Abstract
Low-temperature germinability (LTG) is an important agronomic trait that can affect the planting time, planting area, and grain yield of staple crops, such as rice. However, the genetic mechanism of LTG is still unclear. In this study, a multi-parental permanent population with 208 single segment substitution lines (SSSLs) was used to conduct a genetic dissection for LTG across four cropping seasons. LTG was a typical quantitative trait with a high combined broad-sense heritability of 0.71. By comparison with the recipient parent, Huajingxian74, 24 SSSLs were identified as carrying LTG QTLs, which were further merged into integrated QTLs with shorter genetic distances by substitution mapping. Finally, 14 LTG QTLs were mapped on ten chromosomes, including seven positive-effect and seven negative-effect QTLs, with additive effect contributions ranging from 19.2 to 39.9%. qLTG3a, a main-effect and novel QTL, was confirmed by bulk segregant analysis using an F2 segregating population, and five key recombinants were selected to develop F3 populations for progeny testing. Marker-trait association analysis fine mapped qLTG3a to a 332.7-kb physical region between markers M6026 and M6341. Within this interval, 40 annotated genes were revealed, and three genes (Os03g0213300, Os03g0214400, and Os03g0214600) were considered as pivotal candidate genes for qLTG3a based on their sequence variations and expression patterns. Besides low temperature, qLTG3a can also enhance seed germination under standard temperature and osmotic stress. In summary, this study identified some genetic factors regulating LTG and opened a new window for breeding elite direct-seeded rice varieties. It will help reduce the climate risk in the production process of rice, which is of great significance to ensuring food security.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12284-021-00544-2.
Keywords: Substitution mapping, Bulk segregant analysis, Low temperature, Seed germination vigor, Candidate gene
Introduction
Seed germination, a critical step in the plant life cycle, is a complex process coordinately regulated by internal genetic factors and external environmental cues (Ma et al. 2017; Rajjou et al. 2012). Low temperature is one of the principal restriction factors that limit crop growth and development (Ding et al. 2020). As a thermophilic crop, rice (Oryza sativa L.) is sensitive to chilling stress through its life cycle, especially during the germination stage. However, environmental temperature is frequently below 15 °C in high-latitude and high-altitude rice cultivation regions or early spring seasons, causing a severe rice yield reduction due to low germination rates, delayed seedling emergence, and abnormal seedlings (Liu et al. 2018). Recently, direct seeding cultivation techniques were widely adopted in Asian countries, which increased the risk of encountering cold stress during germination (Mahender et al. 2015; Wang et al. 2018a). High low-temperature germinability (LTG) of rice varieties can extend rice planting areas and growing season to meet the expanding food demand. Therefore, it is of considerable significance to fully understand the genetic basis of LTG in rice.
LTG is a complex quantitative trait controlled by multiple genes. In the past 20 years, researchers have identified more than 100 QTLs for LTG on the 12 rice chromosomes using bi-parental mapping methods based on phenotypic traits such as germination percentage and germination index (Chen et al. 2006; Fujino et al. 2004; Ji et al. 2009; Jiang et al. 2006, 2017; Miura et al. 2001; Wang et al. 2011; Xie et al. 2014). Besides, as an alternative strategy, genome-wide association study (GWAS) has increasingly applied for LTG genetic research to dissect the variation of natural rice accessions (Fujino et al. 2015; Sales et al. 2017; Wang et al. 2018b; Yang et al. 2020).
QTL analysis lays the foundation for fine mapping and map-based cloning of some major LTG QTLs. For example, qLTG-9 was fine mapped using a near-isogenic line population between markers L9-25D and ID-1 to a 72.3-kb physical region on chromosome 9 containing five candidate genes (Li et al. 2013). qSV-5c, a major QTL of radical length and germination rate under optimal and low temperature, was located between SNP3 and CAPS2, a genomic region of approximately 400 kb on chromosome 1 (Xie et al. 2014). By progeny test, qLTG6 was delimited to a 45.8-kb physical interval between M002 and M008, and LOC_Os06g01320 was considered as a possible target gene (Jiang et al. 2020). Currently, only two LTG-related genes, qLTG3-1 and OsSAP16, have been cloned in rice. qLTG3-1 (Os03g0103300) encoded a glycine-rich protein of unknown function and was strongly expressed in the embryo during seed germination (Fujino et al. 2008). Further genome-wide analysis of gene expression and regulation revealed that qLTG3-1 might induce programmed cell death to weaken the tissues covering the embryo and reduce the mechanical resistance of the coleoptile (Fujino and Matsuda 2010; Fujino et al. 2008). OsSAP16 was identified through GWAS; it encodes a stress-associated protein containing two AN1-C2H2 zinc finger domains and acts as an essential LTG regulator (Wang et al. 2018c). These findings of LTG genes have brought some breakthroughs in the genetic control of LTG in rice. However, extra efforts are needed to enrich our knowledge about this critical agronomical trait's genetic and molecular mechanisms.
Single segment substitution lines (SSSLs) contain a single homozygous marker-defined chromosome segment from a donor parent in a recurrent parent background (Zhang et al. 2004). Unlike traditional segregated populations, SSSLs, a permanent advanced mapping population, is a powerful tool for QTL mapping and gene cloning because it can eliminate the interference of genetic background and improve the accuracy of QTL mapping. Recent studies have demonstrated that SSSLs play a crucial role in understanding the genetic mechanisms of important agronomic traits such as seed dormancy, grain shape, and heading date in rice (Wang et al. 2012; Zhou et al. 2017; Zhu et al. 2018).
In this study, the genetic dissection of rice LTG trait was carried out using 208 SSSLs with abundant genetic diversity conferred by 24 donor cultivars in the genetic background of Huajingxian 74 (HJX74) (Li et al. 2021), an elite indica recipient parent, in four cropping seasons. The objects of this research were (1) to identify QTLs controlling LTG using germination percentage at 15 °C as phenotypic data in a rice SSSL population; (2) to integrate and narrow down the QTLs located at similar chromosome locations by substitution mapping; and (3) to validate and fine map a novel major QTL, qLTG3a, and predict the possible candidate genes by sequence and expression analysis. These works will provide new insights into the genetic architecture of LTG and establish the basis for map-based cloning of the casual gene underline qLTG3a.
Materials and Methods
Plant Materials
Genetic analysis of LTG was conducted using 208 SSSLs, which were selected from a 1529 SSSLs library constructed with HJX74 as the recipient and 24 varieties from different countries as donors (Xi et al. 2006; Zhang et al. 2004). Each SSSL contained only one substituted segment from 24 donors (including 14 Indica and 10 japonica varieties) on the 12 chromosomes of rice in the genetic background of HJX74 (Additional file 1: Table S1). The length of substituted segments was 0.4–70.0 cM, the average length was 15.84 cM, and the genome coverage was 68.0% (Additional file 1: Fig. S1).
Field Planting
Rice plants were grown in an experimental field in South China Agricultural University, Guangzhou, China (at approximately 113° E and approximately 23° N). Each SSSL was grown in one block consisting of two rows of 20 plants in 2015E, 2015L, 2016E, or 2016L. "E" represents the early cropping season from March to July, and "L" represents the late cropping season from July to November. Depending on the number of SSSLs, the control line HJX74 was grown in several blocks, and the differences among these blocks were evaluated by one-way ANOVA to assess the effects of the field environment. All rice materials were grown in a wide-narrow row method, 33.33 cm for the wide row and 16.67 cm for the narrow row, with a completely randomized design and regular management. One to two weeks after transplanting to the paddy field, the fresh leaves of rice seedlings were collected and used for genomic DNA extraction. Marker genotyping was conducted with SSR markers to detect the substituted segment of each SSSL in every growing season.
Phenotypic Evaluation of LTG
LTG was evaluated as previously described by Cao et al. (2017) with the following changes. Plants from the middle of each block were tagged for heading date when the first panicle emerged. About 32–35 days after heading, the main panicles of three to five individual plants for each SSSL line were harvested according to the seed maturity status. After threshing and cleaning, the seeds were dried in an oven at 50 °C for seven days to break seed dormancy. About 100–200 well-developed seeds of each plant were randomly distributed into two 9-cm Petri-dishes with 10 ml distilled water per dish. The petri-dishes were then transferred to several large sealed plastic boxes (34.7 × 24.5 × 12.5 cm) with two layers of wet filter paper on the bottom and finally placed in a 15 °C chamber in the dark for ten days. A seed with radicle length longer than 1 mm in 2015E, 2015L, and 2016L, or 2 mm in 2016E, was counted as germinated from the 4th to 10th day after sowing (He et al. 2019). The germination percentages of the 7th day (2015E, 2015L, and 2016L) and 10th day (2016E) showed great variations in the SSSLs and were accordingly used as the phenotypic data of LTG. After germination at 15 °C, the seeds with germination percentages less than 80% were moved into a chamber of 30 °C to eliminate the secondary dormancy induced by low temperature.
QTL Analysis and Substitution Mapping
QTL analysis was performed following the method as previously described (Eshed and Zamir 1995; Zhou et al. 2017). To normalize the variance, the arcsine-transformed germination data were used in the Student's t tests to calculate significant differences between SSSLs and HJX74. When the germination percentage's P value was less than 0.001 in at least two cropping seasons, a putative LTG QTL was considered on the substituted segment of the corresponding SSSL. The nomenclature of identified QTLs follows the general rules in rice. A QTL's name consists of three parts: the abbreviation of the trait, the chromosome number, and the QTL number, which is sequenced from the short arm end to the long arm end of the chromosome. Assuming that each SSSL carried only one QTL, the additive effect (a) of a putative QTL accounted for half of the phenotypic difference in germination percentages between an SSSL and HJX74; the percentage of additive effect contribution (R2) was calculated as the additive effect divided by the mean phenotypic value of HJX74. If a QTL was simultaneously detected in multiple SSSLs with overlapping segments, then substitution mapping method was applied to reduce the region of a QTL as previously described by Wissuwa et al. (2002) and Zhou et al. (2017).
Bulk Segregant Analysis (BSA) Sequencing
In this study, a major QTL, qLTG3a, was simultaneously detected on the overlapping substituted segments of S5, S6, and S7 on chromosome 3. S6 contains a segment derived from Katy, a japonica donor parent, and was selected as a representative of qLTG3a in further analysis. For validation of qLTG3a, S6 was backcrossed with the indica recurrent parent HJX74 to produce an F2 segregating population with 356 individual plants. Young leaves of the 356 F2 individuals were collected separately and stored at -80℃. After evaluation of LTG, the same size leave samples of 30 extremely low-temperature-tolerant and 30 extremely low-temperature-sensitive plants were bulked as LTG-pool and WT-pool, respectively. Total genomic DNA of segregation pools was extracted using the CTAB method. DNA quality was determined by Nanodrop (Thermo Fisher Scientific, Waltham, MA). The two DNA pools, parental cultivars S6 and HJX74 were sequenced with coverage of 30X using Illumina sequencing strategy at Genedenovo (Guangzhou, China). Filtered reads were compared with the R498 (Shuhui498, an indica variety) genome sequence to identify single-nucleotide polymorphisms (SNPs) and Insertion-deletions (InDels) using Burrows-Wheeler Aligner (BWA, v0.7.12). By calculating SNP-index and ΔSNP-index, the distribution of ΔSNP-index was fitted on 12 chromosomes. The 95% confidence level was selected as the threshold to determine the linkage interval. Also, ΔInDel-index was calculated for subsequent sequence variation analysis of candidate genes.
Marker Development and Fine Mapping
New high-density markers were designed on the substituted segment of S6 flanked by RM231 and RM563 based on InDel variations identified by BSA sequencing and the difference between the Nipponbare and 9311 genome sequences. Fourteen polymorphic InDel markers were used for genotyping the 356 F2 individual plants to conduct frequency distribution analysis and select recombinants for the qLTG3a locus. Based on the marker genotypes, five recombinants, named S6_R1 to S6_R5, were detected, and more than 70 progeny lines for each recombinant were genotyped with a marker on the heterozygous region of qLTG3a and used to evaluate LTG. The data of genotype and phenotype were employed in a mark-trait association analysis to fine map qLTG3a. The primer sequence used in fine mapping can be found in supplementary data (Additional file 1: Table S2).
Marker-Trait Association Analysis and Estimation of Genetic Effects
The marker-trait association and genetic effects of qLTG3a on LTG were examined using the SAS program (SAS Institute 2011), as previously reported by Ye et al. (2015). Data from the recombinant-derived progeny populations were analyzed by line correlation analysis to determine the mark-trait associations at the qLTG3a locus. Genotypes for a marker locus were coded as 1, 2, and 3 for HJX74-like homozygote, heterozygote, and S6-like homozygote, respectively, to correlate with the trait value for LTG. Additive and dominance effects of qLTG3a on LTG were assessed using the linear regression model 1: yij = μ + αx + dz + εij, where yij is the phenotypic value for the jth plant of the ith marker genotype; μ is the model mean; x is the dummy variable for the additive component and was coded as -1, 0, and 1 when i = 1, 2, and 3, respectively; z is the dummy variable for the dominance component and was coded as 0, 1, or 0 when i = 1, 2, or 3; a and d are regression coefficients that estimate the additive and dominance effects, respectively; and εij is the error term of the model.
Candidate Gene Identification
Fine mapping and BSA-seq data were comprehensively used to dig the candidate genes underlying qLTG3a. Firstly, the possible candidate genes in the QTL region were chosen according to the functional annotations from the Rice Annotation Project Database (RAP-DB, https://rapdb.dna.affrc.go.jp). Then, based on the whole genome resequencing data by BSA-seq, we further focused on those candidate genes with nonsynonymous SNPs, or InDel mutations in the upstream and exonic regions between the two parents, S6 and HJX74. Finally, those genes that are abundantly or predominantly expressed in seeds were considered as key candidate genes.
RNA Extraction and qRT-PCR Analysis
To verify the temporal and spatial expression patterns of candidate genes, qRT-PCR was conducted with different rice organs of HJX74, including developing seeds at 20 and 30 DAF, mature roots, stems and leaves, and 3-day-old seedlings. Total RNA was isolated from the above tissues by using the RaPure Plant RNA Kit (Magen) according to the manufacturer’s instructions. First-strand cDNA was synthesized from the total RNA with the Evo M-MLV RT Kit with gDNA Clean for qPCR (Accurate biology). qRT-PCR was performed on a CFX 96 (Bio-Rad) in a 20 μl system containing 2 μl of cDNA, 0.4 μl of gene-specific primers (10 μM), 10 μl of SYBR Green Pro Taq HS Premix (Accurate biology), and 7.2 μl of RNase-free water according to the manufacturer’s protocol. The PCR program was 95 °C denaturation for 30 s followed by 40 cycles of 95 °C for 5 s and 60 °C for 30 s. The specificity of the PCR amplification was checked with dissociation curves (65–95 °C) following the final cycle of the PCR. OsActin1 (Os03g0718100) was used as an internal reference for standardization of cDNA amounts. Primers for qRT-PCR are listed in Additional file 1: Table S2. Data represent three biological replicates and three technical replicates.
Germination Assays
To investigate the role of qLTG3a responding to other stresses, the germination performances of S6 and HJX74 were analyzed in various environmental stresses. Each treatment was biologically repeated three times. About 50 seeds for each replicate were placed in 9-cm Petri dishes and cultured under normal temperature (30 °C), high temperature (38 °C), osmotic stress (3% mannitol, 30 °C), and high-salt stress (200 mM NaCl, 30 °C). The germinated seeds were counted daily for one to seven days, depending on germination profiles under different stress conditions.
Results
Phenotypic Variation of LTG in SSSLs
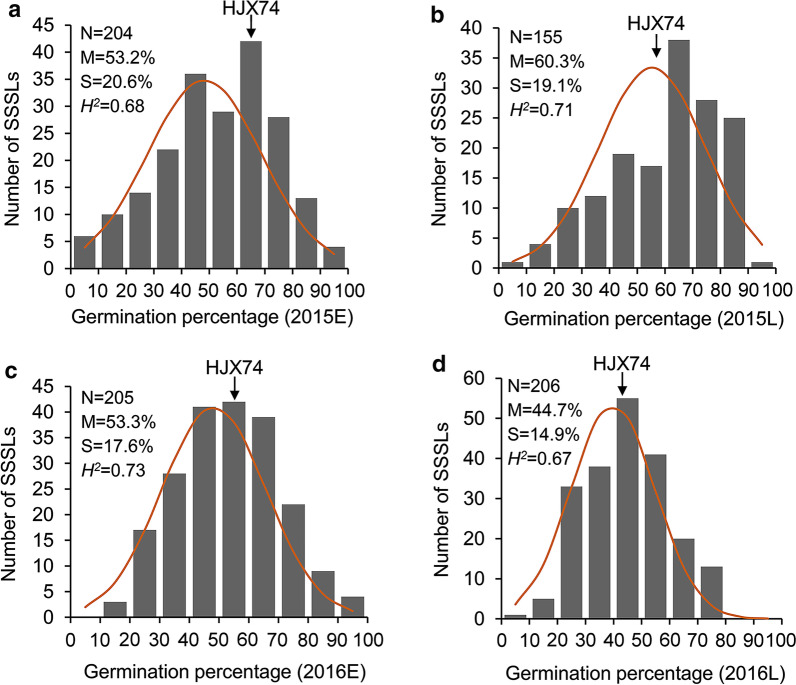
The germination percentages at 15 °C of HJX74 and 208 SSSLs were investigated to evaluate the phenotypic variation of LTG in four growing seasons from 2015 to 2016. The recipient parent HJX74 was planted in 10, 9, 9, and 11 blocks, and the average LTG value was 67.7 ± 1.4, 57.4 ± 1.3, 55.3 ± 1.4, and 44.1 ± 1.2% in 2015E, 2015L, 2016E, and 2016L, respectively (Fig. 1). In addition, the mean LTG values of HJX74 in different blocks of each cropping season were analyzed by one-way ANOVA in 2015 and 2016. The results showed no significant difference in each cropping season, indicating the field environment among different blocks was relatively consistent (Additional file 1: Table S3). On the other hand, the LTG values in SSSLs ranged from 2.8 to 98.1, 9.8–90.8, 12.7–93.4 and 8.7–79.3%, with an average of 53.2, 60.3, 53.3 and 44.7% in 2015E, 2015L, 2016E and 2016L, respectively (Fig. 1). Our data proved that LTG is a typical quantitative trait, as the low-temperature germination percentages in SSSLs displayed wide variations and normal distributions in multiple environments. As shown in Fig. 1, the broad-sense heritability (H2) of LTG was 0.68, 0.71, 0.73, and 0.67 in 2015E, 2015L, 2016E, and 2016L, respectively; the combined broad-sense heritability was 0.71 across four environments. These data indicated that genetic factors mainly contributed to the phenotypic variations observed in SSSLs.
Fig. 1.
Phenotypic distribution of LTG for 208 rice SSSLs in four cropping seasons. a 2015E, b 2015L, c 2016E, d 2016L. Parameters are the number of SSSLs (N) and mean (M) and standard deviation (S) for germination of the freshly harvested seeds at 15 °C. The arrow indicates the germination percentage of the recipient parent HJX74. Broad-sense heritability (H2) of LTG was estimated by the germination rates of SSSLs in each cropping season. 2015E, 2015L, 2016E, and 2016L represent the early (E) and late (L) cropping seasons in 2015 and 2016, respectively
Identification of SSSLs with Putative LTG QTLs
For QTL analysis, the LTG values of each SSSL and the recipient parent HJX74 were analyzed by Student's t test. The results demonstrated that 24 SSSLs displayed significantly different phenotypic values in comparison to HJX74 (P < 0.001), implying that these SSSLs carry putative LTG QTLs on their corresponding substituted segments (Additional file 1: Fig. S2; Table S4).
As shown in Table 1, the 24 SSSLs originated from 10 donor parents, including Tetep (W01), Zihui 100 (W05), Katy (W06), IR64 (W08), Basmati 385 (W09), Basmati 370 (W11), Lianjian33 (W14), Ganxiangnuo (W17), Lemont (W23), and IAPAR9 (W27), of which Katy contributed 12 or half of the SSSLs. The substituted segments of the 24 SSSLs were distributed on ten chromosomes except 10 and 12, and their lengths ranged from 6.5 to 54.5 cM (Table 1). It should be pointed out that some of the 24 SSSLs shared overlapping segments, and they were therefore considered to contain the same QTL of different alleles corresponding to their donor parents. Accordingly, seven positive-effect QTLs, qLTG2, qLTG3a, qLTG5, qLTG6, qLTG8a, qLTG8b, and qLTG11a, were identified on the substituted segments of 14 SSSLs, with additive effects ranging from 0.06 to 0.18 and additive effect contributions ranging from 8.9 to 36.2% (Table 1). In contrast, seven negative-effect QTLs, qLTG1, qLTG3b, qLTG4, qLTG7a, qLTG7b, qLTG9, and qLTG11b, were detected on the substituted segments of 10 SSSLs. The additive effects were between − 0.10 and − 0.24, and additive effect contributions were between 19.2 and 39.9% (Table 1).
Table 1.
Summary of the SSSLs detected with putative LTG QTLs in four field environments
| SSSL code | Donor code | Chr | Substituted segmenta | Length (cM) | Putative QTL | a* | R2 (%)* | Cropping seasonb | Detected seasonc |
|---|---|---|---|---|---|---|---|---|---|
| S1 | W14 | 1 | RM246–RM297-RM212–RM315 | 16.7 | qLTG1 | − 0.18 ± 0.02 | 28.6 ± 7.0 | abcd | ab |
| S2 | W14 | 1 | RM5–RM237-RM212–RM315 | 34.2 | qLTG1 | − 0.17 ± 0.10 | 29.4 ± 11.8 | abcd | acd |
| S3 | W09 | 2 | RM106–RM263-RM530–RM240 | 30.2 | qLTG2 | 0.10 ± 0.04 | 20.0 ± 4.5 | abcd | bd |
| S4 | W08 | 2 | PSM122–RM263-RM166–RM213 | 54.5 | qLTG2 | 0.11 ± 0.06 | 18.9 ± 10.9 | abcd | bc |
| S5 | W06 | 3 | RM175–RM489-PSM428–PSM429 | 16.1 | qLTG3a | 0.16 ± 0.02 | 33.0 ± 1.0 | acd | cd |
| S6 | W06 | 3 | RM231–RM489-RM218–RM563 | 28.7 | qLTG3a | 0.16 ± 0.01 | 29.8 ± 7.8 | acd | acd |
| S7 | W06 | 3 | RM489–RM545-RM218–RM563 | 21.9 | qLTG3a | 0.18 ± 0.01 | 36.2 ± 5.4 | acd | cd |
| S8 | W17 | 3 | PSM127–RM426-RM168–RM186 | 16.4 | qLTG3b | − 0.10 ± 0.01 | 19.2 ± 5.8 | abcd | ad |
| S9 | W06 | 4 | RM119–RM273-RM252–RM241 | 12.9 | qLTG4 | − 0.14 ± 0.01 | 25.8 ± 7.0 | acd | ad |
| S10 | W06 | 4 | PSM194–RM273-RM252–RM241 | 16.2 | qLTG4 | − 0.16 ± 0.08 | 26.9 ± 6.6 | acd | ad |
| S11 | W06 | 4 | PSM358–PSM196-RM252–RM241 | 47.0 | qLTG4 | − 0.20 ± 0.13 | 32.9 ± 13.8 | acd | ad |
| S12 | W11 | 5 | RM480–RM31-PSM386–End | 8.4 | qLTG5 | 0.15 ± 0.02 | 28.0 ± 1.5 | abcd | bcd |
| S13 | W01 | 5 | PSM384–RM178-RM334–End | 22.4 | qLTG5 | 0.14 ± 0.01 | 28.7 ± 2.2 | bcd | cd |
| S14 | W27 | 6 | End–RM508-RM217–RM253 | 16.6 | qLTG6 | 0.06 ± 0.01 | 8.9 ± 0.5 | acd | ac |
| S15 | W23 | 6 | End–RM508-RM121–RM136 | 44.9 | qLTG6 | 0.15 ± 0.02 | 29.9 ± 9.9 | abcd | bd |
| S16 | W06 | 7 | RM481–PSM142–PSM143 | 10.0 | qLTG7a | − 0.22 ± 0.09 | 39.4 ± 3.7 | acd | ad |
| S17 | W05 | 7 | PSM142–RM2-RM10–PSM353 | 45.4 | qLTG7b | − 0.24 ± 0.08 | 39.9 ± 8.9 | abcd | abc |
| S18 | W06 | 8 | PSM155–RM547–PSM393 | 6.5 | qLTG8a | 0.15 ± 0.02 | 26.9 ± 4.4 | abcd | bc |
| S19 | W06 | 8 | RM284–RM556-RM210–RM80 | 9.1 | qLTG8b | 0.17 ± 0.03 | 33.6 ± 1.3 | abcd | cd |
| S20 | W06 | 8 | PSM394–RM515-OSR7–RM447 | 36.4 | qLTG8b | 0.15 ± 0.01 | 28.3 ± 2.4 | abcd | bcd |
| S21 | W27 | 9 | RM105–RM434-RM410–RM257 | 17.1 | qLTG9 | − 0.21 ± 0.01 | 34.2 ± 6.0 | abcd | ac |
| S22 | W06 | 11 | PSM409–PSM412–PSM414 | 28.8 | qLTG11a | 0.16 ± 0.01 | 32.2 ± 4.3 | abcd | bd |
| S23 | W06 | 11 | PSM174–PSM175-RM536–PSM415 | 40.2 | qLTG11a | 0.15 ± 0.03 | 30.3 ± 9.8 | abcd | cd |
| S24 | W17 | 11 | RM21–PSM365-PSM417–PSM460 | 15.4 | qLTG11b | − 0.19 ± 0.07 | 34.1 ± 9.7 | acd | acd |
*a = average additive effect of the QTL in different detected seasons; R2 (%) = average additive effect contribution (%) of the QTL in different detected seasons
aThe single lines in the middle of markers indicate substituted segments while the double lines on the border of markers indicate segments recombination might appear. SSR markers labeled "RM" were selected from public resources, and those labeled "PSM" were developed in our laboratory
b,cThe letters a, c represent early cropping seasons in 2015 and 2016, and b, d represent late cropping seasons in 2015 and 2016, respectively. Detected seasons mean the time when putative QTLs were detected on the SSSLs
Most of these QTLs can be simultaneously detected at the early and late growing seasons, and qLTG1 (S2), qLTG3a (S6), qLTG5 (S12), qLTG7b (S17), qLTG8b (S20), qLTG11b (S24) were repeatedly detected in three seasons, indicating that these QTLs were stable in different environments. Nevertheless, qLTG6 (S14) and qLTG9 (S21) were only identified in the early cropping seasons, while qLTG2 (S3), qLTG6 (S15), and qLTG11a (S22) were specifically detected in the late cropping seasons (Table 1).
Substitution Mapping of LTG QTLs
Five QTLs, qLTG3b, qLTG7a, qLTG8a, qLTG9, and qLTG11b, were detected on a single SSSL. They were located on their corresponding substituted segments with an interval of 6.5–17.1 cM (Table 2). The qLTG7b was detected on a single SSSL (S17), but it was not detected on S28 which overlapped with S17, implying that qLTG7b was located in the non-overlapping interval of S17 and S28 (Fig. 2; Table 2). However, the remaining eight QTLs were detected simultaneously on multiple SSSLs with overlapping fragments. Therefore, we conducted substitution mapping to integrate these QTLs and reduce their location intervals to smaller genetic regions (Fig. 2; Table 2).
Table 2.
Integrated LTG QTLs after substitution mapping
| QTL | Donor code | Chr | Interval (cM) | Length (cM) | a* | R2 (%)* |
|---|---|---|---|---|---|---|
| qLTG1 | W14 | 1 | 123.1–139.8 | 16.7 | − 0.17 ± 0.01 | 29.0 ± 0.6 |
| qLTG2 | W08, W09 | 2 | 102.7–132.9 | 30.2 | 0.11 ± 0.01 | 19.5 ± 0.8 |
| qLTG3a | W06 | 3 | 22.7–29.3 | 6.6 | 0.17 ± 0.01 | 33.0 ± 3.2 |
| qLTG3b | W17 | 3 | 108.6–125.0 | 16.4 | − 0.10 ± 0.01 | 19.2 ± 5.8 |
| qLTG4 | W06 | 4 | 67.7–80.6 | 12.9 | − 0.16 ± 0.03 | 28.5 ± 3.8 |
| qLTG5 | W01, W11 | 5 | 113.9–116.5 | 2.6 | 0.14 ± 0.01 | 28.4 ± 0.5 |
| qLTG6 | W23, W27 | 6 | 0.7–17.3 | 16.6 | 0.10 ± 0.07 | 19.5 ± 15.0 |
| qLTG7a | W06 | 7 | 18.5–28.5 | 10.0 | − 0.22 ± 0.09 | 39.4 ± 3.7 |
| qLTG7b | W05 | 7 | 55.8–80.4 | 24.6 | − 0.24 ± 0.08 | 39.9 ± 8.9 |
| qLTG8a | W06 | 8 | 36.5–43.0 | 6.5 | 0.15 ± 0.02 | 26.9 ± 4.4 |
| qLTG8b | W06 | 8 | 82.6–91.7 | 9.1 | 0.16 ± 0.01 | 31.0 ± 3.7 |
| qLTG9 | W27 | 9 | 48.0–65.1 | 17.1 | − 0.21 ± 0.01 | 34.2 ± 6.0 |
| qLTG11a | W06 | 11 | 39.8–59.3 | 19.5 | 0.15 ± 0.01 | 31.3 ± 1.3 |
| qLTG11b | W17 | 11 | 81.9–97.3 | 15.4 | − 0.19 ± 0.07 | 34.1 ± 9.7 |
*a = average additive effect of the QTL from different SSSLs; R2 (%) = average additive effect contribution (%) of the QTL from different SSSLs
Fig. 2.
Substitution mapping of LTG QTLs in rice. The long horizontal dark bars represent the chromosomes with the molecular markers above them. The short black horizontal bars coded from S1 to S24 represent the SSSLs detected with QTLs in at least two cropping seasons, while the gray ones coded from S25 to S28 represent the SSSLs not detected with a QTL in all cropping seasons. The intervals located LTG QTLs are shown between two vertical dotted lines with the names of QTLs under the substituted segments
qLTG1 was detected simultaneously on S1 and S2; thus, it was located in the overlapping region in the vicinity of RM297-RM212 with an interval length of 16.7 cM. S3 and S4 carried qLTG2, which was delimited to a 30.2 cM overlapping region flanked by RM263 and RM530. qLTG3a was detected simultaneously on S5, S6, and S7, but not on S25 and S26, which shared overlapping fragments with the three former SSSLs. qLTG3a was hence located in the vicinity of RM545 with an interval length of 6.6 cM. qLTG4 was detected simultaneously on S9, S10, and S11. Thus, it was mapped in the overlapping region in the vicinity of RM273 with an interval length of 12.9 cM. qLTG5 was detected simultaneously on S12 and S13, but not on S27, which shared an overlapping region with the other two SSSLs, indicating that qLTG5 was located in the vicinity of RM480 and RM31 with an interval length of 2.6 cM. qLTG6 was detected simultaneously on S14 and S15, and thus it was located in the overlapping region RM508-RM217 with an interval length of 16.6 cM. qLTG8b was detected simultaneously on S19 and S20, and it was located in the overlapping region in the vicinity of RM210 with an interval length of 9.1 cM. S22 and S23 were demonstrated to carry qLTG11a on their overlapping segments in the area of PSM410-PSM412 with an interval length of 19.5 cM.
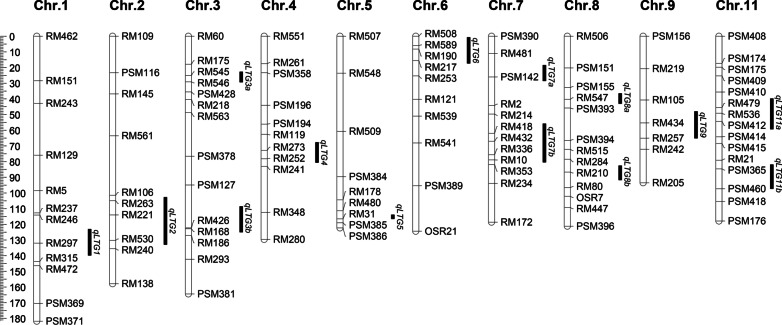
By substitution mapping, 14 integrated LTG QTLs were mapped on ten chromosomes except on chromosomes 10 and 12 (Table 2; Fig. 3). Their locations were narrowed down to shorter intervals ranging from 2.6 to 30.2 cM. The additive effects changed from -0.24 to 0.17, and the additive effect contributions varied from 19.2 to 39.9%. qLTG2, qLTG3a, qLTG5, qLTG6, qLTG8a, qLTG8b, and qLTG11a were positive QTLs with excellent alleles for LTG, while the other seven QTLs were detected with negative effects. It is worth noting that the additive effect contributions of qLTG3a, qLTG8b, and qLTG11a are higher than 30%, so they are the main LTG QTLs with utilization values (Table 2).
Fig. 3.
Chromosome location of 14 QTLs for LTG in rice. The linkage map of QTLs was constructed by using MapChart 2.2 (Voorrips 2002). SSR markers are indicated on the right of the chromosomes. Genetic distance (cM) is shown as rulers on the left margin. Black bars on each chromosome's right are the location intervals of QTLs for LTG with their names on the right. Chr chromosome
Validation and Genetic effect of a Major QTL qLTG3a
It indicated that qLTG3a was a stable and major LTG QTL, and S6 was selected as the representative SSSL of qLTG3a for further study. During seed germination, S6 with the tolerant allele "A" exhibited significantly better low-temperature germination performance than the recipient parent HJX74 carrying the sensitive allele "a", including higher germination rate, faster germination speed, and stronger radicles and plumules (Fig. 4a, b). Moreover, the role of qLTG3a on seed development was investigated at the seed maturation stage. As expected, the tolerant allele "A" could remarkably promote the low-temperature germination ability of seeds developing for 25–40 days compared with the sensitive allele "a" (Fig. 4c).
Fig. 4.
Validate a major LTG QTL, qLTG3a, by an F2 population and BSA sequencing. "aa" represents the HJX74 allele of qLTG3a, and "AA" represents the S6 allele of qLTG3a. Roles of qLTG3a in LTG during seed germination (a) and seed development (c). Dots and bars represent the means and standard deviations of LTG at each time point. Germination data were calculated based on at least four biological replicates during seed germination and nine biological replicates during seed development. b Representative germination images showing different LTG phenotypes between HJX74 and S6. Pictures were taken on the 5th, 7th, and 10th day after sowing. Bar = 1 cm. d Frequency distribution and genetic effects of qLTG3a in a 356-progeny population derived from the cross of HJX74 and S6. The donor parent of S6 is Katy. LTG was evaluated on the 7th day after sowing. The genotypes of individual plants were determined by the InDel P7 marker. The additive effects (a), dominance effects (d), and proportion of the variance explained by the QTL (R2) were estimated based on Model 1. A positive a or d value indicates that the "A" allele increased LTG. * for P < 0.0001 and ns for P ≥ 0.05. e Manhattan plot shows the distribution of SNP-index and ΔSNP-index on 12 chromosomes. The reference genome for BSA sequencing is the indica variety R498 (Shuhui498). The blue and red lines represent 95 and 99% confidence intervals, respectively. The black line shows the ΔSNP-index value of fitting results
To evaluate the genetic effect of qLTG3a, a 356-plants F2 population segregating at the QTL was developed from a cross between S6 and HJX74. Normal distribution was observed in the segregating population (Fig. 4d). The average LTG values of plants with genotypes AA, Aa, and aa were 78, 65, and 52%, respectively, and were significantly different. The numbers of plants with genotypes AA, Aa, and aa were 85, 173, 98, respectively, which accorded with the segregation ratio of 1:2:1 (χ2 = 1.23 < χ20.05,2 = 5.99) (Fig. 4d). A significant positive additive effect of qLTG3a was detected in the segregating population with a phenotypic contribution of 30.6%. However, the dominance effect of qLTG3a was not identified, suggesting it is a purely additive allele for LTG (Fig. 4d). Moreover, qLTG3a was further confirmed by BSA-seq using the above F2 segregation population (Fig. 4e). At the 95% significance level, only one QTL interval was identified on chromosome 3 by the ΔSNP-index method after the whole genome screening. The corresponding position on the R498 genome was Chr.3: 2800001–9600001 bp, a 6.8-Mb region, which is completely located on the substitution segment of S6 (Fig. 4e). Therefore, this QTL detected by BSA-seq was considered as the same one of qLTG3a identified by the conventional method.
Fine Mapping of qLTG3a
Five recombinants selected from the F2 population with crossovers between the flanking markers of qLTG3a were advanced to the F3 generation to conduct a progeny test (Fig. 5). The partial physical map was constructed with 14 new InDel markers, which were applied to determine the recombinants' points of crossovers. Progeny populations (70–86) were genotyped by P1, P8, or P13 segregating in the corresponding populations and phenotyped by testing LTG.
Fig. 5.

Fine mapping of qLTG3a. Five recombinants (S6_R1 to R5) derived from S6_Aa were selected to delimit the locus of qLTG3a to a 332.7-kb region flanked by M6026 and M6341. The dotted vertical lines indicate the target region of qLTG3a. The partial physical maps of chromosome 3 were constructed based on the Nipponbare reference genome (IRGSP-1.0). Black and white bars represent the chromosomal segments from S6 and HJX74, respectively. N, the number of plants in a recombinant-derived progeny population; r, marker-trait correlation coefficients for LTG. A significant r value (* for P < 0.01 and ns for P ≥ 0.05) indicates that the allele of S6 on the marked heterozygous region enhanced LTG
Significant associations between marker genotypes and trait values of LTG were only detected in S6_R1 and S6_R3 progeny populations, with r = 0.56 to 0.70 (Fig. 5). Meanwhile, genetic analysis confirmed that the narrowed qLTG3a-containing interval had significant additive effects in S6_R1 and S6_R3 progeny populations, and also a minor dominance effect in the S6_R3 progeny population, which together accounted for 33% to 53% of the phenotypic variances (Additional file 1: Table S5). Besides, the absence of marker-trait association and the significant additive effect of qLTG3a was observed in the progeny lines of S6_R2, S6_R4, and S6_R5 (Fig. 5; Additional file 1: Table S5). According to the substitution mapping method, qLTG3a was narrowed down to a 332.7-kb heterozygous region of S6_R1 and S6_R3 flanked by M6026 and M6341.
Candidate Genes Underlying qLTG3a
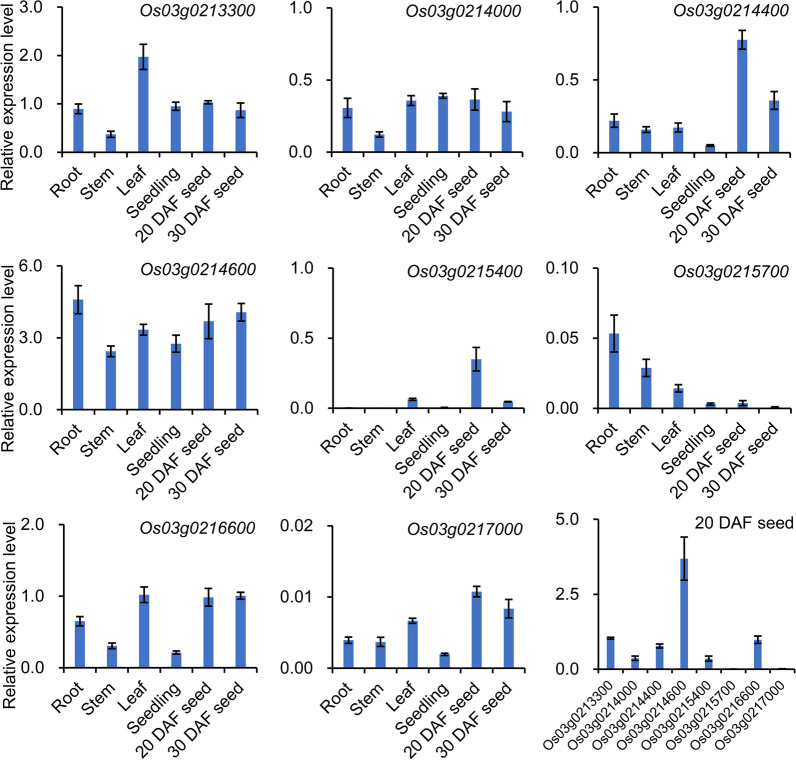
Within the 332.7 kb region of qLTG3a, 40 annotated genes were identified based on the RAP-DB database (https://rapdb.dna.affrc.go.jp) (Table 3). Among these genes, 11 genes were predicted as hypothetical proteins or genes which were excluded from the list of candidate genes, while the other 29 genes showed functional annotations. In addition, BSA-seq data were used to reveal the sequence variations, and only the nonsynonymous SNPs and InDels (ΔSNP/InDel-index > 0.8) in the promoter and coding regions were selected for further analysis. Under this standard, among the 29 genes, a total of 33 sequence variations were detected between the two parents of S6 and HJX74, corresponding to 16 candidate genes (Table 3, Additional file 1: S7, S8). To further reduce the number of candidate genes, we analyzed the temporal and spatial expression patterns of the above 16 genes. Gene expression analysis by RiceXPro indicated that four genes, Os03g0213300, Os03g0214000, Os03g0214400, and Os03g0214600, were highly expressed in seeds (Table 3; Additional file 1: Fig. S3). Moreover, similar expression patterns were obtained after we investigated the expression of eight genes by qRT-PCR (Fig. 6). In mature developing seeds (20-30 DAF), Os03g0213300, Os03g0214400, Os03g0214600, and Os03g0216600 showed high expression levels, Os03g0214000 and Os03g0215400 displayed medium expression levels, while the expression levels of Os03g0215700 and Os03g0217000 were low (Fig. 6). Since Os03g0216600 has no sequence variation, Os03g0213300, Os03g0214400, and Os03g0214600 were considered as the final possible candidate genes responsible for LTG.
Table 3.
Function annotation, sequence variation and gene expression of candidate genes in the qLTG3a region
| Gene RAP IDa | Gene function b | Indel number* | SNP number* | Gene expression level in seeds c |
|---|---|---|---|---|
| Os03g0213300 | Phosphopantetheine attachment site domain containing protein; Hypothetical conserved gene | 1 | 3 | High |
| Os03g0213400 | Similar to RNA helicase | Medium | ||
| Os03g0213500 | Conserved hypothetical protein | High | ||
| Os03g0213600 | Conserved hypothetical protein | 3 | 3 | Medium |
| Os03g0213700 | DUF647 domain containing protein | 1 | Medium | |
| Os03g0213800 | Mitochondrial substrate carrier family protein | 2 | Medium | |
| Os03g0213900 | Conserved hypothetical protein; ROUGH SHEATH2-interacting KH-domain protein | 3 | 3 | Medium |
| Os03g0214000 | Diphosphonucleotide phosphatase 1 precursor; nucleotide pyrophosphatase | 1 | High | |
| Os03g0214050 | Hypothetical protein | no data | ||
| Os03g0214100 | Replication protein A1 | 1 | Low | |
| Os03g0214200 | Mediator of OsbZIP46 deactivation and degradation, Negative regulation of ABA signaling and drought tolerance; Ninja-family protein 1 | Medium | ||
| Os03g0214400 | Digalactosyldiacylglycerol synthase 2 | 4 | High | |
| Os03g0214600 | 26S proteasome subunit RPN9a; 26S proteasome non-ATPase regulatory subunit 13 | 2 | High | |
| Os03g0214900 | Conserved hypothetical protein | 4 | 2 | Medium |
| Os03g0215000 | Integral membrane family protein | High | ||
| Os03g0215200 | Putative transcription factor, Carpel specification, Midrib formation | Low | ||
| Os03g0215400 | MADS-domain-containing protein, sexual reproduction | 1 | Medium | |
| Os03g0215600 | Zinc finger, CCHC retroviral-type domain containing protein | Very low | ||
| Os03g0215700 | Myosin II heavy chain-like family protein | 3 | 1 | Very low |
| Os03g0215800 | Pyridoxal phosphate-dependent enzyme | 1 | Low | |
| Os03g0215900 | 67kD chloroplastic RNA-binding protein, P67 | Low | ||
| Os03g0216000 | Zinc-finger protein KNUCKLES | Very low | ||
| Os03g0216300 | Pentatricopeptide repeat domain containing protein; Hypothetical conserved gene | 1 | Low | |
| Os03g0216400 | Pentatricopeptide repeat domain containing protein | Low | ||
| Os03g0216500 | Conserved hypothetical protein | 1 | Low | |
| Os03g0216600 | Alpha-glucosidase (EC 3.2.1.20) | High | ||
| Os03g0216700 | Citrate transporter, Efficient translocation of Fe | 1 | Medium | |
| Os03g0216733 | Hypothetical protein | no data | ||
| Os03g0216766 | Hypothetical gene | no data | ||
| Os03g0216800 | Polygalacturonase B (Fragment) | 1 | 1 | Very low |
| Os03g0216900 | Prefoldin domain containing protein | Low | ||
| Os03g0217000 | Inhibin beta B chain precursor | 1 | Low | |
| Os03g0217200 | Cyclin-like F-box domain containing protein | Medium | ||
| Os03g0217400 | Conserved hypothetical protein | Low | ||
| Os03g0217801 | Hypothetical gene | no data | ||
| Os03g0217900 | Hypothetical protein | Medium | ||
| Os03g0218100 | Sec63 domain containing protein | no data | ||
| Os03g0218200 | F-box domain containing protein | 1 | Low | |
| Os03g0218300 | Conserved hypothetical protein | 3 | Medium | |
| Os03g0218400 | Similar to Hexose transporter | Low |
*Sequence variation analysis of candidate genes was derived from BSA sequencing data. The number of InDels was derived from the upstream and exonic regions of the candidate genes, and the number of SNPs was derived from the nonsynonymous SNPs in the exonic regions. The ΔSNP-index and ΔInDel-index were greater than 0.8
a,bRAP ID and gene function are based on the Rice Annotation Project Database (RAP-DB): http://rapdb.dna.affrc.go.jp
cThe expression level of candidate genes in seeds was analyzed by the public tool RiceXPro (http://ricexpro.dna.affrc.go.jp/) and divided into four levels
Fig. 6.
Expression patterns of the candidate genes underlying qLTG3a in various tissues. Root, stem, and leaf were collected at the reproductive stage, and seedlings were three days old. The root sample of Os03g0213300 was set as control sample for all genes. Transcript levels were assessed by qRT-PCR using OsActin1 as an internal control. Data are means ± SD (n = three biological replicates). DAF, days after flowering
qLTG3a Enhances Seed Germination Vigor Under Optimal Temperature and Osmotic Stress
To comprehensively reveal the role of qLTG3a, we analyzed the germination performance of S6 with tolerant allele "A" and HJX74 with sensitive allele "a" responding to different environmental conditions in two cropping seasons (Fig. 7). In addition to low-temperature stress, the tolerant allele "A" also promoted seed germination ability under optimal temperature (30℃) and osmotic pressure (3% Mannitol) compared with the sensitive allele "a". At 30℃, the germination rates of S6_AA were significantly higher than that of HJX74_aa at 1 to 1.5 d in 2016E and 1.5 d in 2016L (Fig. 7a, b). Similar germination behavior was also observed after treatment with 3% Mannitol. For example, the germination rates of S6_AA at 1.5 d in 2016E and 2016L were 45.3% and 44.6%, respectively. By contrast, the corresponding germination rates of HJX74_aa were only 19.0% and 23.3%, respectively (Fig. 7g, h). However, S6_AA and HJX74_aa showed similar germination patterns under 38℃ high temperature and 200 mM-NaCl stress, indicating that qLTG3a did not work under these two adversities (Fig. 7c–f).
Fig. 7.
Roles of qLTG3a in various environmental stress tolerance. a, b normal temperature, c, d high temperature, e, f high-salt stress, g, h osmotic stress. "aa" represents the sensitive allele of qLTG3a from HJX74 and "AA" represents the tolerant allele of qLTG3a from S6. At each time point, columns and bars represent the means and standard deviations of germination rates calculated based on three biological replicates. The significance of differences was calculated using Student's t test. *P < 0.05 and **P < 0.01. 2016E, early cropping season in 2016; 2016L, late cropping season in 2016. Bars = 1 cm
Discussion
Low temperatures limit the geographical distribution and planting time of crops, thus affecting food production. Reduced freshwater resources and increasing labor costs have driven the rise of direct-seeded rice in China and worldwide (Lv et al. 2020; Mahender et al. 2015). Genetic improvement of LTG for current rice varieties is a crucial strategy to ensure direct-seeded rice success. As previously reported, LTG is a typical quantitative trait influenced by genetic factors and environmental cues, such as temperature and heading stage (Fujino et al. 2008; Jiang et al. 2017; Li et al. 2013; Wang et al. 2018b, c; Xie et al. 2014). An important prerequisite for QTL mapping is to control environmental factors to improve the accuracy of phenotypic identification. Due to the similar genetic background, the SSSL population showed uniform heading dates, and thereby the seeds can be harvested on the same days after heading to keep the consistent maturity state. Meanwhile, only those SSSLs that can repeatedly detect QTLs in two or more seasons will be selected for subsequent QTL analysis to reduce environmental impact (Table 1).
In this study, 208 multi-donor-derived rice SSSLs were used to conduct an integrated QTL analysis of LTG by substitution mapping over four planting seasons. As expected, a total of 14 LTG QTLs were identified (Table 2; Fig. 3), further demonstrating that SSSL is a powerful tool for the analysis of complex agronomic traits (Wang et al. 2012; Wu et al. 2020; Zhou et al. 2017; Zhu et al. 2018). By comparing chromosome positions and molecular markers, 11 LTG QTLs mapped in this study were similar to those of previous QTLs, which proves the reliability of this experimental system. Most of the QTLs are stably identified in both early and late cropping seasons, especially qLTG1 (S2), qLTG3a (S6), qLTG5 (S12), qLTG7b (S17), qLTG8b (S20), qLTG11b (S24) that were detected in three cropping seasons (Table 1). Interestingly, some QTLs were identified only in early or late cropping seasons, such as qLTG6 (S14), qLTG9 (S21), and qLTG2 (S3), qLTG6 (S15), qLTG11a (S22) separately (Table 1). The season-specific QTLs may correlate with environment temperature and can be applied in specific ecological areas. It is worth noting that qLTG6 derived from S14 or S15 was specifically detected in the early or late planting seasons, respectively, showing distinct expression patterns. This implies that multiple alleles from different donor sources at the qLTG6 locus have an apparent seasonal preference in expression. It is generally believed that japonica rice, mainly distributed in temperate regions, is more tolerant to low temperatures than indica rice widely distributed in tropical and subtropical areas due to habitat adaptability (Jiang et al. 2017; Yang et al. 2019). Consistently, a total of 14 SSSLs with positive-effect QTLs were identified, of which eight SSSLs were derived from a japonica cultivar Katy (W06), corresponding to four QTLs, qLTG3a, qLTG8a, qLTG8b, qLTG11a, respectively (Table 1, Additional file 1: S1). This result indicates that Katy has abundant cold tolerance genes, strengthening our understanding that japonica varieties have excellent cold tolerance.
Four QTLs, qLTG7b, qLTG8b, qLTG9 and qLTG11b, displayed large additive effect contributions of 39.9%, 31.0%, 34.2% and 34.1%, respectively (Table 2). However, only qLTG8b had a positive additive effect, and it corresponds well to qLTG-8-1 (Jiang et al. 2005), which were linked to the same marker RM210. Li et al. (2013) fine-mapped qLTG-9 to a 72.3-kb region between makers L9-25d and ID-1, which was located on the segment of RM105-RM434 of qLTG9 identified in this study (Fig. 3). Noticeably, three novel QTLs, qLTG3a, qLTG7a, and qLTG11a, were identified for the first time in this study. qLTG3a and qLTG11a displayed positive additive effects and large additive effect contributions of 33% and 31%, respectively (Table 2). Also, qLTG3a can be detected repeatedly in different seasons and multiple SSSLs (Table 1). According to the distinctly different chromosomal locations, it is evident that there is no correlation between qLTG3a identified in this study and the first map-based cloned gene qLTG3-1 reported by Fujino et al. (2008). Because the former is located at approximately 25 cM of chromosome 3, closely linked to RM545 (Fig. 3), while the latter is located near the end of chromosome 3 in the vicinity of GBR3001 (Fujino et al. 2004). Therefore, qLTG3a represents a new, main-effect, stably expressed LTG QTL, which can be used as a new candidate gene for breeders to cultivate elite varieties with LTG traits.
To verify qLTG3a, we further examined whether it could enhance LTG during seed germination and development. As expected, S6 carrying the "AA" tolerance gene showed significantly improved seed vigor under low-temperature conditions (Fig. 4a–c). Subsequently, we estimated the genetic effects of qLTG3a through an F2 segregating population and confirmed the location of qLTG3a by BSA-seq (Fig. 4d, e). Finally, qLTG3a was fine mapped to a 332.7-kb physical region by the progeny derived from five recombinants (Fig. 5). Forty candidate genes were revealed based on function annotation, and three most likely candidate genes were identified by gene sequence and expression analysis (Table 3; Fig. 6). Os03g0213300 encodes a phosphopantetheine attachment site domain containing protein, Os03g0214400 encodes a digalactosyldiacylglycerol synthase 2, and Os03g0214600 encodes a 26S proteasome subunit RPN9a. It lays a foundation for dissecting the molecular basis underlying qLTG3a. Currently, only two natural variations related to LTG, qLTG3-1 (Fujino et al. 2008) and OsSAP16 (Wang et al. 2018c), have been cloned by map-based strategy and GWAS, respectively. qLTG3-1 may reduce the mechanical resistance during seed germination by inducing programmed cell death in the covered tissues of the embryo (Fujino et al. 2008).
In addition to low temperature, qLTG3-1 also significantly promoted seed germination at the optimal temperature and responded to various stresses such as high salt, high osmolarity, and high temperature (Fujino et al. 2008). Similarly, we also found that qLTG3a identified in this study could also play a role under normal temperature and high osmotic stress, but did not respond to high temperature and high salt stresses (Fig. 7). Therefore, it is reasonable to speculate that plant cells have established some conservative mechanisms in response to low temperature and osmotic stress. Previous studies have shown that rice accumulates a large number of soluble sugars, including glucose, fructose, sucrose, hexose, raffinose, and trehalose under low-temperature stress, thereby improving the ability of cells to cope with osmotic stress (Liu et al. 2018). In the future, we will explore the possible relationship between qLTG3a and soluble sugars to enrich our understanding of the molecular mechanism of LTG.
In summary, we identified 14 QTLs for LTG by substitution mapping from 208 SSSLs in four seasons, fine mapped qLTG3a and identified three key candidate genes. All these results will help us dissect the genetic mechanisms of LTG and provide elite candidate genes to cultivate new high-vigor rice varieties with low-temperature germination resistance.
Supplementary Information
Additional file 1: Figure S1. Length (a) and genome coverage (b) of substituted segments in 208 rice single segment substitution lines (SSSLs). cM, centimorgan. Figure S2. Phenotypic variation of LTG between recipient parent HJX74 and different SSSLs. S6, S15, S22 were three representative SSSLs with good LTG values. Pictures were taken on the 6th, 8th, and 10th day after sowing. Bars=1 cm. Figure S3. Temporal and spatial expression profiles of candidate genes in qLTG3a region. All expression data were prepared by the public tool RiceXPro (http://ricexpro.dna.affrc.go.jp/). The grey boxes mean no expression data. The heat map was made by TBtools (v1.089). Table S1. Summary the donors and substituted segments distribution of 208 SSSLs. Table S2. The sequence of primers used in this study. Table S3. ANOVA analysis of the mean LTG values of HJX74 in different blocks of each cropping season. Table S4. LTG of HJX74 and 24 SSSLs at 15°C in four cropping seasons. Table S5. Summary of genotypic means for germination and genetic effects of markers segregating in progeny lines derived from selected recombinants for qLTG3a region. Table S6. The RAP ID, MSU ID and R498 (Shuhui498) ID of candidate genes. Table S7. Nonsynonymous SNP variations (ΔSNP-index > 0.8) of candidate genes in the qLTG3a locus. Table S8. Indel variations (ΔInDel-index > 0.8) in the upstream and exonic regions of candidate genes in the qLTG3a locus.
Acknowledgements
Not applicable.
Abbreviations
- BSA
Bulk segregant analysis
- GWAS
Genome-wide association study
- HJX74
Huajingxian 74
- InDels
Insertion-deletions
- LTG
Low-temperature germinability
- SNP
Single-nucleotide polymorphisms
- SSSL
Single segment substitution line
Authors' contributions
YLZ and GQZ designed the experiments. ZYP and GYC performed the experiments with the help of BT, RQZ, ML, RZZ, SKW, HTZ, GMZ, and WC. ZYP and YLZ analyzed the data. HY and GFL assisted in the statistical analysis. YLZ, BT, and ZYP wrote the manuscript. YLZ and GQZ revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31771888, 31301403, 31622041), the Natural Science Foundation of Guangdong Province (S2013040014992, 2017A030310271), and the Specialized Research Fund for the Doctoral Program of Higher Education (20134404120012).
Availability of Data and Materials
The datasets supporting the conclusions of this article are included within the article and its additional files.
Declarations
Ethics Approval and Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Competing interests
The authors declare that there are no conflicts of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zhaoyuan Pan and Bin Tan contributed equally to this work
Contributor Information
Guiquan Zhang, Email: gqzhang@scau.edu.cn.
Yuliang Zhou, Email: zhouyuliang@scau.edu.cn.
References
- Cao W, Wang Y, Tan B, Liu W, Chu L, Pan Z, Zhao G, Cao G, Zhou Y. Evaluation of seed germination vigor of rice cultivars under low temperature or hypoxic condition related with direct seeding rice. Mol Plant Breed. 2017;16:1–13. [Google Scholar]
- Chen L, Lou Q, Sun Z, Xing Y, Yu X, Luo L. QTL mapping of low temperature on germination rate of rice. Rice Sci. 2006;13:93–98. [Google Scholar]
- Ding Y, Shi Y, Yang S. Molecular regulation of plant responses to environmental temperatures. Mol Plant. 2020;13:544–564. doi: 10.1016/j.molp.2020.02.004. [DOI] [PubMed] [Google Scholar]
- Eshed Y, Zamir D. An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics. 1995;141:1147–1162. doi: 10.1093/genetics/141.3.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujino K, Matsuda Y. Genome-wide analysis of genes targeted by qLTG3-1 controlling low-temperature germinability in rice. Plant Mol Biol. 2010;72:137–152. doi: 10.1007/s11103-009-9559-x. [DOI] [PubMed] [Google Scholar]
- Fujino K, Sekiguchi H, Sato T, Kiuchi H, Nonoue Y, Takeuchi Y, Ando T, Lin SY, Yano M. Mapping of quantitative trait loci controlling low-temperature germinability in rice (Oryza sativa L.) Theor Appl Genet. 2004;108:794–799. doi: 10.1007/s00122-003-1509-4. [DOI] [PubMed] [Google Scholar]
- Fujino K, Sekiguchi H, Matsuda Y, Sugimoto K, Ono K, Yano M. Molecular identification of a major quantitative trait locus, qLTG3-1, controlling low-temperature germinability in rice. Proc Natl Acad Sci USA. 2008;105:12623–12628. doi: 10.1073/pnas.0805303105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujino K, Obara M, Shimizu T, Koyanagi KO, Ikegaya T. Genome-wide association mapping focusing on a rice population derived from rice breeding programs in a region. Breed Sci. 2015;65:403–410. doi: 10.1270/jsbbs.65.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Cheng J, He Y, Yang B, Cheng Y, Yang C, Zhang H, Wang Z. Influence of isopropylmalate synthase OsIPMS1 on seed vigour associated with amino acid and energy metabolism in rice. Plant Biotechnol J. 2019;17:322–337. doi: 10.1111/pbi.12979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji S, Jiang L, Wang Y, Zhang W, Liu X, Liu S, Chen L, Zhai H, Wan J. Quantitative trait loci mapping and stability for low temperature germination ability of rice. Plant Breed. 2009;128:387–392. doi: 10.1111/j.1439-0523.2008.01533.x. [DOI] [Google Scholar]
- Jiang X, Li C, Mao T. Mapping of QTL controlling low-temperature germinability in rice. J Wuhan Bot Res. 2005;23:216–220. [Google Scholar]
- Jiang L, Liu S, Hou M, Tang J, Chen L, Zhai H, Wan J. Analysis of QTLs for seed low temperature germinability and anoxia germinability in rice (Oryza sativa L.) Field Crop Res. 2006;98:68–75. doi: 10.1016/j.fcr.2005.12.015. [DOI] [Google Scholar]
- Jiang N, Shi S, Shi H, Khanzada H, Wassan GM, Zhu C, Peng X, Yu Q, Chen X, He X, Fu J, Hu L, Xu J, Ouyang L, Sun X, Zhou D, He H, Bian J. Mapping QTL for seed germinability under low temperature using a new high-density genetic map of rice. Front Plant Sci. 2017;8:1223. doi: 10.3389/fpls.2017.01223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S, Yang C, Xu Q, Wang L, Yang X, Song X, Wang J, Zhang X, Li B, Li H, Li Z, Li W. Genetic dissection of germinability under low temperature by building a resequencing linkage map in japonica rice. Int J Mol Sci. 2020;21:1284. doi: 10.3390/ijms21041284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Liu X, Xie K, Wang Y, Liu F, Lin Q, Wang W, Yang C, Lu B, Liu S, Chen L, Jiang L, Wan J. qLTG-9, a stable quantitative trait locus for low-temperature germination in rice (Oryza sativa L.) Theor Appl Genet. 2013;126:2313–2322. doi: 10.1007/s00122-013-2137-2. [DOI] [PubMed] [Google Scholar]
- Li F, Gao Y, Wu B, Cai Q, Zhan P, Yang W, Shi W, Yang Z, Tan Q, Luan X, Zhang G, Wang S. High-quality de novo genome assembly of Huajingxian 74, a receptor parent of single segment substitution lines. Rice Sci. 2021;28:109–113. doi: 10.1016/j.rsci.2020.09.010. [DOI] [Google Scholar]
- Liu C, Wang W, Mao B, Chu C. Cold stress tolerance in rice: physiological changes, molecular mechanism, and future prospects. Hereditas. 2018;40:171–185. doi: 10.16288/j.yczz.18-007. [DOI] [PubMed] [Google Scholar]
- Lv Y, Shao G, Jiao G, Sheng Z, Xie L, Hu S, Tang S, Wei X, Hu P. Targeted mutagenesis of POLYAMINE OXIDASE 5 that negatively regulates mesocotyl elongation enables the generation of direct-seeding rice with improved grain yield. Mol Plant. 2020 doi: 10.1016/j.molp.2020.11.007. [DOI] [PubMed] [Google Scholar]
- Ma Z, Bykova NV, Igamberdiev AU. Cell signaling mechanisms and metabolic regulation of germination and dormancy in barley seeds. Crop J. 2017;5:459–477. doi: 10.1016/j.cj.2017.08.007. [DOI] [Google Scholar]
- Mahender A, Anandan A, Pradhan SK. Early seedling vigour, an imperative trait for direct-seeded rice: an overview on physio-morphological parameters and molecular markers. Planta. 2015;241:1027–1050. doi: 10.1007/s00425-015-2273-9. [DOI] [PubMed] [Google Scholar]
- Miura K, Lin SY, Yano M, Nagamine T. Mapping quantitative trait loci controlling low temperature germinability in rice (Oryza sativa L.) Breeding Sci. 2001;51:293–299. doi: 10.1270/jsbbs.51.293. [DOI] [Google Scholar]
- Rajjou L, Duval M, Gallardo K, Catusse J, Bally J, Job C, Job D. Seed germination and vigor. Annu Rev Plant Biol. 2012;63:507–533. doi: 10.1146/annurev-arplant-042811-105550. [DOI] [PubMed] [Google Scholar]
- Sales E, Viruel J, Domingo C, Marqués L. Genome wide association analysis of cold tolerance at germination in temperate japonica rice (Oryza sativa L.) varieties. PLoS ONE. 2017;12:e183416. doi: 10.1371/journal.pone.0183416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAS Institute (2011) Base SAS 9.3 procedures guide. SAS Institute, Cary, NC
- Wang Z, Wang F, Zhou R, Wang J, Zhang H. Identification of quantitative trait loci for cold tolerance during the germination and seedling stages in rice (Oryza sativa L.) Euphytica. 2011;181:405–413. doi: 10.1007/s10681-011-0469-z. [DOI] [Google Scholar]
- Wang S, Wu K, Yuan Q, Liu X, Liu Z, Lin X, Zeng R, Zhu H, Dong G, Qian Q, Zhang G, Fu X. Control of grain size, shape and quality by OsSPL16 in rice. Nat Genet. 2012;44:950–954. doi: 10.1038/ng.2327. [DOI] [PubMed] [Google Scholar]
- Wang F, Longkumer T, Catausan SC, Calumpang CLF, Tarun JA, Cattin-Ortola J, Ishizaki T, Tanaka JP, Rose T, Wissuwa M, Kretzschmar T. Genome-wide association and gene validation studies for early root vigour to improve direct seeding of rice. Plant Cell Environ. 2018;41:2731–2743. doi: 10.1111/pce.13400. [DOI] [PubMed] [Google Scholar]
- Wang H, Lee A, Park S, Jin S, Lee J, Ham T, Park Y, Zhao W, Kwon S. Genome-wide association study reveals candidate genes related to low temperature tolerance in rice (Oryza sativa) during germination. 3 Biotech. 2018;8:235. doi: 10.1007/s13205-018-1252-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zou B, Shao Q, Cui Y, Lu S, Zhang Y, Huang Q, Huang J, Hua J. Natural variation reveals that OsSAP16 controls low-temperature germination in rice. J Exp Bot. 2018;69:413–421. doi: 10.1093/jxb/erx413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissuwa M, Wegner J, Ae N, Yano M. Substitution mapping of Pup1: a major QTL increasing phosphorus uptake of rice from a phosphorus-deficient soil. Theor Appl Genet. 2002;105:890–897. doi: 10.1007/s00122-002-1051-9. [DOI] [PubMed] [Google Scholar]
- Wu G, Deng H, Yu M, Cai Y, Zhou D, Tan J, Yu J, Luo X, Tong S, Wang P, Zhang X, Li C, Li C, Wang Y, Cheng Q, He H, Bian J. Genetic analysis of rice seed recovery under low-temperature conditions using a new CSSL population with a high-density genetic map in rice. Mol Breed. 2020;40:109. doi: 10.1007/s11032-020-01189-7. [DOI] [Google Scholar]
- Xi Z, He F, Zeng R, Zhang Z, Ding X, Li W, Zhang G. Development of a wide population of chromosome single-segment substitution lines in the genetic background of an elite cultivar of rice (Oryza sativa L.) Genome. 2006;49:476–484. doi: 10.1139/g06-005. [DOI] [PubMed] [Google Scholar]
- Xie L, Tan Z, Zhou Y, Xu R, Feng L. Identification and fine mapping of quantitative trait loci for seed vigor in germination and seedling establishment in rice. J Integr Plant Biol. 2014;56:749–759. doi: 10.1111/jipb.12190. [DOI] [PubMed] [Google Scholar]
- Yang M, Yang J, Su L, Sun K, Li D, Liu Y, Wang H, Chen Z, Guo T. Metabolic profile analysis and identification of key metabolites during rice seed germination under low-temperature stress. Plant Sci. 2019;289:110282. doi: 10.1016/j.plantsci.2019.110282. [DOI] [PubMed] [Google Scholar]
- Yang T, Zhou L, Zhao J, Dong J, Liu Q, Fu H, Mao X, Yang W, Ma Y, Chen L, Wang J, Bai S, Zhang S, Liu B. The candidate genes underlying a stably expressed QTL for low temperature germinability in rice (Oryza sativa L.) Rice. 2020;13:74. doi: 10.1186/s12284-020-00434-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H, Feng J, Zhang L, Zhang J, Mispan MS, Cao Z, Beighley DH, Yang J, Gu XY. Map-based cloning of Seed Dormancy1-2 identified a gibberellin synthesis gene regulating the development of endosperm-imposed dormancy in rice. Plant Physiol. 2015;169:2152–2165. doi: 10.1104/pp.15.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Zeng R, Zhang Z, Ding X, Li W, Liu G, He F, Tulukdar A, Huang C, Xi Z. The construction of a library of single segment substitution lines in rice (Oryza sativa L.) Rice Genet Newsl. 2004;21:85–87. [Google Scholar]
- Zhou Y, Xie Y, Cai J, Liu C, Zhu H, Jiang R, Zhong Y, Zhang G, Tan B, Liu G, Fu X, Liu Z, Wang S, Zhang G, Zeng R. Substitution mapping of QTLs controlling seed dormancy using single segment substitution lines derived from multiple cultivated rice donors in seven cropping seasons. Theor Appl Genet. 2017;130:1191–1205. doi: 10.1007/s00122-017-2881-9. [DOI] [PubMed] [Google Scholar]
- Zhu H, Li Y, Liang J, Luan X, Xu P, Wang S, Zhang G, Liu G. Analysis of QTLs on heading date based on single segment substitution lines in rice (Oryza sativa L.) Sci Rep. 2018;8:13232. doi: 10.1038/s41598-018-31377-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Length (a) and genome coverage (b) of substituted segments in 208 rice single segment substitution lines (SSSLs). cM, centimorgan. Figure S2. Phenotypic variation of LTG between recipient parent HJX74 and different SSSLs. S6, S15, S22 were three representative SSSLs with good LTG values. Pictures were taken on the 6th, 8th, and 10th day after sowing. Bars=1 cm. Figure S3. Temporal and spatial expression profiles of candidate genes in qLTG3a region. All expression data were prepared by the public tool RiceXPro (http://ricexpro.dna.affrc.go.jp/). The grey boxes mean no expression data. The heat map was made by TBtools (v1.089). Table S1. Summary the donors and substituted segments distribution of 208 SSSLs. Table S2. The sequence of primers used in this study. Table S3. ANOVA analysis of the mean LTG values of HJX74 in different blocks of each cropping season. Table S4. LTG of HJX74 and 24 SSSLs at 15°C in four cropping seasons. Table S5. Summary of genotypic means for germination and genetic effects of markers segregating in progeny lines derived from selected recombinants for qLTG3a region. Table S6. The RAP ID, MSU ID and R498 (Shuhui498) ID of candidate genes. Table S7. Nonsynonymous SNP variations (ΔSNP-index > 0.8) of candidate genes in the qLTG3a locus. Table S8. Indel variations (ΔInDel-index > 0.8) in the upstream and exonic regions of candidate genes in the qLTG3a locus.
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article and its additional files.