Abstract
The expression of c-myc proto-oncogene, a key regulator of cell proliferation and apoptosis, is controlled at different transcriptional and posttranscriptional levels. In particular, the c-myc mRNA contains an internal ribosome entry site (IRES) able to promote translation initiation independently from the classical cap-dependent mechanism. We analyzed the variations of c-myc IRES activity ex vivo in different proliferating cell types, and in vivo in transgenic mice expressing a bicistronic dual luciferase construct. c-myc IRES efficiency was compared to that of encephalomyocarditis virus (EMCV) IRES under the same conditions. The c-myc IRES was active but with variable efficiency in all transiently transfected cell types; it was also active in the 11-day- old (E11) embryo and in some tissues of the E16 embryo. Strikingly, its activity was undetected or very low in all adult organs tested. In contrast, EMCV IRES was very active in most cell types ex vivo, as well as in embryonic and adult tissues. These data suggest a crucial role of IRES in the control of c-myc gene expression throughout development, either during embryogenesis where its activity might participate in cell proliferation or later on, where its silencing could contribute to the downregulation of c-myc expression, whose deregulation leads to tumor formation.
c-myc is a proto-oncogene playing a dual role in the control of cell proliferation-differentiation and apoptosis. Its overexpression contributes to oncogenic transformation of various cell tissues, leading to multiple neoplasms, in particular to hematopoietic tumors in humans and mice (5, 25). Several mechanisms have been shown to generate c-myc activation, including gene amplification, chromosomal translocation, proviral insertion, and retroviral transduction (for reviews see references 25 and 31). However, the correlation between tumor occurrence and increased transcription, marked mRNA stabilization, or elevated level of c-Myc proteins is far from clear. Although it is classically assumed that c-Myc protein abundance is determined by transcriptional control, it has been shown that the c-myc gene is strongly subjected to posttranscriptional regulation (reviewed in reference 12). Numerous studies have demonstrated the role of c-myc mRNA stability in the control of gene expression (for review, see reference 30), and instability determinants have been identified in their 3′ untranslated region (3′ UTR [3, 20, 33]) as well as in coding exons (23, 43, 44). Translational control also plays an important role: the c-myc 5′ UTR has been proposed to be a modulator of translational efficiency (28, 32). It has been recently shown to contain an internal ribosome entry site (IRES) capable of directing the internal initiation of protein synthesis in a cap-independent manner (27, 36). In the human c-myc gene, the IRES lies in the 408 nucleotides (nt) upstream from the AUG start codon of the major c-Myc protein (c-Myc2). It is thus present in the c-myc transcripts initiated from either the P0, P1, or P2 promoter and is able to promote the cap-independent synthesis of the two c-Myc proteins c-Myc1 and c-Myc2, initiated at the two alternative CUG and AUG codons, respectively (15, 27). Various pathologies, including breast cancer (11) and multiple myeloma (41), have been correlated to c-myc aberrant translational upregulation, which is proposed to result from overexpression or activation of the cap-binding protein eIF-4E. Overexpression of this protein relieves the translational repression imposed by highly structured 5′ UTRs (22). However, c-Myc overexpression, at least in multiple myeloma, is clearly eIF-4E independent and would thus most probably result from aberrant activation of the c-myc IRES.
To date, several IRESs have been discovered in cellular mRNAs, most of them encoding proteins that are involved in the control of cell growth and/or apoptosis (1, 16–18, 27, 34, 36, 39). It has been suggested recently that the presence of the IRES might allow translation of these cellular mRNAs in situations where the cap-dependent translation machinery is inactive, for instance in response to stress (34, 40) and apoptosis (16, 17, 35) and during cell cycle G2-to-M transition (6, 29). However, these experiments have mostly been performed with transformed or immortalized cell lines where IRES could be aberrantly upregulated (13, 41), thus precluding a knowledge of their possible regulatory role in physiological conditions.
We have investigated this question by analyzing the activity of c-myc IRES in view of its possible involvement in the well-known role of c-myc in controlling cell proliferation and tumorigenesis. Using the bicistronic vector strategy, we compared activities of c-myc and encephalomyocarditis virus (EMCV) IRESs in various transiently transfected cell lines and transgenic mice during embryogenesis and in adult tissues. Our results revealed that the c-myc IRES was active in all the transiently transfected cells and was subjected to strong developmental control in transgenic mice.
MATERIALS AND METHODS
Plasmid construction.
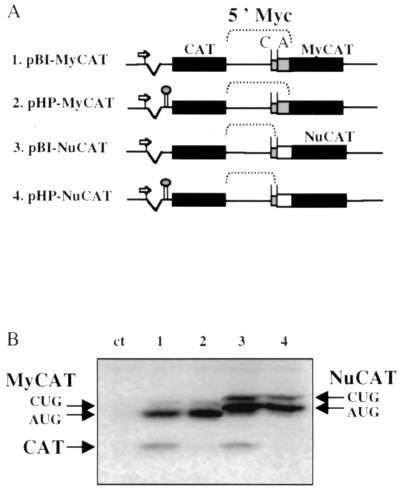
The bicistronic vectors of the MyCAT and NuCAT series were constructed from the previously described plasmids pCVC and pHCVC (containing a hairpin) which have two tandem chloramphenicol acetyltransferase (CAT) genes (18). Part of the c-myc cDNA (551 nt corresponding to the 408 nt of the myc P2 5′ UTR plus the 143 nt downstream from the AUG codon) was introduced into the intercistronic region between the two CAT genes. The two resulting plasmids were called pBI-MyCAT and pHP-MyCAT (Fig. 1A). Two other plasmids were constructed to determine the 3′ border of the IRES. These plasmids contained the myc P2 5′ UTR (408 nt), in which the AUG codon of myc was directly fused to a coding part of the nucleolin gene, resulting in a chimeric NuCAT gene (pSVNC83) (9). The 5′ UTR-NuCAT fusion was obtained by PCR amplification using oligonucleotides T7 and 5′ TTTCCATGGTCGCGGGAGGCTGCTGC 3′ on plasmid pSCT-MyCAT-P2 (27). This PCR fragment was digested at the NcoI site and was fused to the CAT open reading frame in pBI-MyCAT and pHP-MyCAT (in place of MyCAT fusion). These CAT-myc-NuCAT-containing plasmids were called pBI-NuCAT and pHP-NuCAT, respectively (Fig. 1A).
FIG. 1.
Transfection of COS-7 cells with tandem CAT bicistronic vectors. (A) Schematic representation of the tandem CAT bicistronic vectors. The CMV promoter is represented by an arrow, the tandem CAT genes are represented by black boxes, the c-myc sequence fused to CAT is represented by a grey box, and the nucleolin sequence fused to CAT is represented by a white box. The line between the boxes corresponds to the c-myc 5′ UTR (containing the IRES). In the plasmids pHP-MyCAT and pHP-NuCAT, the 5′ hairpin is represented between the intron (broken line) and the first-cistron CAT (left black box). The line between the boxes corresponds to the c-myc 5′ UTR (containing the IRES and the two initiation codons C [CUG] and A [AUG]). (B) Western immunoblotting of transiently transfected COS-7 cells. COS-7 cells were transiently transfected by the bicistronic plasmids described for panel A, and cell extracts were analyzed by Western immunoblotting with anti-CAT antibody as previously described (27). Lane ct corresponds to mock-transfected cells. Lane 1, pBI-MyCAT; lane 2, pHP-MyCAT; lane 3, pBI-NuCAT; lane 4, pHP-NuCAT. The first cistron (CAT) and the second cistron (MyCAT or NuCAT) are indicated by arrows. MyCAT and NuCAT appear as a doublet which corresponds to the initiation of translation at the CUG and AUG codons of the c-myc mRNA, the location of which is shown in panel A.
The plasmids pCRHL and pCREL have already been described (18). The two luciferase genes, Renilla luciferase (LucR) and firefly luciferase (LucF), are under the control of the cytomegalovirus (CMV) promoter and are separated by either a hairpin or the EMCV IRES for pCRHL or pCREL, respectively. The plasmid pCRMyL was constructed by replacing the EMCV IRES with the c-myc IRES between the two luciferase genes. In this plasmid, the 5′ 408 nt of the human c-myc cDNA are fused to the LucF coding sequence.
Cell transfection.
Cell lines were obtained from the American Type Culture Collection (ATCC) or European Collection of Animal Cell Culture (ECACC). NIH 3T3 is a mouse immortalized fibroblast cell line. C2C12 (ECACC no. 91031101) is a mouse muscle myoblast line. ABAE is an adult bovine aortic endothelial primary cell line (7). CHO is a Chinese hamster ovary carcinoma cell line. HeLa (ATCC no. CCL2) is a human uterus carcinoma cell line of epithelial origin. COS-7 (ATCC no. CRL 1654) is a monkey kidney cell line transformed by the simian virus 40 large T antigen. Jurkat (ATCC no. TIB-152) is a human acute leukemia T-cell line. Skin fibroblasts come from a human primary culture cell line (38). ECV304 (ECACC no. 92091712) is a spontaneously transformed human endothelial cell line. 293 (ATCC no. CRL-1573) is a human kidney epithelial cell line transformed with adenovirus 5. SK-Hep-1 (ATCC no. HTB 52) is a human liver adenocarcinoma cell line of endothelial origin; SK-N-AS (ECACC no. 94092302) and SK-N-BE (ECACC no. 95011815) are human neuroblastoma cell lines. Saos2 (ATCC no. HTB-85) is a p53−/− human osteosarcoma cell line of epithelial origin. Cells were cultivated according to ATCC or ECACC instructions.
The different cell types were transfected with 1 μg of plasmid and Fugene 6 reagent (Boehringer-Roche) in 12-well petri dishes. Forty-eight hours after transfection, cell lysates were prepared for luminescence activity as previously described (18).
Generation of transgenic mice.
REL and RMyL transgenic mice were obtained by injecting fragments containing the CMV-LucR-EMCV IRES-LucF and CMV-LucR-c-myc-IRES-LucF sequences, respectively, into one of the pronuclei of (C57BL/6 × CBA)2 fertilized eggs (4). Transgenic embryos and adult mice were identified by PCR and Southern blot analysis of placental or tail DNA using a LucF 500-bp-long probe amplified with the LucF-specific oligonucleotides 5′-CAGTATGAACATTTCGCAGCC-3′ and 5′-CTGAAGGGACTGTAAAAACAGC-3′. Stable lines were maintained by successive crosses with (C57BL/6 × CBA)F1 mice. The transgene copy number was determined by PhosphorImager scanning of Southern blots. The intensity of the bands corresponding to tandem head-to-tail integration was expressed relative to the intensity of the bands corresponding to 5′ and 3′ flanking regions.
Reverse transcription (RT)-PCR analysis.
The cDNAs were synthesized as previously described, using 5 μg of DNase-treated total RNA (38). The PCR was performed with a primer couple (5′-GATTACCAGGGATTTCAGTCG-3′ and 5′-CTGAAGGGACTGTAAAAACAGC-3′ or 5′-CCACATATTGAGCCAGTAGC-3′ and 5′-CCATGATAATGTTGGACGAC-3′) to amplify the LucF or LucR DNA sequence, respectively. The PCRs were carried out using 0.3 U of Goldstar Taq DNA polymerase (Eurogentec) in a final volume of 30 μl, with 1 μl of cDNA. The reaction was performed on a Perkin-Elmer apparatus under the following conditions: 94°C for 3 min and then 25 cycles at 94°C for 30 s, 58°C for 45 s, 72°C for 45 s, and finally 72°C for 5 min. Amplification results (one-third of the reactions) were analyzed on 2% agarose gels (Tris-borate-EDTA), followed by ethidium bromide staining.
Luciferase activity analysis.
The two luciferase activities were measured in cell or tissue extracts, and IRES activity was determined by calculating the LucF-to-LucR ratios. The ratio of IRES efficiency between different cell types was compared ex vivo by calibrating the pCRMyL or pCREL (c-myc or EMCV IRES activity, respectively)-to-pCRHL (background activity without an IRES) ratio. For the in vivo analysis (i) 11-day-old (E11) embryos and placenta; (ii) E16 heart, limb bud, tail, brain, liver, and placenta; and (iii) adult tissue fragments were frozen in liquid nitrogen and stored at −80°C. They were homogenized in 200 μl of passive lysis buffer (Promega) using thurax and were centrifuged for 20 min at 4,500 rpm at 4°C (Biofuge Pico centrifuge; Heraeus). The supernatant was centrifuged for 15 min at 13,000 rpm at 4°C, and the last supernatant was used for luminescence dosage (30 μl). LucR and LucF activities were measured using the Dual Luciferase kit from Promega.
RESULTS
c-myc IRES activity is not influenced by elements located downstream from the AUG start codon.
In a previous report, researchers characterized the human c-myc IRES as requiring RNA elements located between nt −408 and −140 upstream from the c-myc mRNA AUG codon (27). However, the different constructs used in that report contained, in addition to the 5′ UTR, 143 nt of the translated region downstream from the AUG (Fig. 1A, pBI-MyCAT). To evaluate their possible role in the IRES activity, a bicistronic construct with two tandem CAT genes was designed in which the 143 nt of the c-myc coding sequence were replaced by 280 nt from the nucleolin coding sequence (Fig. 1A, pBI-NuCAT). These bicistronic constructs and their counterparts bearing a 5′ hairpin (pHP-MyCAT and pHP-NuCAT) were used for transient transfection of COS-7 cells and expression of the first and second cistrons analyzed by Western immunoblotting with anti-CAT antibodies (Fig. 1B). The results clearly showed that the second cistron was expressed just as efficiently from the constructs pMyCAT and pNuCAT, demonstrating that the coding sequence downstream from the c-myc AUG start codon was not involved in the activity of the c-myc IRES.
c-myc IRES is active in tissue culture cells with varying efficacy.
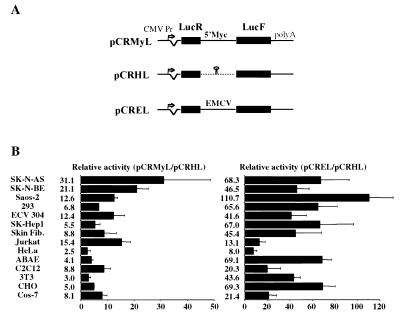
To test the activity of the c-myc IRES in different cell types, we used a bicistronic vector expressing two highly sensitive luciferases, LucR and LucF, under the control of the cytomegalovirus (CMV) promoter. This vector was successfully used in a previous report to demonstrate the existence of the two vascular endothelial growth factor IRESs (18). The human c-myc IRES (the 408-nt-long fragment upstream from the AUG codon) was inserted between the two Luc genes (pCRMyL, Fig. 2A). LucR, the first cistron, provides the level of cap-dependent translation; it is expected to reflect the activity of CMV sequences in the transfected cells and thus to be proportional to the amount of bicistronic mRNA. LucF, the second cistron, is expressed proportionally to the IRES activity. The use of such an assay, based on dual highly sensitive reporters, allows rapid and concomitant quantitation of both reporter gene activities. As positive or negative control, we used similar vectors containing either the EMCV IRES (pCREL) or a hairpin (pCRHL) between the luciferase genes, respectively (18).
FIG. 2.
Analysis of FGF-2 IRES activity in transiently transfected cells. (A) Schematic representation of the bicistronic LucR-I-LucF vectors. Plasmid construction is described in Materials and Methods. They contain a CMV promoter controlling the expression of a bicistronic LucR-LucF mRNA. A synthetic intron is present 5′ of LucR, and a poly(A) site is present 3′ of LucF (18). In the construct pCRMyL, the complete c-myc P2 leader (408 nt) has been fused to the AUG codon of the LucF open reading frame. In the construct pCRHL, a hairpin has been introduced between the two luciferase genes. The construct pCREL contains the EMCV IRES between the two luciferase genes. (B) Fourteen different cell types were transiently transfected with pCRMyL, pCRHL, or pCREL DNA. Cells were harvested 48 h after transfection, and luciferase activities present in cell extracts were measured. c-myc IRES activity was obtained by calculating the ratio 100 × (LucF/LucR). To calibrate the data from the different cell types, the LucF/LucR ratio of pCRMyL was divided by the ratio of the negative control pCRHL. Experiments were repeated three to six times, and results are expressed as means ± standard error (SE). The names of the different cell types (described in Materials and Methods) are indicated. Fib., fibroblast.
Fourteen different cell types of human, monkey, bovine, hamster, or mouse origin were transiently transfected with these constructs. The LucF/LucR ratios obtained in the different cell types for the three constructs pCRMyL, pCREL, and pCRHL are reported in Table 1. As shown by the values obtained using the control without the IRES (pCRHL), the leakage of second-cistron expression strongly varied according to the cell type, indicating that the direct LucF/LucR values were not directly usable for comparing IRES activities from one cell type to the other. Thus, the direct comparison of IRES activity between different cell types was obtained by dividing the LucF/LucR ratios obtained with pCREL and pCRMyL by the LucF/LucR ratios obtained with the negative control pCRHL (Fig. 2B).
TABLE 1.
Measurements of the ratios of LucF to LucR (100 × LucF/LucR) obtained by transient transfection of different cell types with the constructs pCRHL, pCRMyL, and pCREL (Fig. 2A)a
| Cell type | Ratio obtained by use of construct
|
||
|---|---|---|---|
| pCRHL | pCRMyL | pCREL | |
| SK-N-AS | 4.8 | 238.4 | 324.4 |
| SK-N-BE | 4.0 | 65.6 | 135.6 |
| Saos2 | 0.4 | 5.5 | 41.4 |
| 293 | 2.4 | 16.4 | 174.8 |
| ECV304 | 1.1 | 13.4 | 39.5 |
| SK-Hep-1 | 4.4 | 16.4 | 238.0 |
| Skin fibroblast | 5.0 | 47.7 | 256.7 |
| Jurkat | 1.0 | 16.6 | 14.2 |
| HeLa | 7.9 | 12.9 | 75.8 |
| ABAE | 2.1 | 9.9 | 125.6 |
| C2C12 | 29.5 | 292.4 | 500.6 |
| NIH 3T3 | 1.5 | 8.9 | 103.5 |
| CHO | 4.8 | 12.5 | 132.0 |
| COS | 6.3 | 32.9 | 130.4 |
Experiments were performed at least three times, and the values given in this table correspond to a representative experiment.
Both c-myc and EMCV IRESs exhibited a broad spectrum of activity, since they were active in all the different mammalian cell types tested, whatever their origin. However, c-myc IRES activity was globally lower than that of the EMCV IRES. Both IRESs were subjected to cell type-specific regulations, since factors of 11 and 14 were observed between the highest and lowest activities for c-myc and EMCV IRESs, respectively. c-myc IRES activity was high (>20 arbitrary units [AU]) in the two neuroblastoma cell lines SK-N-BE and SK-N-AS and was intermediate (>10 AU) in three other human transformed cell lines originating from osteosarcoma (Saos2), leukemia (Jurkat), or endothelial tumor (ECV304). Low activity was observed in all the nontransformed cells, independently of their origin: murine (3T3 and C2C12), bovine (ABAE), or human (skin fibroblasts). EMCV IRES activity in contrast was very high or high (>50 AU) in most cells, whether transformed or not. The only cell line showing low activity (<10 AU) was HeLa, in which the c-myc IRES is also very weak.
Generation of transgenic mice expressing bicistronic LucR-IRES-LucF constructs.
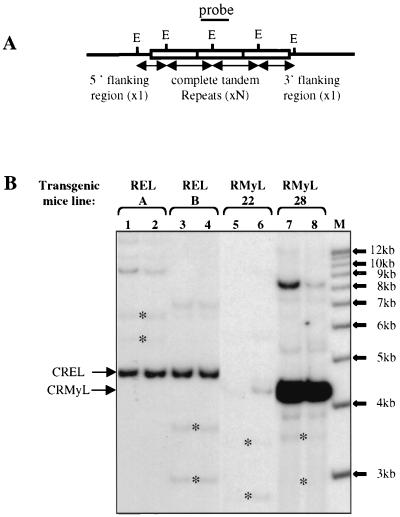
To determine the tissue specificity of the c-myc IRES in the absence of any bias due to cell transformation or cell culture conditions, we decided to study the expression of the LucR-Myc-IRES-LucF construct in vivo. For that purpose, transgenic mice were produced that contained the LucR-IRES-LucF DNA fragment (Fig. 2A, CRMyL, and Materials and Methods). Two independent lines (RMyL-22 and RMyL-28) were established. Southern blots showed that these transgenic lines contained 1 or 2 and 28 to 35 copies of the transgene, respectively (Fig. 3A and B, lanes 5 and 6 and 7 and 8). Two other transgenic lines were derived in parallel, which contained the construct with the viral EMCV IRES in place of c-myc IRES (Fig. 2A, CREL). These two lines, RELA and RELB, both contained 12 to 15 copies of the construct (Fig. 3B, lanes 1 and 2 and 3 and 4).
FIG. 3.
Southern blotting analysis. (A) Schema of the tandem repeat insertion of the bicistronic constructs in the mouse genome. The complete digestion of genomic DNA with the EcoRI enzyme (unique site in the transgene) generates a 4.3-kb (RMyL) or 4.7-kb (REL) fragment corresponding to the complete tandem repeat (N copies) and two fragments of unknown sizes (both hybridizing with the probe which encompasses the EcoRI site) corresponding to the 5′ and 3′ flanking regions. The number of tandem copies has been evaluated by calibrating the signal given by the complete tandem repeat to the signals given by the flanking regions. (B) Twenty micrograms of genomic DNA was digested with the EcoRI enzyme and was hybridized to a LucR-specific PCR fragment (see Materials and Methods). Two offspring of each different strain (RELA and RELB; RMyL-22 and RMyL-28) were tested in parallel. (Only 10 μg of sample 5 was digested.) Arrows indicate the complete tandem repeat fragments corresponding to the CREL and CRMyL transgenes. The flanking region fragments are indicated by asterisks. Other bands of higher molecular weight correspond to partially digested DNA. The intensity of the bands was measured with a phosphorimager, and the copy number of the transgene was evaluated as explained for panel A.
c-myc IRES is active in transgenic embryos but silent in adult tissues.
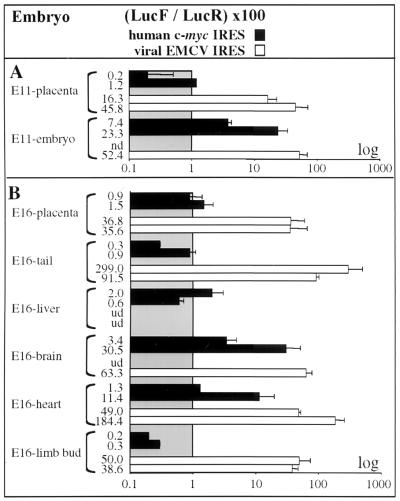
Luciferase activities were measured in total E11 embryos and placenta of the 4 transgenic lines, in 5 E16 tissues and placenta, and in 13 adult organs. As shown in Fig. 4 and Table 2, where the values correspond to representative experiments obtained in the two transgenic lines, the activity of c-myc IRES was high in the E11 embryo, whereas it was hardly detectable in the placenta. In contrast the activity of EMCV IRES was high in both the E11 placenta and embryo (Fig. 4A).
FIG. 4.
Activities of c-myc and EMCV IRESs in transgenic embryos. Embryos from the different transgenic lines were prepared for E11 (A) and E16 (B) embryos, and luciferase activities were measured in different tissues (named on the left), as described in Materials and Methods. IRES activities were measured by calculating the ratios [100 × (LucF/LucR)] and are represented by histograms (black boxes for RMyL-28 and -22 and white boxes for RELB and -A). For values, see Table 2 and reference 8. Experiments were repeated at least three times, and results are expressed as means ± SE. Abbreviations: nd, not determined (the experiment was not performed); ud, undetected activity (LucR activity was not superior to three times the background value in the corresponding organ).
TABLE 2.
Measurement of LucR and LucF activities in whole E11 embryos and in embryonic (E16) and adult tissues of RMyL-22 and RMyL-28 transgenic micea
| Tissue used | LucR and LucF activity values for mice of types
|
|||
|---|---|---|---|---|
| RMyL22
|
RMyL28
|
|||
| LucR (103) | LucF (102) | LucR (103) | LucF (102) | |
| Embryo | ||||
| E11 embryo | 443 | 980 | 4.5 | 3 |
| E11 placenta | 120 | 13 | 40 | 2 |
| E16 limb bud | 45,000 | 1,390 | 2,900 | 63 |
| E16 heart | 280 | 55 | 15 | 1.9 |
| E16 brain | 388 | 220 | 10 | 4.5 |
| E16 liver | 2,650 | 180 | 5.9 | 1.4 |
| E16 tail | 5,050 | 510 | 307 | 12 |
| E16 placenta | 881 | 110 | 885 | 50 |
| Adult | ||||
| Testis | 1,100 | 100 | 14 | 7 |
| Ovary | 189 | 10 | 1.7 | 0.2 |
| Intestine | 380 | 5 | ND | ND |
| Kidney | 1,780 | 150 | 60 | 1 |
| Stomach | 50,000 | 175 | 32.5 | 30 |
| Heart | 1,644 | 51 | 144 | 1.7 |
| Thymus | 75 | 3.5 | 4.7 | 0.2 |
| Tongue | 22,000 | 320 | 274 | 4.5 |
| Brain | 32 | 11 | 4.9 | 3 |
| Muscle | 38,500 | 3,910 | 151 | 45 |
| Skin | 2,600 | 70 | 10.9 | 0.3 |
| Liver | 21 | UD | UD | UD |
| Lung | 9.3 | 0.3 | 3.2 | 0.1 |
These analyses were performed at least three times, and the values given in this table correspond to a representative experiment. ND, not determined (the experiment was not performed); UD, undetected activity (LucR activity was not superior to three times the background value in the corresponding organ). The LucF and LucR activities are measured in luminescence AU. As the sensitivities of the two luciferases are different, these relative values cannot be used to compare the number of LucF and LucR molecules synthesized and consequently do not allow comparison of IRES- versus cap-dependent translation.
For E16 embryos, c-myc IRES was still inactive in extraembryonic tissues. It showed a strong tissue specificity in the embryonic tissues, being active in brain and heart and completely inactive in tail, liver, and limb bud (Fig. 4B). Again, this variation of IRES activity was not observed for EMCV IRES which was very efficient in placenta, tail, heart, and brain. The only organ in which EMCV IRES activity was not detected was the liver.
EMCV IRES exhibited a very broad spectrum of activity in the adult, since 12 out of the 13 tissues tested showed significant IRES activity (Fig. 5). However, as was observed in the tissue-cultured cell lines, there was clear tissue specificity: the highest activities (in AU) (>80) were observed in lung, testis, and skin, and the lowest (<10) were in muscle and liver. The other organs showed high (>40) (tongue, thymus, ovary), moderate (25) (brain), or rather low (<20) (intestine, heart, stomach, and kidney) activity.
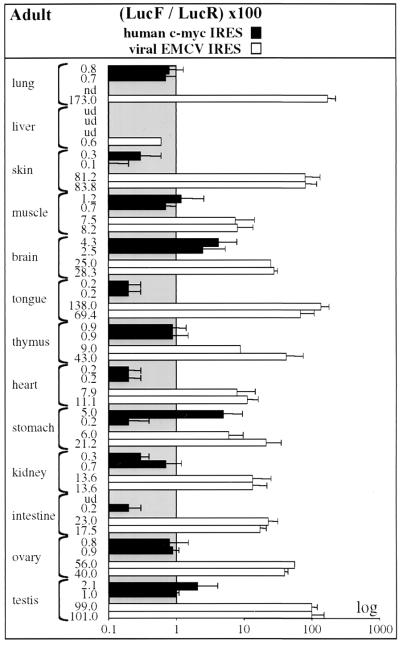
FIG. 5.
c-myc and EMCV IRES activities in transgenic adult tissues. Different organs (named on the left) were prepared from adult transgenic mice, and luciferase activities were measured as described for Fig. 4. IRES activities are represented by histograms corresponding to the equation 100 × (LucF/LucR) (Table 2). These values correspond to representative experiments that were repeated at least three times on the two independent lines expressing each bicistronic construct, pCRMyL (c-myc) and pCREL (EMCV). Abbreviations: nd, not done; ud, undetectable LucR activity.
In contrast to the broad spectrum of activity of EMCV IRES, the c-myc IRES was completely inactive or showed very low activity in all adult organs. In total, the c-myc IRES activity varied from 0.1 AU (most organs) to 5 AU (brain and stomach), whereas that of EMCV IRES varied from 6 to 173 AU (Fig. 5).
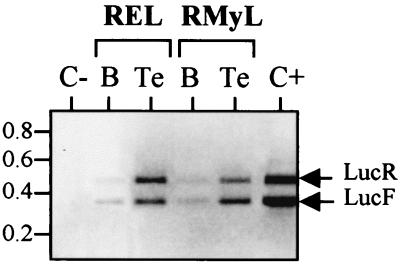
To check that the absence of LucF activity observed with c-myc IRES and not with EMCV IRES was only due to a lack of IRES activity and not to any aberrant event, comparative RT-PCR was used to measure the relative levels of LucF and LucR cistrons. As shown in Fig. 6, the ratio of LucF RNA cistrons to LucR RNA cistrons was similar in RMyL-22 and RELA organs, thus showing that the integrity of the bicistronic mRNAs was comparable in the c-myc IRES and EMCV IRES transgenic mice.
FIG. 6.
Detection of the bicistronic Luc mRNA in transgenic adult tissues. Total RNA was extracted from RMyL-22 or RELA transgenic brain (B) or testis (Te), and the level of LucF and LucR cistrons was analyzed by RT-PCR (as described in Materials and Methods). The negative control (C−) corresponds to a PCR experiment performed using RNA from the brain of a nontransgenic mouse. The positive control (C+) was performed with an in vitro-transcribed bicistronic REL mRNA. The bands corresponding to amplification of LucR (465 nt) and LucF (366 nt) cDNAs are indicated by arrows. This experiment enabled us to check the LucF/LucR ratios at the RNA level in the different organs and to show that they are similar in REL and RMyL transgenic mice.
In summary, these data indicated that, in contrast to EMCV IRES, which is highly efficient in both embryonic and adult mice, the c-myc IRES is active in E11 embryo and tissue specific in E16 embryo, whereas it is strongly silenced in adult tissues.
DISCUSSION
We report here that the IRES of a proto-oncogene, c-myc, is repressed in vivo in adult transgenic mice. In contrast it is active in E11 and E16 embryos (with a strong tissue specificity for E16 embryos) and in all the cultured, proliferating cells tested. Strikingly, the EMCV IRES, albeit with different efficiencies, exhibits similar levels of activity both ex vivo and in vivo in embryos and adult mice. The IRES silencing observed in vivo is thus specific to c-myc IRES and points to a physiological role of this IRES, which, if deregulated, might contribute to tumorigenesis.
We provide here a direct comparison of activities of c-myc and EMCV IRESs in human and nonhuman cell types in a highly calibrated quantitation system. Two previous studies have separately reported cell type-specific variations of these two IRESs (2, 37). First, Borman et al. compared the behavior of seven picornaviral IRESs, either from type I or type II (19), in six primate or nonprimate cell lines (2). The cells could be divided broadly into two groups: cells including HeLa, FRhK4, and SK-HepG2, which were permissive to all the picornaviral IRESs; and cells corresponding to SK-N-BE, BHK21, and Neuro-2A, which were permissive for type II IRESs (such as EMCV) but were refractory to translation from type I IRESs. Accordingly, we have also found that the EMCV IRES is active in both HeLa and SK-N-BE cells. The apparent differences observed between Borman's experiments and ours reflect only the different methods used to measure IRES-dependent translation: whereas in Borman's study, the IRES activities are expressed relative to the cell type in which the IRES is the most efficient (taken as 100%), we calibrate the IRES activity in a certain cell type relative to the background value given by a bicistronic construct devoid of an IRES between the two cistrons.
Second, Stoneley et al., using bicistronic constructs, studied c-myc IRES activity in HeLa cells (37). In agreement with our study, they reported that Hela cells showed a high level of IRES-dependent translation. Furthermore, by comparing CMV- or simian virus 40-initiated bicistronic transcripts, they demonstrated that the level of c-myc IRES activity was dependent upon the amount of bicistronic mRNAs found in the cell: the more they were expressed, the lower the IRES activity. This suggests that noncanonical factors are required to activate IRES-dependent translation and that they might be titrated by an excess of bicistronic mRNAs in the transiently transfected cells.
We have shown by using a large variety of transiently transfected cultured cells that EMCV and c-myc IRESs are both efficient in the five species tested (human, monkey, cow, hamster, and mouse). However, the EMCV IRES is globally more efficient than that of c-myc, with a maximum of 110 AU for EMCV versus 30 AU for the c-myc IRES. This point is well illustrated by the analysis of ABAE, 293, CHO, and 3T3 cell lines, where the EMCV IRES is extremely potent while the c-myc IRES is weak. However, this does not hold true for all cell lines, since in Jurkat lymphoma cells, for instance, both IRESs present similar activities (around 14 AU). These differences, obtained in conditions of mRNA overexpression, reveal the cell types where the trans-acting factors required by the c-myc IRES are the most limiting.
The different activities observed for EMCV and c-myc IRES could rely on particular features due to their viral and cellular origins, respectively. This hypothesis is in agreement with a recent analysis of the activity of another cellular IRES, that of fibroblast growth factor 2 (FGF-2) (8). We found that within the same cell lines, the FGF-2 IRES also presented a broad spectrum of activity, with variations similar to those observed with the c-myc IRES, ranging from 3 to 36 AU. However, the highest efficiency was observed in the p53−/− osteosarcoma Saos2 cell line, in which c-myc IRES was two times less active, and the lowest efficiency was observed in Jurkat cells, where the c-myc IRES was five times more efficient. Taken together, these data point out the tissue specificity of each IRES under study. Strikingly, the c-myc IRES is active in transformed cells and especially in neuroblastoma and leukemia cell lines: as their normal nontransformed counterparts express c-myc mRNA abundantly (26), it is possible that an aberrant activity level of the c-myc IRES in these cells contributes to their progression towards tumorigenesis.
The c-myc IRES shows a much more drastic regulation of its activity in vivo than ex vivo. This contrasts with the EMCV IRES: we show that the viral IRES is active throughout development in the whole E11 embryos as well as in most E16 embryonic and extraembryonic tissues, while c-myc IRES activity is restricted to the E11 embryo proper and to the E16 brain and heart. In adults, the c-myc IRES is silenced, while the EMCV IRES is fully active with a broad spectrum of activity. Whereas the wide activity of EMCV IRES confirms previous data obtained with chicken embryos (14) and transgenic mice (21), the developmental regulation of c-myc IRES activity extends the conclusions reached from analysis of other cellular IRESs, such as the FGF-2 IRES in transgenic mice (8) and Antennapedia and Ultrabithorax IRESs in Drosophila (42). FGF-2 IRES activity was low but significant in most organs of adult transgenic mice and remarkably high in their brains (8). This contrasts with the complete silencing of the c-myc IRES observed in adult tissues and highlights a specific regulatory mechanism for the c-myc IRES. This regulation most probably relies on the presence of spatiotemporally controlled trans-acting factors. Such factors, which are not in limiting amount in vivo when present, since we obtained similar results with two transgenic lines (RMyL-22 and RMyL-28) that express very different levels of pCRMyl transcripts, could be enhancers of IRES activity in embryonic tissues (37, 40). Alternatively, which is not exclusive, they could be inhibitors of c-myc IRES activity in adult tissues.
What could be the physiological relevance of the c-myc IRES regulation observed in vivo? c-myc is widely expressed during mouse embryogenesis, where active cellular proliferation takes place. Its expression is required during development, since c-myc gene invalidation causes lethality during the period between 9.5 and 10.5 days of gestation in homozygotes (10). Strikingly, the pathologic abnormalities include the heart and the neural tube, two developing structures in which we have shown the IRES to be active. Thus the regulation of IRES-dependent translation might play an important role in the control of c-myc expression during development.
c-myc gene expression, active in the embryo, is downregulated in the adult, where most cells are in a nonproliferating state. Aberrant c-myc overexpression in transgenic mice leads to tumor formation (24). Previously, the downregulation of c-myc expression, needed to prevent abnormal cell proliferation and/or differentiation, has been shown to take place mainly at the level of mRNA stability (30). Such a mechanism involves elements located in both the coding and 3′ UTR of the c-myc mRNA sequences (3, 20, 23, 33, 43, 44). The results that we provide in this study indicate that the 5′ UTR of this mRNA, in addition, plays a role in the regulation of c-myc expression in vivo. The translational silencing mediated by the IRES (due to the absence of activating factors or the presence of inhibiting factors) could ensure the complete repression of c-myc expression in fully differentiated tissues. This translational repression could result from the fact that differentiated tissues have reduced levels of proteins required for cell proliferation or increased levels (or activities) of proteins involved in cell growth arrest; such proteins could be involved in the positive or negative control of IRES-mediated translation. Physiologically, the transient activation of the IRES (resulting from a change of IRES-regulating factor activity) would allow quiescent cells to induce rapidly the synthesis of c-Myc proteins in response to various signals inducing proliferation, differentiation, or apoptosis, independently of an increase in the level of mRNA. The transgenic mice that we have described in this study represent a powerful tool to investigate such a hypothesis.
ACKNOWLEDGMENTS
We are grateful to J. Auriol for technical assistance and to D. Warwick for English proofreading. We thank Abderahim Mahfoudi, Cécile Orsini, and Hervé Prats for helpful discussions.
This work was supported by grants from the Association pour la Recherche contre le Cancer, the Ligue Nationale contre le Cancer, the Conseil Régional Midi-Pyrénées, the European Commission BIOTECH program (contract 94199-181), and Rhone-Poulenc-Rorer (now Aventis). L. Créancier was financed first by the European Commission BIOTECH program and then by Retina France.
REFERENCES
- 1.Bernstein J, Sella O, Le S Y, Elroy-Stein O. PDGF2/c-sis mRNA leader contains a differentiation-linked internal ribosome entry site (D-IRES) J Biol Chem. 1997;272:9356–9362. doi: 10.1074/jbc.272.14.9356. [DOI] [PubMed] [Google Scholar]
- 2.Borman A M, Le Mercier P, Girard M, Kean K M. Comparison of picornaviral IRES-driven internal initiation of translation in cultured cells of different origins. Nucleic Acids Res. 1997;25:925–932. doi: 10.1093/nar/25.5.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brewer G, Ross J. Regulation of c-myc mRNA stability in vitro by a labile destabilizer with an essential nucleic acid component. Mol Cell Biol. 1989;9:1996–2006. doi: 10.1128/mcb.9.5.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brinster R L, Chen H Y, Trumbauer M E, Yagle M K, Palmiter R D. Factors affecting the efficiency of introducing foreign DNA into mice by microinjecting eggs. Proc Natl Acad Sci USA. 1985;82:4438–4442. doi: 10.1073/pnas.82.13.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cole M D. The myc oncogene: its role in transformation and differentiation. Annu Rev Genet. 1986;20:361–384. doi: 10.1146/annurev.ge.20.120186.002045. [DOI] [PubMed] [Google Scholar]
- 6.Cornelis S, Bruynooghe Y, Denecker G, Van Huffel S, Tinton S, Beyaert R. Identification and characterization of a novel cell cycle-regulated internal ribosome entry site. Mol Cell. 2000;5:597–605. doi: 10.1016/s1097-2765(00)80239-7. [DOI] [PubMed] [Google Scholar]
- 7.Couderc B, Prats H, Bayard F, Amalric F. Potential oncogenic effects of basic fibroblast growth factor requires cooperation between CUG and AUG-initiated forms. Cell Regul. 1991;2:709–718. doi: 10.1091/mbc.2.9.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Creancier L, Morello D, Mercier P, Prats A C. Fibroblast growth factor 2 internal ribosome entry site (IRES) activity ex vivo and in transgenic mice reveals a stringent tissue-specific regulation. J Cell Biol. 2000;150:275–281. doi: 10.1083/jcb.150.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Creancier L, Prats H, Zanibellato C, Amalric F, Bugler B. Determination of the functional domains involved in nucleolar targeting of nucleolin. Mol Biol Cell. 1993;4:1239–1250. doi: 10.1091/mbc.4.12.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis A C, Wims M, Spotts G D, Hann S R, Bradley A. A null c-myc mutation causes lethality before 10.5 days of gestation in homozygotes and reduced fertility in heterozygous female mice. Genes Dev. 1993;7:671–682. doi: 10.1101/gad.7.4.671. [DOI] [PubMed] [Google Scholar]
- 11.De Benedetti A, Harris A L. eIF4E expression in tumors: its possible role in progression of malignancies. Int J Biochem Cell Biol. 1999;31:59–72. doi: 10.1016/s1357-2725(98)00132-0. [DOI] [PubMed] [Google Scholar]
- 12.DePinho R A, Schreiber-Agus N, Alt F W. myc family oncogenes in the development of normal and neoplastic cells. Adv Cancer Res. 1991;57:1–46. doi: 10.1016/s0065-230x(08)60994-x. [DOI] [PubMed] [Google Scholar]
- 13.Galy B, Maret A, Prats A C, Prats H. Cell transformation results in the loss of the density-dependent translational regulation of the expression of fibroblast growth factor 2 isoforms. Cancer Res. 1999;59:165–171. [PubMed] [Google Scholar]
- 14.Ghattas I R, Sanes J R, Majors J E. The encephalomyocarditis virus internal ribosome entry site allows efficient coexpression of two genes from a recombinant provirus in cultured cells and in embryos. Mol Cell Biol. 1991;11:5848–5859. doi: 10.1128/mcb.11.12.5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hann S R, King M W, Bentley D L, Anderson C W, Eisenman R N. A non-AUG translational initiation in c-myc exon 1 generates an N-terminally distinct protein whose synthesis is disrupted in Burkitt's lymphomas. Cell. 1988;52:185–195. doi: 10.1016/0092-8674(88)90507-7. [DOI] [PubMed] [Google Scholar]
- 16.Henis-Korenblit S, Strumpf N L, Goldstaub D, Kimchi A. A novel form of DAP5 protein accumulates in apoptotic cells as a result of caspase cleavage and internal ribosome entry site-mediated translation. Mol Cell Biol. 2000;20:496–506. doi: 10.1128/mcb.20.2.496-506.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holcik M, Korneluk R G. Functional characterization of the X-linked inhibitor of apoptosis (XIAP) internal ribosome entry site element: role of La autoantigen in XIAP translation. Mol Cell Biol. 2000;20:4648–4657. doi: 10.1128/mcb.20.13.4648-4657.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huez I, Creancier L, Audigier S, Gensac M C, Prats A C, Prats H. Two independent internal ribosome entry sites are involved in translation initiation of vascular endothelial growth factor mRNA. Mol Cell Biol. 1998;18:6178–6190. doi: 10.1128/mcb.18.11.6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jackson R J, Kaminski A. Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA. 1995;1:985–1000. [PMC free article] [PubMed] [Google Scholar]
- 20.Jones T R, Cole M D. Rapid cytoplasmic turnover of c-myc mRNA: requirement of the 3′ untranslated sequences. Mol Cell Biol. 1987;7:4513–4521. doi: 10.1128/mcb.7.12.4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim D G, Kang H M, Jang S K, Shin H S. Construction of a bifunctional mRNA in the mouse by using the internal ribosomal entry site of the encephalomyocarditis virus. Mol Cell Biol. 1992;12:3636–3643. doi: 10.1128/mcb.12.8.3636. . (Erratum, 12:4807.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koromilas A E, Lazaris-Karatzas A, Sonenberg N. mRNAs containing extensive secondary structure in their 5′ non-coding region translate efficiently in cells overexpressing initiation factor eIF-4E. EMBO J. 1992;11:4153–4158. doi: 10.1002/j.1460-2075.1992.tb05508.x. . (Erratum, 11:5138.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lavenu A, Pistoi S, Pournin S, Babinet C, Morello D. Both coding exons of the c-myc gene contribute to its posttranscriptional regulation in the quiescent liver and regenerating liver and after protein synthesis inhibition. Mol Cell Biol. 1995;15:4410–4419. doi: 10.1128/mcb.15.8.4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leder A, Pattengale P K, Kuo A, Stewart T A, Leder P. Consequences of widespread deregulation of the c-myc gene in transgenic mice: multiple neoplasms and normal development. Cell. 1986;45:485–495. doi: 10.1016/0092-8674(86)90280-1. [DOI] [PubMed] [Google Scholar]
- 25.Marcu K B, Bossone S A, Patel A J. myc function and regulation. Annu Rev Biochem. 1992;61:809–860. doi: 10.1146/annurev.bi.61.070192.004113. [DOI] [PubMed] [Google Scholar]
- 26.Morello D, Asselin C, Lavenu A, Marcu K B, Babinet C. Tissue-specific post-transcriptional regulation of c-myc expression in normal and H-2K/human c-myc transgenic mice. Oncogene. 1989;4:955–961. [PubMed] [Google Scholar]
- 27.Nanbru C, Lafon I, Audigier S, Gensac M C, Vagner S, Huez G, Prats A C. Alternative translation of the proto-oncogene c-myc by an internal ribosome entry site. J Biol Chem. 1997;272:32061–32066. doi: 10.1074/jbc.272.51.32061. [DOI] [PubMed] [Google Scholar]
- 28.Parkin N, Darveau A, Nicholson R, Sonenberg N. cis-acting translational effects of the 5′ noncoding region of c-myc mRNA. Mol Cell Biol. 1988;8:2875–2883. doi: 10.1128/mcb.8.7.2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pyronnet S, Pradayrol L, Sonenberg N. A cell cycle-dependent internal ribosome entry site. Mol Cell. 2000;5:607–616. doi: 10.1016/s1097-2765(00)80240-3. [DOI] [PubMed] [Google Scholar]
- 30.Ross J. mRNA stability in mammalian cells. Microbiol Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ryan K M, Birnie G D. Myc oncogenes: the enigmatic family. Biochem J. 1996;314:713–721. doi: 10.1042/bj3140713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saito H, Hayday A C, Wiman K, Hayward W S, Tonegawa S. Activation of the c-myc gene by translocation: a model for translational control. Proc Natl Acad Sci USA. 1983;80:7476–7480. doi: 10.1073/pnas.80.24.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schiavi S C, Belasco J G, Greenberg M E. Regulation of proto-oncogene mRNA stability. Biochim Biophys Acta. 1992;1114:95–106. doi: 10.1016/0304-419x(92)90009-n. [DOI] [PubMed] [Google Scholar]
- 34.Stein I, Itin A, Einat P, Skaliter R, Grossman Z, Keshet E. Translation of vascular endothelial growth factor mRNA by internal ribosome entry: implications for translation under hypoxia. Mol Cell Biol. 1998;18:3112–3119. doi: 10.1128/mcb.18.6.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stoneley M, Chappell S A, Jopling C L, Dickens M, MacFarlane M, Willis A E. c-Myc protein synthesis is initiated from the internal ribosome entry segment during apoptosis. Mol Cell Biol. 2000;20:1162–1169. doi: 10.1128/mcb.20.4.1162-1169.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stoneley M, Paulin F E, Le Quesne J P, Chappell S A, Willis A E. C-Myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene. 1998;16:423–428. doi: 10.1038/sj.onc.1201763. [DOI] [PubMed] [Google Scholar]
- 37.Stoneley M, Subkhankulova T, Le Quesne J P, Coldwell M J, Jopling C L, Belsham G J, Willis A E. Analysis of the c-myc IRES; a potential role for cell-type specific trans-acting factors and the nuclear compartment. Nucleic Acids Res. 2000;28:687–694. doi: 10.1093/nar/28.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Touriol C, Morillon A, Gensac M C, Prats H, Prats A C. Expression of human FGF-2 is post-transcriptionally controlled by a unique destabilization element present in the mRNA 3′ untranslated region between alternative polyadenylation sites. J Biol Chem. 1999;274:21402–21408. doi: 10.1074/jbc.274.30.21402. [DOI] [PubMed] [Google Scholar]
- 39.Vagner S, Gensac M C, Maret A, Bayard F, Amalric F, Prats H, Prats A C. Alternative translation of human fibroblast growth factor 2 mRNA occurs by internal entry of ribosomes. Mol Cell Biol. 1995;15:35–44. doi: 10.1128/mcb.15.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vagner S, Touriol C, Galy B, Audigier S, Gensac M C, Amalric F, Bayard F, Prats H, Prats A C. Translation of CUG- but not AUG-initiated forms of human fibroblast growth factor 2 is activated in transformed and stressed cells. J Cell Biol. 1996;135:1391–1402. doi: 10.1083/jcb.135.5.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.West M J, Sullivan N F, Willis A E. Translational upregulation of the c-myc oncogene in Bloom's syndrome cell lines. Oncogene. 1995;11:2515–2524. [PubMed] [Google Scholar]
- 42.Ye X, Fong P, Iizuka N, Choate D, Cavener D R. Ultrabithorax and Antennapedia 5′ untranslated regions promote developmentally regulated internal translation initiation. Mol Cell Biol. 1997;17:1714–1721. doi: 10.1128/mcb.17.3.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yeilding N M, Lee W M F. Coding elements in exons 2 and 3 target c-myc mRNA downregulation during myogenic differentiation. Mol Cell Biol. 1997;17:2698–2707. doi: 10.1128/mcb.17.5.2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yeilding N M, Rehman M T, Lee W M F. Identification of sequences in c-myc mRNA that regulate its steady-state levels. Mol Cell Biol. 1996;16:3511–3522. doi: 10.1128/mcb.16.7.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]