. (Internet). 27(suppl_6) (2016). Disponible sur: https://academic.oup.com/annonc/article/27/suppl_6/313TiP/2799052. Accessed 17 Feb 2020.
- 27.Di Leo A, Johnston S, Lee KS, Ciruelos E, Lønning PE, Janni W, et al. Buparlisib plus fulvestrant in postmenopausal women with hormone-receptor-positive, HER2-negative, advanced breast cancer progressing on or after mTOR inhibition (BELLE-3): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2018;19(1):87–100. doi: 10.1016/S1470-2045(17)30688-5. [DOI] [PubMed] [Google Scholar]
- 28.Baselga J, Im S-A, Iwata H, Cortés J, De Laurentiis M, Jiang Z, et al. Buparlisib plus fulvestrant versus placebo plus fulvestrant in postmenopausal, hormone receptor-positive, HER2-negative, advanced breast cancer (BELLE-2): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2017;18(7):904–916. doi: 10.1016/S1470-2045(17)30376-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.André F, Ciruelos E, Rubovszky G, Campone M, Loibl S, Rugo HS, et al. Alpelisib for PIK3CA-mutated, hormone receptor-positive advanced breast cancer. N. Engl. J. Med. 2019;380(20):1929–1940. doi: 10.1056/NEJMoa1813904. [DOI] [PubMed] [Google Scholar]
- 30.Clatot F, Perdrix A, Augusto L, Beaussire L, Delacour J, Calbrix C, et al. Kinetics, prognostic and predictive values of ESR1 circulating mutations in metastatic breast cancer patients progressing on aromatase inhibitor. Oncotarget. 2016;7(46):74448–74459. doi: 10.18632/oncotarget.12950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allouchery V, Beaussire L, Perdrix A, Sefrioui D, Augusto L, Guillemet C, et al. Circulating ESR1 mutations at the end of aromatase inhibitor adjuvant treatment and after relapse in breast cancer patients. Breast Cancer Res. 2018;20(1):40. doi: 10.1186/s13058-018-0968-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winn JS, Hasse Z, Slifker M, Pei J, Arisi-Fernandez SM, Talarchek JN, et al. Genetic variants detected using cell-free DNA from blood and tumor samples in patients with inflammatory breast cancer. Int. J. Mol. Sci. 2020;21(4):4. doi: 10.3390/ijms21041290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Atkinson RL, El-Zein R, Valero V, Lucci A, Bevers TB, Fouad T, et al. Epidemiological risk factors associated with inflammatory breast cancer subtypes. Cancer Causes Control CCC. 2016;27(3):359–366. doi: 10.1007/s10552-015-0712-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schairer C, Li Y, Frawley P, Graubard BI, Wellman RD, Buist DSM, et al. Risk factors for inflammatory breast cancer and other invasive breast cancers. JNCI J. Natl. Cancer Inst. 2013;105(18):1373–1384. doi: 10.1093/jnci/djt206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ross JS, Ali SM, Wang K, Khaira D, Palma NA, Chmielecki J, et al. Comprehensive genomic profiling of inflammatory breast cancer cases reveals a high frequency of clinically relevant genomic alterations. Breast Cancer Res. Treat. 2015;154(1):155–162. doi: 10.1007/s10549-015-3592-z. [DOI] [PubMed] [Google Scholar]
- 36.Liang X, Vacher S, Boulai A, Bernard V, Baulande S, Bohec M, et al. Targeted next-generation sequencing identifies clinically relevant somatic mutations in a large cohort of inflammatory breast cancer. Breast Cancer Res. BCR. 2018;20(1):88. doi: 10.1186/s13058-018-1007-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zardavas D, Te Marvelde L, Milne RL, Fumagalli D, Fountzilas G, Kotoula V, et al. Tumor PIK3CA genotype and prognosis in early-stage breast cancer: A pooled analysis of individual patient data. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2018;36(10):981–990. doi: 10.1200/JCO.2017.74.8301. [DOI] [PubMed] [Google Scholar]
- 38.Papaxoinis G, Kotoula V, Alexopoulou Z, Kalogeras KT, Zagouri F, Timotheadou E, et al. Significance of PIK3CA mutations in patients with early breast cancer treated with adjuvant chemotherapy: A hellenic cooperat. PLoS ONE. 2015;10(10):e0140293. doi: 10.1371/journal.pone.0140293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dirican E, Akkiprik M, Özer A. Mutation distributions and clinical correlations of PIK3CA gene mutations in breast cancer. Tumor Biol. 2016;37(6):7033–7045. doi: 10.1007/s13277-016-4924-2. [DOI] [PubMed] [Google Scholar]
- 40.Takeshita T, Yamamoto Y, Yamamoto-Ibusuki M, Inao T, Sueta A, Fujiwara S, et al. Prognostic role of PIK3CA mutations of cell-free DNA in early-stage triple negative breast cancer. Cancer Sci. 2015;106(11):1582–1589. doi: 10.1111/cas.12813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vasan N, Razavi P, Johnson JL, Shao H, Shah H, Antoine A, et al. Double PIK3CA mutations in cis increase oncogenicity and sensitivity to PI3Kα inhibitors. Science. 2019;366(6466):714–723. doi: 10.1126/science.aaw9032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhat-Nakshatri P, Goswami CP, Badve S, Magnani L, Lupien M, Nakshatri H. Molecular insights of pathways resulting from two common PIK3CA mutations in breast cancer. Cancer Res. 2016;76(13):3989–4001. doi: 10.1158/0008-5472.CAN-15-3174. [DOI] [PubMed] [Google Scholar]
- 43.Yang SX, Polley E, Lipkowitz S. New insights on PI3K/AKT pathway alterations and clinical outcomes in breast cancer. Cancer Treat. Rev. 2016;45:87–96. doi: 10.1016/j.ctrv.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Board RE, Wardley AM, Dixon JM, Armstrong AC, Howell S, Renshaw L, et al. Detection of PIK3CA mutations in circulating free DNA in patients with breast cancer. Breast Cancer Res. Treat. 2010;120(2):461–467. doi: 10.1007/s10549-010-0747-9. [DOI] [PubMed] [Google Scholar]
- 45.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014;6(224):224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gautschi O, Bigosch C, Huegli B, Jermann M, Marx A, Chassé E, et al. Circulating deoxyribonucleic acid as prognostic marker in non-small-cell lung cancer patients undergoing chemotherapy. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2004;22(20):4157–4164. doi: 10.1200/JCO.2004.11.123. [DOI] [PubMed] [Google Scholar]
- 47.Zitt M, Müller HM, Rochel M, Schwendinger V, Zitt M, Goebel G, et al. Circulating cell-free DNA in plasma of locally advanced rectal cancer patients undergoing preoperative chemoradiation: A potential diagnostic tool for therapy monitoring. Dis. Markers. 2008;25(3):159–165. doi: 10.1155/2008/598071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park K, Woo M, Kim JE, Ahn J-H, Jung KH, Roh J, et al. Efficacy of assessing circulating cell-free DNA using a simple fluorescence assay in patients with triple-negative breast cancer receiving neoadjuvant chemotherapy: A prospective observational study. Oncotarget. 2017;9(3):3875–3886. doi: 10.18632/oncotarget.23520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Uden DJP, van Maaren MC, Bult P, Strobbe LJA, van der Hoeven JJM, Blanken-Peeters CFJM, et al. Pathologic complete response and overall survival in breast cancer subtypes in stage III inflammatory breast cancer. Breast Cancer Res. Treat. 2019;176(1):217–226. doi: 10.1007/s10549-019-05219-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pierga J-Y, Bidard F-C, Autret A, Petit T, Andre F, Dalenc F, et al. Circulating tumour cells and pathological complete response: Independent prognostic factors in inflammatory breast cancer in a pooled analysis of two multicentre phase II trials (BEVERLY-1 and -2) of neoadjuvant chemotherapy combined with bevacizumab. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2017;28(1):103–109. doi: 10.1093/annonc/mdw535. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.