Abstract
Background
Targeted treatment modalities for non-small cell lung carcinoma (NSCLC) patients are expanding rapidly and demand a constant adaptation of molecular testing strategies. In this regard, broad reflex testing via next-generation sequencing (NGS) might have several advantages. However, real-world data regarding practical feasibility and clinical relevance are scarce, especially for RNA-based NGS.
Methods
We performed a retrospective study comparing NGS use in two consecutive years (2019 and 2020). In 2019, reflex testing mainly consisted of DNA-based NGS for mutations and immunohistochemistry (IHC) for ALK, ROS1, and NTRK fusion products. At the beginning of 2020, our approach has changed, with DNA- and RNA-based NGS panels now being simultaneously performed. This change in protocol allowed us to retrospectively evaluate if broad molecular reflex testing brings additional value to lung cancer patients.
Results
Within the whole cohort (n=432), both DNA- and RNA-based NGS yielded almost always evaluable results. Only in 6 cases, the RNA content was too little for an appropriate analysis. After integrating RNA-based NGS in the reflex testing approach, the number of detected fusions increased significantly (2.6% vs. 8.2%; P=0.0021), but also more patients received targeted therapies. Furthermore, exceedingly rare alterations were more likely to be detected, including the so far undescribed EGFR-NUP160 fusion.
Conclusions
Our study demonstrates that a comprehensive approach to reflex NGS testing is practically feasible and clinically relevant. Including RNA-based panels in the reflex testing approach results in more detected fusions and more patients receiving targeted therapies. Additionally, this broad molecular profiling strategy identifies patients with emerging biomarkers, underscoring its usefulness in the rapidly evolving landscape of targeted therapies.
Keywords: Non-small cell lung carcinoma (NSCLC), reflex testing, next-generation sequencing (NGS), DNA sequencing, RNA sequencing
Introduction
Lung cancer is the leading cause of cancer death worldwide (1). However, treatment modalities for non-small cell lung carcinoma (NSCLC) patients are expanding rapidly with an ever-increasing number of approved targeted therapies. Today, approximately 15 years after applying the first EGFR tyrosine kinase inhibitor (TKI) (2,3), targeted therapies are an established cornerstone in clinical practice. Since detecting the respective molecular alteration is a prerequisite for therapy initiation, pure histologic classification of lung carcinoma is insufficient. It has to be complemented by molecular analyses, providing a complete diagnosis, including predictive and prognostic information.
The increasing number of approved targeted therapies results in a constant need to adapt molecular testing strategies and include more and more genes in testing panels, often leading to insufficient molecular testing (4). The National Comprehensive Cancer Network (NCCN) Guidelines recommend using testing panels that include the following genes: EGFR, ALK, ROS1, BRAF, KRAS, MET, RET, and NTRK 1/2/3. Furthermore, high-level MET amplification and ERBB2 (HER2) mutations are regarded as emerging biomarkers for novel therapies (5). From a practical point of view, several international guidelines are trying to answer the questions of whom, when, and which genes to test and which methods to use (6-8). All these guidelines agree that EGFR, ALK, ROS1, and BRAF should always be included in testing panels. When available, extended testing panels should also have KRAS, MET, RET, and ERBB2. Different guidelines also agree that the optimal diagnostic method is reflex testing and that all advanced stage adenocarcinomas should be tested. Of note, there has been a recent report of a positive double-blind phase 3 trial (ADAURA trial) with osimertinib as adjuvant therapy in patients with stage IB to IIIA EGFR mutation-positive NSCLC (9). This has resulted in a rapid change in NCCN Guidelines, now recommending EGFR molecular testing of all newly diagnosed carcinomas, regardless of the stage (5). National guidelines are in concordance with international guidelines but reflect the local situation regarding drugs and testing availability [for review, see (10,11)]. The 2020 recommendations of the Austrian working group on lung pathology and oncology for the diagnostic workup of NSCLC with a focus on predictive biomarkers recommended reflex testing of all newly diagnosed lung adenocarcinoma for EGFR, ALK, ROS1, KRAS, BRAF, and NTRK 1/2/3 with a recommendation to include also ERBB2, MET and RET, preferably using NGS methods (12).
At our institution (Medical University of Graz, Austria), testing strategies have changed over time, especially between 2019 and 2020. Until the end of 2019, all newly diagnosed lung adenocarcinoma samples were examined via reflex testing using a DNA-based NGS panel, including EGFR, KRAS, BRAF, and ERBB2 genes. ALK was tested using immunohistochemistry (IHC) without additional confirmation if clearly and strongly positive staining was present. For ROS1 and NTRK, IHC was used as a screening method with obligate confirmation via NGS. Since January 2020, we have changed our protocol to optimize tissue management and provide better molecular profiling of tested tumors. All newly diagnosed lung adenocarcinomas are since then examined via reflex testing using DNA- and RNA-based NGS panels. The DNA-based panel (“mutation panel”) comprises 22 genes, among which EGFR, KRAS, BRAF, ERBB2, ALK, and MET. The RNA-based panel (“fusion panel”) encompasses, among other genes, ALK, ROS1, NTRK, RET, and MET.
Although reflex testing might have several advantages for NSCLC patient management, real-world data regarding practical feasibility and clinical relevance are scarce for DNA-based NGS (13-15) and non-existent for RNA-based NGS. Therefore, we performed a retrospective study comparing NGS use in two consecutive years (2019 and 2020), aiming to examine if reflex broad molecular profiling, including DNA- and RNA-based NGS, brings additional value to lung cancer patients.
We present the following article in accordance with the STROBE reporting checklist (available at https://dx.doi.org/10.21037/tlcr-21-570).
Methods
Patient cohort
All lung cancer patients diagnosed at our University Hospital whose tissue specimens were tested via DNA- and/or RNA-based next-generation sequencing between 01.01.2019 and 31.12.2020 were included in the study (n=432). Clinical data were retrospectively obtained from electronic medical records. All patients signed informed consent.
This retrospective study conformed to the principles outlined in the Declaration of Helsinki (as revised in 2013). It was approved by the Ethics Committee of the of the Medical University of Graz (33-066 ex 20/21).
DNA-based NGS
For each case, 2 to 12 FFPE tissue sections (each 4 µm thick) were used for DNA extraction. DNA was extracted from macrodissected tumor areas of FFPE sections. Extraction was performed using the Maxwell 16 instrument (Promega) and the Maxwell RSC DNA FFPE Kit (Promega, CatNr: AS1450). DNA was quantified by Picogreen fluorescence, and 10ng DNA was used for library preparation. NGS libraries were prepared using the AmpliSeq library kit 2.0 (Thermo Fisher Scientific) and the Ion Ampliseq Colon and Lung Cancer Research Panel v2 primer pool covering hotspot mutations in 22 genes (KRAS, EGFR, BRAF, PIK3CA, AKT1, ERBB2, PTEN, NRAS, STK11, MAP2K1, ALK, DDR2, CTNNB1, MET, TP53, SMAD4, FBXW7, FGFR3, NOTCH1, ERBB4, FGFR1, FGFR2). Sequencing was performed on an Ion S5XL benchtop sequencer (Thermo Fisher Scientific) to a length of 200 base pairs. Initial data analysis was done using the Ion Torrent Suite Software Plug-ins (Thermo Fisher Scientific, open-source, GPL, https://github.com/iontorrent/). Briefly, this included base calling, alignment to the reference genome (HG19) using the TMAP mapper, and variant calling by a modified diBayes approach considering the flow space information. Called variants were annotated using open source software ANNOVAR (16) and SnpEff (17). All coding, nonsynonymous mutations were further evaluated and visually inspected in IGV (http://www.broadinstitute.org/igv/), and variant calls resulting from technical read errors or sequence effects were excluded from the result.
RNA-based NGS
For each case, five to eight FFPE tissue sections (each 10 µm thick) were used for RNA-based NGS. After microdissection, RNA extraction was performed via the Maxwell RSC RNA FFPE kit (Promega, Mannheim, Germany). After RNA quantification via ribogreen fluorescence on a Qubit fluorometer (Life Tech Austria, Vienna, Austria), 250 ng of total RNA were utilized for further analyses using the Archer FusionPlex Expanded Lung 18090 v1.0 primer pool (ArcherDX, Boulder, CO, USA). Sequencing was performed on the S5XL benchtop sequencer (Ion Torrent, Thermo Fischer, Waltham, MA) using the Ion S5 Sequencing 200 kit (Thermo Fischer, Waltham, MA, USA) and Ion 550 chip kit. Acquired sequencing data were analyzed via the ArcherDX Analysis software Version 5.1.3. (ArcherDX, Boulder, CO, USA). Translocations called with more than 10 individual reads by the Archer analysis software were included in the final reports.
Validation of assays for DNA and RNA-based analyzes
All analyses were performed in the diagnostic context on the Ion Torrent platform in conjunction with Ampliseq and Archer chemistry, neither is CE-IVD. However, a thorough validation of all in-house NGS assays has been performed. In detail, assays were tested for specificity, sensitivity, the limit of detection, as well as repeatability and reproducibility to ascertain concordance to the general safety and performance requirements described in Annex I of the IVDR (EU 2017/746). For the validation, we used a mix of commercial known-truth samples and patient samples previously analyzed with alternative technologies (qPCR, Pyrosequencing, or FISH) at our Institute. Furthermore, our Institute participates regularly in EQA schemes for all diagnostic analyses.
Statistical analyses
Categorical data are reported as absolute frequencies (%), numerical data as medians (range). All statistical analyses (Fisher’s exact test, Mann Whitney test, Spearman rank correlation test, as appropriate) were performed with GraphPad Prism version 9.1.0 for Mac, GraphPad Software, San Diego, California, USA, www.graphpad.com. Data are expressed as box-and-whisker plots indicating a median and interquartile range (boxes) as well as minimum and maximum (whiskers) unless otherwise specified. P values ≤0.05 were considered statistically significant.
Results
Patient characteristics
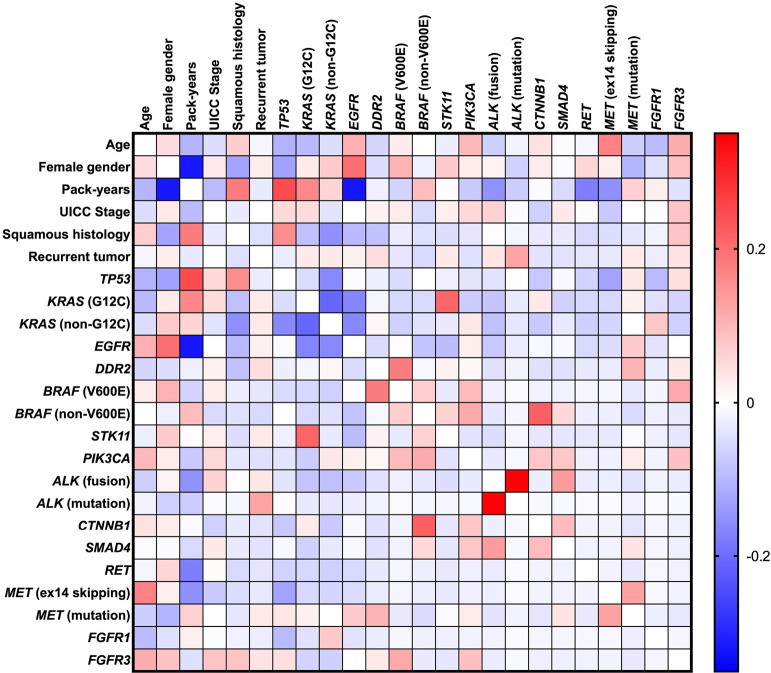
Within the whole cohort (n=432), the median age was 66 years (range, 32–88 years), and 194 (44.9%) patients were female (Table 1). Most patients were smokers (n=322; 74.5%), while only 87 (20.1%) were non-smokers. In 23 (5.3%) patients, the smoking status could not be determined. The median number of pack-years was 30 (range, 0–150). Female gender was associated with lower pack-years (r=−0.318; P<0.0001; Figure 1). Histologically, the most common subtype was adenocarcinoma (AC) (n=368; 85.2%), followed by squamous cell carcinoma (SCC) (n=36; 8.3%). The high amount of adenocarcinoma in this population is explained by the fact that reflex testing in SCC is not officially recommended. Only certain patients are tested (e.g., younger patients). SCC histology was associated with higher numbers of pack-years (r=0.181; P=0.001) and was less common in women (r=−0.128; P=0.010). In most cases (n=406; 94%), molecular testing was performed on the primary tumor at the time of initial diagnosis, while in 26 (6%) cases, molecular testing was performed on tumors that recurred, both locally (n=11; 2.5%) and distantly (n=15; 3.5%). It is important to note that there were no significant differences in these characteristics between 2019 (n=189) and 2020 (n=243).
Table 1. Patient characteristics of the study cohort.
| Clinical/pathological characteristics | All (n=432) | 2019 (n=189) | 2020 (n=243) | P value |
|---|---|---|---|---|
| Basic characteristics | ||||
| Age - median (range) | 66 [32–88] | 66 [35–88] | 66 [32–87] | 0.733 |
| Female gender, n (%) | 194 (44.9) | 87 (46.0) | 107 (44.4) | 0.697 |
| Male gender, n (%) | 238 (55.1) | 102 (54.0) | 136 (55.6) | 0.697 |
| Smoking status, n (%) | ||||
| Smoker | 322 (74.5) | 145 (76.7) | 177 (72.8) | 0.375 |
| Current smoker | 145 (33.6) | 61 (32.3) | 84 (34.6) | 0.681 |
| Former smoker | 177 (41.0) | 84 (44.4) | 93 (38.3) | 0.202 |
| Non-smoker | 87 (20.1) | 34 (18.0) | 53 (21.8) | 0.336 |
| Smoking status not known | 23 (5.3) | 10 (5.3) | 13 (5.3) | >0.9999 |
| Pack-years, median (range) | 30 (0–150) | 35 (0–150) | 30 (0–125) | 0.179 |
| Histology, n (%) | ||||
| Adenocarcinoma | 368 (85.2%) | 168 (88.9%) | 200 (82.3%) | 0.058 |
| Squamous cell carcinoma | 36 (8.3%) | 11 (5.8%) | 25 (10.3%) | 0.115 |
| Other | 28 (6.5%) | 10 (5.3%) | 18 (7.4%) | 0.434 |
| Staging, n (%) | ||||
| Initial tumor | 406 (94.0) | 180 (95.2) | 226 (93.0) | 0.416 |
| UICC 2017 stage | 0.411 | |||
| IA1 | 15 (3.5) | 9 (4.8) | 6 (2.5) | |
| IA2 | 46 (10.6) | 24 (12.7) | 22 (9.1) | |
| IA3 | 37 (8.6) | 15 (7.9) | 22 (9.1) | |
| IB | 28 (6.5) | 12 (6.3) | 16 (6.6) | |
| IIA | 13 (3.0) | 6 (3.2) | 7 (2.9) | |
| IIB | 26 (6.0) | 10 (5.3) | 16 (6.6) | |
| IIIA | 42 (9.7) | 23 (12.2) | 19 (7.8) | |
| IIIB | 21 (4.9) | 6 (3.2) | 15 (6.2) | |
| IIIC | 4 (0.9) | 2 (1.1) | 2 (0.8) | |
| IVA | 57 (13.2) | 19 (10.1) | 38 (15.6) | |
| IVB | 117 (27.1) | 54 (28.6) | 63 (25.9) | |
| Recurrent tumor | 26 (6.0) | 9 (4.8) | 17 (7.0) | 0.416 |
| Local recurrence | 11 (2.5) | 4 (2.1) | 7 (2.9) | 0.762 |
| Distant recurrence | 15 (3.5) | 5 (2.6) | 10 (4.1) | 0.443 |
Figure 1.
Correlation matrix of patient characteristics and corresponding NGS data. Positive correlation coefficients are marked red, and negative correlation coefficients are observed blue (Spearman rank correlation test). NGS, next-generation sequencing.
DNA-based NGS
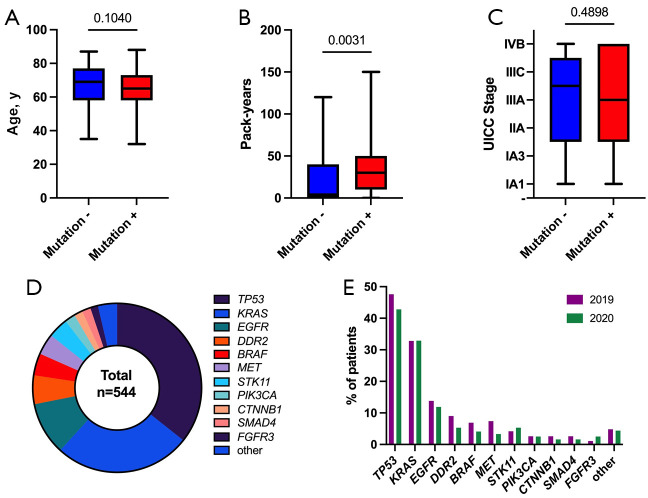
In all cases where DNA-based NGS was performed (n=407; 94.2%), also evaluable results could be obtained, although in 4 (0.9%) cases, the tumor cell content was <10%. Analyzed sample types in 2019 and 2020 included resection specimens (31.1% and 34.4%, respectively), biopsies (61.2% and 60.3%, respectively) and cytologic samples (7.7% and 5.4%, respectively). Most of the samples originated from the primary tumor in the lung (79.8% and 75.4%, in 2019 and 2020, respectively) (Table S1). A genetic alteration could be detected in 348 (80.6%) cases (Table S1). The presence of a detectable mutation was significantly associated with higher numbers of pack-years (Figure 2). The distribution of detected alterations in our cohort is depicted in Figure 2. The most frequent mutations occurred within the TP53 gene (n=194; 44.9%), followed by KRAS mutations (n=142; 32.9%). The targetable G12C mutation accounted for almost half of all KRAS mutations (n=62; 14.4%). EGFR mutations occurred in 55 (12.7%) cases, with exon 19 deletions (n=22; 5.1%) and L858R mutations (n=21; 4.9%) being by far the most common ones. A T790M mutation was detected in 1 (0.2%) patient with recurrence of a tumor initially treated with afatinib due to the presence of the exon 19 deletion. As depicted in Figure 1, EGFR mutations occurred more commonly in women (r=0.199; P<0.0001) and were associated with lower numbers of pack-years (r=0.321; P<0.0001), while TP53 mutations occurred less commonly in women (r=−0.129; P=0.009) and were associated with higher numbers of pack-years (r=0.246; P<0.0001). Furthermore, there was a significant negative correlation between EGFR and KRAS mutations (r=-0.253; P<0.0001), both being well-known driver mutations and therefore most often exclusively present. In 23 (5.3%) cases, mutations within the BRAF gene could be detected, with almost a third of them being V600E mutations (n=7; 1.6%). In 2 (0.5%) patients with a recurrent tumor, a point mutation within the ALK gene was detected, both of them concurrently harbored an EML4-ALK fusion, which had been treated with a TKI before. In 15 (3.5%) cases with SCC histology, DNA-based NGS was performed, and in 14 (3.2%) cases, a genetic alteration could be detected. Almost all tested SCC cases harbored a TP53 mutation (n=13; 3%), followed by mutations within FGFR3 (n=2; 0.5%), PTEN (n=1; 0.2%), MET (n=1; 0.2%), and EGFR (n=1; 0.2%).
Figure 2.
DNA-based NGS results. (A-C) In contrast to age and UICC stage, the number of pack-years is significantly positively correlated with the detection of at least one mutation (Mann-Whitney test); (D) Overall distribution of detected mutations; (E) Comparison of detected mutations between both subgroups (patients from 2019 and 2020). NGS, next-generation sequencing.
The distribution of described mutations according to different years (2019 vs. 2020) is presented in Table S1.
There is a discrepancy between the total number of patients tested (189 in 2019 and 243 in 2020) and the number of patients with DNA-based NGS performed (183 in 2019 and 224 in 2020). The reason is that some samples were only tested with an RNA-based panel based on clinicians’ wishes or in search of eventual mutation in samples already tested for DNA (in previous years).
RNA-based NGS
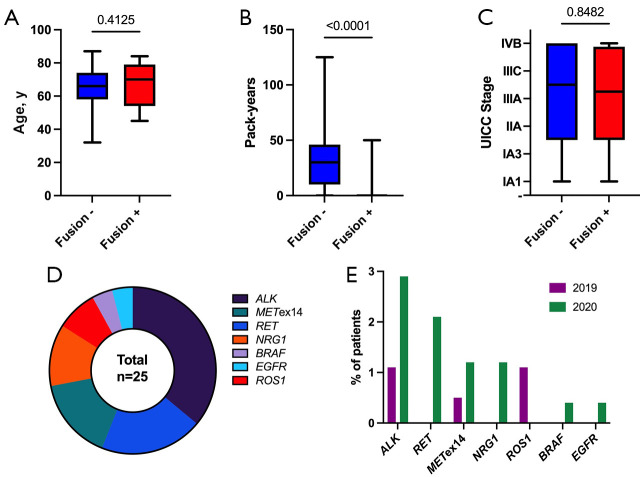
RNA-based NGS was performed in 269 (62.3%) cases and almost always yielded evaluable results (Table S2). Only in 6 (1.4%) cases, the RNA content within the sample was too low or of poor quality for an appropriate analysis. Analyzed sample types in 2019 and 2020 included resection specimens (34.2% and 35.9%, respectively), biopsies (60.5% and 59.3%, respectively) and cytologic samples (5.3% and 4.8%, respectively). Out of 6 “failed” samples, two were resection specimens and 4 biopsies. All cytologic samples were satisfactory for analysis. Most of the samples originated from the primary tumor in the lung (76.3% and 77.9%, in 2019 and 2020, respectively) (Table S2). A genetic alteration could be detected in 25 (5.8%) cases. Contrary to DNA-based NGS, the presence of a detectable alteration in RNA-based NGS was significantly associated with lower numbers of pack years (Figure 3). The introduction of reflex testing in 2020 resulted in a significantly higher number of detected alterations (Table S2). The distribution of detected alterations in our cohort is depicted in Figure 3. Overall, the most frequent alterations detected via RNA-based NGS were ALK fusions (n=9; 2.1%), with EML4-ALK fusions being by far the most common ones (n=8; 1.9%). Rearrangements involving the RET proto-oncogene could be detected in 5 (1.2%) cases and its fusion partners were CCDC6 (n=3; 0.7%) or KIF5B (n=2; 0.5%). MET exon 14 (METex14) skipping was detectable in 4 (0.9%) cases and was more common in the elderly (r=0.173; P=0.005). Fusions involving NRG1 or ROS1 were seen in 3 (0.7%) and 2 (0.5%) cases, respectively. Furthermore, exceedingly rare rearrangements involving otherwise commonly mutated genes (BRAF, EGFR) were also observed. RET fusions, NRG1 fusions, BRAF fusions, and EGFR fusions could only be detected in the year 2020 (Figure 3), demonstrating the importance of reflex testing in detecting rare but increasingly targetable gene rearrangements. In 30 (6.9%) cases with SCC histology, RNA-based NGS was performed, and in 2 (0.5%) cases, a genetic alteration could be detected, one harboring an ALK-KRT6A, the other an EGFR-NUP160 fusion.
Figure 3.
RNA-based NGS results. (A-C) In contrast to age and UICC stage, the number of pack-years is significantly negatively correlated with the detection of a fusion (Mann-Whitney test); (D) Overall distribution of detected fusions; (E) Comparison of detected fusions between both subgroups (patients from 2019 and 2020). NGS, next-generation sequencing.
The distribution of detected fusions according to different years (2019 vs. 2020) is presented in Table S2.
There is also a discrepancy between the total number of patients tested in 2020 [243] and the number of patients with RNA-based NGS performed [231]. The reason is that only adenocarcinomas were always tested with both panels, while non-adenocarcinoma samples were sometimes not tested with RNA-based panel.
Detection of targetable genetic alterations
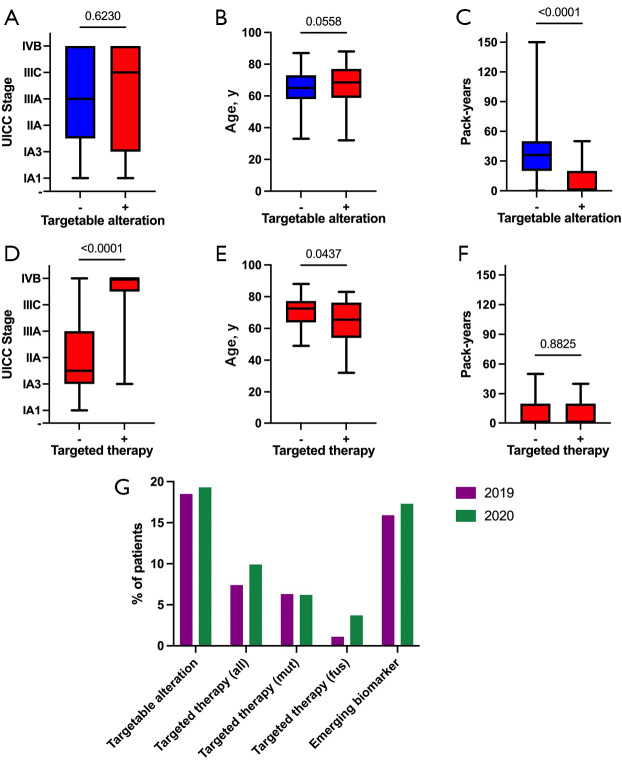
There are several targeted therapies approved for NSCLC patients harboring specific genetic alterations. These currently targetable alterations include EGFR mutations, ALK fusions, the BRAF V600E mutation, RET fusions, the METex14 skipping mutation, ROS1 fusions, and NTRK fusions. At least one of these alterations could be detected in 82 (19%) patients (Table 2). In 51 (11.8%) of those cases, an additional synchronous genetic alteration was observed, with 7 (1.6%) of them harboring a second targetable alteration. Interestingly, detecting a targetable alteration was significantly more likely in patients with lower numbers of pack-years but did not significantly correlate with UICC tumor stage or age (Figure 4). Two SCC cases (0.5%) harbored a targetable genetic alteration, one with an EGFR mutation, the other with an ALK-KRT6A fusion.
Table 2. Clinical relevance of detected genetic alterations.
| Patients with genetic alterations | All, n=432 (%) | 2019, n=189 (%) | 2020, n=243 (%) | P value |
|---|---|---|---|---|
| Alteration detected (all) | 364 (84.3) | 163 (86.2) | 201 (82.7) | 0.353 |
| Targetable alteration detected | 82 (19.0) | 35 (18.5) | 47 (19.3) | 0.902 |
| In early stage (I−IIIA) | 36 (8.3) | 17 (9) | 19 (7.8) | 0.727 |
| In late stage (IIIB−IV) | 40 (9.3) | 16 (8.5) | 24 (9.9) | 0.738 |
| In recurrent tumor | 6 (1.4) | 2 (1.1) | 4 (1.6) | 0.7 |
| With co-alteration (all) | 51 (11.8) | 26 (13.8) | 25 (10.3) | 0.294 |
| With targetable co-alteration | 7 (1.6) | 3 (1.6) | 4 (1.6) | >0.9999 |
| Targeted therapy received (all) | 38 (8.8) | 14 (7.4) | 24 (9.9) | 0.397 |
| In early stage (I−IIIA) | 3 (0.7) | 2 (1.1) | 1 (0.4) | 0.584 |
| In late stage (IIIB−IV) | 29 (6.7) | 10 (5.3) | 19 (7.8) | 0.337 |
| In recurrent tumor | 6 (1.4) | 2 (1.1) | 4 (1.6) | 0.7 |
| Therapy based on detected mutation1 | 27 (6.3) | 12 (6.3) | 15 (6.2) | >0.9999 |
| Osimertinib | 19 (4.4) | 9 (4.8) | 10 (4.1) | 0.815 |
| Afatinib | 11 (2.5) | 5 (2.6) | 6 (2.5) | >0.9999 |
| Trametinib/Dabrafenib | 4 (0.9) | 2 (1.1) | 2 (0.8) | >0.9999 |
| Gefitinib | 2 (0.5) | 2 (1.1) | 0 (0) | 0.191 |
| Mobocertinib | 1 (0.2) | 0 (0) | 1 (0.4) | >0.9999 |
| Therapy based on detected fusion/skipping1 | 11 (2.5) | 2 (1.1) | 9 (3.7) | 0.123 |
| Alectinib | 6 (1.4) | 0 (0) | 6 (2.5) | 0.038 |
| Brigatinib | 4 (0.9) | 0 (0) | 4 (1.6) | 0.135 |
| Selpercatinib | 3 (0.7) | 0 (0) | 3 (1.2) | 0.26 |
| Capmatinib | 1 (0.2) | 1 (0.5) | 0 (0) | 0.438 |
| Lorlatinib | 1 (0.2) | 1 (0.5) | 0 (0) | 0.438 |
| Emerging biomarker2 detected | 72 (16.7) | 30 (15.9) | 42 (17.3) | 0.795 |
| In early stage (I−IIIA) | 29 (6.7) | 15 (7.9) | 14 (5.8) | 0.439 |
| In late stage (IIIB−IV) | 38 (8.8) | 13 (6.9) | 25 (10.3) | 0.235 |
| In recurrent tumor | 5 (1.2) | 2 (1.1) | 3 (1.2) | >0.9999 |
Results of both subgroups (patients from 2019 and 2020, respectively) were compared using Fisher’s exact test. 1, some patients received more than one substance; 2, NRG1 fusion, KRAS G12C, ERBB2, FGFR1 [according to (10)].
Figure 4.
Clinical relevance of detected genetic alterations. (A,D) UICC stage significantly correlates with the application of targeted therapy, but not with the presence of a targetable alteration (Mann-Whitney test); (B,E) Age significantly negatively correlates with the application of targeted therapy, but not with the presence of a targetable alteration (Mann-Whitney test); (C,F) The number of pack-years significantly negatively correlates with the presence of a targetable alteration, but not with the application of targeted therapy (Mann-Whitney test); (G) Comparison of detected targetable alterations, applied targeted therapy (mutation and fusion specific), and detected emerging biomarkers between both subgroups (patients from 2019 and 2020).
In addition to well-established biomarkers for already approved targeted therapies, many further genetic alterations are on the horizon for implementation in clinical practice. In a recent comprehensive overview about the evolving landscape of biomarker testing in Europe, Kerr and colleagues regarded ERBB2 mutations, KRAS G12C mutations, NRG1 fusions, and FGFR1 mutations as emerging biomarkers (10). At least one of these alterations could be detected in 72 (16.7%) patients in our cohort, laying the groundwork for future targeted therapy in this subgroup (Table 2). However, no SCC cases harbored such an alteration.
Of note, both established and emerging biomarkers were more frequently detected in 2020 (Table 2 and Figure 4).
Therapeutic consequences
The detection of a targetable genetic alteration via NGS resulted in the application of targeted therapy in 38 (8.8%) patients, 14 (7.4%) in 2019, and 24 (9.9%) in 2020. Almost all of these patients (n=35; 8.1%) were in late-stage (IIIB-IV) or in recurrence (Table 2). In addition to patients with AC, also one SCC patient harboring an ALK-fusion received targeted therapy. Overall, in patients with a targetable alteration, consecutive administration of targeted therapy was highly associated with an advanced tumor stage (Figure 4), reflecting current therapy guidelines. Younger patients with a targetable alteration were more likely to receive targeted therapy than older patients (Figure 4). Furthermore, patients with lower numbers of pack-years were significantly more likely to harbor a targetable alteration, mainly caused by the presence of EGFR mutations that are far more common in non-smokers (Figure 1). Importantly, the higher frequency of targeted therapies in 2020 can be clearly attributed to much higher fusion detection rates, as shown in Figure 4. The most commonly applied substances were Osimertinib (n=19; 4.4%), Afatinib (n=11; 2.5%), Alectinib (n=6; 1.4%), Brigatinib (n=4; 0.9%), and Trametinib/Dabrafenib (n=4; 0.9%). Especially substances targeting a genetic rearrangement were more frequently applied in 2020, mirroring the higher fusion detection rates after introducing reflex testing via RNA-based NGS (for comparisons, see Table 2 and Table S2).
Very important issue is also the turnaround time. Introducing both NGS panels in 2020 prolonged turnaround time for 2 working days, from 8 in 2019 to 10 in 2020.
Discussion
Comparing NGS-based NSCLC testing in two consecutive years in a real-life setting, we showed that both DNA- and RNA-based analysis could be performed in a remarkably high number of patients, identifying a plethora of targetable genetic alterations, resulting in targeted therapy for many patients. More specifically, we demonstrated that RNA-based analysis, when performed in a reflex manner at the same time as DNA-based analysis, reliably identifies patients with targetable genetic rearrangements who would otherwise be missed.
However, to have such a high yield, adequate tissue management is essential. One of the most fundamental challenges in the diagnostic process of NSCLC is tissue availability, as more and more biomarkers should be examined within small histologic (18) or cytologic specimens (19). Therefore, proper tissue handling of samples from lung cancer patients is critical. Saving tissue is actually one of the major arguments for reflex testing. At our institute, we have established a workflow that guarantees the economical use of all tiny lung cancer specimens. First, a certified pulmonary pathologist evaluates if a malignant tumor is present and orders a specific tissue re-cutting program for lung cancer specimens if that is the case. This means that the paraffin block will be cut on the same microtome by the same person, preventing the additional “leveling” steps and saving tissue. The tumor sample will be cut on up to 15 blank slides. The first six have one 4 µm thick slice (primarily reserved for immunohistochemistry), and the last nine up to three 4 µm thick slices per slide (used mainly for molecular analysis). Furthermore, the usage of diagnostic immunohistochemical stains (if needed) is restricted to thyroid transcription factor 1 (TTF1) and deltaNp63 (p40) [as recommended in (20)], as well as to programmed death-ligand 1 (PD-L1) for predictive purposes [as recommended in (21)]. Since the recent introduction of the entity “NSCLC—not otherwise specified (NOS)” in biopsy/cytologic specimens, no further immunohistochemical staining are required in cases where the amount of tumor tissue is small (22), facilitating economic tissue handling even more. In other words, if there is a very small amount of NSCLC in a sample, for the patient it is more important to have an adequate molecular profile than the exact histologic diagnosis.
Our study cohorts, looking at both years separately, were well balanced concerning all clinical and histologic parameters. Interestingly, although adenocarcinoma was the most predominant histologic type, there is still a very high proportion of males and smokers. This fact reflects still the high number of smokers in the Austrian population, and the only recently introduced smoking-ban law in Austria. SCC patients have been rarely tested (5.8% and 10% in 2019 and 2020, respectively), and as previously explained, only on the request from clinicians (e.g., for younger patients). The increase of tested SCC in 2020 is due to reflex screening of all lung carcinomas with the pan-TRK antibody, with obligate confirmation of any positive reaction with NGS.
It is known that the frequency of mutations in NSCLC is dependent on multiple factors, including ethnicity. For example, EGFR mutations are more common in Asians (23), while the KRAS G12C mutation is more common in Caucasians (24). In our cohort, frequencies of detected mutations were mostly comparable with other studies examining Caucasian lung cancer patients, although slight differences could be encountered (25-28). In comparison to the study by Volckmar et al., for example, we detected more mutations in MET (5.1% vs. 3%) and DDR2 (6.9% vs. 2%), while mutations in ERBB2 (0.7% vs. 3%), STK11 (4.9% vs. 10%), PIK3CA (2.5% vs. 6%), FGFR3 (1.9% vs. 4%) and PTEN (0.5% vs. 2%) were less frequent in our cohort (27). Within the SCC subgroup, our NGS results are largely consistent with other studies that included SCC cases. For example, both in our cohort and in the CRISP study, TP53 mutations were by far the most frequent alterations in SCC (93.3% and 69.1%, respectively) (26). Furthermore, 2 (5.3%) of our SCC patients harbored a targetable alteration (EGFR mutation and ALK fusion, respectively), with the latter even receiving targeted therapy. These findings again correspond well to the CRISP study, where targetable alterations could also be detected in SCC patients, including 4.4% with an EGFR mutation and 0.5% with an ALK fusion (26).
A major argument for multiple gene testing using DNA- and RNA-based NGS is the rapidly evolving landscape of targetable alterations in NSCLC. KRAS mutations, for example, have long been regarded as strictly undruggable and are now in the spotlight of drug development (29). Especially drugs targeting the KRAS G12C mutation (e.g., Sotrasib and Adagrasib) show promising data in clinical trials (30,31). The same is true for targeted therapies against MET, RET, and Her2 (32-34). Furthermore, STK11 and KEAP mutations are associated with an impaired response to anti-PD(L)1 agents, expanding the role of NGS in detecting biomarkers for immunotherapy (35). In general, routine testing for emerging biomarkers even before drug approval has several advantages. First, characterizing clinical and pathological parameters of a patient subgroup harboring a specific molecular alteration is paramount for upcoming clinical studies and clinical decision-making after drug approval. Two recent studies have comprehensively done this in large cohorts of lung cancer patients harboring the KRAS G12C mutation (36,37), which would not have been possible without previous routine testing. This need for testing before drug approval also becomes evident when looking at the early years of EGFR inhibitors, where their clinical benefit in a fraction of patients was attributed to factors like gender, smoking status, or histological subtype. Only after the correlation of these factors with sequencing data, it became clear that they are confounders of EGFR mutation status, which subsequently influenced treatment indications (38). Another crucial argument for routine testing of emerging biomarkers before drug approval is that patients with the detected alteration can potentially be treated in early access programs and/or immediately after drug approval.
Recently, several papers have investigated the effects of reflex testing in NSCLC, most of them confirming its benefits (13-15). However, these studies were limited to DNA-based NGS, although gene rearrangements in NSCLC are increasingly recognized and targetable. In our cohort, the inclusion of RNA-based NGS into our reflex testing approach led to much higher detection rates of the respective gene alterations. The frequencies of detected ALK fusions and METex14 skipping increased nearly threefold (1.1% to 2.9% and 0.5% to 1.2%, respectively), resulting in a concomitant increase of patients receiving targeted therapy (0.5% to 2.5%). Furthermore, most other detected fusions could only be recognized after introducing reflex RNA-based NGS. RET fusions, for example, were detected in 2.1% of patients in 2020 (with 1.2% receiving targeted therapy), while none was detected in 2019. A very peculiar finding is that in both 2019 and 2020, there is an unexpectedly low number of samples with ROS1 fusion (1.1% and 0%, respectively) compared to the published data [ranging from 1–2% (26,27) to 3–5% (25)]. One possible explanation could be that in the Austrian population, these mutations are lower than in other published data. The second possibility is that NGS might not be the ideal method for ROS1 fusion detection.
In addition to overall higher numbers of detected fusions, the introduction of RNA-based NGS into our reflex testing approach also enabled us to recognize exceedingly rare alterations. EGFR fusions, for example, do only rarely occur but are potentially targetable, with several reports showing a clinical benefit for these patients when treated with a TKI (39-42). We have also detected the so far unrecognized EGFR-NUP160 fusion in an SCC patient and could therefore expand the known spectrum of EGFR rearranged NSCLC with the help of our comprehensive reflex testing approach.
It is important to stress again that parallel, reflex testing with both DNA and RNA based NGS panels prolonged turnaround time for only 2 days, which is in our opinion very good, and still in the frame of international recommendations.
Another very important issue is immunotherapy, which is out of the scope of this manuscript. However, in a real-life setting, as previously mentioned, it is very important to test for PD-L1, and to have it included in a reflex protocol. Combination of these results (PD-L1 immunohistochemistry and NGS) provides better information and can help in identification patients that could potentially be treated effectively with immunotherapy.
In conclusion, our study demonstrated that a comprehensive approach to reflex NGS testing in NSCLC is practically feasible and clinically relevant. Including RNA-based panels in the reflex testing approach results in more detected fusions and more patients receiving targeted therapies. Additionally, this broad molecular profiling strategy identifies patients with emerging biomarkers, providing some of them with the possibility of early drug access. Furthermore, a pool of patients positive for different biomarkers is known, and no additional testing will be needed after the new drugs are approved. Finally, with clinical studies increasingly using targeted therapy as adjuvant/neoadjuvant treatment, our comprehensive reflex testing approach will become even more relevant in the future.
Supplementary
The article’s supplementary files as
Acknowledgments
We would like to thank Iris Halbwedl from the Medical University of Graz for her technical assistance.
Funding: All financials have been obtained from the institutional budget of the Diagnostic and Research Institute of Pathology, Medical University of Graz. No external funding has been received.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This retrospective study conformed to the principles outlined in the Declaration of Helsinki (as revised in 2013). It was approved by the Ethics Committee of the of the Medical University of Graz (33-066 ex 20/21). All patients signed informed consent.
Footnotes
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at https://dx.doi.org/10.21037/tlcr-21-570
Data Sharing Statement: Available at https://dx.doi.org/10.21037/tlcr-21-570
Peer Review File: Available at https://dx.doi.org/10.21037/tlcr-21-570
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tlcr-21-570). GA received payment for lectures and participated in advisory boards from Lilly, AstraZeneca, Roche, MSD, Merck, BMS, Pfizer, Novartis, Takeda, Janssen, Böhringer Ingelheim, Amgen, Sanofi; KK received grants from AstraZeneca; he also received payment for lectures and participated in advisory boards from AstraZeneca, Roche, MSD, BMS, Novartis and Thermo Fisher; RW received payment for presentations and participated in advisory boards for Takeda, Roche, Astra Zeneca, BMS, MSD, Janssen, Sanofi, Amgen and Boehringer Ingelheim; AT received grants from Sanofi, AstraZeneca, BMS, Roche and MSD; she also received payment for lectures and support for attending meetings and participated in advisory boards from Eli-Lilly, AstraZeneca, Roche, MSD, Merck, BMS, Pfizer, Novartis, Janssen and LB received grants from Takeda, AstraZeneca, BMS and Roche; he also received payment for lectures and participated in advisory boards form Invitae, Eli-Lilly, AstraZeneca, Roche, MSD, Merck, BMS, Pfizer, Novartis, Takeda, Janssen; support for attending meeting from Pfizer. He is Int. Secretary-Austrian Society of Pathology; PPS Membership and Awards Committee; Member of the Mesothelioma Committee of IASLC. The other authors have no conflicts of interest to declare.
References
- 1.Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129-39. 10.1056/NEJMoa040938 [DOI] [PubMed] [Google Scholar]
- 3.Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004;304:1497-500. 10.1126/science.1099314 [DOI] [PubMed] [Google Scholar]
- 4.Pennell NA, Arcila ME, Gandara DR, et al. Biomarker Testing for Patients With Advanced Non-Small Cell Lung Cancer: Real-World Issues and Tough Choices. Am Soc Clin Oncol Educ Book 2019;39:531-42. 10.1200/EDBK_237863 [DOI] [PubMed] [Google Scholar]
- 5.NCCN Clinical Practice Guidlines in Oncology- Non-Small Cell Lung Cancer. Available online: https://www.nccn.org/professionals/physician_gls/pdf/nscl.pdf. Accessed 2021 March 3, 2021.
- 6.Mosele F, Remon J, Mateo J, et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: a report from the ESMO Precision Medicine Working Group. Ann Oncol 2020;31:1491-505. 10.1016/j.annonc.2020.07.014 [DOI] [PubMed] [Google Scholar]
- 7.Metastatic non-small cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Available online: https://www.esmo.org/content/download/347819/6934778/1/ESMO-CPG-mNSCLC-15SEPT2020.pdf. Accessed 15 September 2020.
- 8.Lindeman NI, Cagle PT, Aisner DL, et al. Updated Molecular Testing Guideline for the Selection of Lung Cancer Patients for Treatment With Targeted Tyrosine Kinase Inhibitors: Guideline From the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. J Mol Diagn 2018;20:129-59. 10.1016/j.jmoldx.2017.11.004 [DOI] [PubMed] [Google Scholar]
- 9.Wu YL, Tsuboi M, He J, et al. Osimertinib in Resected EGFR-Mutated Non-Small-Cell Lung Cancer. N Engl J Med 2020;383:1711-23. 10.1056/NEJMoa2027071 [DOI] [PubMed] [Google Scholar]
- 10.Kerr KM, Bibeau F, Thunnissen E, et al. The evolving landscape of biomarker testing for non-small cell lung cancer in Europe. Lung Cancer 2021;154:161-75. 10.1016/j.lungcan.2021.02.026 [DOI] [PubMed] [Google Scholar]
- 11.Thunnissen E, Weynand B, Udovicic-Gagula D, et al. Lung cancer biomarker testing: perspective from Europe. Transl Lung Cancer Res 2020;9:887-97. 10.21037/tlcr.2020.04.07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Popper HH, Gruber-Mösenbacher U, Pall G, et al. The 2020 update of the recommendations of the Austrian working group on lung pathology and oncology for the diagnostic workup of non-small cell lung cancer with focus on predictive biomarkers. Memo 2020;13:11-26. 10.1007/s12254-019-00565-0 [DOI] [Google Scholar]
- 13.Lassalle S, Hofman V, Heeke S, et al. Targeted Assessment of the EGFR Status as Reflex Testing in Treatment-Naive Non-Squamous Cell Lung Carcinoma Patients: A Single Laboratory Experience (LPCE, Nice, France). Cancers (Basel) 2020;12:955. 10.3390/cancers12040955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anand K, Phung TL, Bernicker EH, et al. Clinical Utility of Reflex Ordered Testing for Molecular Biomarkers in Lung Adenocarcinoma. Clin Lung Cancer 2020;21:437-42. 10.1016/j.cllc.2020.05.007 [DOI] [PubMed] [Google Scholar]
- 15.Miller TE, Yang M, Bajor D, et al. Clinical utility of reflex testing using focused next-generation sequencing for management of patients with advanced lung adenocarcinoma. J Clin Pathol 2018;71:1108-15. 10.1136/jclinpath-2018-205396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164. 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cingolani P, Platts A, Wang le L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80-92. 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirsch FR, Wynes MW, Gandara DR, et al. The tissue is the issue: personalized medicine for non-small cell lung cancer. Clin Cancer Res 2010;16:4909-11. 10.1158/1078-0432.CCR-10-2005 [DOI] [PubMed] [Google Scholar]
- 19.Lozano MD, Echeveste JI, Abengozar M, et al. Cytology Smears in the Era of Molecular Biomarkers in Non-Small Cell Lung Cancer: Doing More With Less. Arch Pathol Lab Med 2018;142:291-8. 10.5858/arpa.2017-0208-RA [DOI] [PubMed] [Google Scholar]
- 20.Yatabe Y, Dacic S, Borczuk AC, et al. Best Practices Recommendations for Diagnostic Immunohistochemistry in Lung Cancer. J Thorac Oncol 2019;14:377-407. 10.1016/j.jtho.2018.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lantuejoul S, Sound-Tsao M, Cooper WA, et al. PD-L1 Testing for Lung Cancer in 2019: Perspective From the IASLC Pathology Committee. J Thorac Oncol 2020;15:499-519. 10.1016/j.jtho.2019.12.107 [DOI] [PubMed] [Google Scholar]
- 22.Thoracic Tumours: WHO Classification of Tumours. 5 ed. World Health Organization; 2021. [Google Scholar]
- 23.Zhang YL, Yuan JQ, Wang KF, et al. The prevalence of EGFR mutation in patients with non-small cell lung cancer: a systematic review and meta-analysis. Oncotarget 2016;7:78985-93. 10.18632/oncotarget.12587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nassar AH, Adib E, Kwiatkowski DJ. Distribution of KRAS G12C Somatic Mutations across Race, Sex, and Cancer Type. N Engl J Med 2021;384:185-7. 10.1056/NEJMc2030638 [DOI] [PubMed] [Google Scholar]
- 25.Trédan O, Wang Q, Pissaloux D, et al. Molecular screening program to select molecular-based recommended therapies for metastatic cancer patients: analysis from the ProfiLER trial. Ann Oncol 2019;30:757-65. 10.1093/annonc/mdz080 [DOI] [PubMed] [Google Scholar]
- 26.Griesinger F, Eberhardt W, Nusch A, et al. Biomarker testing in non-small cell lung cancer in routine care: Analysis of the first 3,717 patients in the German prospective, observational, nation-wide CRISP Registry (AIO-TRK-0315). Lung Cancer 2021;152:174-84. 10.1016/j.lungcan.2020.10.012 [DOI] [PubMed] [Google Scholar]
- 27.Volckmar AL, Leichsenring J, Kirchner M, et al. Combined targeted DNA and RNA sequencing of advanced NSCLC in routine molecular diagnostics: Analysis of the first 3,000 Heidelberg cases. Int J Cancer 2019;145:649-61. 10.1002/ijc.32133 [DOI] [PubMed] [Google Scholar]
- 28.Cohen D, Hondelink LM, Solleveld-Westerink N, et al. Optimizing Mutation and Fusion Detection in NSCLC by Sequential DNA and RNA Sequencing. J Thorac Oncol 2020;15:1000-14. 10.1016/j.jtho.2020.01.019 [DOI] [PubMed] [Google Scholar]
- 29.Salgia R, Pharaon R, Mambetsariev I, et al. The improbable targeted therapy: KRAS as an emerging target in non-small cell lung cancer (NSCLC). Cell Rep Med 2021;2:100186. 10.1016/j.xcrm.2020.100186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hong DS, Fakih MG, Strickler JH, et al. KRASG12C Inhibition with Sotorasib in Advanced Solid Tumors. N Engl J Med 2020;383:1207-17. 10.1056/NEJMoa1917239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jänne PA, Rybkin II, Spira AI, et al. KRYSTAL-1: Activity and Safety of Adagrasib (MRTX849) in Advanced/ Metastatic Non–Small-Cell Lung Cancer (NSCLC) Harboring KRAS G12C Mutation. Eur J Cancer 2020;138:S1-S2. 10.1016/S0959-8049(20)31076-5 [DOI] [Google Scholar]
- 32.Drilon A, Oxnard GR, Tan DSW, et al. Efficacy of Selpercatinib in RET Fusion-Positive Non-Small-Cell Lung Cancer. N Engl J Med 2020;383:813-24. 10.1056/NEJMoa2005653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolf J, Seto T, Han JY, et al. Capmatinib in MET Exon 14-Mutated or MET-Amplified Non-Small-Cell Lung Cancer. N Engl J Med 2020;383:944-57. 10.1056/NEJMoa2002787 [DOI] [PubMed] [Google Scholar]
- 34.Smit EF, Nakagawa K, Nagasaka M, et al. Trastuzumab deruxtecan (T-DXd; DS-8201) in patients with HER2-mutated metastatic non-small cell lung cancer (NSCLC): Interim results of DESTINY-Lung01. J Clin Oncol 2020;38:9504. 10.1200/JCO.2020.38.15_suppl.9504 [DOI] [Google Scholar]
- 35.Codima A, Monteiro G, Costa I, et al. 122P STK11 and/or KEAP1 mutations and outcomes in non-small cell lung cancer patients treated with immune checkpoint inhibitors: A systematic literature review. J Thorac Oncol 2021;16:S764. 10.1016/S1556-0864(21)01964-X [DOI] [Google Scholar]
- 36.Arbour KC, Rizvi H, Plodkowski AJ, et al. Treatment Outcomes and Clinical Characteristics of Patients with KRAS-G12C-Mutant Non-Small Cell Lung Cancer. Clin Cancer Res 2021;27:2209-15. 10.1158/1078-0432.CCR-20-4023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sebastian M, Eberhardt WEE, Hoffknecht P, et al. KRAS G12C-mutated advanced non-small cell lung cancer: A real-world cohort from the German prospective, observational, nation-wide CRISP Registry (AIO-TRK-0315). Lung Cancer 2021;154:51-61. 10.1016/j.lungcan.2021.02.005 [DOI] [PubMed] [Google Scholar]
- 38.Camidge DR, Doebele RC, Kerr KM. Comparing and contrasting predictive biomarkers for immunotherapy and targeted therapy of NSCLC. Nat Rev Clin Oncol 2019;16:341-55. 10.1038/s41571-019-0173-9 [DOI] [PubMed] [Google Scholar]
- 39.Konduri K, Gallant JN, Chae YK, et al. EGFR Fusions as Novel Therapeutic Targets in Lung Cancer. Cancer Discov 2016;6:601-11. 10.1158/2159-8290.CD-16-0075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raez LE, Pinto JA, Schrock AB, et al. EGFR-RAD51 Fusion: A Targetable Partnership Originated from the Tumor Evolution? J Thorac Oncol 2018;13:e33-4. 10.1016/j.jtho.2017.10.005 [DOI] [PubMed] [Google Scholar]
- 41.Xu H, Shao C. KIF5B-EGFR Fusion: A Novel EGFR Mutation in Lung Adenocarcinoma. Onco Targets Ther 2020;13:8317-21. 10.2147/OTT.S263994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu YC, Wang WX, Xu CW, et al. EGFR-RAD51 fusion variant in lung adenocarcinoma and response to erlotinib: A case report. Lung Cancer 2018;115:131-4. 10.1016/j.lungcan.2017.12.001 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as