Abstract
Fifty-five Shiga toxin (Stx)-producing Escherichia coli (STEC) O26:H11 and O26:H− strains isolated from humans between 1965 and 1999 in Germany and the Czech Republic were investigated for their chromosomal and plasmid characteristics. All motile (n = 23) and nonmotile (n = 32) STEC O26 strains were shown to possess the identical flagellin subunit-encoding gene (fliC). We observed a striking recent shift of the stx genotype from stx1 to stx2 among the STEC O26 isolates. While stx1 was the exclusive genotype identified in our collection until 1994, 94% of the isolates obtained after 1997 possessed stx2 either alone (71%) or together with stx1 (23%). Plasmid profiling demonstrated a remarkable heterogeneity with respect to plasmid sizes and combinations. Southern blot analysis of plasmid DNA with probes specific to potential accessory virulence genes revealed considerable additional variability in gene composition and arrangement. Pulsed-field gel electrophoresis (PFGE) differentiated 16 subgroups among the 55 STEC O26 strains. Using these techniques we demonstrate the emergence of a new clonal subgroup characterized by PFGE pattern A and a unique combination of virulence markers including stx2 and a single, approximately 90-kb plasmid harboring the enterhemorrhagic E. coli hlyA and etp genes. The proportion of PFGE subgroup A strains among STEC O26 isolates rose from 30% in 1996 to more than 50% in 1999. Four clusters of infections with the clonal subgroup A were identified. We conclude that the STEC serogroup O26 is diverse and that pathogenic clonal subgroups can rapidly emerge during short intervals. The extensive genetic diversity of STEC O26 provides a basis for molecular subtyping of this important non-O157 STEC serogroup.
Shiga toxin (Stx)-producing Escherichia coli (STEC) cause a broad spectrum of diseases in humans ranging from diarrhea to hemolytic uremic syndrome (HUS) (46). In addition to STEC O157:H7/H−, particular non-O157 STEC serogroups, especially O26, have emerged as significant causes of human diseases (2, 6, 46).
Five years before the identification of E. coli O157:H7 as a pathogen, STEC strains belonging to the serotype O26:H11 were recognized as causes of diarrhea (21). In 1987, Levine et al. (23) classified a variety of Stx-producing E. coli O26:H11 strains as enterohemorrhagic E. coli (EHEC) because they were associated with hemorrhagic colitis and HUS and hybridized to the CVD 419 probe, which is complementary to sequences of the plasmid-encoded EHEC hemolysin gene (EHEC-hly) of E. coli O157:H7 (35).
STEC O26 strains produce Stx1, Stx2, or both (6, 31, 41). In the prototype STEC O26 strain H19, the stx1 gene was shown to be localized to the lambdoid prophages H19B (43) and H19J (25). STEC O26 also possess the eae gene (6) which, in STEC O157:H7 and enteropathogenic E. coli (EPEC), is located within the pathogenicity island LEE (locus of enterocyte effacement) (16). Another gene, designated pas (protein associated with secretion), has recently been identified in the LEE of STEC O157 and of an STEC O26:H− strain of bovine origin (22). The pas gene product is required for the secretion of Esp proteins (22). Moreover, we have recently shown that the genome of human STEC O26, but not of STEC O157:H7, contains a region also found in pathogenic yersiniae and termed a high-pathogenicity island (HPI) (19). The HPI contains genes encoding the pesticin receptor FyuA and the siderophore yersiniabactin (40).
STEC O26 possess large plasmids (9, 18) that contain genes encoding additional potential virulence factors originally identified in STEC O157:H7. In addition to the EHEC hemolysin, which acts as a pore-forming cytolysin (35), the bifunctional catalase-peroxidase (KatP) (7), and a serine protease (EspP) which cleaves human coagulation factor V (8) have been detected (9). Another closely related serine protease, termed PssA (protease secreted by STEC), has been identified on the large plasmid of a bovine STEC O26:H− strain (13). The etp gene cluster that presumably encodes a type II secretion pathway system in STEC O157:H7 (36) has not been found in STEC O26 isolates (37).
In the present study we analyzed 55 STEC O26 strains isolated between 1965 and 1999 from patients in Germany and the Czech Republic for their chromosomal and plasmid characteristics to determine their clonal structure and to create a basis for subtyping of STEC O26 isolates in epidemiological studies.
MATERIALS AND METHODS
Bacterial strains.
Fifty-five STEC strains of serotypes O26:H11 and O26:H−, originating from stools of patients with HUS (n = 30) or diarrhea (n = 24) and of one asymptomatic carrier were investigated (Table 1). The strains were isolated between 1965 and 1999 at the Institute for Hygiene and Microbiology of the University of Würzburg, Würzburg, Germany (48 isolates), and at the Institute for Medical Microbiology of the Charles University, Prague, Czech Republic (7 isolates) by previously described procedures (2, 6, 44). The isolates obtained from 1965 to 1982 were detected by slide agglutination with O26 antiserum and identified as STEC retrospectively based on their cytotoxicity for Vero cells. Eleven strains originated from four clusters of STEC O26-associated diarrhea and HUS (Table 1). The remaining 44 strains were from sporadic infections without apparent geographical and temporal linkage. Twenty-three of the isolates from Germany were described previously (9, 19, 37, 38). STEC O26:H11 strain H19 (21, 42) from our collection was included as a reference strain. E. coli O157 strains used as controls for fliC PCR included EDL 933 (O157:H7), 702/88 (O157:H−), 693/91 (O157:H19), 241/88 (O157:H43), and 1083/87 (O157:H45) (39). E. coli G5244 (28) was obtained from the Centers for Disease Control and Prevention, Atlanta, Ga.
TABLE 1.
STEC O26 strains used in the study and their chromosomal and plasmid characteristics
| Strain/yr of isolation | Diseasea | Serotype | Chromosomal characteristics
|
Plasmid characteristics
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| PFGE patternb | stx genotypen | Plasmid profilec [size(s) in kb] | Sizes (kb) found by Southern blot hybridization with probed:
|
||||||
| EHEC-hlyA | espP | katPe | etp | ||||||
| 1226/65*m | D | O26:H11 | G | 1 | 90, 6, 4f | 18 | 9.5 | 8.5, 7.5 | – |
| 3608/71* | D | O26:H11 | O | 1 | 57, 6, 4, 2.8, 1 | – | – | – | – |
| 1557/77 | D | O26:H11 | G | 1 | 12, 8, 6, 4 | – | – | – | – |
| 147/82* | D | O26:H11 | Q | 1 | 90, 57, 6, 4, 1 | 17 | 9.5 | 8.5, 4.2 | – |
| 2350/85 | HUS | O26:H11 | G | 1 | 12, 8, 6, 4 | – | – | – | – |
| 37/89* | HUS | O26:H11 | Q | 1 | 90, 6 | 17 | 9.5 | 8.5, 4.2 | – |
| 144/91* | HUS | O26:H11 | Q | 1 | 90, 6 | 18 | 9.5 | 8.5, 4.2 | – |
| 3909/94 | D | O26:H−g | G | 1 | 90, 12, 6, 4 | 18 | 9.5 | 8.5, 7.5 | – |
| 4166/94 | D | O26:H− | G | 1 | 90, 57, 13, 6 | 17 | 10.5 | 8.5, 7.5 | – |
| 107/95 | D | O26:H− | M | 1 | 90, 6, 2 | 17 | 11.6 | 8.5, 7.5 | – |
| 1247/95 | HUS | O26:H− | E | 1+2 | 90, 110, 6, 2.8 | 18 | 13 | 8.5, 4.2 | – |
| 5953/95 | HUS | O26:H− | C | 2 | 6, 4 | – | – | – | – |
| 7015/95 | HUS | O26:H11 | B | 2 | 90, 6, 3.3 | 18 | 9.2 | 8.5, 7.5 | – |
| 7161/95 | A | O26:H11 | G | 1 | 90, 4 | 18 | 10.5 | 8.5, 7.5 | – |
| 3967/96 | D | O26:H− | B | 1 | 90, 6 | 18 | 9.5 | 8.5, 20 | – |
| 5157/96 | D | O26:H11 | I | 1 | 90, 4 | 17 | 9.5, 13e | 8.5, 20, 7.5 | – |
| 5236/96 | D | O26:H11 | K | 1 | 90, 6, 2.8 | 18 | 9.5 | 8.5, 10.5 | – |
| 5720/96 | HUS | O26:H− | A | 2 | 90 | 12.4 | – | – | 4 |
| 6061/96 | HUS | O26:H11 | A | 2 | 90 | 12.4 | – | 4 | |
| 6068/96 | HUS | O26:H− | A | 2 | 90 | 12.4 | – | – | 4 |
| 6105/96 | D | O26:H− | E | 1 | 90, 6, 4 | 18 | 11.4 | 8.5, 4.2 | – |
| 7661/96 | D | O26:H− | C | 2 | 90, 4 | 22 | 11.6 | 8.5, 4.2 | – |
| 7662/96 | HUS | O26:H− | C | 2 | 90, 6, 4 | 18 | 13 | 8.5, 4.2 | – |
| 8574/96 | D | O26:H11 | H | 1 | 90, 57 | 16 | 9.2 | 8.5, 4.2 | – |
| 1448/97 | D | O26:H11 | B | 2 | 90, 6, 1 | 18 | 9.5 | 8.5, 6.2 | – |
| 1705/97 | HUS | O26:H− | B | 2 | 90, 4 | 18 | 10.5 | 8.5, 6.2 | – |
| 2574/97 | HUS | O26:H11 | A | 2 | 90 | 12.4 | – | – | 4 |
| 3901/97 | HUS | O26:H− | D | 1+2 | 90, 6 | 24 | 10.5 | 8.5, 7.5 | – |
| 3905/97 | D | O26:H− | D | 1+2 | 90, 57, 6, 3.3 | 18 | 13 | 8.5, 7.5 | – |
| 3584/97 | HUS | O26:H− | E | 2 | 90 | 18 | 10.5 | 8.5, 4.2 | – |
| 4038/97 | D | O26:H11 | J | 2 | 90, 6 | 18 | 13 | 8.5, 6.2 | – |
| 4104/97 | HUS | O26:H11 | B | 2 | 90, 4 | 18 | 10.5 | 8.5, 6.2 | – |
| 5080/97 | HUS | O26:H− | F | 1+2 | 90, 6, 3.3 | 18 | 10.5 | 8.5, 7.5 | – |
| 5917/97h | HUS | O26:H− | A | 2 | 90 | 12.4 | – | – | 4 |
| 5918/97h | D | O26:H− | A | 2 | 90 | 12.4 | – | – | 4 |
| 1833/98 | HUS | O26:H− | B | 2 | 90, 6, 4 | 18 | 9.5 | 8.5, 20 | – |
| 2150/98 | HUS | O26:H− | L | 1+2 | 90, 6 | 18 | 9.2 | 8.5, 4.2 | – |
| 2245/98 | HUS | O26:H− | L | 1 | 90, 6 | 18 | 9.5 | 8.5, 4.2 | – |
| 2569/98 | D | O26:H− | A | 2 | 90 | 12.4 | – | – | – |
| 0379/99* | HUS | O26:H11 | A | 2 | 90 | 12.4 | – | – | 4 |
| 0573/99 | D | O26:H− | P | 1+2 | 90, 110, 57, 6, 4 | 18 | 11.6 | 8.5, 4.2 | – |
| 1530/99i | HUS | O26:H11 | A | 2 | 90 | 12.4 | – | – | 4 |
| 1531/99i | HUS | O26:H11 | A | 2 | 90 | 12.4 | – | – | 4 |
| 1532/99i | HUS | O26:H11 | A | 2 | 90 | 12.4 | – | – | 4 |
| 1655/99j | HUS | O26:H− | A | 2 | 90 | 12.4 | – | – | 4 |
| 1656/99j | HUS | O26:H− | A | 2 | 90 | 12.4 | – | – | 4 |
| 2160/99 | HUS | O26:H− | A | 2 | 90 | 12.4 | – | – | 4 |
| 2162/99k | D | O26:H− | A | 2 | 90 | 12.4 | – | – | 4 |
| 2163/99k | D | O26:H− | A | 2 | 90 | 12.4 | – | – | 4 |
| 2164/99k | D | O26:H− | A | 2 | 90 | 12.4 | – | – | 4 |
| 2165/99k | D | O26:H− | A | 2 | 90 | 12.4 | – | – | 4 |
| 2185/99 | HUS | O26:H11 | A | 2 | 90 | 12.4 | – | – | 4 |
| 2640/99 | HUS | O26:H11 | D | 1+2 | 90, 6, 3.3 | 18 | 9.2 | 8.5, 7.5 | – |
| 2959/99 | D | O26:H− | I | 1 | 90, 110, 6, 2 | 17 | 13 | 8.5, 4.2 | – |
| 2971/99 | HUS | O26:H− | I | 1+2 | 90, 110, 6, 2 | 17 | 13 | 8.5, 7.5 | – |
| H19 reference strainl | D | O26:H11 | N | 1 | 90, 57, 6, 4 | 17 | 9.5 | 8.5, 20 | – |
D, diarrhea; A, asymptomatic.
PFGE patterns designated by the same letters differed in four bands at the maximum level and indicate clonal subgroups.
Size of plasmids are given in kilobases (the sizes of large plasmids are approximately 90 and 110 kb.
Sizes in kilobases of BamHI restriction fragments hybridizing with the respective probes; –, no signal obtained.
Two or more fragments of different sizes hybridized.
Plasmids of different sizes as indicated were detected.
H−, nonmotile strains.
Strains isolated from an HUS patient and his mother.
Strains isolated from three children admitted to the same hospital in Jena in April 1999.
Strains isolated from two children admitted to the University hospital in Hamburg in April, 1999
Strains isolated from four members of the same family.
Isolated in 1967 (40).
Strains from the Czech Republic are indicated by an asterisk. All other strains were from Germany.
Genotypes: 1, stx1; 2, stx2; 1+2, stx1 and stx2.
Phenotypic methods.
Following isolation, the strains were serotyped (5) and stored on nutrient agar slants. For experiments, bacteria were inoculated on fresh MacConkey agar plates and subsequently cultured as required. Stx production was tested by the Vero cell cytotoxicity assay (37). The enterohemolytic phenotype was determined on blood agar plates containing 5% of defibrinated and washed human red blood cells and 10 mM CaCl2 (35, 37).
fliC PCR-restriction fragment length polymorphism (PCR-RFLP) and sequencing.
The flagellin-encoding fliC gene was amplified with primers F-FLIC1 and R-FLIC2 and the PCR products were digested with RsaI (Gibco BRL, Eggenstein, Germany) as described by Fields et al. (14). Restriction fragments were separated on a 2% (wt/vol) agarose gel and visualized by staining with ethidium bromide. The sequencing of fliC PCR products was performed with an automated 377 DNA Sequencer (Perkin-Elmer Applied Biosystems, Weiterstadt, Germany) using a fluorescence procedure with the Taq Prism Ready Reaction DyeDeoxy Terminator Cycle Sequencing Kit (Perkin-Elmer Applied Biosystems) according to the manufacturer's instructions. Nucleotide sequence analysis was performed with the HUSAR program package (Heidelberg Unix Sequence Analysis Resources; German Cancer Research Center, Heidelberg, Germany) and with the DNASIS program (Hitachi Software).
PFGE.
Genomic DNA was prepared and embedded in agarose as described earlier (48), and digested with XbaI (Roche Diagnostics, Mannheim, Germany). Pulsed-field gel electrophoresis (PFGE) was performed with the CHEF DRII equipment (Bio-Rad, Munich, Germany) in 1.2% agarose gels at 14°C for 42 h at a constant voltage of 200 V. Pulse times were ramped with 12.6 s at the beginning and 40 s at the end. Chromosomal DNA of E. coli G5244 (28) digested with XbaI was used as a molecular size marker. Restriction fragment patterns of genomic DNA were determined using the GelScan System (Scanalytics; CSP, Inc.) and analyzed with the RFLPScan software (Scanalytics) (12). The patterns differing in up to four bands were considered related in accordance with the criteria of Tenover et al. (47). Isolates with related PFGE patterns were defined as clonal subgroups.
PCR.
PCRs for detecting STEC-specific sequences were performed in the GeneAmp PCR System 9600 (Perkin-Elmer Applied Biosystems) in a volume of 50 μl containing 5 μl of bacterial suspension (104 bacteria), 200 μM deoxynucleoside triphosphates (dATP, dCTP, dGTP, and dTTP), 30 pmol of each primer, 5 μl of 10-fold-concentrated polymerase synthesis buffer, 1.5 mM MgCl2, and 2.0 U of AmpliTaq DNA polymerase (Perkin-Elmer Applied Biosystems).
To detect stx genes, primer pairs KS7-KS8 (stx1B) and GK3-GK4 (stx2B, stx2cB) were used as described previously (15, 33). stx2 and stx2c were differentiated by restriction analysis of GK3-GK4 amplification products using HaeIII or FokI (32).
eae and pas genes were detected using primer pairs SK1-SK2 and ANK49-ANK50, respectively, as described earlier (17, 22). The eae gene was further characterized using primers LP2 (34), LP3 (34), LP4 (27), or LP5 (27) in combination with SK1 as a forward primer. Primer pairs SK1-LP2, SK1-LP3, SK1-LP4, and SK1-LP5 amplify eae types α, γ, β, and ɛ, respectively (27, 34). The irp2 and fyuA genes that were used as markers for the HPI of pathogenic yersiniae were detected with primer pairs irp2 FP-irp2 RP and fyuA FP-fyuA RP, respectively, as described by Schubert et al. (40).
The EHEC-hlyA gene was detected with primer hlyA1 and hlyA4 (35).
Plasmid analysis.
Plasmids were isolated with the Nucleobond AX100 preparation kit (Macherey-Nagel, Düren, Germany) using the protocol for low-copy plasmids. Plasmid DNA was separated by agarose (0.6%) gel electrophoresis, and bands were visualized by staining with ethidium bromide (17). Reference plasmids pBR325 (5.5 kb), pRK290 (20 kb), pRP4 (57 kb), and pR1 (93 kb) were used as molecular size markers.
Southern blot hybridization.
Plasmids were digested with BamHI (Gibco BRL), and the resulting fragments were separated in a 0.6% agarose gel. The DNA was transferred to a nylon membrane (Zeta-Probe GT; Bio-Rad) and UV cross-linked (Stratalinker UV Crosslinker 1800; Stratagene, Heidelberg, Germany). Stringent hybridization was achieved with the DIG DNA Labelling and Detection Kit (Boehringer GmbH, Mannheim, Germany) according to the manufacturer's instructions. The EHEC-hlyA, katP and etp gene probes were prepared by incorporating digoxigenin-11-deoxy-uridine-triphosphate (Boehringer GmbH) during PCR (37). Primers hlyA1 and hlyA4 (35) were used to amplify a 1,551-bp fragment of the EHEC-hlyA gene of E. coli O157:H7 strain EDL 933. A 2,125-bp fragment of the katP gene was amplified by using primers wkat-B and wkat-F (7). A 1,062-bp fragment of the etpD gene was amplified with primers D1 and D13R (36). PCR products were purified with the QIAquick PCR purification kit (Qiagen, Hilden, Germany). The espP probe, a 4.4-kb HindIII/SmaI fragment derived from plasmid pB9-5 (8), was purified from 0.6% agarose gel with the Prep-a-Gene kit (Bio-Rad) and labeled with the DIG DNA Labelling and Detection Kit (Boehringer GmbH).
Nucleotide sequence accession numbers.
The nucleotide sequences for the fliC genes of E. coli O26:H− (strain 5720/96) and O26:H11 (strain 6061/96) have been entered into the EMBL database library under accession numbers AJ243795 and AJ243796, respectively.
RESULTS
PCR-RFLP of fliC gene and fliC sequence analysis.
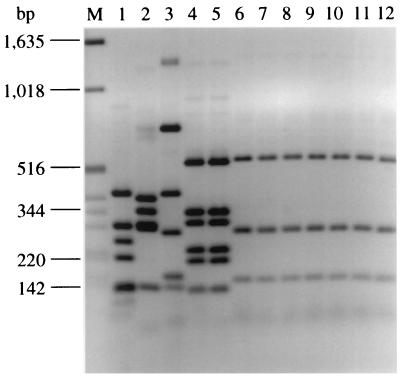
To assess whether nonmotile STEC O26 isolates possess fliC and to characterize the gene, the fliC PCR-RFLP analysis was performed with all STEC O26 strains. Representative STEC O26:H− and O26:H11 isolates are shown and compared with E. coli O157 strains in Fig. 1. A typical fliC PCR-RFLP pattern of three bands of 520, 280, and 150 bp (Fig. 1, lanes 6 to 10) was found in all 32 STEC O26:H− isolates. An identical banding pattern was displayed by the STEC O26:H11 isolates (Fig. 1, lanes 11 and 12). In contrast, the fliC-RFLP patterns of both STEC O26:H− and STEC O26:H11 isolates differed markedly from those of control E. coli O157 strains with flagellar antigens H7, H19, H43, and H45 (Fig. 1, lanes 5, 3, 2, and 1, respectively), and nonmotile E. coli O157:H− in the STEC O157:H7 lineage (Fig. 1, lane 4).
FIG. 1.
Agarose gel electrophoresis of fliC PCR products of representative STEC O26:H− and O26:H11 isolates and control E. coli O157 strains after restriction with RsaI. Lane M, molecular weight marker (1-kb DNA ladder; Gibco BRL). In lanes 1 to 12, the following E. coli strains are shown: lane 1, 1083/87 (O157:H45); lane 2, 241/88 (O157:H43); lane 3, 693/91 (O157:H19); lane 4, 702/88 (O157:H−); lane 5, EDL933 (O157:H7); lane 6, 5720/96 (O26:H−); lane 7, 6068/96 (O26:H−); lane 8, 7662/96 (O26:H−); lane 9, 1705/97 (O26:H−); lane 10, 5917/97 (O26:H−); lane 11, 6061/96 (O26:H11); and lane 12, 2574/97 (O26:H11). Based on the data derived from nucleotide sequence analysis of fliC, bands of 280 and 150 bp each consist of two fragments of similar size (269 and 276 bp and 141 and 142 bp, respectively) that could not be separated on the gel. Three additional fragments of 41, 31, and 14 bp could not be detected on the gel.
Nucleotide sequence analysis of the fliC PCR products derived from STEC O26:H− strain 5720/96 (EMBL number AJ243795) and STEC O26:H11 strain 6061/96 (EMBL number AJ243796) showed that the sequences were identical. Hence, nonmotile STEC O26 isolates possessed the H11-encoding fliC gene; however, nonmotility of such strains cannot be attributed to mutations in fliC.
PFGE analysis.
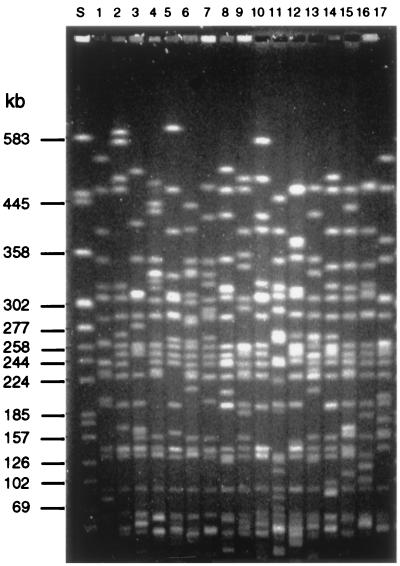
Sixteen different PFGE patterns indicating 16 clonal subgroups were identified among 55 isolates (Table 1). Representative PFGE patterns of the STEC O26 clonal subgroups and of reference strain H19 are shown in Fig. 2. Thirty-one isolates clustered in one of three main PFGE subgroups: A (19 isolates), B (6 isolates), or G (6 isolates). The remaining 24 isolates showed 13 different PFGE patterns, 7 of which were represented by a single isolate (Table 1). When banding patterns of all STEC O26 isolates were compared by the RFLPScan program, they showed more than 80% similarity, indicating that the STEC O26 strains belong to one clone complex (data not shown).
FIG. 2.
Representative PFGE patterns of XbaI-digested genomic DNA of STEC O26. Each PFGE pattern indicates a clonal subgroup. Lane S, molecular weight standard (DNA from E. coli G5244 restricted with XbaI). In lanes 1 to 17, the following STEC O26 strains (with PFGE patterns given in parentheses) are depicted: lane 1, 2162/99 (A); lane 2, 1448/97 (B); lane 3, 7662/96 (C); lane 4, 3905/97 (D); lane 5, 3584/97 (E); lane 6, 5080/97 (F); lane 7, 4166/94 (G); lane 8, 8574/96 (H); lane 9, 2971/99 (I); lane 10, 4038/97 (J); lane 11, 5236/96 (K); lane 12, 2150/98 (L); lane 13, 107/95 (M); lane 14, H19 reference strain (N); lane 15, 3608/71 (O); lane 16, 0573/99 (P); and lane 17, 37/89 /Q).
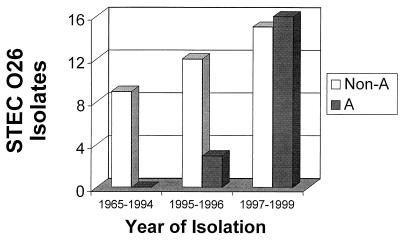
All 19 PFGE subgroup A strains were isolated between 1996 and 1999 (Table 1, Fig. 3). The strains of PFGE subgroup B were obtained between 1995 and 1998 (Table 1). In contrast, PFGE pattern G was observed in 6 of 14 strains isolated between 1965 and 1995 (Table 1). Remarkably, the subgroup G persisted for over a 30-year period. Three additional isolates obtained between 1982 and 1991 had PFGE pattern Q. Neither PFGE pattern G nor PFGE pattern Q were observed in strains isolated after 1995 (Table 1). The PFGE pattern of STEC O26 reference strain H19 differed from the patterns found in German and Czech O26 STEC isolates (Table 1, Fig. 2).
FIG. 3.
Isolation of PFGE subgroup A strains and STEC O26 isolates belonging to the other PFGE subgroups during 1965 to 1999.
stx genotypes.
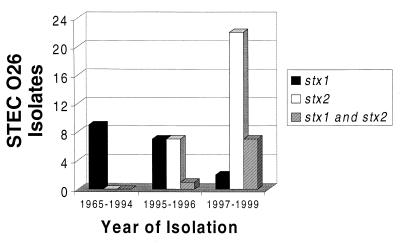
stx genotypes of 55 STEC O26 isolates were investigated by PCR using primer pairs KS7-KS8 and GK3-GK4. Of 18 strains showing the stx1 genotype, 13 were isolated from patients with diarrhea. A total of 20 of 29 isolates with the stx2 genotype and 6 of 8 strains that harbored both stx1 and stx2 originated from HUS patients (Table 1). Interestingly, 16 of 18 strains containing the stx1 gene only were isolated between 1965 and 1996, but only two such strains were identified since 1997 (Table 1, Fig. 4). In contrast, all 37 strains harboring the stx2 gene, either alone (n = 29) or in combination with stx1(n = 8), were isolated between 1995 and 1999 (Table 1, Fig. 4). No isolate contained stx2c (Table 1). All strains were cytotoxic for Vero cells indicating that the stx genes were expressed.
FIG. 4.
stx genotypes of STEC O26 strains isolated during 1965 to 1999.
Pathogenicity islands LEE and HPI.
All 55 STEC O26 strains were found to be positive by PCR with primer pair SK1-SK2, which amplifies a conserved region of eae. All strains yielded an amplification product of the expected size of 2,287 bp with primer pair SK1-LP4 demonstrating the presence of the β eae gene; no amplification products were obtained with primer pairs specific for eae types α, γ, and ɛ, respectively. All 55 STEC O26 isolates and the reference strain H19 carried the pas gene, as evidenced by specific PCR products. In addition, the pesticin receptor-encoding gene fyuA and one of the components of yersiniabactin-encoding gene cluster (irp2) were identified in all STEC O26 isolates, including the H19 reference strain.
Plasmid profiles.
All but four STEC O26 strains harbored a large plasmid of approximately 90 kb (Table 1). In addition, another large plasmid of approximately 110 kb was present in four strains. Altogether, 20 different plasmid patterns were observed among 55 STEC O26 isolates (Table 1). A total of 36 strains belonged to four major plasmid patterns characterized by the presence of a 90-kb plasmid alone (20 strains) or in combination with either a 6-kb plasmid (7 strains) or a 4-kb plasmid (5 strains) or both the small plasmids (4 strains). Thirteen plasmid patterns were represented by a single isolate (Table 1). The plasmid pattern of H19 reference strain was distinct from those of the 55 German and Czech STEC O26 isolates (Table 1).
Plasmid-encoded genes.
Plasmid-encoded accessory virulence genes were only detected in the 51 STEC O26 isolates that contained the large ca. 90-kb plasmid (Table 1). The four strains that lacked this plasmid did not contain any of these genes. The 51 strains that possessed EHEC-hlyA displayed the enterohemolytic phenotype.
The EHEC-hlyA probe hybridized to a single fragment in all 51 strains. The size of this fragment was 18 or 12.4 kb in most instances, but fragments of four different sizes also hybridized (Table 1). The espP probe elicited signals from 32 strains. The signals were localized on single fragments of various sizes (9.2 to 13 kb) except for one strain showing probe-specific sequences on two different fragments (Table 1). The katP probe demonstrated the presence of homologous sequences in 32 isolates, all of which were also positive for espP. In 31 strains, katP-specific sequences were localized on two fragments of different sizes. In addition to a 8.5-kb fragment that was present in all katP-positive strains, fragments of 4.2, 6.2, 7.5, 10.5, or 20 kb demonstrated homology (Table 1). In one strain, three different fragments hybridized to the katP probe (Table 1). The etp probe hybridized to a single 4-kb fragment of BamHI-restricted plasmid DNA in 18 isolates; etp-specific sequences were only observed in STEC O26 strains isolated after 1995 (Table 1).
In summary, three combinations of plasmid-encoded genes were detected by Southern blot analysis: 32 strains contained EHEC-hlyA, espP, and katP, and 18 strains contained EHEC-hlyA and etp; one additional strain contained only the EHEC-hlyA gene (Table 1). Interestingly, espP and katP were always found together, while etp was never observed in the presence of espP and katP. The presence of EHEC-hlyA in all 51 isolates that hybridized with the respective probe was confirmed by PCR.
Evidence for the emergence of a new clonal subgroup of STEC O26.
All 19 isolates of PFGE subgroup A harbored stx2 but not stx1 and had identical plasmid profiles (Table 1). Moreover, all but one of these strains shared a combination of the EHEC-hlyA and etp genes that was not observed in any of the other strains. The EHEC-hlyA gene was localized to the same 12.4-kb BamHI restriction fragment in all cases (Table 1). Thirteen of these strains were isolated from HUS patients, and six strains were from patients with diarrhea. Potential epidemiological links between some of these isolates are shown in Table 1. Eight cases (seven with HUS and one with diarrhea) had no apparent epidemiological linkage. The patients were from six German cities and two Czech cities. Six other patients with HUS and five patients with diarrhea were identified in four clusters of infection (Table 1). The relative frequency of isolation of PFGE subgroup A strains rose from 30% in 1996 when this clonal subgroup was identified for the first time (Table 1) to more than 50% during 1997 to 1999 (Fig. 3).
DISCUSSION
The incidence of STEC O26-mediated disease is probably underestimated because of diagnostic limitations (20), and the epidemiology and ecological niche of these pathogens are not well characterized. STEC O26 have been isolated from the feces of healthy cattle (4, 11) and pigs (31), calves with diarrhea (4, 11) and slaughter cattle (1), suggesting that these animal species may be reservoirs of this pathogen. However, routes of transmission and the minimal infectious dose remain unknown and may differ from those of STEC O157:H7. For example, STEC O26 were not detected in more than 500 food samples from various countries (29, 31). To identify the sources and modes of spreading of STEC O26 infection, isolates need to be subtyped to provide adequate strain discrimination. For STEC O157 strains, a useful phage typing scheme has been introduced for the initial characterization of isolates during outbreaks (45). To our knowledge, no such system has been proposed and evaluated for STEC O26. In the present work, we analyzed genotypic markers of 55 clinical STEC O26:H11 and O26:H− isolates obtained between 1965 and 1999 from different parts of Germany and the Czech Republic. We show that STEC O26 strains display considerable genetic diversity which can be applied to molecular typing to investigate the epidemiology of STEC O26 infections. It is of note that the STEC O26 isolate from 1965 represents, to our knowledge, the first well-characterized STEC O26 strain back to the past.
Among the 55 STEC O26 isolates investigated in our study, a total of 32 strains were nonmotile. Although nonmotility is frequent within E. coli O26 serogroup, the molecular basis leading to this phenomenon remains unresolved. In contrast to the intact fliC gene in STEC O26:H− strain 5720/96 sequenced here, nonmotile sorbitol-fermenting STEC O157 strains that have been reported to possess H7-encoding fliC (14, 39) have two insertions in the 5′ conserved region of the flagellin gene that produce a shift in the reading frame, thus introducing a premature stop codon (30).
We observed a striking, recent shift of the stx genotype from stx1 to stx2 among STEC O26 isolates. While in our collection the stx1 was the exclusive genotype identified in STEC O26 until 1994, its relative frequency decreased substantially during 1995 and 1996 and was only rarely detected in STEC O26 isolates obtained between 1997 and 1999 (Fig. 4). STEC O26 strains with the stx2 genotype first appeared in 1995 and prevailed since 1997 (Fig. 4). These figures are in agreement with data reported from the United Kingdom (41) and from Czechoslovakia (44) during the early 1990s that showed mostly Stx1-producing STEC O26 isolates. The reason for this apparent shift from the stx1 to stx2 genotype in STEC O26 isolates during 1995 to 1996 is not yet known. Our data suggest that it has occurred because of the emergence of the stx2-harboring clonal subgroup A, perhaps due to better adaptation to the human host or to changes in food consumption. Mechanistically, the STEC O26 stx genotype switch might also result from the infection of endemic E. coli O26 strains with stx2-encoding bacteriophages. Indeed, such bacteriophages have been shown to be abundant in sewage (24) and we have recently demonstrated that an Stx2-converting phage isolated from E. coli O157:H7 was able to infect and lysogenize various enteric E. coli strains, including EPEC and STEC O26 (38).
Multiple studies have documented the preponderance of Stx2 production—alone or in combination with Stx1—both in STEC O157 and non-O157 isolates from patients with HUS (6, 26). The basic N-glycosidase activity of the Stx1 and Stx2 A subunit is identical, and the findings on differential effects, for example in cultured endothelial cells, are subtle (3). Yet, experimental data from animal models using Stx2-producing strains or purified Stx proteins clearly demonstrated different biological effects (10). We believe that the presence of the stx2 gene in strains of PFGE A clonal subgroup contributes to the increased isolation rate of this clonal subgroup from patients with HUS.
All 55 STEC O26 isolates contained the fyuA and irp2 genes, indicating the presence of the HPI of pathogenic yersiniae (40). This extends our observation of the HPI in all 31 STEC O26 strains investigated recently (19) and demonstrates that the HPI is a common component of the genome of STEC O26 clonal lineage. The presence of this element in all early STEC O26 isolates in our collection, including the strain isolated 35 years ago, suggests a high degree of genetic stability of the HPI in STEC O26. This might result from the deletion in the integrase gene of the STEC O26 HPI that we observed in our previous study (19) and that may lead to a nonfunctional integrase gene and thus fixation of the HPI in the genome of this STEC clonal lineage (19). Similarly, as we reported previously (19), all 55 STEC O26 isolates harbored the pathogenicity island LEE in addition to HPI, as demonstrated by the presence of the eae gene. The finding of eae β type in all STEC O26 isolates investigated in this study is in agreement with the presence of this eae variant in STEC O26 strains investigated by Oswald et al. (27).
In contrast to the large plasmids of STEC O157:H7 that uniformly harbor EHEC-hlyA, katP, espP, and etp (18), none of the STEC O26 isolates investigated in this study demonstrated the full complement of these genes. Instead, two combinations of plasmid genes, either EHEC-hlyA, espP, and katP or EHEC-hlyA and etp, were observed, suggesting the existence of two closely related but distinct large plasmids among STEC O26. The observed polymorphism of the EHEC-hlyA, espP, and katP genes as demonstrated by varied BamHI restriction patterns and Southern blot hybridization further indicates that the large plasmids of STEC O26 are variable elements with considerable heterogeneity in gene composition and arrangement. This confirms and extends observations by Brunder et al. (9), who investigated large plasmids of several non-O157 STEC serogroups, including O26 using espP as a marker. Many STEC O26 isolates possess one or more additional, smaller plasmids. The extensive plasmid variability in strains of the same serotype is probably due to the lateral transfer of these mobile elements resulting in the loss, acquisition or exchange of plasmid DNA. Four of the STEC O26 strains investigated here lacked the 90-kb plasmid. All of these strains were patient isolates. This suggests that the plasmid may be dispensable as a virulence factor or that it has been lost during infection or storage.
Based on PFGE analysis we distinguished 16 separate clonal subgroups among the 55 STEC O26 isolates. PFGE has been proven a powerful tool for clonal definition and discrimination of STEC O157:H7/H− isolates (28). Here we demonstrate its potential usefulness for subtyping strains within one of the most important non-O157 STEC serogroups. Strains within some PFGE subgroups could be further discriminated by their plasmid characteristics. This suggests that plasmid profiling, and especially Southern blot analysis of the plasmid-encoded virulence genes, could be a useful adjunct to PFGE for subtyping of STEC O26 isolates in epidemiological studies.
Whittam et al. (49) examined 93 E. coli O26 isolates by multilocus enzyme electrophoresis. There were 20 electrophoretic types and two main clusters: one was E. coli O26:H32 and the other was E. coli O26:H11. Most STEC O26:H11 isolates belonged to two electrophoretic types termed DEC9 and DEC10. The methods used in our study are more sensitive for epidemiological typing and allow subtyping of the STEC O26 clone. We demonstrate a remarkable heterogeneity among STEC O26 isolates at the plasmid and chromosomal DNA level. This allowed us to identify 19 strains of a distinct clonal subgroup that shared PFGE pattern A, harbored the stx2 gene only, and with one exception (strain 2569/98 that probably lost etp) were identical in the structure and gene composition of their large plasmids (Table 1). The widespread distribution of the clonal subgroup A in Germany since 1996 and its spread into the Czech Republic in 1999 may be explained by two mechanisms. First, all 19 PFGE pattern A isolates may have been part of a diffuse outbreak. Alternatively, these strains indeed represent a new clonal subgroup of STEC O26 with high pathogenic potential for humans. The latter interpretation is supported by the fact that such strains were not identified among 21 STEC O26 isolates from Germany and two other countries analyzed in our previous studies (37; H. Karch, unpublished data), and by their strong association with HUS and with clusters of infections that clearly demonstrates the potential of the clonal subgroup A to cause outbreaks. These characteristics make this STEC O26 clonal subgroup similar to the prototypic STEC O157:H7 and warrant that its further spread be monitored carefully. Further investigations are necessary to evaluate the significance of STEC O26 clonal subgroup A as a cause of human disease, to monitor its appearance in other countries, to identify its reservoir(s) and mode(s) of transmission, and to understand its pathogenic mechanism(s).
ACKNOWLEDGMENTS
This study was supported by Bundesministerium für Bildung und Forschung Verbundprojekt, Forschungsnetzwerk “Emerging foodborne pathogens in Germany” grants 01KI 9903 and 01KI 9901. Investigation of the Czech isolates was supported by grant IGA 4563-3 from the Ministry of Health of the Czech Republic. Wen-Lan Zhang is a recipient of a scholarship 97834013 from the China Scholarship Council.
We thank Phillip I. Tarr and Thomas S. Whittam for critical reading of the manuscript and for helpful discussions.
REFERENCES
- 1.Beutin L, Zimmermann S, Gleier K. Zur Epidemiologie und Diagnostik von Infektionen durch enterohämorrhagische E. coli (EHEC) in der Bundesrepublik Deutschland. Bundesgesundheitsblatt. 1996;9:326–331. [Google Scholar]
- 2.Bielaszewska M, Janda J, Blahova K, Feber J, Potuznik V, Souckova A. Verocytotoxin-producing Escherichia coli in children with hemolytic uremic syndrome in the Czech Republic. Clin Nephrol. 1996;46:42–44. [PubMed] [Google Scholar]
- 3.Bitzan M M, Wang Y, Lin J, Marsden P. Verotoxin and ricin have novel effects on preproendothelin-1 expression but fail to modify nitric oxide synthase (eeNOS) expression and NO production in vascular endothelium. J Clin Investig. 1998;101:372–382. doi: 10.1172/JCI522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanco M, Blanco J, Blanco J E, Gonzales E A, Gomes T A T, Zerbini L F, Yano T, Pestana de Castro A F. Genes coding for Shiga-like toxins in bovine verotoxin-producing Escherichia coli (VTEC) strains belonging to different O:K:H serotypes. Vet Microbiol. 1994;42:105–110. doi: 10.1016/0378-1135(94)90010-8. [DOI] [PubMed] [Google Scholar]
- 5.Bockemühl J, Aleksic S, Karch H. Serological and biochemical properties of Shiga-like toxin (verocytotoxin)-producing strains of Escherichia coli, other than O-group 157, from patients in Germany. Zentbl Bakteriol. 1992;276:189–195. doi: 10.1016/s0934-8840(11)80005-8. [DOI] [PubMed] [Google Scholar]
- 6.Bockemühl J, Karch H, Tschäpe H. Zur Situation der Infektionen des Menschen durch enterohämorrhagische Escherichia coli (EHEC) in Deutschland, 1997. Bundesgesundheitsblatt. 1998;Suppl. (October):2–5. doi: 10.1007/s00103-002-0458-4. [DOI] [PubMed] [Google Scholar]
- 7.Brunder W, Schmidt H, Karch H. KatP, a novel catalase-peroxidase encoded by the large plasmid of enterohaemorrhagic Escherichia coli O157:H7. Microbiology. 1996;142:3305–3315. doi: 10.1099/13500872-142-11-3305. [DOI] [PubMed] [Google Scholar]
- 8.Brunder W, Schmidt H, Karch H. EspP, a novel extracellular serine protease of enterohaemorrhagic Escherichia coli O157:H7 cleaves human coagulation factor V. Mol Microbiol. 1997;24:767–778. doi: 10.1046/j.1365-2958.1997.3871751.x. [DOI] [PubMed] [Google Scholar]
- 9.Brunder W, Schmidt H, Frosch M, Karch H. The large plasmids of Shiga-toxin-producing Escherichia coli (STEC) are highly variable genetic elements. Microbiology. 1999;145:1005–1014. doi: 10.1099/13500872-145-5-1005. [DOI] [PubMed] [Google Scholar]
- 10.Brunton J L. Animal models: pathogenesis, animal infections. In: Karmali M A, Goglio A G, editors. Recent advances in verocytotoxin-producing Escherichia coli infections. Amsterdam, The Netherlands: Elsevier Science; 1984. pp. 211–220. [Google Scholar]
- 11.Caprioli A, Nigrelli A, Gatti R, Zavanella M, Blado A M, Minelli F, Donelli G. Characterisation of verocytotoxin-producing Escherichia coli isolated from pigs and cattle in northern Italy. Vet Rec. 1993;133:323–324. doi: 10.1136/vr.133.13.323. [DOI] [PubMed] [Google Scholar]
- 12.Claus H, Cuny C, Pasemann B, Witte W. A database system for fragment patterns of genomic DNA of Staphylococcus aureus. Zentbl Bakteriol. 1998;287:105–116. doi: 10.1016/s0934-8840(98)80154-0. [DOI] [PubMed] [Google Scholar]
- 13.Djafari S, Ebel F, Deibel C, Krämer S, Hudel M, Chakraborty T. Characterization of an exported protease from Shiga toxin-producing Escherichia coli. Mol Microbiol. 1997;25:771–784. doi: 10.1046/j.1365-2958.1997.5141874.x. [DOI] [PubMed] [Google Scholar]
- 14.Fields P I, Blom K, Hughes H J, Helsel L O, Feng P, Swaminathan B. Molecular characterization of the gene encoding H antigen in Escherichia coli and development of a PCR-restriction fragment length polymorphism test for identification of E. coli O157:H7 and O157:NM. J Clin Microbiol. 1997;35:1066–1070. doi: 10.1128/jcm.35.5.1066-1070.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gunzer F, Böhm H, Rüssmann H, Bitzan M, Aleksic S, Karch H. Molecular detection of sorbitol-fermenting Escherichia coli O157 in patients with hemolytic-uremic syndrome. J Clin Microbiol. 1992;30:1807–1810. doi: 10.1128/jcm.30.7.1807-1810.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaper J B, Elliot S, Sperandio V, Perna N T, Mayhew G F, Blattner F R. Attaching-and-effacing intestinal histopathology and the locus of enterocyte effacement. In: Kaper J B, O'Brien A D, editors. Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. Washington, D.C.: American Society for Microbiology; 1998. pp. 163–182. [Google Scholar]
- 17.Karch H, Böhm H, Schmidt H, Gunzer F, Aleksic S, Heesemann J. Clonal structure and pathogenicity of Shiga-like toxin-producing, sorbitol-fermenting Escherichia coli O157:H−. J Clin Microbiol. 1993;31:1200–1205. doi: 10.1128/jcm.31.5.1200-1205.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karch H, Schmidt H, Brunder W. Plasmid-encoded determinants of Escherichia coli O157:H7. In: Kaper J B, O'Brien A D, editors. Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. Washington, D.C.: American Society for Microbiology; 1998. pp. 183–194. [Google Scholar]
- 19.Karch H, Schubert S, Zhang D, Zhang W, Schmidt H, Ölschlager T, Hacker J. A genomic island, termed “high pathogenicity island,” is present in certain non-O157 Shiga toxin-producing Escherichia coli clonal lineages. Infect Immun. 1999;67:5994–6001. doi: 10.1128/iai.67.11.5994-6001.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karch H, Bielaszewska M, Bitzan M, Schmidt H. Epidemiology and diagnosis of Shiga toxin-producing Escherichia coli infections. Diagn Microbiol Infect Dis. 1999;34:229–243. doi: 10.1016/s0732-8893(99)00031-0. [DOI] [PubMed] [Google Scholar]
- 21.Konowalchuk J, Speirs J I, Stavric S. Vero response to a cytotoxin of Escherichia coli. Infect Immun. 1977;18:775–779. doi: 10.1128/iai.18.3.775-779.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kresse A, Schulze K, Deibel C, Ebel F, Rohde M, Chakraborty T, Guzman C A. Pas, a novel protein required for protein secretion and attaching and effacing activities of enterohemorrhagic Escherichia coli. J Bacteriol. 1998;180:4370–4379. doi: 10.1128/jb.180.17.4370-4379.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levine M M, Xu J G, Kaper J B, Lior H, Prado V, Tall B, Nataro J, Karch H, Wachsmuth K. A DNA probe to identify enterohemorrhagic Escherichia coli of O157:H7 and other serotypes that cause hemorrhagic colitis and hemolytic uremic syndrome. J Infect Dis. 1987;156:175–182. doi: 10.1093/infdis/156.1.175. [DOI] [PubMed] [Google Scholar]
- 24.Muniesa M, Jofre J. Abundance in sewage of bacteriophages that infect Escherichia coli O157:H7 and that carry the Shiga toxin 2 gene. Appl Environ Microbiol. 1998;64:2443–2448. doi: 10.1128/aem.64.7.2443-2448.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Brien A D, Newland J W, Miller S F, Holmes R K, Smith H W, Formal S B. Shiga-like toxin-converting phages from Escherichia coli strains that cause hemorrhagic colitis or infantile diarrhea. Science. 1984;226:694–696. doi: 10.1126/science.6387911. [DOI] [PubMed] [Google Scholar]
- 26.Ostroff S M, Tarr P I, Neill M A, Lewis J H, Hargrett-Bean N, Kobayashi J M. Toxin genotypes and plasmid profiles as determinants of systemic sequelae in Escherichia coli O157:H7 infections. J Infect Dis. 1989;160:994–998. doi: 10.1093/infdis/160.6.994. [DOI] [PubMed] [Google Scholar]
- 27.Oswald E, Schmidt H, Morabito S, Karch H, Marches O, Caprioli A. Typing of intimin genes in human and animal enterohemorrhagic and enteropathogenic Escherichia coli: characterization of a new intimin variant. Infect Inmun. 2000;68:64–71. doi: 10.1128/iai.68.1.64-71.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.PulseNet. Standardized molecular subtyping of foodborne bacterial pathogens by pulsed-field gel electrophoresis. Atlanta, Ga: Centers for Disease Control and Prevention; 1998. [Google Scholar]
- 29.Read S C, Gyles C L, Clarke R C, Lior H, McEwen S. Prevalence of verocytotoxigenic Escherichia coli in ground beef, pork and chicken in southwestern Ontario. Epidemiol Infect. 1990;105:11–20. doi: 10.1017/s0950268800047592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reid S D, Selander R K, Whittam T S. Sequence diversity of flagellin (fliC) alleles in pathogenic Escherichia coli. J Bacteriol. 1999;181:153–169. doi: 10.1128/jb.181.1.153-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rios M, Prado V, Trucksis M, Arellano C, Borie C, Alexandre M, Fica A, Levine M M. Clonal diversity of Chilean isolates of enterohemorrhagic Escherichia coli from patients with hemolytic-uremic syndrome, asymptomatic subjects, and food products. J Clin Microbiol. 1999;37:778–781. doi: 10.1128/jcm.37.3.778-781.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rüssmann H, Schmidt H, Heesemann J, Caprioli A, Karch H. Variants of Shiga-like toxin II constitute a major toxin component in Escherichia coli O157 strains from patients with haemolytic uraemic syndrome. J Med Microbiol. 1994;40:338–343. doi: 10.1099/00222615-40-5-338. [DOI] [PubMed] [Google Scholar]
- 33.Rüssmann H, Kothe E, Schmidt H, Franke S, Harmsen D, Caprioli A, Karch H. Genotyping of Shiga-like toxin genes in non-O157 Escherichia coli strains associated with haemolytic uraemic syndrome. J Med Microbiol. 1995;42:404–410. doi: 10.1099/00222615-42-6-404. [DOI] [PubMed] [Google Scholar]
- 34.Schmidt H, Rüssmann H, Karch H. Virulence determinants in nontoxigenic Escherichia coli O157 strains that cause infantile diarrhea. Infect Immun. 1993;61:4894–4898. doi: 10.1128/iai.61.11.4894-4898.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmidt H, Beutin L, Karch H. Molecular analysis of the plasmid-encoded hemolysin of Escherichia coli O157:H7 strain EDL 933. Infect Immun. 1995;63:1055–1061. doi: 10.1128/iai.63.3.1055-1061.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmidt H, Henkel B, Karch H. A gene cluster closely related to type II secretion pathway operons of gram-negative bacteria is located on the large plasmid of enterohaemorrhagic Escherichia coli O157 strains. FEMS Microbiol Lett. 1997;148:265–272. doi: 10.1111/j.1574-6968.1997.tb10299.x. [DOI] [PubMed] [Google Scholar]
- 37.Schmidt H, Geitz C, Tarr P I, Frosch M, Karch H. Non-O157 pathogenic Shiga toxin-producing Escherichia coli: phenotypic and genetic profiling of virulence traits and evidence for clonality. J Infect Dis. 1999;179:115–123. doi: 10.1086/314537. [DOI] [PubMed] [Google Scholar]
- 38.Schmidt H, Bielaszewska M, Karch H. Transduction of enteric Escherichia coli isolates with a derivative of Shiga toxin 2-encoding bacteriophage φ3538 isolated from Escherichia coli O157:H7. Appl Environ Microbiol. 1999;65:3855–3861. doi: 10.1128/aem.65.9.3855-3861.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmidt H, Scheef J, Huppertz H I, Frosch M, Karch H. Escherichia coli O157:H7 and O157:H− strains that do not produce Shiga toxin: phenotypic and genetic characterization of isolates associated with diarrhea and hemolytic uremic syndrome. J Clin Microbiol. 1999;37:3491–3496. doi: 10.1128/jcm.37.11.3491-3496.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schubert S, Rakin A, Karch H, Carniel E, Heesemann J. Prevalence of the “high pathogenicity island” of Yersinia species among Escherichia coli strains that are pathogenic to humans. Infect Immun. 1998;66:480–485. doi: 10.1128/iai.66.2.480-485.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scotland S M, Willshaw G A, Smith H R, Rowe B. Properties of strains of Escherichia coli O26:H11 in relation to their enteropathogenic or enterohemorrhagic classification. J Infect Dis. 1990;162:1069–1074. doi: 10.1093/infdis/162.5.1069. [DOI] [PubMed] [Google Scholar]
- 42.Smith H W, Linggood M A. The transmissible nature of enterotoxin production in human enteropathogenic strain of Escherichia coli. J Med Microbiol. 1971;4:301–305. doi: 10.1099/00222615-4-3-301. [DOI] [PubMed] [Google Scholar]
- 43.Smith H W, Green P, Parsell Z. Vero cell toxin in Escherichia coli and related bacteria: transfer by phage and conjugation and toxic action in laboratory animals, chicken and pigs. J Gen Microbiol. 1983;129:3121–3137. doi: 10.1099/00221287-129-10-3121. [DOI] [PubMed] [Google Scholar]
- 44.Sramkova L, Bielaszewska M, Janda J, Blahova K, Hausner O. Verocytotoxin-producing strains of Escherichia coli in children with haemolytic uraemic syndrome and diarrhoea in Czechoslovakia. Infection. 1990;18:204–209. doi: 10.1007/BF01643386. [DOI] [PubMed] [Google Scholar]
- 45.Strockbine N A, Wells J G, Bopp C A, Barrett T J. Overview of detection and subtyping methods. In: Kaper J B, O'Brien A D, editors. Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. Washington, D.C.: American Society for Microbiology; 1998. pp. 331–356. [Google Scholar]
- 46.Tarr P I, Neill M A. Perspective: the problem of non-O157 Shiga toxin (verocytotoxin)-producing Escherichia coli. J Infect Dis. 1996;174:1136–1139. doi: 10.1093/infdis/174.5.1136. [DOI] [PubMed] [Google Scholar]
- 47.Tenover F C, Arbeit R D, Goering R V, Mickelsen P M, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tschäpe H, Prager R, Streckel W, Fruth A, Tietze E, Böhme B. Verotoxigenic Citrobacter freundii associated with severe gastroenteritis and cases of haemolytic urameic syndrome in a nursery school: green butter as the infection source. Epidemiol Infect. 1995;114:441–450. doi: 10.1017/s0950268800052158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Whittam T S, Wolfe M L, Wachsmuth I K, Orskov F, Orskov I, Wilson R. Clonal relationships among Escherichia coli strains that cause hemorrhagic colitis and infantile diarrhea. Infect Immun. 1993;61:1619–1629. doi: 10.1128/iai.61.5.1619-1629.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]