Abstract
Modern biomarker and translational research as well as personalized health care studies rely heavily on powerful omics’ technologies, including metabolomics and lipidomics. However, to translate metabolomics and lipidomics discoveries into a high-throughput clinical setting, standardization is of utmost importance. Here, we compared and benchmarked a quantitative lipidomics platform. The employed Lipidyzer platform is based on lipid class separation by means of differential mobility spectrometry with subsequent multiple reaction monitoring. Quantitation is achieved by the use of 54 deuterated internal standards and an automated informatics approach. We investigated the platform performance across nine laboratories using NIST SRM 1950–Metabolites in Frozen Human Plasma, and three NIST Candidate Reference Materials 8231–Frozen Human Plasma Suite for Metabolomics (high triglyceride, diabetic, and African-American plasma). In addition, we comparatively analyzed 59 plasma samples from individuals with familial hypercholesterolemia from a clinical cohort study. We provide evidence that the more practical methyl-tert-butyl ether extraction outperforms the classic Bligh and Dyer approach and compare our results with two previously published ring trials. In summary, we present standardized lipidomics protocols, allowing for the highly reproducible analysis of several hundred human plasma lipids, and present detailed molecular information for potentially disease relevant and ethnicity-related materials.
Introduction
Lipids are involved in a plethora of (patho-)physiological functions and therefore their accurate identification and quantification are important for the study and diagnosis of numerous diseases. Lipids and their corresponding metabolic enzymes can serve as endogenous bioactive mediators,1 potential disease markers,2 future drug targets,3 or precision medicine markers for future development of personalized drug treatment.4 These developments have contributed to the field of lipidomics5 and generated substantial interest in the scientific community during the last decade.6 However, as recently argued by Liebisch et al., lipidomics needs standardization.7 The complexity of the human lipidome not only results in challenges related to lipid annotation8 but also identification9 and quantification.10 These issues are crucial to address if the success of lipidomics is to be translated from experimental in vitro and in vivo studies to the clinical space. Ultimately, the goal is to obtain accurate and precise quantitative measurements from lipids with standardized names across large cohort studies. Moving toward this goal, successful interlaboratory comparison of lipidomics studies and data will be key to further expanding the application of lipidomics into a clinical setting. Present-day clinical lipidomics applies diverse analytical approaches leading to vastly deviating results. The most prominent platforms applied in preclinical lipidomics include nuclear magnetic resonance-11 and liquid chromatography–mass spectrometry (LC–MS)-based approaches, either involving triple quadrupole or high-resolution mass spectrometers,12 as well as flow-injection based approaches, with some now including ion-mobility13,14 technologies. To this end, only a small number of studies have investigated the necessary analytical parameters mandatory for a broader application of lipidomics in the clinic. Cajka et al. compared several untargeted LC–MS-based lipidomics platforms in an interlaboratory study under identical experimental conditions and found largely comparable results.11 In contrast, Thompson et al. compared a kit-based metabolomics and lipidomics workflow in an international interlaboratory setting and found high coefficients of variation (CV) of up to 306% for glycerolipids and 181% for glycerophospholipids, even under identical analytical settings.15 Another study by Bowden et al.10 compared the results obtained using different approaches at different sites and presented consensus location estimates (e.g., consensus means) with associated uncertainties in the NIST Standardized Reference Material 1950–Metabolites in frozen human plasma (SRM 1950). The authors compared these results with the concentration values noted from the LIPID MAPS consortium16 and found that the participating laboratories measured phospholipids (particularly LPC, PC, PE, and PI) and, in general, the most abundant lipids more consistently.
In the current study, we evaluated the applicability of the Lipidyzer platform for generating high-throughput quantitative lipidomics data in multiple laboratories. Both traditional mass spectrometry-based shotgun lipidomics and the Lipidyzer platform measure and quantify comparable numbers of lipid species. However, shotgun lipidomics suffers from inaccurate quantification caused by overlapping isomeric and isobaric lipid species. The Lipidyzer platform overcomes this issue by employing differential mobility spectrometry (DMS) and multiple reaction monitoring (MRM) on a QTRAP mass spectrometer and by applying 54 isotopically labeled internal standards. At present, the platform can quantify >1000 lipid species across 13 lipid classes (CE, cholesterylester; CER, ceramide; DAG, diacylglycerol; DCER, dihydroceramide; FFA, free fatty acid; HCER, hexosylceramide; LCER, lactosylceramide; LPC, lysophosphatidylcholine; LPE, lysophosphatidylethanolamine; PC, phosphatidylcholine; PE, phosphatidylethanolamine; SM, sphingomyelin; and TAG, triacylglycerol).17 However, while some studies have compared the Lipidyzer performance with conventional platforms,18,19 an evaluation of its performance across different laboratories in a preclinical setting is still needed. Moreover, this further extends to evaluate the use of either the classic Bligh and Dyer (BD) protocol,20 with which the platform has originally been validated against, or the potentially more practical methyl tert-butyl ether (MTBE) protocol.21 Ultimately, such comparisons are an important prerequisite for successfully applying quantitative lipidomics in clinical research studies to ensure standardized and highly reproducible data. In the current study, we (i) compared two different lipid extraction methods using NIST SRM 1950 in nine laboratories, (ii) determined consensus concentration values of lipid species and lipid classes in four NIST pooled plasma reference materials (nine laboratories measured NIST SRM 1950 and seven laboratories measured the remaining NIST candidate 8231 (RM) reference materials), and (iii) compared the lipid concentration among three laboratories in plasma samples from individuals identified with familial hypercholesterolemia (FH) and known to suffer from cardiovascular disease (CVD). We were able to assign consensus concentration values for hundreds of lipid species in the NIST SRM 1950 and NIST candidate RM 8231 derived of diabetic (DB) individuals, high triglyceride (HTG) individuals, and Young, African-American (AA) donors, using a single analytical technique.
In summary, we investigated the use of standardized extraction protocols and the reproducibility for an extensive array of lipids in human plasma materials of different metabolic health states and ethnicity, as well as conducted a comparative analysis of a clinical cohort study.
Experimental Section
Study Design
The study was conducted in four different phases, starting with organization and development of the study protocol and workflow. Nine out of 12 invited laboratories (five from Europe and four from the United States) agreed to participate in the study. Each partner was assigned a unique site number for anonymous data sharing. All participating laboratories were provided with the NIST standards and patient samples and the study protocol. A detailed description of the study design can be found in Supporting Information S1.
Samples
The NIST candidate RM suite was provided by NIST (Gaithersburg, Maryland, US) and consisted of the following materials: (i) Pooled Plasma 1: Diabetic Plasma (DB) (ii) Pooled Plasma 2: High Triglyceride Plasma (HTG) (iii) Pooled Plasma 3: Young, African American Plasma (AA), and (iv) Pooled Plasma 4: SRM 1950 Plasma (SRM1950).
Along with the NIST RM, three laboratories analyzed 59 plasma samples from a clinical trial provided by the Erasmus MC, Rotterdam, The Netherlands (Eric Sijbrands). More details are provided in Supporting Information S1.
Chemicals and Consumables
The laboratories were asked to use chemicals and consumables of LC–MS grade or higher. A reference list, including the Lipidyzer internal standard (IS) kits and Lipidyzer control plasma standard, was provided. It was not mandatory to use the same lot# for IS between the laboratories (further details can be found in Supporting Information S1).
Sample Preparation
Two lipid extraction methods were evaluated in this study. The BD extraction protocol20 (modified to use dichloromethane) is the standard protocol provided with the Lipidyzer platform. The MTBE extraction protocol21 provided a faster and more facile alternative (for further details, please refer to the detailed sample preparation protocols provided in Supporting Information S1).
Lipidyzer Platform Analysis
The Lipidyzer platform consists of a SCIEX QTRAP 5500 mass spectrometer equipped with a SelexION differential ion mobility (DMS) interface and a Nexera X2 ultrahigh-performance liquid chromatography/tandem mass spectrometry system operated with Analyst software (detailed information can be found in Supporting Information S1).
Initial Screening and Selection Criteria
Data were received as xls result files. For each day and for each lipid species, three out of five replicates had to be reported to consider the lipid concentration from the measurement day. Furthermore, for SRM 1950 plasma, only lipids reported by at least seven out of nine sites were considered in the study. In the remaining NIST plasma standards, the lipids were reported if at least five out of seven sites provided data for them. The mean concentration from each site was used to calculate the consensus value. The mean concentration of each measurement day from each site was used to calculate the CV.
Calculation of Consensus Values and Associated Uncertainties
Among various approaches22−24 available to calculate consensus values, the median of means (MEDM) approach25 was applied in this study as it has been used previously in related investigations.10,26 A detailed description of the calculations and criteria can be found in Supporting Information S1.
Nomenclature
Although the output format of the Lipidyzer platform does not strictly adhere to the lipid short-hand notation described by Liebisch et al.,27 we have kept this format to facilitate data handling and comparison in this multicenter study. Nevertheless, Table S1A in Supporting Information S1 exemplifies how the Lipidyzer format translates into the lipid short-hand annotation as described by Liebisch et al.27 Additionally, it must be noted that for TAG lipids, the Lipidyzer platform can only define one of the three FA side chains. Hence, a TAG lipid specified as, for example, TAG 54: 6-FA 18:1 would refer to a TAG lipid with 54 carbons, 6 double bonds, and 1 side chain being FA 18:1. This also results in the fact that one concatenated TAG lipid can be reported with various combinations of fatty acyl tails (e.g., TAG 54:6-FA 16:0, TAG 54:6-FA 20:4, and so on). Consequently, this leads to an overestimation of the number of measured TAG species and TAG 54:6 will be reported three times, once for each acyl side chain. While this annotation is advantageous for fatty acid composition analysis, it overestimates the number of TAGs reported. When reporting the TAG lipid class concentration, the LWM software corrects for this by dividing the summed concentration of all TAG combinations by a factor of three.
Results and Discussion
The main goal of our study was to investigate the interlaboratory performance of the Lipidyzer platform in a preclinical setting. For this purpose, we started our investigation with SRM 1950 and compared the BD20 and MTBE21 extraction methods, based on the observed CV and COD values as an essential first step toward harmonization among the Lipidyzer community. Subsequently, we compared the interlaboratory repeatability using SRM 1950 plasma in combination with the MTBE extraction protocol. As a next step, we investigated the interlaboratory performance using more clinically relevant materials, such as three NIST candidates RM 8231 consisting of DB, HTG plasma, and AA plasma. The latter may help shed light on possible analytical challenges across ethnic group differences, which is an important aspect of personalized health care. Ultimately, we investigated the correlation of the observed data analyzing a clinical cohort study related to FH between three participating laboratories.
Comparison of Extraction Methods
The BD20 method is the most widely used lipid extraction method and is also provided as the standard extraction protocol with the Lipidyzer platform. However, for practical purposes, the MTBE21 method has an advantage as the lipid fraction is portioned into the upper layer during extraction, facilitating automation and miniaturization. To compare the two extraction methods, we used the SRM 1950 data from four measurement days for both methods from all participating laboratories and calculated consensus values after applying the exclusion criteria as described in the method section.
In total, we were able to quantify 856 and 854 lipids in SRM 1950 using the BD and MTBE extractions, respectively. After applying the exclusion criteria, we selected 730 lipids from the BD extraction and 724 lipids from the MTBE extraction for the consensus value calculations. Ultimately, we were able to assign consensus values to 685 and 696 lipid species from BD and MTBE extraction, respectively (a complete list of consensus valued of lipid species and lipid classes can be found in Tables S2–S5 in Supporting Information S2). Both methods resulted in comparable numbers of lipid species for consensus concentration values with COD ≤ 20% except for FFA, PC, and PE lipid classes, where slightly higher differences were observed. A total of 21 FFAs and 49 PCs were selected with the MTBE protocol compared to 16 FFAs and 34 PCs with BD. Similarly, 50 PEs were selected with the BD protocol compared to 43 PEs with MTBE. However, both methods performed almost equally well when investigating the number of lipid species for which a consensus value could be assigned. Ultimately, we compared the distribution of COD values among the lipid classes, confirming that both methods worked equally well, as the average COD of lipid species for all lipid classes was largely comparable between the methods.
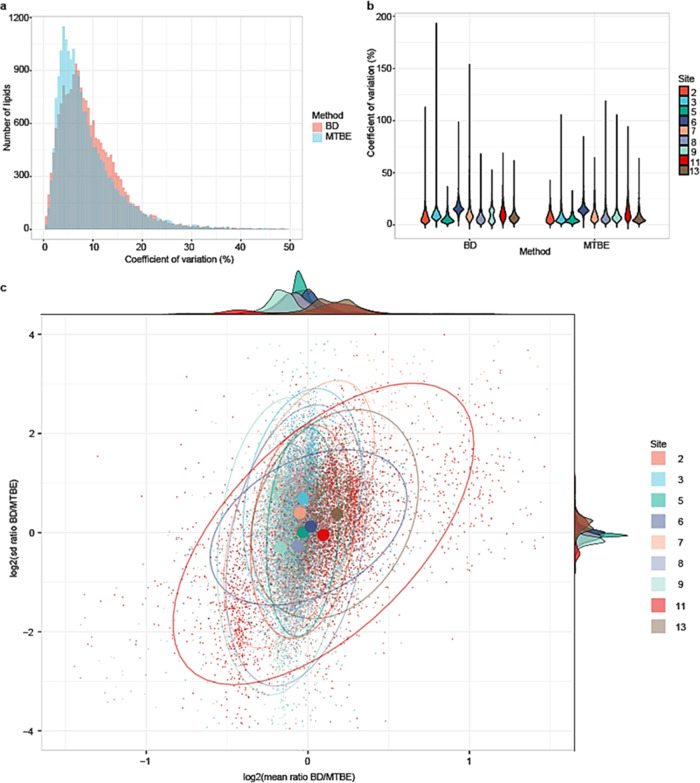
To further investigate the interlaboratory performance of the two extraction methods, we next compared the distribution of CV values between the two methods in all participating laboratories (Figure 1). Prior to the CV calculation, we included lipids from a measurement day of a laboratory only if three out of five replicates were reported successful. Next, we calculated the mean concentration of each lipid for each measurement day to calculate the CV. As can be seen from Figure 1a, improved repeatability was observed using the MTBE extraction protocol. Comparing the performance of the individual sites (Figure 1b), we found that all sites reported similar or better results employing the MTBE method during the full four-day SRM 1950 analysis trial. A total of 704 lipid species were measured with a CV ≤ 20%, with the average CV across the 13 lipid classes below 15% (Figure S2 in Supporting Information S1). To further study and compare the performance of the two methods, we investigated the relationships between the mean ratio and standard deviation of the two methods per lipid species per laboratory (Figure 1c). Three important aspects became evident. First, the two methods reported similar mean concentrations, that is, the data points (representing lipid species) were close to zero on the x-axis with randomness representing inaccuracies between the two methods. Ideally, the two methods are of similar precision for identical lipids, that is, the data points are centered around zero on the y-axis. Systematic bias on the y-axis indicates that one method is more precise. Furthermore, in an ideal situation, the mean and standard deviation ratios are independent, that is, the major axis of the oval representing the data distribution is parallel to the y-axis (or the x-axis, in case the precision is better than accuracy). Overall, the methods are in better agreement when the distribution represented by the oval is small and vice versa. Most sites had comparable method performance. Site 13 reported, on average, higher concentrations with lower precision for BD. Site 11 reported worse agreement between the two methods with the largest spread in the distribution. In summary, the MTBE extraction protocol was not only more practical but also resulted in more precise interlaboratory results. For that purpose, we opted to use the MTBE method throughout the remainder of the study.
Figure 1.
Comparison between BD and MTBE lipid extraction methods. (a) Histogram of overlapping lipids between the two methods with CV values on the x-axis and counts of lipid species on the y-axis. (b) Violin plot to compare the performance of sites between the two methods. (c) Scatter plot to summarize the performance of each laboratory between the method: the x-axis represents the ratio of mean concentration of the BD extraction method to the MTBE method using the base-2 logarithmic scale and the y-axis represents the ratio of standard deviation of the two methods. Larger dots represent the center of the distribution of the data points for each laboratory calculated from the fitted distribution (represented by the ovals). The ovals represent 95% of the data entries using the fitted multivariate t-distribution.
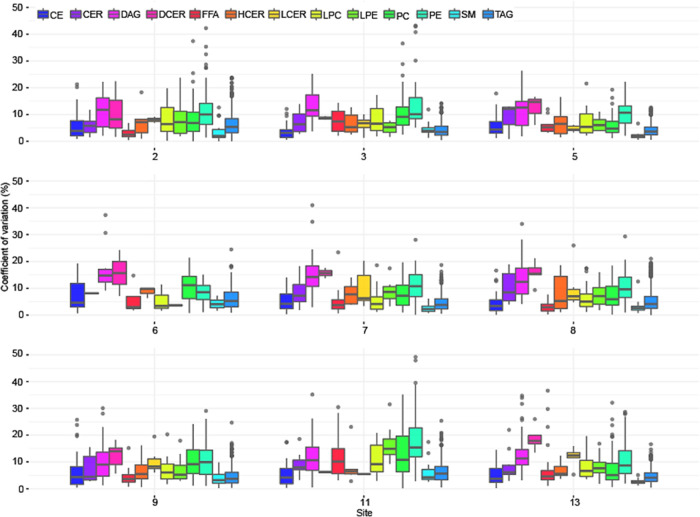
Furthermore, we evaluated the technical repeatability (i.e., repeated injection of pooled materials) between the sites by comparing the results obtained from the analysis of pooled samples across four consecutive days (pooled BD and MTBE aliquots). As can be seen from Figure 2 all sites reported acceptable CVs across all lipid classes. The overall technical repeatability across all lipid classes had a mean CV of 6.6% and third quartile at 8.6%, with each lipid class reporting a CV below 15% on average. It is important to mention that the CVs of the DCER lipid class are relatively high due to its very low abundance in plasma. The highest mean CV value of 8.3% was observed by site 11 and the lowest of 5.5% by site 5.
Figure 2.
Distribution of CV across the sites and lipid classes of all repeated measurements of the pooled sample aliquots. The median value is marked by a horizontal line inside the box. The whiskers extending from top and bottom of the box represent the largest and smallest non-outlier values, respectively.
Consensus Values and Breakdown of the SRM 1950 Plasma Lipidome
As the next step in our investigation, we aimed for consensus value determination and comparison with earlier studies using the SRM 1950. We were able to quantify 854 lipids representing 13 major lipid classes and assigned consensus values to more than 80% of the quantified lipids. Given that the participating laboratories applied identical workflows, we assigned final consensus values to lipids with a strict exclusion criterion of COD > 20%. In total, 696 out of 724 selected lipids satisfied the COD ≤ 20% criterion to assign consensus values. The average COD of all lipid classes was <15%. The top five lipid classes with the lowest average COD were CER (5%), HCER (5%), LCER (6%), TAG (7%), and DAG (8%). The top 100 most abundant and least abundant lipids had an average COD of 8.8 and 9.5%, respectively, indicating excellent repeatability at both the upper and lower ends of the concentration range.
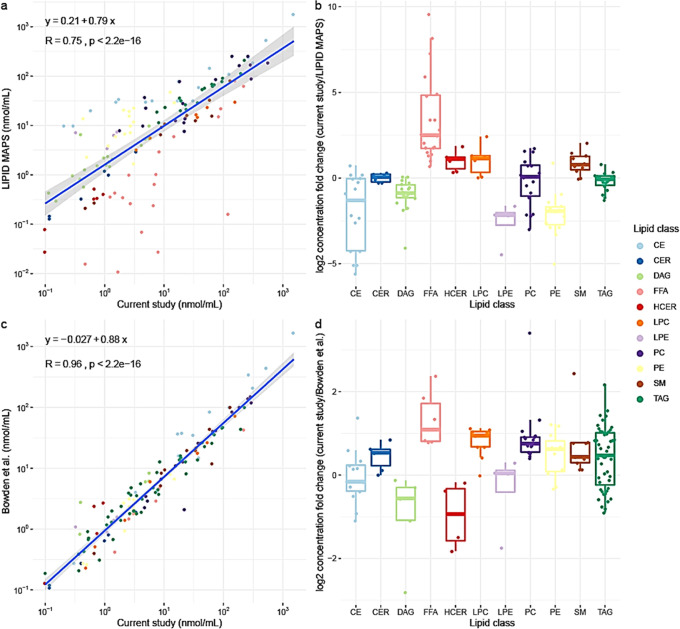
Finally, we compared the consensus values for SRM 1950 from this study with the values reported by earlier reports.10,16 The two earlier reports determined consensus values for SRM 1950 plasma using either triple quadrupole technology (LIPID MAPS) or diverse analytical methods and platforms (Bowden et al.10). LIPID MAPS and Bowden et al. reported consensus concentrations of 588 and 339 lipid species, respectively. In total, 138 overlapping lipid species were found between our study and LIPID MAPS and 131 lipid species between our study and Bowden et al. A comparison of the reported concentrations of overlapping lipids is shown in Figure 3. For comparison, the lipid species of TAG, DAG, PC, and PE lipid classes from the current study were reported in the concatenated form. As seen in the correlation plots (Figure 3), the reported lipid concentrations from the current study showed a higher correlation with Bowden et al. compared to LIPID MAPS, while our method appears to be more sensitive than both as can be judged from the slope of the regression equations (a detailed comparison of lipid species concentrations is listed in Table S6 of Supporting Information S2). Given that Bowden et al. reported on measurements performed on different platforms, it is remarkable how good the correlation was. This is another confirmation that lipidomics, as a field, is converging toward the higher standards of repeatability. Furthermore, appropriate normalization methods can be used to correct quantitative biases and harmonize lipidomics data across different platforms.28 Moreover, isotope correction for increased quantitative accuracy is gaining attention in the field.29 However, the Lipidyzer platform was initially developed without isotope correction, and both previous ring trials (LIPID MAPS and Bowden et al.10,16) did not apply any isotope correction. Consequently, when comparing with previous works, we refrained from isotope correction. Nevertheless, using our recently published algorithm for isotopically correcting the Lipidyzer data,30 we provide a comparison between corrected and uncorrected values for all investigated NIST materials in Tables S7 and S8 in Supporting Information S2. As can be seen, roughly 85% of all reported lipids present with a deviation of <20%.
Figure 3.
Comparison of reported consensus values of the current study with LIPID MAPS16 and Bowden et al.10 (a) and (b) Correlation and boxplot representing the fold change comparison between the reported consensus values and Lipid MAPS. (c) and (d) Correlation and boxplot representing the fold change comparison between the reported consensus values of the current study and Bowden et al.
NIST Candidate Reference Material 8231–Diabetic, High Triglyceride, and African-American Plasma
For the next step in evaluating the interlaboratory performance of the Lipidyzer platform, we compared the interlaboratory performance of the plasma materials using disease and ethnicity-related categories. Recently, semiquantitative values for complex lipids in candidate RM 8231 have been reported using a non-targeted UHPLC-MS/MS method.31 A breakdown of these lipidomes from our study is summarized in Figure S6 in Supporting Information S1. We quantified 849, 843, and 827 lipids in DB, HTG, and AA plasma, respectively. After applying our exclusion criteria, we selected 760, 744, and 714 lipids for consensus value calculations. Subsequently, we assigned consensus values to 714, 692, and 683 lipids in DB, HTG, and AA plasma samples. A list of consensus values of lipid species can be found in Tables S9 and S10 in Supporting Information S2. Consensus values were again calculated with a strict exclusion criterion of COD > 20% to represent the most probable interval of true concentration values in the investigated plasma lipidomes. The average COD of the selected lipids in each lipid class was ≤17% in all plasma samples. The highest average COD was observed for the PC (17%) and lowest for the CE (4%) lipid class. The PC lipid class also had the highest number of excluded lipids, with COD > 20% for up to 40% of the reported lipids. A list of consensus values of excluded lipids is shown in Table S11 in Supporting Information S2.
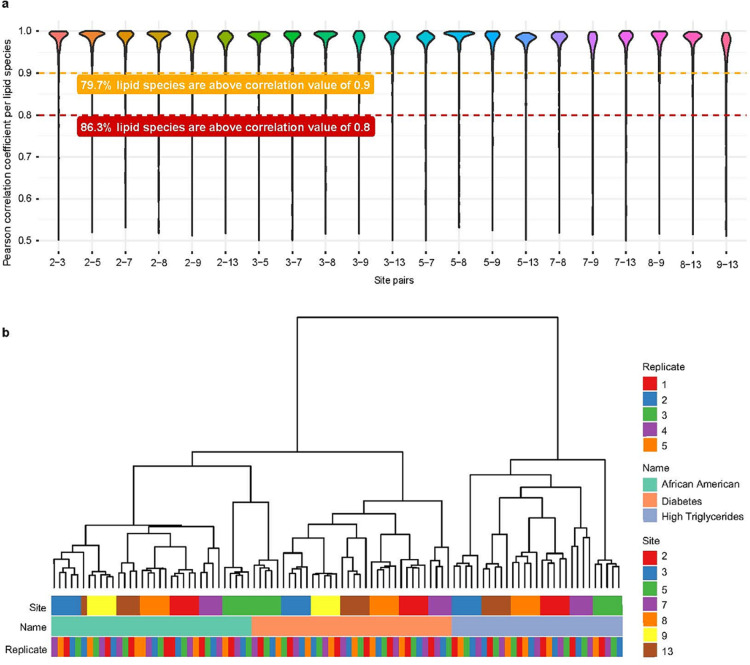
The main goal of analyzing these plasma samples was to test the robustness of the platform measuring plasma from different clinical and ethnic backgrounds. This is an important step toward the standardization and validation of the platform for its use in preclinical studies and biomarker research. For this evaluation, we considered the 704 lipids selected from our previous analysis of the SRM 1950 material with CV ≤ 20%. First, we calculated pairwise Pearson correlations of lipid concentrations reported between all seven participating laboratories (Figure 4a). A significant number of lipid species had correlations close to 1. This proved that the reported lipid species concentrations were in very good agreement between the sites. Next, we performed hierarchical clustering (Figure 4b) to further compare the obtained results. We observed separate clusters for each of the investigated plasma materials showing that all laboratories could distinguish the different plasma samples. The cluster analysis also revealed a site-specific separation for each analyzed plasma.
Figure 4.
Interlaboratory performance of the Lipidyzer platform on NIST candidate RM 8231. (a) All Pearson correlations of determined lipid species in NIST candidate RM 8231 between sites. (b) Cluster analysis of selected lipids. Only those lipids selected in SRM 1950 (measured by at least seven sites with minimum three replicates in a day and CV ≤ 20%) were considered. Site 9 did not measure high triglyceride plasma.
Familial Hypercholesterolemia Cohort Samples
As a final step of our investigation, three sites analyzed 59 plasma samples from individuals with FH. We considered the same 704 lipids as selected in SRM 1950 plasma for these cohort samples. The main goal of this analysis was to compare whether different laboratories would obtain similar results when analyzing a “real” clinical study.
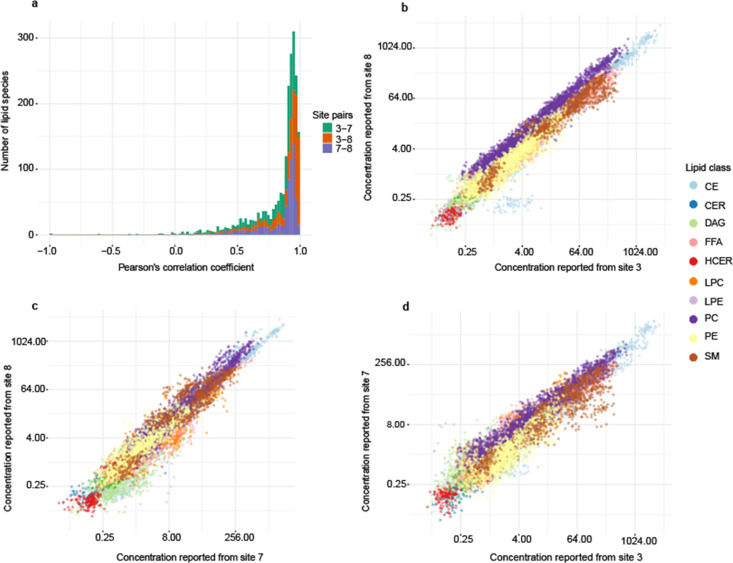
To evaluate the concordance across laboratories, we calculated pairwise correlations between sites for the monitored lipid species concentrations. A histogram of the Pearson’s correlation coefficient (Figure 5a) showed very good agreement between sites, with a significant number of measurements correlated with values above 80%. The agreement was the highest between sites 3 and 8 and the lowest between sites 7 and 8. Figure 5b–d shows that different classes were correlated between sites (TAGs were removed to visualize the other classes and are reported in Figure S4 in Supporting Information S1). Lipid classes correlated best between sites 7 and 8, showing the narrowest spread of data points along the diagonal. It is also worth mentioning the visible systematic bias in one class, PC, at site 3, reported lower concentration values compared to both sites 7 and 8. Site 3 also reported the lowest concentrations for PC in SRM 1950 among the participating laboratories. However, such a systematic difference should not significantly bias the overall interpretation in most instances.
Figure 5.
Correlation of reported concentration of lipids in 59 plasma samples related to familial hypercholesterolemia between three sites. (a) Histogram of pairwise Pearson’s correlation coefficient between three sites. (b–d) Correlation of the reported lipid concentrations between sites 3 and 8, 7 and 8, and 3 and 7, respectively.
Overall, our analysis showed congruent results considering several additional sources of variation when comparing a “real” clinical trial with the analysis of a standardized material such as SRM 1950. Aliquotation of small sample volumes, 50 μL in our case, as well as sample thawing, shipment conditions, and individual processing are inevitable sources of variation, which can largely be prevented when analyzing the SRM 1950 material. Considering these circumstances, three sites robustly measured more than 500 lipid species in the cohort plasma samples. A recent study on reproducibility of targeted lipidome analyses in plasma and erythrocytes using the Lipidyzer platform also reported that 491 lipid measurements in plasma were reproduced well over a 6 week period.32 This promising number of robustly measured lipids puts the platform in a strong position to be used in important clinical research and biomarker discovery studies. At present, it is still a challenging task to reproduce clinical metabolomics findings between laboratories, with the major impediment being the limited methodological and practical agreement among laboratories. Although harmonization will remain a significant challenge for the entire lipidomics community, we showed here that the Lipidyzer platform can provide robust and clinically relevant results by using harmonized analysis protocols.
Conclusions
Our study analytically compared two well-established lipid extraction protocols and incorporated clinically relevant plasma materials to prove that a harmonized analysis protocol can result in excellent interlaboratory lipidomics data. Moreover, we have assigned an unprecedented number of lipid consensus values not only for pooled SRM 1950 plasma but also for clinically relevant disease-related HTG, DB, as well as for AA plasma materials, and provided a quantitative lipidomics overview of these materials. The robust measurement of lipids was possible because the Lipidyzer platform standardizes all steps of the analysis process: sample extraction, standard preparation, acquisition method, and data processing and reporting. Furthermore, using a validated targeted method provides consistent results compared to untargeted methods across multiple laboratories, instruments, and operators. The entire lipidomics community can utilize the reported reference values to extend their quality control activities and serve as a quantitative benchmark for several hundred lipid species in the investigated human plasma materials. At present, the Lipidizer platform misses out on some important lipid classes such as phosphatidylinositol and phosphatidylserine. However, just recently an open access version of the LWM has been published not only enabling workflow adaptations such as the addition of additional classes or the definition and assignment of internal standards but also isotope correction.30 We believe that this new approach together with the here provided evidence, that standardized methods and protocols lead to reproducible and comparable data, will bring quantitative, comprehensive lipidomics closer to the bedside.
Acknowledgments
The authors would like to acknowledge Tracy Schock, Dan Bearden, and Yamil Simón-Manso for their help with material design and procurement. Niek Blomberg is acknowledged for assistance with Lipidyzer analysis. Part of the TOC graphic was created with BioRender.com. D.R. thanks the NIH (grants P30CA015704 and S10OD021562) for financial support. Mohan Ghorasaini (M.G.) is an early-stage researcher supported by the H2020 ITN consortium ArthritisHeal (#812890). Martin Giera (M.G.) was partially supported by the NWO XOmics project #184.034.019. This research was supported in part by the Intramural Research Program of the National Institute on Aging, NIH. This research was supported in part by NIH grants 5U54HG010426-03 (M.P.S.) and 1U2CCA233311-01 (M.P.S.). The FIMM Metabolomics Unit was supported by HiLIFE and Biocenter Finland.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.analchem.1c02826.
Study design; samples; calculations of consensus values and associated uncertainties; experimental section; lipid annotations by Lipidyzer and LIPID MAPS; complete overview of study design and workflow of the inter-laboratory comparison study; CV distribution along 13 lipid classes; scatter plot to summarize the performance of each laboratory between the methods; correlation of reported concentration of lipids; comparison of COD distribution; and breakdown of plasma lipidome of NIST plasma standards (PDF)
Consensus values; method details; and isotope correction comparison (XLSX)
Author Contributions
○○ Mohan Ghorasaini (M.G.) and Y.M. contributed equally. Mohan Ghorasaini (M.G.), Y.M., and Martin Giera (M.G.) wrote the manuscript. Y.d.R. and E.S. contributed the clinical cohort. Martin Giera (M.G.) conducted the overall study design. C.H., Mohan Ghorasaini (M.G.), and Martin Giera (M.G.) were responsible for study management and planning. C.J. and J.B. contributed NIST SRM1950 and RM8231 reference materials. All authors contributed to the study and either analyzed study samples, provided data, and/or critically assessed the manuscript.
The authors declare no competing financial interest.
Notes
Certain commercial equipment, instruments, or materials are identified in this paper to adequately specify the experimental procedures. Such identification does not imply recommendation or endorsement by the National Institute of Standards and Technology nor does it imply that the materials or equipment identified are necessarily the best for the purpose. Furthermore, the content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Standards and Technology. Products mentioned by name are for research use only and not for use in diagnostic procedures.
Supplementary Material
References
- Serhan C. N.; Levy B. D. Resolvins in inflammation: emergence of the pro-resolving superfamily of mediators. J. Clin. Invest. 2018, 128, 2657–2669. 10.1172/jci97943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K.; Han X. Lipidomics: Techniques, Applications, and Outcomes Related to Biomedical Sciences. Trends Biochem. Sci. 2016, 41, 954–969. 10.1016/j.tibs.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Körner A.; Zhou E.; Müller C.; Mohammed Y.; Herceg S.; Bracher F.; Rensen P. C.; Wang Y.; Mirakaj V.; Giera M. Inhibition of Δ24-dehydrocholesterol reductase activates pro-resolving lipid mediator biosynthesis and inflammation resolution. Proc. Natl. Acad. Sci. U.S.A. 2019, 116, 20623–20634. 10.1073/pnas.1911992116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell V. B.; Ekroos K.; Liebisch G.; Wakelam M. Lipidomics: Current state of the art in a fast moving field. Wiley Interdiscip. Rev.: Syst. Biol. Med. 2020, 12, e1466 10.1002/wsbm.1466. [DOI] [PubMed] [Google Scholar]
- Quehenberger O.; Dennis E. A. The human plasma lipidome. N. Engl. J. Med. 2011, 365, 1812–1823. 10.1056/nejmra1104901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenk M. R. The emerging field of lipidomics. Nat. Rev. Drug Discov. 2005, 4, 594–610. 10.1038/nrd1776. [DOI] [PubMed] [Google Scholar]
- Lipidomics needs more standardization. Nat. Metab. 2019, 1, 745–747. 10.1038/s42255-019-0094-z. [DOI] [PubMed] [Google Scholar]
- Liebisch G.; Vizcaíno J. A.; Köfeler H.; Trötzmüller M.; Griffiths W. J.; Schmitz G.; Spener F.; Wakelam M. J. O. Shorthand notation for lipid structures derived from mass spectrometry. J. Lipid Res. 2013, 54, 1523–1530. 10.1194/jlr.m033506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebisch G.; Ejsing C. S.; Ekroos K. Identification and Annotation of Lipid Species in Metabolomics Studies Need Improvement. Clin. Chem. 2015, 61, 1542–1544. 10.1373/clinchem.2015.244830. [DOI] [PubMed] [Google Scholar]
- Bowden J. A.; Heckert A.; Ulmer C. Z.; Jones C. M.; Koelmel J. P.; Abdullah L.; Ahonen L.; Alnouti Y.; Armando A. M.; Asara J. M.; Bamba T.; Barr J. R.; Bergquist J.; Borchers C. H.; Brandsma J.; Breitkopf S. B.; Cajka T.; Cazenave-Gassiot A.; Checa A.; Cinel M. A.; Colas R. A.; Cremers S.; Dennis E. A.; Evans J. E.; Fauland A.; Fiehn O.; Gardner M. S.; Garrett T. J.; Gotlinger K. H.; Han J.; Huang Y.; Neo A. H.; Hyötyläinen T.; Izumi Y.; Jiang H.; Jiang H.; Jiang J.; Kachman M.; Kiyonami R.; Klavins K.; Klose C.; Köfeler H. C.; Kolmert J.; Koal T.; Koster G.; Kuklenyik Z.; Kurland I. J.; Leadley M.; Lin K.; Maddipati K. R.; McDougall D.; Meikle P. J.; Mellett N. A.; Monnin C.; Moseley M. A.; Nandakumar R.; Oresic M.; Patterson R.; Peake D.; Pierce J. S.; Post M.; Postle A. D.; Pugh R.; Qiu Y.; Quehenberger O.; Ramrup P.; Rees J.; Rembiesa B.; Reynaud D.; Roth M. R.; Sales S.; Schuhmann K.; Schwartzman M. L.; Serhan C. N.; Shevchenko A.; Somerville S. E.; St. John-Williams L.; Surma M. A.; Takeda H.; Thakare R.; Thompson J. W.; Torta F.; Triebl A.; Trötzmüller M.; Ubhayasekera S. J. K.; Vuckovic D.; Weir J. M.; Welti R.; Wenk M. R.; Wheelock C. E.; Yao L.; Yuan M.; Zhao X. H.; Zhou S. Harmonizing lipidomics: NIST interlaboratory comparison exercise for lipidomics using SRM 1950-Metabolites in Frozen Human Plasma. J. Lipid Res. 2017, 58, 2275–2288. 10.1194/jlr.m079012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajka T.; Smilowitz J. T.; Fiehn O. Validating Quantitative Untargeted Lipidomics Across Nine Liquid Chromatography-High-Resolution Mass Spectrometry Platforms. Anal. Chem. 2017, 89, 12360–12368. 10.1021/acs.analchem.7b03404. [DOI] [PubMed] [Google Scholar]
- Chen X.; Chen H.; Dai M.; Ai J.; Li Y.; Mahon B.; Dai S.; Deng Y. Plasma lipidomics profiling identified lipid biomarkers in distinguishing early-stage breast cancer from benign lesions. Oncotarget 2016, 7, 36622–36631. 10.18632/oncotarget.9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höring M.; Ejsing C. S.; Hermansson M.; Liebisch G. Quantification of cholesterol and cholesteryl ester by direct flow injection high-resolution Fourier transform mass spectrometry utilizing species-specific response factors. Anal. Chem. 2019, 91, 3459–3466. 10.1021/acs.analchem.8b05013. [DOI] [PubMed] [Google Scholar]
- Loef M.; Faquih T. O.; von Hegedus J. H.; Ghorasaini M.; Ioan-Facsinay A.; Kroon F. P. B.; Giera M.; Kloppenburg M. The lipid profile for the prediction of prednisolone treatment response in patients with inflammatory hand osteoarthritis: the HOPE study. Osteoarthritis Cartilage Open 2021, 100167. 10.1016/j.ocarto.2021.100167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. W.; Adams K. J.; Adamski J.; Asad Y.; Borts D.; Bowden J. A.; Byram G.; Dang V.; Dunn W. B.; Fernandez F.; Fiehn O.; Gaul D. A.; Hühmer A. F.; Kalli A.; Koal T.; Koeniger S.; Mandal R.; Meier F.; Naser F. J.; O’Neil D.; Pal A.; Patti G. J.; Pham-Tuan H.; Prehn C.; Raynaud F. I.; Shen T.; Southam A. D.; St. John-Williams L.; Sulek K.; Vasilopoulou C. G.; Viant M.; Winder C. L.; Wishart D.; Zhang L.; Zheng J.; Moseley M. A. International Ring Trial of a High Resolution Targeted Metabolomics and Lipidomics Platform for Serum and Plasma Analysis. Anal. Chem. 2019, 91, 14407–14416. 10.1021/acs.analchem.9b02908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quehenberger O.; Armando A. M.; Brown A. H.; Milne S. B.; Myers D. S.; Merrill A. H.; Bandyopadhyay S.; Jones K. N.; Kelly S.; Shaner R. L.; Sullards C. M.; Wang E.; Murphy R. C.; Barkley R. M.; Leiker T. J.; Raetz C. R. H.; Guan Z.; Laird G. M.; Six D. A.; Russell D. W.; McDonald J. G.; Subramaniam S.; Fahy E.; Dennis E. A. Lipidomics reveals a remarkable diversity of lipids in human plasma. J. Lipid Res. 2010, 51, 3299–3305. 10.1194/jlr.m009449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubhi B. K.Direct Infusion-Tandem Mass Spectrometry (DI-MS/MS) Analysis of Complex Lipids in Human Plasma and Serum Using the Lipidyzer Platform. In Clinical Metabolomics; Springer, 2018; pp 227–236. [DOI] [PubMed] [Google Scholar]
- Contrepois K.; Mahmoudi S.; Ubhi B. K.; Papsdorf K.; Hornburg D.; Brunet A.; Snyder M. Cross-Platform Comparison of Untargeted and Targeted Lipidomics Approaches on Aging Mouse Plasma. Sci. Rep. 2018, 8, 17747. 10.1038/s41598-018-35807-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z.; Schmitt T. C.; Varma V.; Sloper D.; Beger R. D.; Sun J. Evaluation of the Performance of Lipidyzer Platform and Its Application in the Lipidomics Analysis in Mouse Heart and Liver. J. Proteome Res. 2020, 19, 2742–2749. 10.1021/acs.jproteome.9b00289. [DOI] [PubMed] [Google Scholar]
- Bligh E. G.; Dyer W. J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Matyash V.; Liebisch G.; Kurzchalia T. V.; Shevchenko A.; Schwudke D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J. Lipid Res. 2008, 49, 1137–1146. 10.1194/jlr.d700041-jlr200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rukhin A. L.; Vangel M. G. Estimation of a common mean and weighted means statistics. J. Am. Stat. Assoc. 1998, 93, 303–308. 10.1080/01621459.1998.10474111. [DOI] [Google Scholar]
- Vangel M. G.; Rukhin A. L. Maximum likelihood analysis for heteroscedastic one-way random effects ANOVA in interlaboratory studies. Biometrics 1999, 55, 129–136. 10.1111/j.0006-341x.1999.00129.x. [DOI] [PubMed] [Google Scholar]
- DerSimonian R.; Laird N. Meta-analysis in clinical trials. Controlled Clin. Trials 1986, 7, 177–188. 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- CCQM, CCQM guidance note . Estimation of a Consensus KCRV and Associated Degrees of Equivalence, 2013. [Google Scholar]
- Mainka M.; Dalle C.; Pétéra M.; Dalloux-Chioccioli J.; Kampschulte N.; Ostermann A. I.; Rothe M.; Bertrand-Michel J.; Newman J. W.; Gladine C.; Schebb N. H. Harmonized procedures lead to comparable quantification of total oxylipins across laboratories. J. Lipid Res. 2020, 61, 1424–1436. 10.1194/jlr.ra120000991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebisch G.; Fahy E.; Aoki J.; Dennis E. A.; Durand T.; Ejsing C. S.; Fedorova M.; Feussner I.; Griffiths W. J.; Köfeler H.; Merrill A. H. Jr.; Murphy R. C.; O’Donnell V. B.; Oskolkova O.; Subramaniam S.; Wakelam M. J. O.; Spener F. Update on LIPID MAPS classification, nomenclature, and shorthand notation for MS-derived lipid structures. J. Lipid Res. 2020, 61, 1539–1555. 10.1194/jlr.s120001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triebl A.; Burla B.; Selvalatchmanan J.; Oh J.; Tan S. H.; Chan M. Y.; Mellet N. A.; Meikle P. J.; Torta F.; Wenk M. R. Shared reference materials harmonize lipidomics across MS-based detection platforms and laboratories. J. Lipid Res. 2020, 61, 105–115. 10.1194/jlr.d119000393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L.; Ji S.; Burla B.; Wenk M. R.; Torta F.; Cazenave-Gassiot A. LICAR: An Application for Isotopic Correction of Targeted Lipidomic Data Acquired with Class-Based Chromatographic Separations Using Multiple Reaction Monitoring. Anal. Chem. 2021, 93, 3163–3171. 10.1021/acs.analchem.0c04565. [DOI] [PubMed] [Google Scholar]
- Su B.; Bettcher L. F.; Hsieh W.-Y.; Hornburg D.; Pearson M. J.; Blomberg N.; Giera M.; Snyder M. P.; Raftery D.; Bensinger S. J.; Williams K. J. A DMS Shotgun Lipidomics Workflow Application to Facilitate High-Throughput, Comprehensive Lipidomics. J. Am. Soc. Mass Spectrom. 2021, 32, 2655–2663. 10.1021/jasms.1c00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aristizabal-Henao J. J.; Jones C. M.; Lippa K. A.; Bowden J. A. Nontargeted lipidomics of novel human plasma reference materials: hypertriglyceridemic, diabetic, and African-American. Anal. Bioanal. Chem. 2020, 412, 7373–7380. 10.1007/s00216-020-02910-3. [DOI] [PubMed] [Google Scholar]
- Loef M.; von Hegedus J. H.; Ghorasaini M.; Kroon F. P. B.; Giera M.; Ioan-Facsinay A.; Kloppenburg M. Reproducibility of Targeted Lipidome Analyses (Lipidyzer) in Plasma and Erythrocytes over a 6-Week Period. Metabolites 2021, 11, 26. 10.3390/metabo11010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.