Abstract
Background
Lurbinectedin recently received FDA accelerated approval as a second line treatment option for metastatic small cell lung cancer (SCLC). However, there are currently no established biomarkers to predict SCLC sensitivity or resistance to lurbinectedin or preclinical studies to guide rational combinations.
Methods
Drug sensitivity was assayed in proliferation assays and xenograft models. Baseline proteomic profiling was performed by reverse-phase protein array. Lurbinectedin-induced changes in intracellular signaling pathways were assayed by Western blot.
Results
Among 21 human SCLC cell lines, cytotoxicity was observed following lurbinectedin treatment at a low dose (median IC50 0.46 nM, range, 0.06–1.83 nM). Notably, cell lines with high expression of Schlafen-11 (SLFN11) protein, a promising biomarker of response to other DNA damaging agents (e.g., chemotherapy, PARP inhibitors), were more sensitive to single-agent lurbinectedin (FC =3.2, P=0.005). SLFN11 was validated as a biomarker of sensitivity to lurbinectedin using siRNA knockdown and in xenografts representing SLFN11 high and low SCLC. Replication stress and DNA damage markers (e.g., γH2AX, phosphorylated CHK1, phosphorylated RPA32) increased in SCLC cell lines following treatment with lurbinectedin. Lurbinectedin also induced PD-L1 expression via cGAS-STING pathway activation. Finally, the combination of lurbinectedin with the ataxia telangiectasia and Rad3-related protein (ATR) inhibitors ceralasertib and berzosertib showed a greater than additive effect in SLFN11-low models.
Conclusions
Together our data confirm the activity of lurbinectedin across a large cohort of SCLC models and identify SLFN11 as a top candidate biomarker for lurbinectedin sensitivity. In SLFN11-low SCLC cell lines which are relatively resistance to lurbinectedin, the addition of an ATR inhibitor to lurbinectedin re-sensitized otherwise resistant cells, confirming previous observations that SLFN11 is a master regulator of DNA damage response independent of ATR, and the absence of SLFN11 leads to synthetic lethality with ATR inhibition. This study provides a rationale for lurbinectedin in combination with ATR inhibitors to overcome resistance in SCLC with low SLFN11 expression.
Keywords: Small cell lung cancer (SCLC), lurbinectedin, ataxia telangiectasia and Rad3-related protein (ATR), SLFN11, replication stress
Introduction
Small cell lung cancer (SCLC) is an aggressive form of high-grade neuroendocrine lung cancer with dismal patient outcome of a 5-year survival of less than 10% (1). It is characterized by loss of function of the tumor suppressor genes TP53 and RB1, and accounts for around 13–15% of all lung cancers (1). For decades the first line treatment for extensive-stage SCLC, has been platinum-based chemotherapy combinations, with the incorporation of PD-L1 checkpoint blockade as part of the frontline regimen only since 2018 (2,3). In the second line setting, topotecan, a topoisomerase I inhibitor was the only US Food and Drug Administration (FDA) approved treatment (4) until the FDA recently granted accelerated approval to lurbinectedin for the treatment of patients with metastatic SCLC with disease progression on or after frontline platinum-based chemotherapy (4). In a single-arm, open-label, phase 2 basket trial overall response by investigator assessment was seen in 37 of 102 patients enrolled (35.2%; 95% CI: 26.2–45.2) with an acceptable and manageable safety profile (5). Lurbinectedin (PM01183) is a synthetic analog of the natural marine-based tetrahydroisoquinoline, trabectedin which is derived from the sea squirt species Ecteinascidia turbinate (6,7). Lurbinectedin blocks the activity of RNA-polymerase-II and induces its degradation by the ubiquitin/proteasome machinery, also inducing DNA damage (8). By hampering the transcription processes within tumor-associated macrophages lurbinectedin also modifies tumor microenvironment (9). To date, no biomarkers of response to lurbinectedin have been reported in SCLC, or any other cancer type. Furthermore, little is known about how intra-cellular signaling is altered following treatment with lurbinectedin.
Schlafen-11 (SLFN11) has been identified as a biomarker of response to cisplatin, and other DNA damaging agents such as PARP inhibitors, in multiple cancer types. In SCLC pre-clinical models, SLFN11 expression assayed by RNASeq, RPPA, and immunohistochemistry predicted response to both cisplatin and PARP inhibitors (10,11). In a phase II clinical trial testing temozolomide with or without veliparib, a PARP inhibitor, in relapsed extensive stage SCLC, patients with SLFN11 positive tumors had significantly longer overall survival (12). The mechanisms by which SLFN11 expression levels are regulated are as yet not fully understood, but its expression is believed to be controlled epigenetically, and by the cGAS-STING-Interferon γ innate immune response pathway (13).
In this preclinical study, we investigate the therapeutic efficacy of lurbinectedin in large panel of profiled cell lines representing the molecular heterogeneity of SCLC to identify candidate predictive biomarkers of response. We then characterized changes in DNA damage response pathway and immunogenic cell death associated pathway in SCLC cells upon treatment with lurbinectedin. Finally, we demonstrate a significantly greater than additive effect when lurbinectedin is combined with an ataxia telangiectasia and Rad3-related protein (ATR) inhibitor, ceralasertib. We present the following article in accordance with the MDAR checklist (available at https://dx.doi.org/10.21037/tlcr-21-437).
Methods
Cell lines and cell culture
Human SCLC cell lines (21 cell lines) were purchased from ATCC. Cell lines were tested and authenticated by short tandem repeat profiling (DNA fingerprinting) and not cultured for greater than 6-month and were routinely tested for Mycoplasma contamination. Cell lines were cultured in either RPMI or HITES medium with 10% fetal bovine serum and 1% pen-strep, at 37 °C in a humidified chamber with 5% CO2.
Chemical compounds
Lurbinectedin was purchased from MedChemExpress (NJ, USA) or acquired from the MD Anderson pharmacy. Ceralasertib (AZD6738), was provided by Astra Zeneca. Berzosertib (VX-970) was purchased from Selleck Chemicals (Houston, TX, USA). All compounds were dissolved in dimethyl sulfoxide (DMSO) for in vitro treatments.
Cell proliferation assay
Cells were seeded in 96-well plates at 2,000 cells per well in triplicate for each cell line. After 24 hours, the cells in each well were treated with a lurbinectedin and/or AZD6738 and VX-970 or with vehicle control. Four days later, proliferation was assayed by Cell Titer Glo (Promega, Fitchburg, WI, USA). For single drug treatments, median inhibitory concentration (IC50) values were estimated by the Drexplorer software using date from duplicate experiments (14). For drug combinations, the area under the curve (AUC) of the observed (or experimental) effect of the combination was compared to the predicted additive effect. Data was subsequently presented as a ratio of the experimental effect relative to the predicted additive effect based on the Bliss Independence model, as previously reported (15). Using 10% above or below the predicted additive effect as a cut-off, we then assigned the following groups: observed/predicted <−0.1= greater than additive; observed/predicted <0.1= less than additive; observed/predicted <0.1 and >−0.1= additive.
siRNA based knockdown of SLFN11
For SLFN11 gene silencing, pooled small interfering RNAs (siRNAs) targeting SLFN11 (L-016764–01-0005) or its corresponding scramble control (D-001810–10-05; GE Dharmacon) were transfected into DMS79 and H209 cells for 72 hours using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. The cells were then plated for 96 h proliferation assays. Knock down efficiency was validated by Western blotting.
Western blot
Protein lysates were isolated from SCLC cells treated with 0.9 nM single agent lurbinectedin or DMSO for 24 and 48 hours and for the combination experiment 0.6 nM of lurbinectedin with 0.3 µM of ATR inhibitor. Nitrocellulose membranes were incubated with primary antibodies (1:1,000) for Human SLFN11 (Santa Cruz Biotechnology), phospho-(S345) CHK1 Cell Signaling Technologies (CST), phospho-(S4/S8) RPA32 (Bethyl laboratories), phospho-(S139) γH2AX (CST), cleaved caspase-3 (CST), phospho-(S366) STING (SAB Biotech), cGAS (CST), PD-L1 (CST), GAPDH (Santa Cruz Biotechnology) and Vinculin (Sigma-Aldrich) overnight. Secondary anti-goat, anti-mouse (Santa Cruz Biotechnology) and anti-rabbit HRP linked antibodies (CST) were used and detected using the Chemidoc imaging system, image captured with Image Studio Version 3.1 software (BioRad).
Biomarker analysis
Differences in IC50 values between subgroups were tested using ANOVA. Differences between SLFN11 high and low groups, defined using the bi-modality index (16), were tested using an un-paired t-test. Spearman rank correlation was used to assess the association between IC50 values and RPPA protein markers, The Benjamini & Hochberg [1995] method (17) was used to control false discovery rate (FDR). RPPA generates a quantitative measurement of expression of over 200 total and phosphorylated proteins, these values can be correlated to other variables such as IC50 values, and be compared between biomarker defined groups using standard statistical techniques. All analyses were performed using the R software.
Mouse strains
For the flank xenograft models, 6-week-old female athymic nude mice (Envigo) were used. Animals were maintained in accordance with the Institutional Animal Care and Use Committee (IACUC) of The University of Texas MD Anderson Cancer Center and the NIH Guidelines for the Care and Use of Laboratory Animals. Experiments were performed under protocol: 00001191-RN03.
Establishment of flank xenografts and efficacy studies in nude mice
High SLFN11 protein expressing SCLC cell line DMS79 and low SLFN11 protein expressing cell line H865 were used for generation of xenografts. For subcutaneous injections, 1×106 SCLC cells were injected into one flank of each mouse with PBS: Matrigel (1:1, BD Biosciences). At an average tumor volume of 100 mm3, mice were randomly allocated to three groups (n=5). Mice received either Vehicle (0.9% saline), lurbinectedin (0.1 mg/kg i.v. once per week), or cisplatin (6 mg/kg i.p. once per week). Tumor volumes were measured every other day using digital calipers.
Statistical analysis
Differences in IC50 values between subgroups were tested using ANOVA. Differences between SLFN11 high and low groups, defined using the bi-modality index were tested using an un-paired t-test. Spearman rank correlation was used to assess the association between IC50 values and RPPA protein markers, The Benjamini & Hochberg (1995) method was used to control FDR. Differences in vivo drug sensitivity were tested by comparing tumor control ratios using an un-paired t-test.
Results
Human SCLC cells show sensitivity to lurbinectedin both in vitro and in vivo
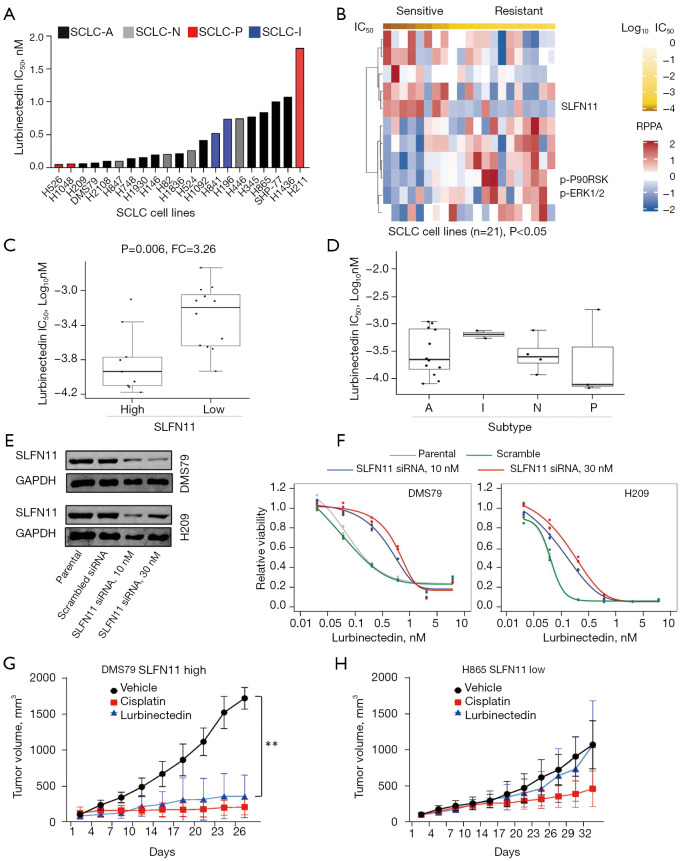
To assess the sensitivity of SCLC to lurbinectedin we screened a panel of 21 human SCLC cell lines, representing all the four SCLC subtypes (A, N, P and I) in cell proliferation assays. Though majority of SCLC cell lines were sensitive to lurbinectedin at a nanomolar range, they showed a range of sensitivities (Figure 1A). Notably, all IC50 values are lower than the peak plasma concentration of lurbinectedin of 148.2–153.8 ng/mL (188.8–196.0 nM) reported in clinical testing (5). Next, we sought to identify potential protein biomarkers of response leveraging baseline reverse phase protein array (RPPA) proteomic profiling data of our panel of cell lines, which quantifies expression of 209 total and phosphorylated proteins. To identify novel biomarkers of response we correlated IC50 values with the RPPA expression dataset—SLFN11 was identified as the strongest biomarkers of sensitivity to lurbinectedin (R=−0.618, P=0.003), with phosphorylated P90RSK, phosphorylated ERK1/2 and phosphorylated PI3K amongst the top biomarkers of resistance (R=0.664, P=0.001, R=0.474, P=0.030, and R=0.433, P=0.049, respectively) (Figure 1B, Table S1). Additionally activated mTOR (p-mTOR) was associated with relative resistance to lurbinectedin, but failed to reach statistical significance (P=0.067). As SLFN11 is a known biomarker of sensitivity to chemotherapy and other DNA damaging agents, we performed a secondary analysis dichotomizing the cell lines based on SLFN11 expression which is bimodal in SCLC (10). As shown in Figure 1C, cell lines with higher SLFN11 expression (bimodal separation by RPPA) were significantly more sensitive to lurbinectedin (P=0.006, FC=3.26). We then compared lurbinectedin cytotoxicity based on our recently published molecularly-defined subtypes of SCLC (SCLC-A, SCLC-N, SCLC-P, SCLC-I) (18), there was, however, no significant difference in the in vitro activity of lurbinectedin between the subtypes (Figure 1D), a not unsurprising observation as all four subtypes contain models with high SLFN11 expression. To validate SLFN11 as a biomarker of response to lurbinectedin, we first knocked down SLFN11 expression using siRNA in high SLFN11 protein expressing cell lines-DMS79 and H209 (Figure 1E) and performed proliferation assays. In both DMS79 and H209, SLFN11 knockdown reduced sensitivity to lurbinectedin as compared to their respective parental and scrambled siRNA controls (Figure 1F). Second, we compared the in vivo efficacy of lurbinectedin in models representing SLFN11-high and -low disease. In DMS79 xenografts that have high SLFN11 expression, lurbinectedin treatment significantly reduced tumor growth as compared to vehicle (Figure 1G). However, H865 xenografts, with lower SLFN11 expression, were significantly less sensitive to lurbinectedin, supporting SLFN11 as a biomarker of response to lurbinectedin (Figure 1H). Since SCLC-A is the most prevalent SCLC subtype (18) and showed a range of in vitro sensitivities to lurbinectedin (Figure 1D), we repeated our biomarker analysis in SCLC-A cell lines. Again, SLFN11 was identified as a top biomarker of sensitivity, and phosphorylated P90RSK as biomarker of resistance (Figure S1, Table S2).
Figure 1.
SLFN11 predicts sensitivity to lurbinectedin in SCLC. (A) Sensitivity of a panel of 21 human SCLC cell lines to lurbinectedin (ranked by IC50) and color coded for SCLC subtypes. (B) Spearman correlation of RPPA protein markers and IC50 values in panel of 21 cell lines (P<0.05). (C) Comparison of lurbinectedin IC50 values between cell lines with high and low SLFN11 expression by RPPA (bi-modal separation, mean ± SEM, P value by t-test). (D) Comparison of lurbinectedin IC50 values between cell lines from the four SCLC subtypes SCLA-A (n=12), -N (n=4), -P (n=3) and -I (n=2) (P values by t-test). (E) Western-blot showing knockdown of SLFN11 following siRNA DMS79 and H209 cell lines. (F) Sensitivity of cell lines following SLFN11 knockdown. (G,H) Tumor growth curves of SLFN11 high xenografts (DMS79) and SLFN11 low xenografts (H865) treated with vehicle, lurbinectedin (0.1 mg/kg), or cisplatin (6 mg/kg) (**, P=0.01 by un-paired t-test).
These observations confirm SLFN11 as a biomarker of response to lurbinectedin, in agreement with previous data describing SLFN11 as a biomarker of response to a number of DNA damaging agents (10,12,13,19). Activation of the PI3K/mTOR pathway has also been identified as a biomarker of resistance to DNA damaging agents such as cisplatin and PARP inhibitors (15,20), further suggesting overlapping mechanisms of resistance between DNA damaging agents in SCLC.
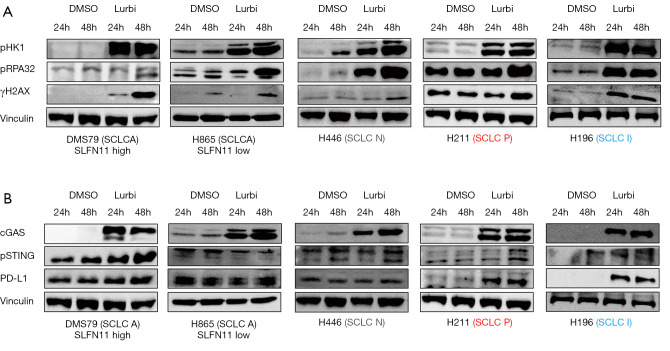
Lurbinectedin induces DNA damage and replication stress.
Based on its mechanism of action, we predicted that lurbinectedin would lead to increased replication stress and DNA damage in SCLC models. To test this, SCLC cells from all four subtypes were treated with lurbinectedin (0.9 nM/L) for 24 and 48 hours and markers of DNA damage and replication stress were assayed by Western blot. All cell lines tested showed increases in markers of replication stress—phospho CHK1 (S345) and phospho RPA32 (S4/S8)—as compared to DMSO control (Figure 2A and Figure S2A). Similar increases in phospho γH2AX were also observed, indicating DNA double-strands breaks in SCLC cells upon treatment with lurbinectedin (Figure 2A and Figure S2A).
Figure 2.
Induction of DNA damage and PD-L1 expression following lurbinectedin treatment. (A) Western-blot showing changes in replication stress markers (pCHK1 and pRPA32) and DNA damage (pγH2Ax) in SCLC cell lines following 24- and 48-hour treatment with DMSO or lurbinectedin (Lurbi, 0.9 nM). (B) Treatment with lurbinectedin activates cGAS, phosphorylates STING and increases PD-L1 expression in SCLC-P and SCLC-I cell lines.
Lurbinectedin treatment activates the cGAS-STING pathway and increases PD-L1 expression
As our group has shown that DNA damage induces release of cytosolic DNA and increases PD-L1 level via activation of the cGAS-STING pathway in pre-clinical models of SCLC (21,22), we sought to test if lurbinectedin also induces PD-L1 expression via activation of cGAS-STING signaling. Interestingly, treatment of SCLC cells with lurbinectedin for 24 and 48 hours induced an increase in protein levels of cGAS in all cell lines tested, as compared to DMSO control (Figure 2B and Figure S2B). However, a corresponding increase in phospho STING and PD-L1 was only observed in cell lines from the SCLC-P and SCLC-I subtypes (Figure 2B and Figure S2C). These data suggest potential for a synergistic interaction between lurbinectedin and anti-PD-L1 treatment, but that further testing is required to identify potential biomarkers for this interaction to explain the variation seen in the cell lines tested, and to test the combination in immune competent models of SCLC.
As treatment with other DNA damaging agents (cisplatin, PARP inhibitors) can reduce expression of SLFN11, and reports have shown that the activation of the cGAS-IFN axis can induce SLFN11 expression, we were interested in understanding the effect of lurbinectedin on SLFN11. In three cell lines with high SLFN11 expression—DMS-79, H209, H1048—lurbinectedin caused variable changes in SLFN11 levels ranging from a minimal decrease (DMS-79), to reduced to an undetectable level (H1048) (Figure S2B). We hypothesize that these variable changes are reflective of a balance between epigenetic down-regulation related to treatment, and up-regulation via activation of the cGAS-IFN pathway —further testing of this relationship, including in immune competent systems, will be required to fully understand this relationship (13).
ATR inhibition potentiates the cytotoxic activity of lurbinectedin in SLFN11 low SCLC cell lines
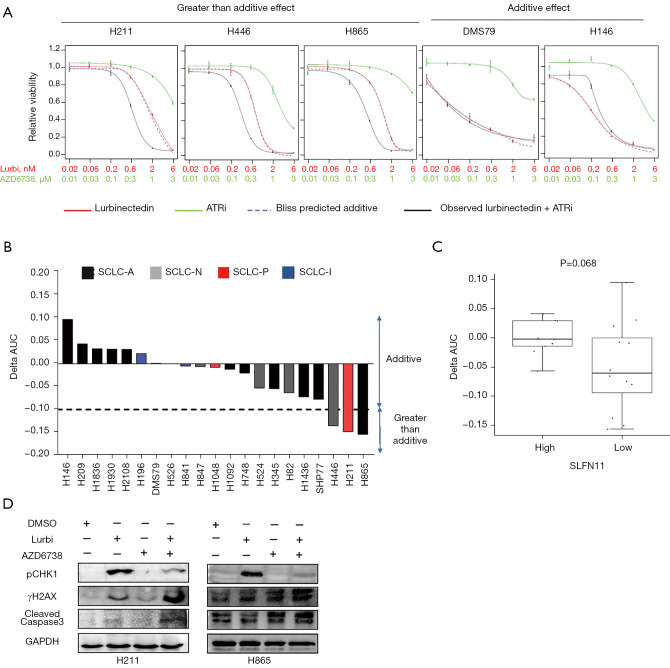
Since we observed an increase in phosphorylated CHK1 in SCLC cell lines upon treatment with lurbinectedin, and that SLFN11 deficient cells are reliant on the ATR/CHK1-mediated replication stress and DNA damage repair pathway (23-26), we sought to investigate the combination of an ATR inhibitor, ceralasertib (AZD6738), with lurbinectedin in SCLC cell lines. Using the Bliss Delta AUC (ΔAUC) method, which compares the observed and predicted additive interaction between two drugs, we observed a range of interactions (Figure 3A, Figure S3A), both additive (ΔAUC >−0.1 and <0.1) and greater than additive (Δ <−0.1). The three cell lines with a greater than additive response to the combination with ceralasertib—H865, H211, and H446—are members of subsets A, P, and N respectively (Figure 3B) and as expected, are amongst the cell lines with low SLFN11 level and least sensitive to single agent lurbinectedin. To account for potential off-target effects, we also tested lurbinectedin with a second ATR inhibitor, berzosertib (VX-970). Similar to the combination with ceralasertib, the lurbinectedin/berzosertib combination showed a range of interactions, with the three cell lines with a greater than additive response -SHP77, H211 and H446 represent the A, P, and N subtypes respectively (Figure S3B). To explore potential biomarkers of response to the combination, we compared ΔAUC between the subtypes and between SLFN11 high and low cell lines. Opposite to single agent lurbinectedin (Figure 1C), cell lines with lower SLFN11 expression had better responses to the combination of lurbinectedin and ceralasertib (i.e., smaller ΔAUC values; P=0.068, Figure 3C) or berzosertib (P=0.101, Figure S3C). All three cell lines in which a greater than additive response to the combination of lurbinectedin and ceralasertib or berzosertib was observed were in the SLFN11 low group, and were relatively resistant to single agent ceralasertib or berzosertib. To understand the mechanism of action when we treated H211 and H865 cell lines with the combination of lurbinectedin and ceralasertib or berzosertib for 48 h, Western blotting results showed marked increase in γH2AX and cleaved caspase 3 levels following the combination treatment as compared to either single agent lurbinectedin and ceralasertib (Figure 3D) and for berzosertib (Figure S3D). Reduction in pCHK1 in single agent ATR inhibitor and in combination treatment group in compared to lurbinectedin represents a halt in the DNA damage repair system (Figure 3D). Similar to single agent lurbinectedin, there appear to be no differences in effectiveness of combining lurbinectedin with ceralasertib or berzosertib between the SCLC-A, -N, -P, and -I subtypes (Figure S3E).
Figure 3.
Combination of lurbinectedin with ATR inhibitor is greater than additive in SLFN11 low cell lines. (A) Relative proliferation of cell lines representing greater than additive, and additive responses following 96 h treatment with lurbinectedin (Lurbi), ceralasertib and their combination at indicated concentrations (mean ± SEM). (B) Bar-graph representing ∆AUC for all 21 cell lines tested, color coded by SCLC subtype. ∆AUC value denotes difference in area under the dose-response curve of the observed drug combination and the predicted additive effect of the single agents was calculated using the BLISS independence model. ∆AUC <−0.1 indicates a greater than additive effect of the combination. (C) Comparison of ∆AUC values between cell lines with high and low SLFN11 expression by RPPA (bi-modal separation, mean ± SEM, P values by t-test) shows a better response to the combination in SLFN11 low cell lines. (D) Western-blot showing changes in pCHK1, pγH2Ax and cleaved caspase3 in H211 and H865 cell lines treated with 0.6 nM of lurbinectedin, 0.3 µM of ceralasertib and their combination.
Discussion
The lack of effective therapies and predictive biomarkers are major challenges in the management of advanced SCLC. For decades, topotecan was the only FDA approved treatment for relapsed SCLC, until the recent accelerated FDA approval of lurbinectedin in the second line setting for metastatic SCLC. In the current study we explored the therapeutic efficacy of lurbinectedin in a large number of proteomically profiled SCLC cell lines as well as in cell line xenograft models as a single-agent, and in combination with ATR inhibitors (ceralasertib and berzosertib). Lurbinectedin showed robust cytotoxic effect in SCLC cell lines with IC50 values ranging from 0.06 to 1.83 nM (Figure 1A), all comfortably below the reported clinical plasma concentration (5).
In this study, we also found that SCLC models with high levels of SLFN11 protein were more sensitive to lurbinectedin. Previous studies from our group and others have shown that SLFN11 predicts sensitivity to some DNA damaging agents (e.g., cisplatin and PARP inhibitors), but not others (e.g., temozolomide) (10,12,25,27-31). Recently, our group developed a CLIA-certified immunohistochemistry (IHC) assay (12) for SLFN11 for a cooperative group trial testing the activity of PARP inhibitors in a SLFN11-positive patient population (SWOG1929/NCT04334941). Based on our findings here, we propose that SLFN11 warrants further investigation in the clinical setting to determine whether it may help identify patients who may receive relatively greater benefit from lurbinectedin.
Related to this, prior studies by our group and others in SCLC have shown that SLFN11 is downregulated in preclinical models with platinum resistance and that fewer SLFN11 positive circulating tumor cells are observed in patients with relapsed SCLC (as compared to treatment naïve) (10,12,32,33). This may help to explain the higher response rates observed with single agent lurbinectedin in platinum-sensitive disease (ORR 45%) as compared to platinum-resistant SCLC (22%) (5). Lurbinectedin not only blocks RNA-polymerase-II but also causes DNA damage (8,34). Our observations of increases in DNA damage and DNA damage response markers in SCLC cell lines after lurbinectedin treatment support that notion. In all cell lines tested—representing both the four SCLC subtypes, and SLFN11 high/low SCLC—lurbinectedin treatment induced a robust increase in pγH2AX, pCHK1, pRPA32 (Figure 2A). Based upon these molecular changes that indicated activation of the ATR pathway, and previous studies demonstrating ATR inhibition could re-sensitize otherwise resistant SLFN11-low cells to chemotherapy and PARP inhibitors (23-25), we further studied whether an ATR inhibitor (ceralasertib/berzosertib) mediated induction of replication stress could overcome single agent resistance to lurbinectedin. Combining ceralasertib/berzosertib with lurbinectedin showed a greater than additive effect in SCLC cell lines with higher IC50 values for lurbinectedin (Figure 3A, Figure S3A). Importantly, SLFN11 low SCLC cells which were less sensitive to single-agent lurbinectedin (and relatively resistant to single agent ATR inhibition) became sensitive when treated with combination of lurbinectedin and ceralasertib or berzosertib.
We have previously reported that treatment with DNA damage response inhibitors (DDRi) increases protein expression of PD-L1 in SCLC cells via activation of the cGAS-STING pathway and combination of DDRi with anti-PD-L1 showed robust anti-tumor activity (13,21,22). Here we also observed significant increase in cGAS, phosphorylated STING, and PD-L1 expression in SCLC-P and SCLC-I subtype cells following lurbinectedin treatment (Figure 2B, Figure S2C). While lurbinectedin treatment failed to increase PD-L1 expression in all models tested, the potential for an approved second line treatment to induce such an increase, and therefore to synergize with immune therapy warrants further exploration.
SLFN11 has recently emerged as a promising predictive biomarker in SCLC, predicting response to a wide range of DNA-damaging chemotherapies and PARPi. SLFN11 detection by IHC has been validated and evaluated in several clinical trials retrospectively (12,27). The first clinical trial in SCLC selecting patients for SLFN11-positive tumors is currently ongoing (SWOG1929/NCT04334941), which uses a CLIA-certified SLFN11 IHC assay. Given the availability of SLFN11 IHC testing, this biomarker could be rapidly adopted in clinical trials, both to be explored in ongoing lurbinectedin studies, and to select for SLFN11 status in prospective trials.
In conclusion, our findings demonstrate that SLFN11 is a top candidate predictive biomarker of response to single-agent lurbinectedin. Notably lurbinectedin in combination with an ATR inhibitor had greater than additive effects and re-sensitized otherwise resistant SLFN11-low cells. SLFN11 IHC could easily be translated into clinical setting and be immediately leveraged in ongoing and future clinical trials studying lurbinectedin and combination treatment strategies for patients with SCLC.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This work was supported by: The NIH/NCI CCSG P30-CA016672 (Bioinformatics Shared Resource); NIH/NCI R01-CA207295 (LAB); University of Texas SPORE in Lung Cancer P5-CA070907 (KRC, CMG, JW, LAB); NIH/NCI U01-CA213273 (LAB); NIH/NCI T32 CA009666 (CMG); NIH/NCI R50-CA243698 (CAS); IALSC ILCF Fellowship (KRC); ASCO Young Investigator Award (CMG); the Khalifa Bin Zayed Al Nahyan Foundation (CMG); through generous philanthropic contributions to The University of Texas MD Anderson Lung Cancer Moon Shot Program (JW, LAB) and to the Byers Lab; The Andrew Sabin Family Fellowship (LAB); CMG and LAB were supported by the Abell Hangar Foundation, the LUNGevity Foundation Career Development Award (CMG), and the Rexanna Foundation for Fighting Lung Cancer (CMG, LAB).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Animals were maintained in accordance with the Institutional Animal Care and Use Committee (IACUC) of the University of Texas MD Anderson Cancer Center and the NIH Guidelines for the Care and Use of Laboratory Animals. Experiments were performed under protocol: 00001191-RN03.
Footnotes
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/tlcr-21-437
Data Sharing Statement: Available at https://dx.doi.org/10.21037/tlcr-21-437
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tlcr-21-437). Dr. CMG serves on advisory committees for Jazz Pharmaceuticals and AstraZeneca. Dr. LAB serves on advisory committees for Jazz Pharmaceuticals, AstraZeneca, Pharma Mar SA, and Merck and has research support from AstraZeneca. The other authors have no conflicts of interest to declare.
References
- 1.Rudin CM, Brambilla E, Faivre-Finn C, et al. Small-cell lung cancer. Nat Rev Dis Primers 2021;7:3. 10.1038/s41572-020-00235-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Horn L, Mansfield AS, Szczęsna A, et al. First-Line Atezolizumab plus Chemotherapy in Extensive-Stage Small-Cell Lung Cancer. N Engl J Med 2018;379:2220-9. 10.1056/NEJMoa1809064 [DOI] [PubMed] [Google Scholar]
- 3.Paz-Ares L, Dvorkin M, Chen Y, et al. Durvalumab plus platinum-etoposide versus platinum-etoposide in first-line treatment of extensive-stage small-cell lung cancer (CASPIAN): a randomised, controlled, open-label, phase 3 trial. Lancet 2019;394:1929-39. 10.1016/S0140-6736(19)32222-6 [DOI] [PubMed] [Google Scholar]
- 4.Available online: NCCN. http://www.nccn.org/professionals/physician_gls/pdf/sclc.pdf
- 5.Trigo J, Subbiah V, Besse B, et al. Lurbinectedin as second-line treatment for patients with small-cell lung cancer: a single-arm, open-label, phase 2 basket trial. Lancet Oncol 2020;21:645-54. 10.1016/S1470-2045(20)30068-1 [DOI] [PubMed] [Google Scholar]
- 6.Soares DG, Machado MS, Rocca CJ, et al. Trabectedin and its C subunit modified analogue PM01183 attenuate nucleotide excision repair and show activity toward platinum-resistant cells. Mol Cancer Ther 2011;10:1481-9. 10.1158/1535-7163.MCT-11-0252 [DOI] [PubMed] [Google Scholar]
- 7.Vidal A, Muñoz C, Guillén MJ, et al. Lurbinectedin (PM01183), a new DNA minor groove binder, inhibits growth of orthotopic primary graft of cisplatin-resistant epithelial ovarian cancer. Clin Cancer Res 2012;18:5399-411. 10.1158/1078-0432.CCR-12-1513 [DOI] [PubMed] [Google Scholar]
- 8.Santamaría Nuñez G, Robles CM, Giraudon C, et al. Lurbinectedin Specifically Triggers the Degradation of Phosphorylated RNA Polymerase II and the Formation of DNA Breaks in Cancer Cells. Mol Cancer Ther 2016;15:2399-412. 10.1158/1535-7163.MCT-16-0172 [DOI] [PubMed] [Google Scholar]
- 9.Belgiovine C, Bello E, Liguori M, et al. Lurbinectedin reduces tumour-associated macrophages and the inflammatory tumour microenvironment in preclinical models. Br J Cancer 2017;117:628-38. 10.1038/bjc.2017.205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allison Stewart C, Tong P, Cardnell RJ, et al. Dynamic variations in epithelial-to-mesenchymal transition (EMT), ATM, and SLFN11 govern response to PARP inhibitors and cisplatin in small cell lung cancer. Oncotarget 2017;8:28575-87. 10.18632/oncotarget.15338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lok BH, Gardner EE, Schneeberger VE, et al. PARP Inhibitor Activity Correlates with SLFN11 Expression and Demonstrates Synergy with Temozolomide in Small Cell Lung Cancer. Clin Cancer Res 2017;23:523-35. 10.1158/1078-0432.CCR-16-1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pietanza MC, Waqar SN, Krug LM, et al. Randomized, Double-Blind, Phase II Study of Temozolomide in Combination With Either Veliparib or Placebo in Patients With Relapsed-Sensitive or Refractory Small-Cell Lung Cancer. J Clin Oncol 2018;36:2386-94. 10.1200/JCO.2018.77.7672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang B, Ramkumar K, Cardnell RJ, et al. A wake-up call for cancer DNA damage: the role of Schlafen 11 (SLFN11) across multiple cancers. Br J Cancer 2021. [Epub ahead of print]. doi: . 10.1038/s41416-021-01476-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tong P, Coombes KR, Johnson FM, et al. drexplorer: A tool to explore dose-response relationships and drug-drug interactions. Bioinformatics 2015;31:1692-4. 10.1093/bioinformatics/btv028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cardnell RJ, Feng Y, Mukherjee S, et al. Activation of the PI3K/mTOR Pathway following PARP Inhibition in Small Cell Lung Cancer. PLoS One 2016;11:e0152584. 10.1371/journal.pone.0152584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J, Wen S, Symmans WF, et al. The bimodality index: a criterion for discovering and ranking bimodal signatures from cancer gene expression profiling data. Cancer Inform 2009;7:199-216. 10.4137/CIN.S2846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. JRSS (B) 1995;57:289-300. 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- 18.Gay CM, Stewart CA, Park EM, et al. Patterns of transcription factor programs and immune pathway activation define four major subtypes of SCLC with distinct therapeutic vulnerabilities. Cancer Cell 2021;39:346-360.e7. 10.1016/j.ccell.2020.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murai J, Thomas A, Miettinen M, et al. Schlafen 11 (SLFN11), a restriction factor for replicative stress induced by DNA-targeting anti-cancer therapies. Pharmacol Ther 2019;201:94-102. 10.1016/j.pharmthera.2019.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cardnell RJ, Feng Y, Diao L, et al. Proteomic markers of DNA repair and PI3K pathway activation predict response to the PARP inhibitor BMN 673 in small cell lung cancer. Clin Cancer Res 2013;19:6322-8. 10.1158/1078-0432.CCR-13-1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sen T, Della Corte CM, Milutinovic S, et al. Combination Treatment of the Oral CHK1 Inhibitor, SRA737, and Low-Dose Gemcitabine Enhances the Effect of Programmed Death Ligand 1 Blockade by Modulating the Immune Microenvironment in SCLC. J Thorac Oncol 2019;14:2152-63. 10.1016/j.jtho.2019.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sen T, Rodriguez BL, Chen L, et al. Targeting DNA Damage Response Promotes Antitumor Immunity through STING-Mediated T-cell Activation in Small Cell Lung Cancer. Cancer Discov 2019;9:646-61. 10.1158/2159-8290.CD-18-1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jo U, Murai Y, Chakka S, et al. SLFN11 promotes CDT1 degradation by CUL4 in response to replicative DNA damage, while its absence leads to synthetic lethality with ATR/CHK1 inhibitors. Proc Natl Acad Sci U S A 2021;118:e2015654118. 10.1073/pnas.2015654118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murai J, Feng Y, Yu GK, et al. Resistance to PARP inhibitors by SLFN11 inactivation can be overcome by ATR inhibition. Oncotarget 2016;7:76534-50. 10.18632/oncotarget.12266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murai J, Tang SW, Leo E, et al. SLFN11 Blocks Stressed Replication Forks Independently of ATR. Mol Cell 2018;69:371-384.e6. 10.1016/j.molcel.2018.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murai J, Pommier Y. PARP Trapping Beyond Homologous Recombination and Platinum Sensitivity in Cancers. Annu Rev Cancer Biol 2019;3:131-50. 10.1146/annurev-cancerbio-030518-055914 [DOI] [Google Scholar]
- 27.Byers LA, Bentsion D, Gans S, et al. Veliparib in Combination with Carboplatin and Etoposide in Patients with Treatment-Naïve Extensive-Stage Small Cell Lung Cancer: A Phase 2 Randomized Study. Clin Cancer Res 2021;27:3884-95. 10.1158/1078-0432.CCR-20-4259 [DOI] [PubMed] [Google Scholar]
- 28.Kang MH, Wang J, Makena MR, et al. Activity of MM-398, nanoliposomal irinotecan (nal-IRI), in Ewing's family tumor xenografts is associated with high exposure of tumor to drug and high SLFN11 expression. Clin Cancer Res 2015;21:1139-50. 10.1158/1078-0432.CCR-14-1882 [DOI] [PubMed] [Google Scholar]
- 29.Nogales V, Reinhold WC, Varma S, et al. Epigenetic inactivation of the putative DNA/RNA helicase SLFN11 in human cancer confers resistance to platinum drugs. Oncotarget 2016;7:3084-97. 10.18632/oncotarget.6413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takashima T, Sakamoto N, Murai J, et al. Immunohistochemical analysis of SLFN11 expression uncovers potential non-responders to DNA-damaging agents overlooked by tissue RNA-seq. Virchows Arch 2021;478:569-79. 10.1007/s00428-020-02840-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zoppoli G, Regairaz M, Leo E, et al. Putative DNA/RNA helicase Schlafen-11 (SLFN11) sensitizes cancer cells to DNA-damaging agents. Proc Natl Acad Sci U S A 2012;109:15030-5. 10.1073/pnas.1205943109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gardner EE, Lok BH, Schneeberger VE, et al. Chemosensitive Relapse in Small Cell Lung Cancer Proceeds through an EZH2-SLFN11 Axis. Cancer Cell 2017;31:286-99. 10.1016/j.ccell.2017.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lheureux S, Oaknin A, Garg S, et al. EVOLVE: A Multicenter Open-Label Single-Arm Clinical and Translational Phase II Trial of Cediranib Plus Olaparib for Ovarian Cancer after PARP Inhibition Progression. Clin Cancer Res 2020;26:4206-15. 10.1158/1078-0432.CCR-19-4121 [DOI] [PubMed] [Google Scholar]
- 34.Tumini E, Herrera-Moyano E, San Martín-Alonso M, et al. The Antitumor Drugs Trabectedin and Lurbinectedin Induce Transcription-Dependent Replication Stress and Genome Instability. Mol Cancer Res 2019;17:773-82. 10.1158/1541-7786.MCR-18-0575 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as