Abstract
The correlation between the prototype AMPLICOR CMV MONITOR test (Roche Molecular Systems), a quantitative PCR assay, and the cytomegalovirus (CMV) pp65 antigenemia assay was evaluated in transplant recipients. Sequential blood specimens were collected on 29 patients (491 specimens), the leukocyte fraction was tested by CMV antigenemia, and quantitative PCR was performed on plasma specimens. None of the 15 patients (242 specimens) who were antigenemia negative were positive for CMV DNA by PCR, and none of these patients developed active CMV disease. There were 14 antigenemia-positive patients, 8 of whom developed active CMV disease. In all patients, there was a good association between the antigenemia and PCR assays. Ganciclovir-resistant virus was isolated from three patients with active CMV disease. These three patients had persistently elevated levels of antigenemia and CMV DNA by PCR when resistance to ganciclovir developed. This standardized, quantitative CMV PCR assay on plasma has clinical utility for the diagnosis of active disease and in monitoring the response to antiviral therapy in transplant recipients.
Cytomegalovirus (CMV) is the most important pathogen affecting transplant recipients (11), causing significant morbidity and mortality (30). The effects of CMV infection in transplant recipients may be classified as direct and indirect (11). Direct effects are seen clinically as manifestations of active CMV disease including, for example, fever, leukopenia, hepatitis, colitis, and retinitis. Indirect effects are more subtle and are believed to lead to allograft injury and loss (8, 27, 31) and increased susceptibility to infections with other organisms, as well as to decreased patient survival (6, 10, 38, 39).
With effective anti-CMV treatments available, a sensitive and reliable diagnostic test that will rapidly confirm or exclude active CMV infection is of practical clinical relevance. The introduction of the CMV antigenemia assay (40, 41) and the PCR assay (5, 17) marked a new era in our ability to rapidly detect CMV infection and have led to the use of anti-CMV therapy in a preemptive mode (4, 15, 29, 32). Hence, these assays have been used for both diagnosis and surveillance of CMV infection in solid-organ and bone marrow transplant recipients (1, 3, 7, 12, 18, 20, 22, 24, 26, 28, 35, 36, 42). The clinical sensitivity of the antigenemia assay is better than shell vial and conventional culture for the early diagnosis of CMV infection (23, 36). Moreover, culture assays have been found to be insufficiently sensitive for the timely initiation of preemptive therapy (9, 13). This has prompted many laboratories to use the antigenemia assay as their standard of care for detecting CMV in the blood of transplant recipients. While studies have shown that CMV infection can be detected even earlier using the PCR assay for the detection of CMV DNA in blood leukocytes (12, 24), there is concern that the assay may not correlate with active disease (12, 19, 36). That is, the increased analytical sensitivity of the PCR assays leads to a lower clinical specificity. Studies by Spector and coworkers (33, 44) have shown that detection of CMV DNA in blood plasma by PCR may correlate with disease better than assays using leukocytes or whole blood. Other studies have found that CMV DNA detected by PCR in plasma correlated more closely than that in leukocytes or whole blood when compared with the CMV pp65 antigenemia assay (3, 14, 37, 43, 45).
Many quantitative CMV PCR assays used in clinical studies have been developed in-house (7, 28) and, as a result, the methods are not well standardized. In-house PCR assays may differ in the target sequence, primer pairs, nucleic acid extraction methods, and detection methods, which result in differences in the limit of sensitivity and linear range of the assays. This makes it difficult to compare studies performed at different institutions or to make general comments on the clinical utility of quantitative CMV PCR. The clinical utility of a standardized qualitative CMV PCR assay has been shown in transplant recipients (2, 37). We have therefore assessed the prototype AMPLICOR CMV MONITOR test, a commercially available quantitative PCR assay for the detection of CMV DNA in plasma and evaluated the extent to which it correlated with the CMV antigenemia assay in transplant recipients.
MATERIALS AND METHODS
Patients and specimens.
A total of 491 specimens from 29 patients were tested by both the CMV antigenemia and quantitative CMV PCR assays at either the Massachusetts General Hospital (MGH) Clinical Microbiology Laboratory (Virology Section) or the University of Pittsburgh Medical Center (UPMC) Clinical Virology Laboratory. Specimens were collected from the MGH January 1995 through October 1997 and from the UPMC August 1995 through April 1997. Antigenemia assays were performed in real time, and results were used for patient management decisions. Plasma samples were stored frozen at −70°C and batch tested retrospectively. The patient population included individuals that had undergone either kidney, lung, liver, heart, or bone marrow transplantation. Chart review was done to obtain information regarding clinical symptoms and treatment.
Each study was approved by the corresponding institutional review board. At both the MGH and the UPMC the antigenemia test is the standard test used on blood specimens for the detection of CMV infection.
Definition of active CMV disease.
CMV infection was defined as a positive CMV antigenemia assay. Active CMV disease was defined as a positive CMV antigenemia assay and any of the following: the presence of appropriate symptoms (fever, malaise, and diarrhea) or signs (leukopenia and transaminitis), the presence of retinitis on ophthalmologic exam, or a tissue biopsy CMV positive by either culture or immunohistochemical staining.
CMV antigenemia assay.
The CMV antigenemia assays were done by standard procedures in the MGH and UPMC Clinical Virology Laboratories (23, 34). Results were reported as the number of positive staining cells per 200,000 leukocytes.
Quantitative CMV PCR assay.
The prototype AMPLICOR CMV MONITOR test, a quantitative microtiter-based PCR assay, was developed by Roche Molecular Systems (Alameda, Calif.). CMV viral DNA in the specimen was quantitated by coamplifying a region of the CMV DNA polymerase gene in the presence of a known quantity of quantitative standard. The primers used were specific for the CMV polymerase gene and amplified a 362-bp fragment of the gene (21). The test is designed with an internal quantitation standard (QS) that was constructed with the same primer sequence as the target DNA but with different intervening sequences. The unique probe sequence allows for the differentiation of QS DNA and target DNA. To ensure a similar amplification efficiency, the QS is the same size and has a similar G+C content as the CMV target region. The QS is added at a known concentration during specimen processing so that extraction and recovery of DNA, in addition to amplification and detection, can be monitored. The assay was performed according to the manufacturer's recommendations. The lower limit of sensitivity of the assay is 400 copies/ml of plasma.
Prevention of PCR contamination.
Contamination precautions included the use of aerosol barrier pipette tips; the use of separate areas of the laboratory for master mix preparation, specimen extraction, and detection; the use of UTP and uracil-N-glycosylase in the reaction mixture; and the inclusion of multiple negative controls in each run.
RESULTS
Sequential specimens obtained from 29 transplant recipients were tested in both the CMV antigenemia and the quantitative CMV PCR assays. Of the 29 patients, 14 developed CMV infection as evidenced by positive antigenemia. The remaining 15 patients never exhibited a positive antigenemia test during the posttransplant surveillance period. Additional demographic information is given in Table 1. The number of specimens collected from each patient ranged from 4 to 35, with a median of 12 specimens. Of the 491 plasma specimens tested by the quantitative PCR assay, 7 (1.4%) showed evidence of inhibition and, therefore, were not quantifiable. None of the 15 patients (242 specimens) that were antigenemia negative were found to have detectable levels of CMV DNA by the quantitative PCR assay. In addition, none of these patients developed active CMV disease.
TABLE 1.
Demographic information
| Patient parameter | Patient group
|
|
|---|---|---|
| Infectiona | No infection | |
| No. of patients | 14 | 15 |
| Age (yr) | ||
| Mean | 45 | 39 |
| Range | 5–63 | 2–65 |
| Sex (no. male/no. female) | 9/5 | 9/6 |
| No. with each type of transplant | ||
| Liver | 2 | 0 |
| Kidney | 8 | 5 |
| Lung | 2 | 8 |
| Heart | 1 | 0 |
| Bone marrow | 1 | 2 |
| No. donor seropositive | 13 | 8b |
| No. recipient seropositive (no. with equivocal serology) | 0 (1) | 4c (0) |
| No. D+/R−d | 12 | 6 |
Infection was defined by positive antigenemia status during posttransplant monitoring, and no infection was defined by negative antigenemia status during posttransplant monitoring.
One patient received lung tissue from two different donors, one seropositive and the other seronegative; for two patients the donor serostatus was unknown.
The serostatus of one recipient was unknown.
D+/R−, donor CMV seropositive and recipient CMV seronegative.
The patients that had CMV infection (positive antigenemia assay) were similar to those who did not have infection with regard to age, sex, and type of organ transplanted (Table 1). As expected, there were differences in the CMV serostatus of the recipients (R) and donors (D) in the two groups. In the group with infection, 86% (12 of 14) were D+/R−, that is, the donor was seropositive for CMV and the recipient was seronegative for CMV prior to transplant. In the group without infection, 40% (6 of 15) were D+/R− (two-sided exact test, P = 0.021).
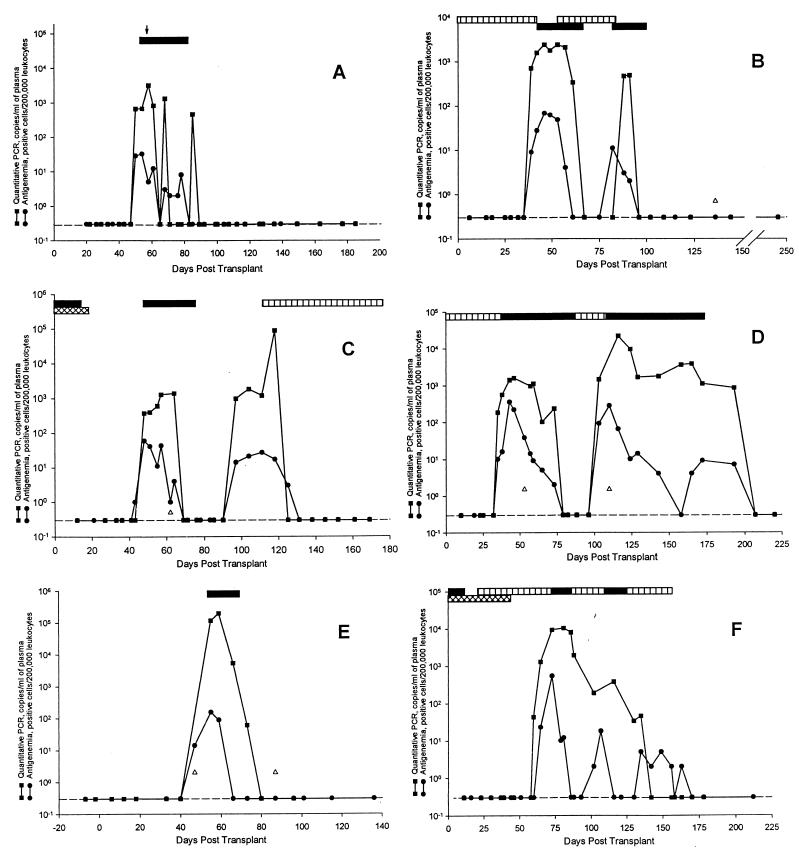
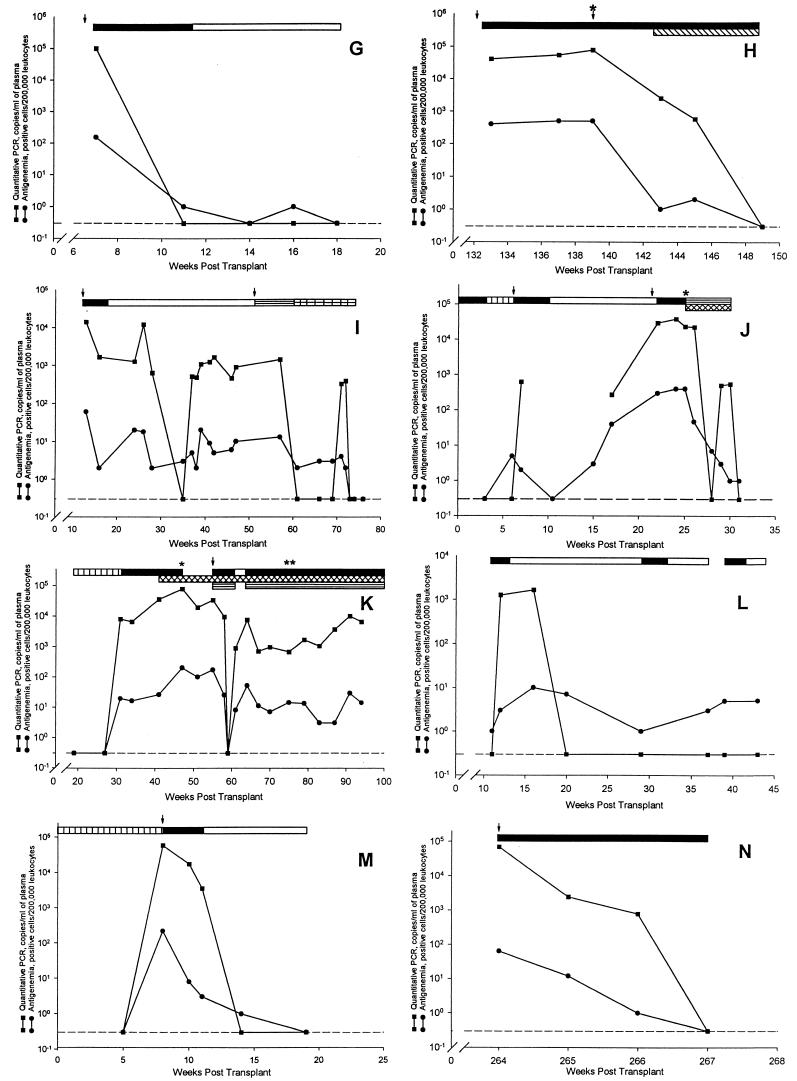
The results of the 14 antigenemia-positive patients are shown in Fig. 1. There was a good association between the PCR and antigenemia assays in all of the patients. When the initial antigenemia test was positive and the quantitative PCR was negative, the antigenemia values were ≤11 positive cells (Fig. 1C, J, and L). Two patients (Fig. 1D and F) had time points when the antigenemia returned to negative while CMV DNA remained detectable by PCR. Since subsequent specimens from both patients were found to be positive by antigenemia and PCR, this may indicate a better sensitivity for the detection of unresolved infections by PCR.
FIG. 1.
Comparison of CMV antigenemia and quantitative PCR
results for the 14 patients who were found to be positive by
antigenemia testing. The dashed line (---) represents the negative
antigenemia results (no positive cells per 200,000 leukocytes) or a CMV
DNA copy number below the limit of detection of the assay (<400
copies/ml). Symbols: ↓, an episode of symptomatic CMV disease; ▵, a
specimen that failed to amplify due to the presence of inhibitors; ∗,
detection of ganciclovir-resistant virus; ∗∗, detection of
foscarnet resistant virus. Patients: A, renal transplant, 25-year-old
male, ↓ gastritis; B, renal transplant, 52-year-old male; C, renal
transplant, 43-year-old female; D, renal transplant, 62-year-old male;
E, bone marrow transplant, 5-year-old male; F, renal transplant,
29-year-old male; G, renal transplant, 49-year-old male, ↓
fever and liver transaminitis; H, renal transplant, 52-year-old female,
↓ (first) fever, fatigue, and leukopenia, ↓ (second) retinitis; I,
lung transplant, 40-year-old female, ↓ (first) fever, abdominal pain,
liver transaminitis, ↓ (second) retinitis; J, liver transplant,
63-year-old female, ↓ (first) fever and malaise, ↓ (second)
leukopenia, low-grade fever and malaise; K, lung transplant,
41-year-old female, ↓ pneumonitis and colitis; L, heart transplant,
56-year-old male; M, liver transplant, 60-year-old male, ↓ fever and
fatigue; N, renal transplant, 58-year-old male, ↓ low-grade fever,
leukopenia, and liver transaminitis. Bars: □, oral ganciclovir;
, intravitreal ganciclovir; ▤, intravenous foscarnet; ▥, oral
acyclovir; ■, intravenous ganciclovir; ▧, intravitreal foscarnet;
 ,
intravenous acyclovir;
,
intravenous acyclovir;
 ,
cytogam.
,
cytogam.
Of the 14 CMV antigenemia-positive patients that were studied, 8 developed active CMV disease (Fig. 1A, G to K, M, and N); 3 of the 8 patients had two episodes of active disease. The 11 episodes of active disease included gastritis (one patient); fever with leukopenia, fatigue, or liver transaminitis (seven patients); retinitis (two patients); and colitis (one patient). In these patients both the antigenemia and PCR assays were positive when the patient presented with signs and symptoms of acute infection.
Of the 14 antigenemia-positive patients, 3 developed ganciclovir-resistant CMV while receiving therapy (50% inhibitory concentrations [IC50s] of 4.9, 10.9, and 3.7 μg/ml; assay cutoff, >3.0 μg/ml). One patient received a liver transplant (Fig. 1J), another received a lung transplant (Fig. 1K), and the final patient received a renal transplant (Fig. 1H); in each case the recipient was CMV seronegative, and the donor was CMV seropositive. The patients were found to be positive by the antigenemia assay and received courses of intravenous and oral ganciclovir. Eventually the patients developed persistent positive CMV antigenemia, with between 300 to 400 positive cells, while on therapy. PCR testing revealed a similar pattern as seen with the antigenemia assay, with the CMV DNA copy number remaining elevated at between 20,000 to 70,000 copies per ml of plasma, while the patient was on therapy. Persistently elevated antigenemia or CMV DNA levels during therapy may therefore be a marker of resistant virus.
Of the 14 CMV antigenemia-positive patients, there was 1 patient in whom the CMV DNA levels remained at <400 copies/ml. A total of 14 specimens from this patient were tested by PCR that were found to have between 1 and 100 positive cells by antigenemia. Only one of the specimens showed evidence of inhibition. When the plasma specimens were tested in a second laboratory with the PCR assay, the CMV DNA values ranged at between 372 and 1,920 copies/ml of plasma (Fig. 1E). Again, one of the specimens was inhibitory to amplification. Other patient samples tested by both laboratories were concordant. The reason for the differences in results for this patient between the two laboratories is unclear. The patient did not develop active CMV disease.
DISCUSSION
There have been many studies evaluating the clinical utility of PCR-based assays for the detection of CMV DNA (1, 3, 7, 18, 22, 24, 28, 35, 36, 44). Many of these investigations have shown that, although the assays may be sensitive, they often lack clinical specificity, detecting CMV DNA in patients that do not develop active CMV disease. The quantitative PCR assay studied here was designed with a reduced level of sensitivity (400 copies/ml of plasma) in an effort to improve the disease specificity. This approach appears to have been effective, since none of the 242 specimens from 15 transplant recipients that did not have a positive antigenemia or develop active CMV disease posttransplantation were found to have detectable CMV DNA levels by quantitative PCR. To further assess the specificity of the CMV PCR assay, plasma specimens were tested from 100 CMV seronegative and 100 seropositive healthy blood donors, and all were found to be negative using the qualitative version of the assay (data not shown).
There was good association between the patterns of positive CMV antigenemia and CMV DNA levels. These 14 patients represent a wide range of clinical presentations and antigenemia results. There were patients with a single antigenemia peak and patients with multiple antigenemia peaks, and patients who developed active CMV disease shortly after receiving their transplant and patients who developed active CMV disease many months after transplantation. In all of these patients, there was a strong temporal relationship between antigenemia and CMV DNA as quantitated by PCR. Three patients with initial low positive antigenemia results had undetectable CMV DNA levels. In all of these patients the antigenemia level was ≤11 cells, and the subsequent specimen was found to be positive by both the PCR and antigenemia assays. Two of these three patients went on to develop active CMV disease. Performing quantitative CMV PCR using whole blood or WBC specimens usually improves the sensitivity of the assay, but it also decreases the clinical specificity. To further evaluate this issue, we are currently doing studies comparing cellular and plasma CMV DNA levels with the development of clinical disease in transplant recipients.
Though this study was not designed to test the utility of the quantitative PCR assay in a posttransplant surveillance protocol, we have been able to demonstrate a good association between CMV DNA measurements obtained using a commercial quantitative PCR assay and the antigenemia results. In addition, it appears that the development of active CMV disease is very unlikely when CMV DNA levels remain undetectable (<400 copies/ml). Results from this retrospective analysis suggest that this quantitative PCR assay may have clinical utility for monitoring CMV disease in much the same way that the antigenemia test is currently used. This concept is supported by other studies (1, 7, 28) which suggest that quantitative PCR assays will accurately predict the occurrence of symptomatic disease in transplant recipients. One such study with this commercial assay has recently been reported for liver transplant recipients with encouraging results (16). Further studies in other transplant and patient groups are needed.
As with the antigenemia assay, there are patients who have detectable CMV DNA who do not develop clinical CMV disease. A factor contributing to the difficulty of correlating quantities of CMV DNA with the development of active disease is the wide use of preemptive therapy. However, 8 of the 14 (69%) patients with detectable CMV DNA levels did develop active CMV disease, a finding which supports the use of the quantitative test as a screening test to assess when to initiate preemptive therapy. The clinical utility of this quantitative PCR assay in monitoring response to therapy was evident for the three patients that developed ganciclovir-resistant virus. For these patients, inadequate suppression of CMV DNA levels (between 20,000 and 70,000 copies/ml) eventually led to recrudescence of disease and the isolation of ganciclovir-resistant virus.
The quantitative CMV PCR assay described here has several advantages over the antigenemia assay. The test requires a small volume of plasma (200 μl) compared to 3 to 5 ml of whole blood required for antigenemia testing. Since PCR testing is performed on plasma specimens, the test can be done on patients with low leukocyte counts and the specimen processing is much simpler compared to methods for leukocyte preparation for antigenemia testing. Furthermore, CMV DNA has been found to be stable in whole blood for up to 5 days when stored at 4°C (data not shown). This study was performed on an early version of the assay, which used a microtiter plate format. The test has now been adapted to the semiautomated COBAS AMPLICOR format (25), so the technical time required to perform the PCR test is much less than that required for antigenemia assay. For example, testing 12 specimens by the antigenemia test requires about 6 h of technical time, whereas the semiautomated, quantitative CMV PCR testing requires about 2 h. An important technical component of the PCR assay is the quantitation standard, which allows for the detection of substances in the clinical specimen that may be inhibitory to amplification.
One challenge in interpreting the clinical utility of quantitative CMV DNA has been the variability in assay methods and the resulting variation in assay sensitivity and specificity. A major advantage of the prototype AMPLICOR CMV MONITOR test described here is that it provides clinical laboratories with a standardized assay. This will allow the results of studies to be compared between laboratories and may allow for the determination of broadly applicable viral load cutoffs for preemptive therapy. In addition, this commercial assay is simple to perform, allowing it to be used in laboratories without extensive experience with PCR testing.
ACKNOWLEDGMENTS
We thank Arlene Bullotta for preparation of the graphs, Richard Day for advice with statistical analysis, and the staffs of the Clinical Virology Laboratories at the MGH and the UPMC.
This work was supported in part by Roche Molecular Systems.
REFERENCES
- 1.Abecassis M M, Koffron A J, Kaplan B, Buckingham M, Muldoon J P, Cribbins A J, Kaufman D B, Fryer J P, Stuart J, Stuart F P. The role of PCR in the diagnosis and management of CMV in solid organ transplant recipients. Transplantation. 1997;63:275–279. doi: 10.1097/00007890-199701270-00017. [DOI] [PubMed] [Google Scholar]
- 2.Aitken C, Barrett-Muir W, Millar C, Templeton K, Thomas J, Sheridan F, Jefferies D, Yaqoob M, Breuer J. Use of molecular assays in diagnosis and monitoring of cytomegalovirus disease following renal transplantation. J Clin Microbiol. 1999;37:2804–2807. doi: 10.1128/jcm.37.9.2804-2807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boeckh M, Gallez-Hawkins G M, Myerson D, Zaia J, Bowden R A. Plasma polymerase chain reaction for cytomegalovirus DNA after allogeneic bone marrow transplantation. Transplantation. 1997;64:108–113. doi: 10.1097/00007890-199707150-00020. [DOI] [PubMed] [Google Scholar]
- 4.Brennan D C, Garlock K A, Lippmann B A, Buller R S, Gaudreault-Keener M, Lowell J A, Miller S B, Shenoy S, Howard T K, Storch G A. Control of cytomegalovirus-associated morbidity in renal transplant recipients using intensive monitoring and either preemptive or deferred therapy. J Am Soc Nephrol. 1997;8:118–125. doi: 10.1681/ASN.V81118. [DOI] [PubMed] [Google Scholar]
- 5.Cassol S A, Poon M C, Pal R, Naylor M J, Culver-James J, Bowen T J, Russell J A, Krawetz S A, Pon R T, Hoar D I. Primer-mediated enzymatic amplification of CMV DNA. J Clin Investig. 1989;83:1109–1115. doi: 10.1172/JCI113990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collins L A, Samore M H, Roberts M S, Luzzati R, Jenkins R L, Lewis W D, Karchmer A W. Risk factors for invasive fungal infection complicating orthotopic liver transplantation. J Infect Dis. 1994;170:644–652. [PubMed] [Google Scholar]
- 7.Cope A V, Sabin C, Burroughs A, Rolles K, Griffiths P D, Emery V C. Interrelationships among quantity of human cytomegalovirus (HCMV) DNA in blood, donor-recipient serostatus, and administration of methylprednisolone as risk factors for HCMV disease following liver transplantation. J Infect Dis. 1997;176:1484–1490. doi: 10.1086/514145. [DOI] [PubMed] [Google Scholar]
- 8.de Otero J, Gavalda J, Murio E, Vargas V, Calico I, Llopart L, Rossello J, Margarit C, Pahissa A. Cytomegalovirus disease as a risk factor for graft loss and death after orthotopic liver transplantation. Clin Infect Dis. 1998;26:865–870. doi: 10.1086/513949. [DOI] [PubMed] [Google Scholar]
- 9.Einsele H, Ehninger G, Hebart H, Wittkowski K M, Schuler U, Jahn G, Mackes P, Herter M, Klingebiel T, Loffler J, Wagner S, Muller C A. Polymerase chain reaction monitoring reduces the incidence of cytomegalovirus disease and the duration and side effects of antiviral therapy after bone marrow transplantation. Blood. 1995;86:2815–2820. [PubMed] [Google Scholar]
- 10.Falagas M E, Snydman D R, Griffith J, Werner B G the Boston Center for Liver Transplantation CMVIG Study Group. Exposure to cytomegalovirus from the donated organ is a risk factor for bacteremia in orthotopic liver transplant recipients. Clin Infect Dis. 1996;23:468–474. doi: 10.1093/clinids/23.3.468. [DOI] [PubMed] [Google Scholar]
- 11.Fishman J A, Rubin R H. Infection in organ-transplant recipients. N Engl J Med. 1998;338:1741–1751. doi: 10.1056/NEJM199806113382407. [DOI] [PubMed] [Google Scholar]
- 12.Gerna G, Zipeto D, Parea M, Revello M G, Silini E, Percivalle E, Zavattoni M, Grossi P, Milanesi G. Monitoring of human cytomegalovirus infections and ganciclovir treatment in heart transplant recipients by determination of viremia, antigenemia and DNAemia. J Infect Dis. 1991;164:488–498. doi: 10.1093/infdis/164.3.488. [DOI] [PubMed] [Google Scholar]
- 13.Goodrich J M, More M, Gleaves C A, DuMond C, Cays M, Ebeling D F, Buhles W C, DeArmand B, Meyer J D. Early treatment with ganciclovir to prevent cytomegalovirus disease after allogeneic bone marrow transplantation. N Engl J Med. 1991;325:1601–1607. doi: 10.1056/NEJM199112053252303. [DOI] [PubMed] [Google Scholar]
- 14.Hebart H, Muller C, Loffler J, Jahn G, Einsele H. Monitoring of CMV infection: a comparison from whole blood, plasma PCR, pp65-antigenemia and virus culture in patients after bone marrow transplantation. Bone Marrow Transplant. 1996;17:861–868. [PubMed] [Google Scholar]
- 15.Hibberd P L, Tolkoff-Rubin N E, Conti D, Stuart F, Thistlethwaite J R, Neylan J F, Suydman D R, Freeman R, Lorber M I, Rubin R H. Preemptive ganciclovir therapy to prevent cytomegalovirus disease in cytomegalovirus-antibody-positive renal transplant recipients. Ann Intern Med. 1995;123:18–26. doi: 10.7326/0003-4819-123-1-199507010-00002. [DOI] [PubMed] [Google Scholar]
- 16.Humar A, Gregson D, Caliendo A M, McGeer A, Malkan G, Krajden M, Corey P, Greig P, Walmsley S, Levy G, Mazzulli T. Clinical utility of quantitative cytomegalovirus viral load determination for predicting cytomegalovirus disease in liver transplant recipients. Transplantation. 1999;68:1305–1311. doi: 10.1097/00007890-199911150-00015. [DOI] [PubMed] [Google Scholar]
- 17.Jiwa N M, van Gemert G W, Raap A K, van de Rijke F M, Mulder A, Lens P F, Salimans M M M, Zwann F E, van Dorp W, van der Ploeg M. Rapid detection of human cytomegalovirus DNA in peripheral blood leucocytes of viremic transplant recipients by the polymerase chain reaction. Transplantation. 1989;48:72–76. doi: 10.1097/00007890-198907000-00017. [DOI] [PubMed] [Google Scholar]
- 18.Kidd I M, Fox J C, Pillay D, Charman H, Griffiths P D, Emery V C. Provision of prognostic information in immunocompromised patients by routine application of the polymerase chain reaction for cytomegalovirus. Transplantation. 1993;56:867–871. doi: 10.1097/00007890-199310000-00018. [DOI] [PubMed] [Google Scholar]
- 19.Koehler M, St. George K, Ehrlich G D, Mirro J, Neudorf S M, Rinaldo C. Prevention of CMV disease in allogeneic BMT recipients by CMV antigenemia-guided preemptive therapy. J Pediatr Hematol Oncol. 1997;19:43–47. doi: 10.1097/00043426-199701000-00006. [DOI] [PubMed] [Google Scholar]
- 20.Koskinen P T, Markku S N, Mattila S P, Hayry P J, Lautenschlager I T. The correlation between symptomatic CMV infection and CMV antigenemia in heart allograft recipients. Transplantation. 1993;55:547–551. doi: 10.1097/00007890-199303000-00017. [DOI] [PubMed] [Google Scholar]
- 21.Kouzarides T, Banjier A T, Satchwell S C, Weston K, Tomlinson P, Barell B G. Sequence and transcription analysis of the human cytomegalovirus DNA polymerase gene. J Virol. 1987;61:125–133. doi: 10.1128/jvi.61.1.125-133.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Landry M L, Ferguson D. Comparison of quantitative cytomegalovirus antigenemia assay with culture methods and correlation with clinical disease. J Clin Microbiol. 1993;31:2851–2856. doi: 10.1128/jcm.31.11.2851-2856.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mazzulli T, Rubin R H, Ferraro M J, D'Aquila R T, Doveikis S A, Smith B R, The T H, Hirsch M S. Cytomegalovirus antigenemia: clinical correlations in transplant recipients and in persons with AIDS. J Clin Microbiol. 1993;31:2824–2827. doi: 10.1128/jcm.31.10.2824-2827.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nolte F S, Emmens R K, Thurmond C, Mitchell P S, Pascuzzi C, Devine S M, Saral R, Wingard J R. Early detection of human cytomegalovirus viremia in bone marrow transplant recipients with DNA amplification. J Clin Microbiol. 1995;33:1263–1266. doi: 10.1128/jcm.33.5.1263-1266.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pasternack R, Vuorinen P, Pitkajarvi T, Koskela M, Miettinen A. Comparison of manual Amplicor PCR, Cobas Amplicor PCR, and Lcx assays for detection of Chlamydia trachomatisinfection in women by using urine specimens. J Clin Microbiol. 1997;35:402–405. doi: 10.1128/jcm.35.2.402-405.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patel R, Smith T F, Espy M, Portela D, Wiesner R H, Krom R A F, Paya C V. A prospective comparison of molecular diagnostic techniques for the early detection of cytomegalovirus in liver transplant recipients. J Infect Dis. 1995;171:1010–1014. doi: 10.1093/infdis/171.4.1010. [DOI] [PubMed] [Google Scholar]
- 27.Reinke P, Fietze E, Ode-Hakim S, Prosch S, Lippert J, Ewert R, Volk H-D. Late-acute renal allograft rejection and symptomless cytomegalovirus infection. Lancet. 1994;344:1737–1738. doi: 10.1016/s0140-6736(94)92887-8. [DOI] [PubMed] [Google Scholar]
- 28.Roberts T C, Brennan D C, Buller R S, Gaudreault-Keener M, Schnitzler M A, Sternhell K E, Garlock K A, Singer G G, Storch G A. Quantitative polymerase chain reaction to predict occurrence of symptomatic cytomegalovirus infection and assess response to ganciclovir therapy in renal transplant recipients. J Infect Dis. 1998;178:626–635. doi: 10.1086/515383. [DOI] [PubMed] [Google Scholar]
- 29.Rubin R H. Preemptive therapy in immunocompromised hosts. N Engl J Med. 1991;324:1057–1059. doi: 10.1056/NEJM199104113241509. [DOI] [PubMed] [Google Scholar]
- 30.Rubin R H. Infection in the organ transplant recipient. New York, N.Y: Plenum Medical Book Co.; 1994. [Google Scholar]
- 31.Rubin R H. Cytomegalovirus disease and allograft loss after organ transplantation. Clin Infect Dis. 1998;26:871–873. doi: 10.1086/513948. [DOI] [PubMed] [Google Scholar]
- 32.Rubin R H, Tolkoff-Rubin N E. Antimicrobial strategies in the care of organ transplant recipients. Antimicrob Agents Chemother. 1993;37:619–624. doi: 10.1128/aac.37.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spector S A, Hsia K, Wolf D, Shinkai M, Smith I. Molecular detection of human cytomegalovirus and determination of genotypic ganciclovir resistance in clinical specimens. Clin Infect Dis. 1995;21(Suppl. 2):S170–S173. doi: 10.1093/clinids/21.supplement_2.s170. [DOI] [PubMed] [Google Scholar]
- 34.St. George K, Rinaldo C R. Comparison of commercially available antibody reagents for the cytomegalovirus pp65 antigenemia assay. Clin Diagn Virol. 1997;7:147–152. doi: 10.1016/s0928-0197(96)00264-4. [DOI] [PubMed] [Google Scholar]
- 35.Storch G A, Buller R S, Bailey T C, Ettinger N A, Langlois T, Gaudreault-Keener M, Welby P L. Comparison of PCR and pp65 antigenemia assay with quantitative shell vial culture for detection of cytomegalovirus in blood leukocytes from solid-organ transplant recipients. J Clin Microbiol. 1994;32:997–1003. doi: 10.1128/jcm.32.4.997-1003.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanabe K, Tokumoto T, Ishikawa N, Koyama I, Takahashi K, Fuchinoue S, Kawai T, Koga S, Yagisawa T, Toma H, Ota K, Nakajima H. Comparative study of CMV antigenemia assay, polymerase chain reaction, serology, and shell vial assay in the early diagnosis and monitoring of CMV infection after renal transplantation. Transplantation. 1997;64:1721–1725. doi: 10.1097/00007890-199712270-00016. [DOI] [PubMed] [Google Scholar]
- 37.Tong C Y, Cuevas L, Williams H, Bakran A. Use of laboratory assays to predict cytomegalovirus disease in renal transplant recipients. J Clin Microbiol. 1998;36:2681–2685. doi: 10.1128/jcm.36.9.2681-2685.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van den Berg A P, Klompmaker I J, Haagsma E B, Peeters P M, Meerman L, Verwer R, The T H, Slooff M J. Evidence for an increased rate of bacterial infections in liver transplant recipients with cytomegalovirus infection. Clin Transplant. 1996;10:224–231. [PubMed] [Google Scholar]
- 39.van den Berg A P, Meyaard L, Otto S A, van Son W J, Klompmaker I J, Mesander G, de Leij L H F M, Miedema F, The T H. Cytomegalovirus infection associated with decreased proliferative capacity and increased rate of apoptosis of peripheral blood lymphocytes. Transplant Proc. 1995;27:936–938. [PubMed] [Google Scholar]
- 40.van der Bij W, Schirm J, Torensma R, van Son W J, Tegzess A M, The T H. Comparison between viremia and antigenemia for the detection of cytomegalovirus in blood. J Clin Microbiol. 1988;26:2531–2535. doi: 10.1128/jcm.26.12.2531-2535.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van der Bij W, Torensma R, van Son W J, Anema J, Schirm J, Tegzess A M, The T H. Rapid immunodiagnosis of active cytomegalovirus infection by monoclonal antibody staining of blood leucocytes. J Med Virol. 1988;25:179–188. doi: 10.1002/jmv.1890250208. [DOI] [PubMed] [Google Scholar]
- 42.van Dorp W T, Vlieger A, Jiwa N M, van Es L A, van der Ploeg M, van Saase J L C M, van der Woude F J. The polymerase chain reaction, a sensitive and rapid technique for detecting cytomegalovirus infection after renal transplantation. Transplantation. 1992;54:661–664. doi: 10.1097/00007890-199210000-00019. [DOI] [PubMed] [Google Scholar]
- 43.Wirgart B Z, Claesson K, Eriksson B M, Brundin M, Tufveson G, Totterman T, Grillner L. Cytomegalovirus (CMV) DNA amplification from plasma compared with CMV pp65 antigen (ppUL83) detection in leukocytes for early diagnosis of symptomatic CMV infection in kidney transplant recipients. Clin Diagn Virol. 1996;7:99–110. doi: 10.1016/s0928-0197(96)00258-9. [DOI] [PubMed] [Google Scholar]
- 44.Wolf D G, Spector S A. Early diagnosis of human cytomegalovirus disease in transplant recipients by DNA amplification in plasma. Transplantation. 1993;56:330–334. doi: 10.1097/00007890-199308000-00014. [DOI] [PubMed] [Google Scholar]
- 45.Wolff C, Skourtopoulos M, Hornschemeyer D, Wolff D, Korner M, Hufert F, Korfer R, Kleesiek K. Significance of human cytomegalovirus DNA detection in immunocompromised heart transplant patients. Transplantation. 1996;61:750–757. doi: 10.1097/00007890-199603150-00014. [DOI] [PubMed] [Google Scholar]