Abstract
Oral squamous cell carcinoma (OSCC), which is the most common epithelial malignant neoplasm in the head and neck, is characterized by local infiltration and metastasis of lymph nodes. The five-year survival rate of OSCC remains low despite the advances in clinical methods. miR-141-3p has been shown to activate or inhibit tumorigenesis. However, the effects of miR-141-3p on invasion and migration of OSCC remain unclear. The present study aimed to evaluate the effects of miR-141-3p on invasion, proliferation, and migration in oral squamous cell carcinoma (OSCC). Reverse transcription quantitative PCR, western blotting and immunohistochemistry were used to detect microRNA(miR)-141-3p and pre-B-cell leukaemia homeobox-1 (PBX1) expression in OSCC tissues and cell lines. The luciferase reporter assay was used to detect targets of miR-141-3p in OSCC. MTT, Transwell and wound healing assays were used to determine the cell proliferation and invasive and migratory abilities, respectively. Expression of constitutive phosphorylated (p)-Janus kinase 2 (JAK2) and p-signal transducer and activator of transcription 3 (STAT3) was detected using western blotting in tissues and cells. miR-141-3p expression was decreased in OSCC tissues and cells, while PBX1 protein expression was increased compared with non-cancerous controls. The result from the dual-luciferase reporter assay revealed that PBX1 was the direct target of miR-141-3p in OSCC tissues. Furthermore, miR-141-3p overexpression and PBX1 knockdown could reduce cell invasion, proliferation and migration, and inhibit the JAK2/STAT3 pathway; however, miR-141-3p downregulation had the opposite effects. In addition, silencing of PBX1 using small interfering RNA could weaken the effects of miR-141-3p inhibitor on JAK2/STAT3 pathway and cell progression in CAL27 cells. In summary, the findings from this study indicated that miR-141-3p upregulation could inhibit OSCC cell invasion, proliferation and migration, by targeting PBX1 via the JAK2/STAT3 pathway.
Keywords: pre-B-cell leukaemia homeobox-1, microRNA-141-3p, oral squamous cell carcinoma, Janus kinase 2/signal transducer and activator of transcription 3 signaling pathway
Introduction
Oral cancer is a general term for various malignant tumors of the mouth. Oral squamous cell carcinoma (OSCC) is one the most common types of oral cancer, accounting for ~90% of all oral cancers found in the mouth and lips (1). In addition to being one of the most common types of oral tumor, is also the 8th leading cause of cancer-associated mortality worldwide (1,2). The current treatment of OSCC has been significantly improved and includes radiotherapy, chemotherapy and surgical resection (3). However, some patients with OSCC are ineligible for radiation or chemotherapy (4) and their survival is not better than that of patients with OSCC. At present, the 5-year survival rate is ~50% (5,6). Further investigation on the mechanism of OSCC tumorigenesis is therefore crucial.
Pre-B-cell leukemia homeobox protein 1 (PBX1) is a member of the PBX family. PBX1 can form hetero-oligomeric complexes with other homeodomain transcription factors, such as Hox and engrailed, to prevent the development of OSCC (7,8). Increasing evidence showed that dysregulated expression of PBX1 contributes to the proliferation, survival and metastasis of various types of cancer, such as breast, lung, gastric and ovarian cancers. For example, in gastric carcinomas (9) and non-small cell lung cancers (10), PBX1 can promote the transformation between the epithelial and mesenchymal cells associated with chemoresistance. Furthermore, it has recently been found that PBX1 is upregulated in several OSCC cell lines (8).
Signal transducer and activator of transcription 3 (STAT3) is a cytoplasmic transcription factor, which expression can be regulated by various cytokines and growth factors, such as IL6, IL10 and TNF-α (11-14).
Previous studies have demonstrated that STAT3 is an important protein in tumorigenesis and progression of different malignancies, such as hepatocellular carcinoma (HCC), clear cell renal cell carcinoma and non-small cell lung cancer (15-18).
The phosphorylation of janus kinase 2 (JAK2) protein can activate STAT3 protein (19). Furthermore, the JAK2/STAT3 signaling pathway plays crucial roles in cell proliferation and apoptosis of various cancers (20). Activating the JAK2/STAT3 signaling pathway can promote the stemness of OSCC (21) and certain microRNAs (miRs/miRNAs) might regulate the expression of JAK/STAT, such as miR-203, miR-409 and miR-365a-3p (20,22-24).
miRs are small non-coding RNAs of ~22 nucleotides in length. They regulate mRNA translation and deterioration, and gene expression is modulated following mRNA transcription (25). Some miRNAs associated with cancer can influence multiple biological processes, such as cell survival, differentiation, proliferation, apoptosis and migration (26). Certain miRNAs have been characterized as tumor suppressors and others as oncogenic (27,28). Previous studies reported that OSCC progression is affected by numerous miRNAs, including miR-141, miR-186(29), miR-145 and miR-429 (30,31). miR-141-3p is a member of the miR-141 family. Previous studies have demonstrated that miR-141-3p expression is related to several types of tumors, such as nasopharyngeal carcinoma, esophageal squamous cell carcinoma and HCC (32-36). However, the effects of miR-141-3p on OSCC have been rarely reported, and the function of miR-141-3p and the relationship between miR-141-3p and JAK2/STAT3 pathway remain unclear in OSCC.
The present study aimed to investigate the possible modulation of JAK2/STAT3 pathway by miR-141-3p via targeting PBX1, and to determine the potential effects of miR-141-3p in OSCC.
Materials and methods
Clinical specimens
A total of 30 pairs of tumor and adjacent normal tissues were collected from clinical surgical cases of OSCC between March 2018 and December 2019 from Lishui People's Hospital (Lishui, China). Adjacent tissues were collected at least 1.5 cm from the tumor tissue margin. The clinical specimen information of the 30 patients is presented in Table I. The study complied with the ethics committee regulations and was approved by the Clinical Ethical Committee of Lishui University (approval no. 201803). Written informed consent was provided by all the patients or their relatives for the use of their tissues in experiments. Following collection, tissue samples were immediately frozen in liquid nitrogen and preserved at -80˚C for subsequent experiments.
Table I.
Clinicopathological characteristics of patients with oral squamous cell carcinoma.
| Variable | Number (%) |
|---|---|
| Age, years | |
| <60 | 11 (36.7) |
| ≥60 | 19 (63.3) |
| Sex | |
| Male | 14 (46.7) |
| Female | 16 (53.3) |
| BMI | |
| <22.5 | 12 (40.0) |
| ≥22.5 | 18 (60.0) |
| Differentiation | |
| Poorly differentiated | 3 (10.0) |
| Moderate differentiation | 6 (20.0) |
| Well differentiated | 21 (70.0) |
| Clinical stage | |
| I-II | 16 (53.3) |
| III-IV | 14 (46.7) |
| Tumor size, cm | |
| <2.9 | 13 (43.3) |
| ≥2.9 | 17 (56.7) |
| T stage | |
| T1-2 | 16 (53.3) |
| T3-4 | 14 (46.7) |
| Infiltration depth | |
| Primary tissue layer | 7 (23.3) |
| Adjacent tissue layer | 19 (63.3) |
| Surrounding tissue layer | 4 (13.3) |
| Smoking status | |
| No | 22 (73.3) |
| Yes | 8 (26.7) |
| Underlying disease | |
| No | 17 (56.7) |
| Yes | 13 (43.3) |
BMI, body mass index.
Cell lines and culture
The OSCC cell lines CAL27, SCC-9, SCC-4 and normal human oral keratinocytes (NHOK; cat. no. PCS-200-014 TM) cells were purchased from The Cell Bank of Type Culture Collection of The Chinese Academy of Sciences. All OSCC cells were cultured in DMEM medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 1% penicillin-streptomycin and 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) placed at 37˚C in a humidified incubator containing 5% CO2.
Cell transfection
CAL27 cells were seeded in a six-well plate at 1x105 cells/well for incubation. After 24 h, the miR-141-3p inhibitor, miR-141-3p mimic, mimic/inhibitor negative control (NC; 50 nM; Guangzhou RiboBio Co., Ltd.) and small interfering (si)-PBX1 and si-NC (2 µM; Guangzhou RiboBio Co., Ltd.), were transfected into the cells using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Subsequently, the cells were incubated in a humidified atmosphere of 5% CO2 at 37˚C. After 72 h, all transfected cells were collected, and subsequent experiments were immediately carried out. CAL27 cells were also co-transfected with 50 nM miR-141-3p inhibitor and 2 µM si-PBX1. The sequences were as follows: si-NC sense, 5'-AAUUCUCCGAACGUGUCACGU-3' and antisense, 5'-ACGUGACACGUUCGGAGAAUU-3'; si-PBX1 sense, 5'-AGCUGUCACUGCUACCAAUGU-3' and antisense, 5'-ACAUUGGUAGCAGUGACAGCU-3'; mimic NC sense, 5'-UUCUCCGAACGUGUCACUGUU-3' and antisense, 5'-AACAGUGACACGUUCGGAGAA-3'; miR-141-3p mimic sense, 5'-AACACUGUCUGGUAAAGAUGG-3' and antisense, 5'-CCAUCUUUACCAGACAGUGUU-3' inhibitor NC sense, 5'-CAGUACUUUUGUGUAGUACAA-3' and antisense, 5'-UUGUACUACACAAAAGUACUG-3'; miR-141-3p inhibitor sense, 5'-CCAUCUUUACCAGACAGUGUUA-3' and antisense, 5'-UAACACUGUCUGGUAAAGAUGG-3'.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from tumor, adjacent tissues and cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. RNA was reverse transcribed into cDNA using the PrimeScript RT reagent kit (Takara Biotechnology Co., Ltd.). The thermocycling conditions were the following: Pre-denaturation at 95˚C for 10 min; 40 cycles of denaturation at 95˚C for 10 sec, annealing at 60˚C for 20 sec. The reaction system contained SYBR Premix Ex Taq™ II 10 µl, PCR Forward Primer (10 µM) 0.8 µl, PCR Reverse Primer (10 µM) 0.8 µl, ROX Reference Dye 0.4 µl, DNA template (2.0 µl) and sterile distilled water (6.0 µl). U6 served as an internal control. The 2-ΔΔCq method was used to calculate expression of the relative mRNA (37). The sequences of the primers were as follows: miR-141-3p forward, 5'-ACACTCCAGCTGGGTAACACTGTCTGGTAA-3' and reverse, 5'-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCCATCTTT-3'; U6 forward, 5'-ATTGGAACGATACAGAGAAGATT-3' and reverse, 5'-GGAACGCTTCACGAATTTG-3'. RT-qPCR reactions were performed using a RealTime PCR System (Applied Biosystems).
Immunohistochemistry (IHC)
Tissues were fixed in 4% paraformaldehyde at 4˚C for 24 h. Then, the tissues were dehydrated in graded dilutions of ethanol at room temperature (70% ethanol for 3-4 h; 80% ethanol for 3-4 h; 90% ethanol for 2-3 h; 95% ethanol for 2-3 h, 100% ethanol I for 1.5-2 h; 100% ethanol II for 1.5-2 h). Then, the tissues were made transparent in xylene I and xylene II for 0.5-1 h each time at room temperature. Next, a mixture of low melting point wax (melting point, 56-58˚C) pre-heated at 60˚C with an equivalent volume of fresh xylene was prepared in a glass bottle. The tissues were dipped into the mixture and incubated overnight at room temperature. Then, the tissues were dipped in low melting point wax and incubated in a 60-˚C oven for 0.5-1 h three times. Following this, the tissues were embedded in low melting point wax at room temperature until the wax concreted. Finally, the waxes of containing tissue were trimmed and marked at room temperature. Wax-embedded oral tissues were cut into 5-µm slices. Xylene and graded dilutions of ethanol (100% I; 100% II; 95% I; 95% II; 85%; 75%) were used to remove wax and rehydrate sections, respectively. Slices were heated in an autoclave with citric acid buffer (pH 7.0) for antigen retrieval for 10 min, and then cooled down at room temperature; they were next incubated with 3% H2O2 for 30 min at 37˚C. Slices were washed three times with PBS for 5 min and subsequently incubated with 10% goat serum (Beyotime Institute of Biotechnology) for 30 min at 37˚C, without subsequent washing. Sections were incubated with an anti-rabbit PBX1 primary antibody (1:100; cat. no. ab104247; Abcam) overnight at 4˚C. Sections were then washed three times with PBS for 5 min. Subsequently, slices were incubated with an HRP-conjugated goat anti-rabbit IgG secondary antibody (1:1,000; cat. no. ab6721; Abcam) at room temperature for 30 min at 37˚C and with an avidin-biotin HRP complex for 30 min at 37˚C, followed by three washes with PBS for 5 min. Slices incubated with only an HRP-conjugated goat anti-rabbit IgG secondary antibody (1:1,000; cat. no. ab6721; Abcam) at room temperature for 30 min at 37˚C served as a negative control. The slices were subsequently stained with 3,3'-diaminobenzidine avoid light for 15 min at room temperature and counterstained with hematoxylin for 15 min at room temperature. Sections were observed using a light microscope (Eclipse Ni-U; Nikon Corporation) at x20 magnification and ImageJ v1.8.0 software (National Institutes of Health) was used to evaluate the positive area of PBX1 staining. PBX1 expression was calculated as follows: Average Optical Density = Integrated Optical Density/Area (Area of target protein distribution).
Western blotting
Tissues and cells were lysed using RIPA buffer (Thermo Fisher Scientific, Inc.) supplemented with protease and phosphatase inhibitors (Thermo Fisher Scientific, Inc.) on ice for 20 min. Samples were centrifuged at 1,500 x g at 4˚C for 5 min, and a BCA protein assay kit (Thermo Fisher Scientific, Inc.) was used to determine the protein concentration after collecting the supernatants. A total of 20 µg of total proteins were separated by 8-12% SDS-PAGE and transferred onto a PVDF membrane. Membranes were blocked with 5% skimmed milk at room temperature for 1 h and were incubated with primary antibodies anti-rabbit JAK2 (Abcam; cat. no. ab245303; 1:1,000), anti-rabbit p-JAK2 (Abcam; cat. no. ab195055; 1:1,000), anti-rabbit STAT3 (Abcam; cat. no. ab226942; 1:1,000), anti-rabbit p-STAT3 (Abcam; 1:1,000), anti-rabbit PBX1 (Abcam; cat. no. ab104247; 1:1,000), and anti-mouse GAPDH (Abcam; cat. no. ab181602; 1:1,000) at 4˚C overnight. Membranes were washed three times with TBST (0.2% Tween) and were incubated with secondary antibodies: HRP-conjugated goat anti-rabbit IgG (Abcam; cat. no. ab6721; 1:5,000) and HRP-conjugated goat anti-mouse IgG (Abcam; cat. no. ab205719; 1:5,000) for 2 h at 37˚C. Membranes were washed three times with TBST and enhanced chemiluminescence reagent (Thermo Fisher Scientific, Inc.) was used to detect the signal on the membrane. The data were analyzed via densitometry using ImageJ v1.8.0 software (National Institutes of Health) and normalized to expression of the internal control GAPDH.
MTT assay
CAL27 cells were seeded at the density of 2x103 per well in 96-well plates. MTT solution (20 µl; Sigma-Aldrich; Merck KGaA) was added at the concentration of 5 g/l. After 4 h, the supernatant from each well was discarded and 150 µl/well DMSO was added. The samples reacted for 10 min in the dark at room temperature. A microplate reader (Beckman Coulter, Inc.) was used to detect the optical density at 490 nm and the cell proliferation curve was plotted.
Luciferase assay
TargetScanHuman v7.2 (www.targetscan.org) online prediction software was used to predict the binding sites of miR-141-3p and PBX1. We first constructed two plasmids wild type (Wt) and mutant type (Mut), and then we constructed two vectors, Wt-PBX1 and Mut-PBX1. Subsequently, miR-141-3p mimic, miR-141-3p inhibitor and mimic/inhibitor-NC were transfected into CAL27 cells using Lipofectamine® 2000. After 48 h, the cells were collected and lysed and used for luminescence detection. The luciferase activity was detected using a dual-luciferase reporter system (Promega Corporation) which includes the use of two reporters, firefly and Renilla luciferases. The activity of the co-transfected control reporter (Renilla luciferase) provided an internal control to normalize the results.
Transwell assay
Transwell chambers were used for the Transwell assay (Corning). Matrigel (cat. no. E1270; Sigma-Aldrich; Merck KGaA) and serum-free DMEM medium were used to solidify the upper chamber. Matrigel was thawed at 4˚C overnight. Matrigel was then diluted with serum-free DMEM medium to a Matrigel concentration of 300 µl/ml mixture at 4˚C. Upper chambers were pre-coated with the mixture, then pre-warmed at 37˚C. After 1 h, 200 µl cell suspension (2x104) containing 1% FBS was added to the upper chamber, whereas 500 µl DMEM medium containing 10% FBS was added to the lower chamber. After 24 h, the non-invading cells were removed from the upper chamber and cells were fixed with 4% formaldehyde for 10 min at 4˚C and stained with 0.05% crystal violet for 30 min at room temperature. Five fields were randomly chosen and observed under an inverted light microscope (Eclipse Ts2; Nikon Corporation) at x20 magnification and the number of invasive cells was calculated.
Wound healing assay
Cells were seeded onto 6-well plates at the density of 5x105 cells per well. Once the cell density reached ~80%, a 200-µl pipette tip was used to create a vertical wound on the cell layer, and detached cells were washed away with PBS. Images were captured at 0 h and at 48 h under a light microscope (Eclipse Ni-U; Nikon Corporation) at x40 magnification and the cell migration distances was calculated.
Statistical analysis
Data were presented as the means ± standard deviation of three independent experiments. Data were analyzed using SPSS 22.0 software (SPSS, Inc.). Paired t-test was used to compare tumor group and adjacent normal tissue group and others significant difference between two groups was compared with unpaired t-test. Multiple comparison was performed using one-way ANOVA followed by Bonferroni's post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of miR-141-3p and PBX1 in OSCC tissues and cell lines
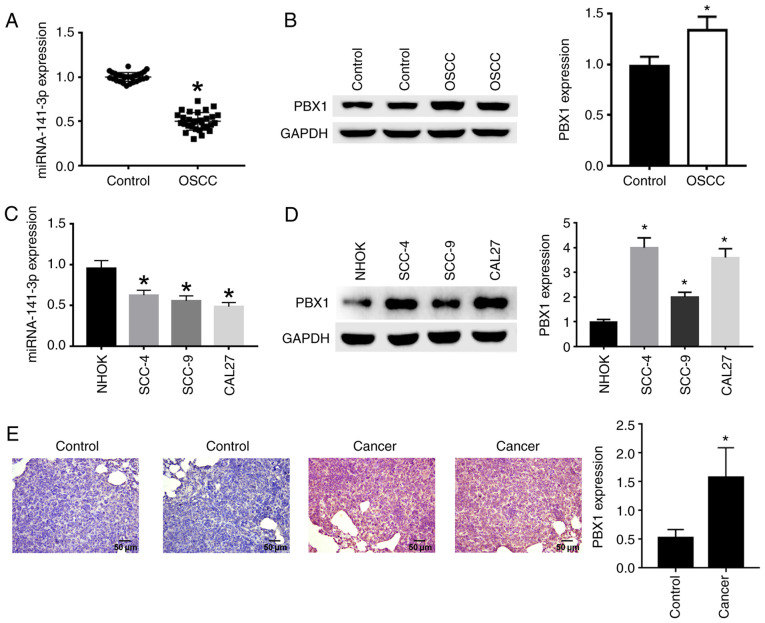
RT-qPCR, western blotting and IHC were used to analyze the expression of miR-141-3p and the protein expression of PBX1. The results demonstrated that miR-141-3p expression in OSCC tissues (Fig. 1A) and cell lines (Fig. 1C) was significantly decreased compared with non-cancerous tissues and NHOK cell line, respectively. In addition, PBX1 protein expression was significantly increased in OSCC tissues (Fig. 1B) and cell lines (Fig. 1D) compared with non-cancerous tissues and NHOK cell line. The expression of PBX1 was significantly higher in CAL27 and SCC-4 cells compared with the other cell lines (Fig. 1D), and miR-141-3p expression levels were the lowest in CAL27 cells compared with the other cell lines (Fig. 1C). Thus, CAL27 cell line was selected for PBX1 knockdown in subsequent experiments to confirm the effects of PBX1 in OSCC. Immunohistochemical staining with anti-PBX1 primary antibody showed that cancer tissues were stained brown in cytoplasm, corresponding to a positive staining. Control tissues were stained blue, corresponding to a negative staining (Fig. 1E). From the above results, it can be seen that miR-141-3p was downregulated, while PBX1 was upregulated in OSCC tissues and cell lines.
Figure 1.
Detection of miR-141-3p and PBX1 expression in OSCC tissues and cell lines. (A) RNA expression level of miR-141-3p and (B) protein expression level of PBX1 in OSCC tissues were detected by RT-qPCR and by western blotting, respectively. (C) RNA expression level of miR-141-5p and (D) protein expression level of PBX1 in OSCC cell lines were detected by RT-qPCR and by western blotting, respectively. (E) Expression level of PBX1 in OSCC tissues was detected by IHC assays. Scale bar=50 µm. *P<0.05 vs. control or NHOK. RT-qPCR, reverse transcription-quantitative PCR; IHC, immunohistochemistry; OSCC, oral squamous cell carcinoma; miRNA/miR, microRNA; PXB1, pre-B-cell leukaemia homeobox-1.
PBX1 is a potential target of miR-141-3p in OSCC
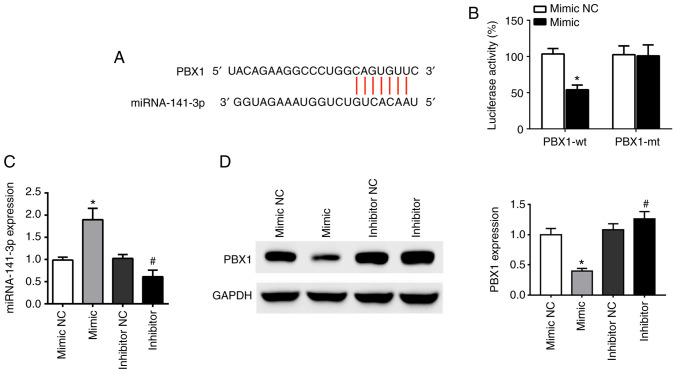
TargetScanHuman v7.2 online software was used to analyze the binding sites of miR-141-3p and PBX1. The results demonstrated that the sequence of the 3'-untranslated region (UTR) of the PBX1 gene was complementary to the sequence of miR-141-3p (Fig. 2A). Furthermore, the co-existence of PBX1-Wt and miR-141-3p mimic in the cells significantly inhibited luciferase activity, while the co-existence of PBX1-Mut and miR-141-3p mimic or co-existence of PBX1-Wt and miR-141-3p mimic NC had no inhibition effect (Fig. 2B).
Figure 2.
PBX1 is a direct target of miR-141-3p in CAL27 cells. (A) Predicted duplex formation of PBX1-wt-3'-UTR and miR-141-3p. (B) Luciferase gene reporter assay results in cells co-transfected with PBX1-wt and miR-141-3p mimic or mimic NC, or co-transfected with PBX1-mt and miR-141-3p mimic or mimic NC. (C) Expression levels of miR-141-3p in cells transfected with miR-141-3p mimic, mimic NC, miR-141-3p inhibitor or inhibitor NC was analyzed by RT-qPCR. (D) PBX1 expression in cells transfected with miR-141-3p mimic, mimic NC, miR-141-3p inhibitor or inhibitor NC was analyzed by western blotting. *P<0.05 vs. mimic NC. #P<0.05 vs. inhibitor NC. wt, wild type; mt, mutant; miRNA/miR, microRNA; NC, negative control; UTR, untranslated region; PXB1, pre-B-cell leukaemia homeobox-1.
To determine the relationship between miR-141-3p and PBX1, the expression of miR-141-3p mRNA and PBX1 protein was detected using RT-qPCR and western blotting, respectively, following CAL27 cell transfection miR-141-3p mimic or inhibitor. The results demonstrated that transfections with miR-mimic and inhibitor were successful in CAL27 cells. The level of miR-141-3p was upregulated by miR-141-3p mimic and downregulated by miR-141-3p inhibitor, compared with corresponding NC (Fig. 2C). Furthermore, PBX1 protein expression was significantly decreased following transfection with miR-141-3p mimics, whereas miR-141-3p inhibitor had the opposite effect (Fig. 2D). These findings indicated that miR-141-3p could negatively regulate PBX1 protein expression in CAL27 cells, suggesting that PBX1 may be a target of miR-141-3p in OSCC cells.
miR-141-3p inhibits CAL27 cell invasion, proliferation and migration
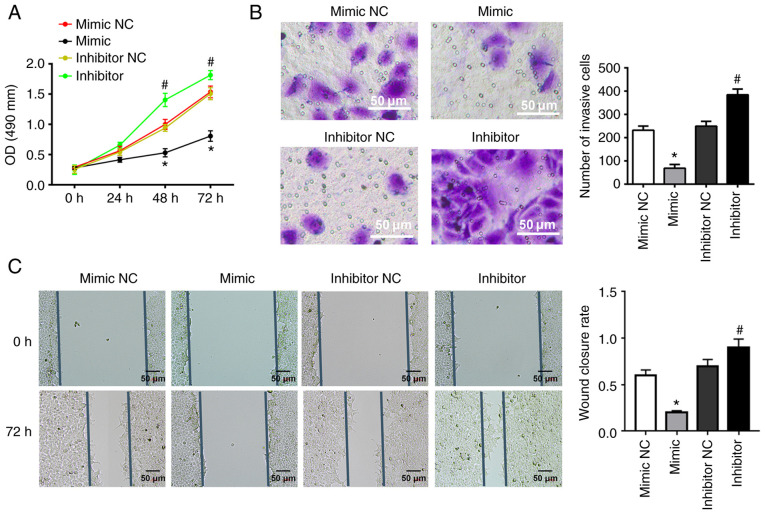
To determine the effects of miR-141-3p on CAL27 cells, miR-141-3p mimic or inhibitor and corresponding NC were transfected into cells. miR-141-3p mimic significantly inhibited the proliferation of CAL27 cells, as determined by MTT assay; however, miR-141-3p inhibitor had the opposite effect (Fig. 3A). In addition, miR-141-3p mimic significantly inhibited the invasion and migration of CAL27 cells, while miR-141-3p inhibitor exhibited the opposite effects, according to results from Transwell and wound healing assays (Fig. 3B and C).
Figure 3.
miR-141-3p mimic and inhibitor inhibit or enhance CAL27 cell invasion, proliferation and migration. Cells were transfected with miR-141-3p mimic, mimic NC, miR-141-3p inhibitor, or inhibitor NC. (A) Cell proliferation assay. (B) Cell invasion assay. Scale bar=50 µm. (C) Wound healing assay. Scale bar=50 µm. *P<0.05 vs. mimic NC. #P<0.05 vs. inhibitor NC. miR, microRNA; NC, negative control; OD, optical density.
miRNA-141-3p inhibitor activates the JAK2/STAT3 pathway
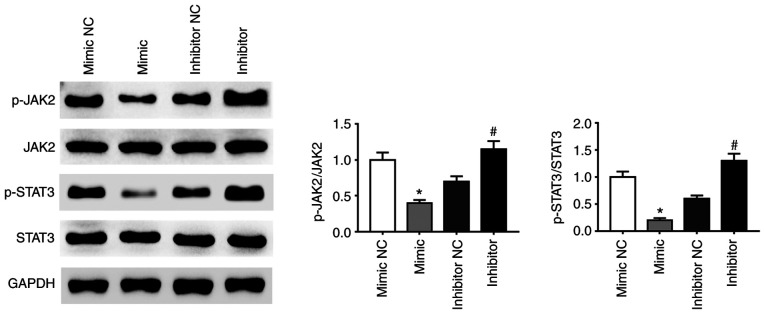
A previous study demonstrated that activation of the JAK2-STAT3 signaling pathway enhanced proliferation, invasion and metastasis of HCC cells (38). The present study demonstrated that the phosphorylation of JAK2 and STAT3 proteins was affected by miR-141-3p mimic and inhibitor. Expression of p-JAK2 and p-STAT3 was decreased in CAL27 cells transfected with miRNA-141-3p mimic compared with NC whereas miR-141-3p inhibitor had the opposite effect (Fig. 4). These findings suggested that the JAK2/STAT3 signaling pathway may be activated following inhibition of miR-141-3p expression in CAL27 cells.
Figure 4.
miR-141-3p inhibits the activation of the JAK2/STAT3 signaling pathway. Expression of STAT3 and p-STAT3 and of JAK2 and p-JAK2 in CAL27 cells was determined by western blotting. *P<0.05 vs. mimic NC. #P<0.05 vs. inhibitor NC. miR, microRNA; NC, negative control; JAK2, janus kinase 2; STAT3, signal transducer and activator of transcription 3 signaling pathway.
si-PBX1 weakens the effects of miRNA-141-3p inhibitor on JAK2/STAT3 pathway and cell behavior in CAL27 cells
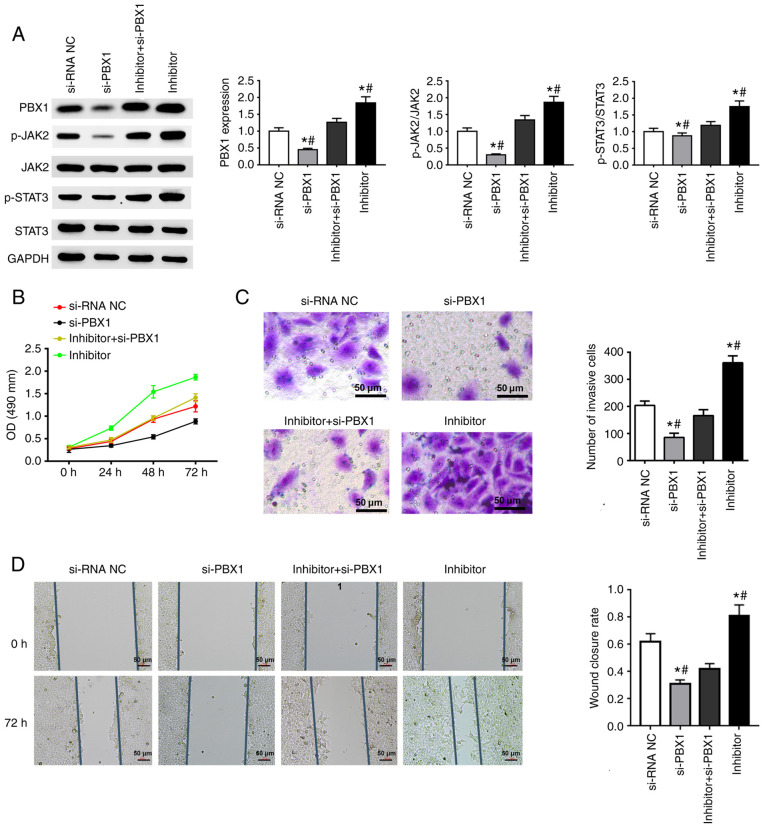
si-PBX1, miR-141-3p inhibitor and the combination of PBX1-siRNA + miR-141-3p inhibitor were transfected into CAL27 cells. The results from western blotting confirmed the successful downregulation of PBX1 in the si-PBX1 group in comparison with the si-NC group, and also a decreased expression of p-JAK2 and p-STAT3. The combined transfection of miR-141-3p inhibitor and si-PBX1 increased the expression of p-JAK2, p-STAT3 and PBX1, whereas si-PBX1 transfection attenuated the increase in constitutive levels of p-JAK2, p-STAT3 and PBX1 expression induced by miR-141-3p inhibitor (Fig. 5A).
Figure 5.
Effect of si-PBX1 and miRNA-141-3p inhibitor on JAK2/STAT3 signaling pathway and CAL27 cell invasion, proliferation and migration. (A) Western blotting was used to determine the expression of PBX1, JAK2, STAT3, p-JAK2 and p-STAT3. Quantitative data are shown for PBX1, p-JAK2/JAK2 and p-STAT3/STAT3 in CAL27 cells transfected with si-PBX1 or miR-141-3p inhibitor. (B) Cell proliferation assay was used to detected CAL27 cells transfected with si-PBX1, siRNA NC, inhibitor + si-PBX1 or inhibitor. (C) Invasion assay results of CAL27 cells transfected with si-PBX1, siRNA NC, inhibitor + si-PBX1 or inhibitor. Scale bar=50 µm. (D) Wound healing assay results of CAL27 cells transfected with si-PBX1, siRNA NC, inhibitor + si-PBX1 or inhibitor. Scale bar=50 µm. *P<0.05 vs. si-NC. #P<0.05 vs. miRNA-141-3p inhibitor + si-PBX1. miR, microRNA; NC, negative control; si, small interfering; JAK2, janus kinase 2; STAT3, signal transducer and activator of transcription 3 signaling pathway; PXB1, pre-B-cell leukaemia homeobox-1; OD, optical density.
The results from MTT, Transwell and wound healing assays suggested that miR-141-3p inhibitor could significantly enhance the cell proliferation, migration and invasion of CAL27 cells. Furthermore, PBX1 downregulation inhibited the invasion, proliferation and migration of CAL27 cells. In addition, in the combination group, si-PBX1 notably weakened the promoting effects of miR-141-3p inhibitor on cell invasion, proliferation and migration (Fig. 5B-D). Taken together, these results suggested that si-PBX1 could weaken the effects of miRNA-141-3p inhibitor on the JAK2/STAT3 pathway and CAL27 cell behavior.
Discussion
OSCC is considered as a common malignant tumor and its incidence rate is continuously rising in developing countries (31). In OSCC, invasion occurs before metastasis, which is an important reason why OSCC leads to mortality (39). Determining novel targeted treatments for OSCC is therefore crucial, and this can be achieved by exploring the molecular properties of OSCC. The dysregulation of miRs contributes to the development and metastasis of many types of tumor, such as lung, breast, gastric, ovarian and liver cancers (40). Increasing evidence indicates that certain miRs can regulate the metastasis and invasion of OSCC cells, such as microRNA-22, microRNA-205-5p and miRNA-491-5p (41-43). miR-141-3p is a member of the miR-200 family and exists in two clusters on chromosomes 1 and 12, named miRs-200b/a/429 and miRs-200c/141. The five miRs form the miR-200 family (44). miR-141 has the same sequence as miR-200a, with only one nucleotide difference between miR-141 and miR-200a (45). Previous studies have reported that miR-141 is involved in ovarian tumorigenesis, colon cancer, small cell lung cancer and renal cell carcinoma (46-48).
However, the effects of miR-141-3p on OSCC remain unclear. Thus, the effects of miR-141-3p on OSCC were analyzed in the present study. The results from the present study demonstrated that miR-141-3p was downregulated in OSCC tissues and cells compared with non-cancerous controls, indicating its potential role in OSCC tumorigenesis. Furthermore, consistent with a previous study, PBX1 expression was increased in OSCC cells and tissues (8).
miRs are short endogenous single chain RNA molecules that can bind to 3'UTR to regulate the post-transcriptional expression of miR of target genes (49). In addition, target genes can be activated or inhibited by miRs binding to their promoters after transcription (50,51). The result from luciferase reported assay demonstrated that miR-141-3p mimic significantly suppressed the activity of PBX1-Wt, whereas the activity of PBX1-Mut was not modified by miR-141-3p mimic. PBX1 was predicted to be a direct target of miR-141-3p. miR-141-3p has been reported to inhibit the occurrence and development of numerous types of tumors. For example, miR-141-3p was shown to inhibit the migration and invasion of HCC cells (52). The present results demonstrated that miR-141-3p overexpression could inhibit the invasion, proliferation and migration of OSCC cells.
The present study demonstrated that miR-141-3p expression was decreased in OSCC cells, thereby promoting cell invasion, proliferation and migration. It was previously reported that the JAK2/STAT3 signaling pathway is related to tumor metastasis (53,54). In OSCC, the JAK2/STAT3 signaling pathway has also been found to mediate the process of cancer metastasis (55). Therefore, we hypothesized that miR-141-3p could inhibit the invasion, proliferation and migration of OSCC cells via the JAK2/STAT3 signaling pathway. The results demonstrated that miR-141-3p inhibitor could activate the JAK2/STAT3 signaling pathway in OSCC cells, indicating that miR-141-3p and the JAK2/STAT3 signaling pathway may have a role in OSCC tumorigenesis. Furthermore, PBX1 expression has been found to be abnormally high in a variety of human tissues, and PXB1 gene was shown to play a key role in tumor proliferation (56). The results from the present study demonstrated that PBX1 downregulation could attenuate OSCC cell invasion, proliferation and migration. It has been previously reported that PBX1 downregulation can inhibit the expression of STAT3 and p-STAT3 in esophageal squamous cancer (42). The present study thus speculated that miR-141-3p could be related to PBX1, and the interaction between PBX1 and miR-141-3p was subsequently investigated. The results demonstrated that miR-141-3p exerted its function in OSCC cells by targeting PBX1, when PBX1 knockdown attenuated the invasion, proliferation and migration caused by miR-141-3p knockdown. Taken together, the findings from our study revealed that miR-141-3p may inhibit the invasion, proliferation and migration of OSCC cells through the JAK2/STAT3 signaling pathway by targeting PBX1.
In summary, the present study demonstrated that PBX1 may be considered as a direct target of miR-141-3p, and that miR-141-3p may interact with JAK2 and its downstream signaling kinases, such as STAT3, by inhibiting the catalytic activities of kinases, thus inhibiting the invasion, proliferation and migration of OSCC cells. This study provided a novel target that may be useful for monitoring the progression of OSCC.
Acknowledgements
Not applicable.
Funding Statement
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
MC and WS designed the study and performed experiments. KT and WS performed experiments, confirmed the authenticity of all the raw data, interpreted the results and wrote the manuscript. JX and YT interpreted the results and contributed to the preparation, review and revision of the manuscript. SW provided research funding, modified the study design, provided guidance for the experiments and revised the manuscript. All authors agreed to be accountable for the content of the work. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
The study was approved by the Clinical Ethical Committee of Lishui University (approval no. 201803). Written informed consent was provided by all the patients or their relatives for the use of their tissues in experiments.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Markopoulos AK. Current aspects on oral squamous cell carcinoma. Open Dent J. 2012;6:126–130. doi: 10.2174/1874210601206010126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wei J, Wu J, Xu W, Nie H, Zhou R, Wang R, Liu Y, Tang G, Wu J. Salvianolic acid B inhibits glycolysis in oral squamous cell carcinoma via targeting PI3K/AKT/HIF-1α signaling pathway. Cell Death Dis. 2018;9(599) doi: 10.1038/s41419-018-0623-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adjuvant bisphosphonate treatment in early breast cancer: Meta-analyses of individual patient data from randomised trials. Lancet. 2015;386:1353–1361. doi: 10.1016/S0140-6736(15)60908-4. Early Breast Cancer Trialists' Collaborative Group (EBCTCG) [DOI] [PubMed] [Google Scholar]
- 4.Kim SM, Jeong D, Min KK, Sang SL, Lee SK. Two different protein expression profiles of oral squamous cell carcinoma analyzed by immunoprecipitation high-performance liquid chromatography. World J Surg Oncol. 2017;15(151) doi: 10.1186/s12957-017-1213-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Radhika T, Jeddy N, Nithya S, Muthumeenakshi RM. Salivary biomarkers in oral squamous cell carcinoma-an insight. J Oral Biol Craniofac Res. 2016;6 (Suppl 1):S51–S54. doi: 10.1016/j.jobcr.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sasahira T, Kirita T, Kuniyasu H. Update of molecular pathobiology in oral cancer: A review. Int J Clin Oncol. 2014;19:431–436. doi: 10.1007/s10147-014-0684-4. [DOI] [PubMed] [Google Scholar]
- 7.Liu N, Zhang Z, Li L, Shen X, Sun B, Wang R, Zhong H, Shi Q, Wei L, Zhang Y, et al. MicroRNA-181 regulates the development of ossification of posterior longitudinal ligament via Epigenetic modulation by targeting PBX1. Theranostics. 2020;10:7492–7509. doi: 10.7150/thno.44309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Platais C, Radhakrishnan R, Niklander Ebensperger S, Morgan R, Lambert DW, Hunter KD. Targeting HOX-PBX interactions causes death in oral potentially malignant and squamous carcinoma cells but not normal oral keratinocytes. BMC Cancer. 2018;18(723) doi: 10.1186/s12885-018-4622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He C, Wang Z, Zhang L, Yang L, Li J, Chen X, Zhang J, Chang Q, Yu Y, Liu B, Zhu Z. A hydrophobic residue in the TALE homeodomain of PBX1 promotes epithelial-to-mesenchymal transition of gastric carcinoma. Oncotarget. 2017;8:46818–46833. doi: 10.18632/oncotarget.17473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Risolino M, Mandia N, Iavarone F, Dardaei L, Longobardi E, Fernandez S, Talotta F, Bianchi F, Pisati F. Transcription factor PREP1 induces EMT and metastasis by controlling the TGF-β-SMAD3 pathway in non-small cell lung adenocarcinoma. Proc Natl Acad Sci USA. 2014;111:E3775–E3784. doi: 10.1073/pnas.1407074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE Jr. Stat3 as an Oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 12.Degboé Y, Rauwel B, Baron M, Boyer JF, Ruyssen-Witrand A, Constantin A, Davignon JL. Polarization of rheumatoid macrophages by TNF targeting through an IL-10/STAT3 mechanism. Front Immunol. 2019;10(3) doi: 10.3389/fimmu.2019.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson DE, O'Keefe RA, Grandis JR. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat Rev Clin Oncol. 2018;15:234–248. doi: 10.1038/nrclinonc.2018.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu J, Niu P, Zhao Y, Cheng Y, Chen W, Lin L, Lu J, Cheng X, Xu Z. Impact of miR-223-3p and miR-2909 on inflammatory factors IL-6, IL-1β, and TNF-α, and the TLR4/TLR2/NF-κB/STAT3 signaling pathway induced by lipopolysaccharide in human adipose stem cells. PLoS One. 2019;14(e0212063) doi: 10.1371/journal.pone.0212063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: New and unexpected biological functions. Nat Rev Cancer. 2014;14:736–746. doi: 10.1038/nrc3818. [DOI] [PubMed] [Google Scholar]
- 16.Wang B, Liu T, Wu JC, Luo SZ, Chen R, Lu LG, Xu MY. STAT3 aggravates TGF-β1-induced hepatic epithelial-to-mesenchymal transition and migration. Biomed Pharmacother. 2018;98:214–221. doi: 10.1016/j.biopha.2017.12.035. [DOI] [PubMed] [Google Scholar]
- 17.Li YL, Wu LW, Zeng LH, Zhang ZY, Wang W, Zhang C, Lin NM. ApoC1 promotes the metastasis of clear cell renal cell carcinoma via activation of STAT3. Oncogene. 2020;39:6203–6217. doi: 10.1038/s41388-020-01428-3. [DOI] [PubMed] [Google Scholar]
- 18.Chen B, Ling CH. Long noncoding RNA AK027294 acts as an oncogene in non-small cell lung cancer by up-regulating STAT3. Eur Rev Med Pharmacol Sci. 2019;23:1102–1107. doi: 10.26355/eurrev_201902_17000. [DOI] [PubMed] [Google Scholar]
- 19.Devarajan E, Huang S. STAT3 as a central regulator of tumor metastases. Curr Mol Med. 2009;9:626–633. doi: 10.2174/156652409788488720. [DOI] [PubMed] [Google Scholar]
- 20.Zhou W, Bi X, Gao G, Sun L. miRNA-133b and miRNA-135a induce apoptosis via the JAK2/STAT3 signaling pathway in human renal carcinoma cells. Biomed Pharmacother. 2016;84:722–729. doi: 10.1016/j.biopha.2016.09.074. [DOI] [PubMed] [Google Scholar]
- 21.Chen Y, Shao Z, Jiang E, Zhou X, Wang L, Wang H, Luo X, Chen Q, Liu K, Shang Z. CCL21/CCR7 interaction promotes EMT and enhances the stemness of OSCC via a JAK2/STAT3 signaling pathway. J Cell Physiol. 2020;235:5995–6009. doi: 10.1002/jcp.29525. [DOI] [PubMed] [Google Scholar]
- 22.Lin XM, Chen H, Zhan XL. miR-203 regulates JAK-STAT pathway in affecting pancreatic cancer cells proliferation and apoptosis by targeting SOCS3. Eur Rev Med Pharmacol Sci. 2019;23:6906–6913. doi: 10.26355/eurrev_201908_18730. [DOI] [PubMed] [Google Scholar]
- 23.Zhang CS, Lin Y, Sun FB, Gao J, Han B, Li SJ. miR-409 down-regulates Jak-Stat pathway to inhibit progression of liver cancer. Eur Rev Med Pharmacol Sci. 2019;23:146–154. doi: 10.26355/eurrev_201901_16758. [DOI] [PubMed] [Google Scholar]
- 24.Hong YG, Xin C, Zheng H, Huang ZP, Yang Y, Zhou JD, Gao XH, Hao L, Liu QZ, Zhang W, Hao LQ. miR-365a-3p regulates ADAM10-JAK-STAT signaling to suppress the growth and metastasis of colorectal cancer cells. J Cancer. 2020;11:3634–3644. doi: 10.7150/jca.42731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 26.Yates L, Norbury C, Gilbert RC. The long and short of microRNA. Cell. 2013;153:516–519. doi: 10.1016/j.cell.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 27.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 28.Kozaki K, Imoto I, Mogi S, Omura K, Inazawa J. Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Res. 2008;68:2094–2105. doi: 10.1158/0008-5472.CAN-07-5194. [DOI] [PubMed] [Google Scholar]
- 29.Cai Z, Hao XY, Liu FX. MicroRNA-186 serves as a tumor suppressor in oral squamous cell carcinoma by negatively regulating the protein tyrosine phosphatase SHP2 expression. Arch Oral Biol. 2018;89:20–25. doi: 10.1016/j.archoralbio.2018.01.016. [DOI] [PubMed] [Google Scholar]
- 30.Arunkumar G, Deva Magendhra Rao AK, Manikandan M, Prasanna Srinivasa Rao H, Subbiah S, Ilangovan R, Murugan AK, Munirajan AK. Dysregulation of miR-200 family microRNAs and epithelial-mesenchymal transition markers in oral squamous cell carcinoma. Oncol Lett. 2018;15:649–657. doi: 10.3892/ol.2017.7296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shao Y, Qu Y, Dang S, Yao B, Ji M. miR-145 inhibits oral squamous cell carcinoma (OSCC) cell growth by targeting c-Myc and Cdk6. Cancer Cell Int. 2013;13(51) doi: 10.1186/1475-2867-13-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Della Vittoria Scarpati G, Calura E, Di Marino M, Romualdi C, Beltrame L, Malapelle U, Troncone G, De Stefano A, Pepe S, De Placido S, et al. Analysis of differential miRNA expression in primary tumor and stroma of colorectal cancer patients. Biomed Res Int. 2014;2014(840921) doi: 10.1155/2014/840921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verrando P, Capovilla M, Rahmani R. Trans-nonachlor decreases miR-141-3p levels in human melanocytes in vitro promoting melanoma cell characteristics and shows a multigenerational impact on miR-8 levels in Drosophila. Toxicology. 2016;368-369:129–141. doi: 10.1016/j.tox.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 34.Li M, Huang H, Cheng F, Hu X, Liu J. miR-141-3p promotes proliferation and metastasis of nasopharyngeal carcinoma by targeting NME1. Adv Med Sci. 2020;65:252–258. doi: 10.1016/j.advms.2020.03.005. [DOI] [PubMed] [Google Scholar]
- 35.Ishibashi O, Akagi I, Ogawa Y, Inui T. miR-141-3p is upregulated in esophageal squamous cell carcinoma and targets pleckstrin homology domain leucine-rich repeat protein phosphatase-2, a negative regulator of the PI3K/AKT pathway. Biochem Biophys Res Commun. 2018;501:507–513. doi: 10.1016/j.bbrc.2018.05.025. [DOI] [PubMed] [Google Scholar]
- 36.Hou X, Yang L, Jiang X, Liu Z, Li X, Xie S, Li G, Liu J. Role of microRNA-141-3p in the progression and metastasis of hepatocellular carcinoma cell. Int J Biol Macromol. 2019;128:331–339. doi: 10.1016/j.ijbiomac.2019.01.144. [DOI] [PubMed] [Google Scholar]
- 37.Qiu X, Ye Q, Sun M, Wang L, Tan Y, Wu G. Saturated hydrogen improves lipid metabolism disorders and dysbacteriosis induced by a high-fat diet. Exp Biol Med (Maywood) 2020;245:512–521. doi: 10.1177/1535370219898407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu Y, Yuan T, Wang WW, Ge PL, Gao ZQ, Zhang G, Tang Z, Dang XW, Zhao YF, Zhang JY, Jiang GZ. Long noncoding RNA HOST2 promotes epithelial-mesenchymal transition, proliferation, invasion and migration of hepatocellular carcinoma cells by activating the JAK2-STAT3 signaling pathway. Cell Physiol Biochem. 2018;51:301–314. doi: 10.1159/000495231. [DOI] [PubMed] [Google Scholar]
- 39.Steeg PS. Tumor metastasis: Mechanistic insights and clinical challenges. Nat Med. 2006;12:895–904. doi: 10.1038/nm1469. [DOI] [PubMed] [Google Scholar]
- 40.Xu W, Sun D, Wang Y, Zheng X, Li Y, Xia Y, Teng Y. Inhibitory effect of microRNA-608 on lung cancer cell proliferation, migration, and invasion by targeting BRD4 through the JAK2/STAT3 pathway. Bosn J Basic Med Sci. 2019;20:347–356. doi: 10.17305/bjbms.2019.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang WC, Chan SH, Jang TH, Chang JW, Ko YC, Yen TC, Chiang SL, Chiang WF, Shieh TY, Liao CT, et al. miRNA-491-5p and GIT1 serve as modulators and biomarkers for oral squamous cell carcinoma invasion and metastasis. Cancer Res. 2014;74:751–564. doi: 10.1158/0008-5472.CAN-13-1297. [DOI] [PubMed] [Google Scholar]
- 42.Nagai H, Hasegawa S, Uchida F, Terabe T, Ishibashi Kanno N, Kato K, Yamagata K, Sakai S, Kawashiri S, Sato H, et al. MicroRNA-205-5p suppresses the invasiveness of oral squamous cell carcinoma by inhibiting TIMP-2 expression. Int J Oncol. 2018;52:841–850. doi: 10.3892/ijo.2018.4260. [DOI] [PubMed] [Google Scholar]
- 43.Feng X, Luo Q, Wang H, Zhang H, Chen F. MicroRNA-22 suppresses cell proliferation, migration and invasion in oral squamous cell carcinoma by targeting NLRP3. J Cell Physiol. 2018;233:6705–6713. doi: 10.1002/jcp.26331. [DOI] [PubMed] [Google Scholar]
- 44.Korpal M, Kang Y. The emerging role of miR-200 family of microRNAs in epithelial-mesenchymal transition and cancer metastasis. RNA Biol. 2008;5:115–119. doi: 10.4161/rna.5.3.6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Renthal NE, Williams KC, Mendelson CR. MicroRNAs-mediators of myometrial contractility during pregnancy and labour. Nat Rev Endocrinol. 2013;9:391–401. doi: 10.1038/nrendo.2013.96. [DOI] [PubMed] [Google Scholar]
- 46.Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- 47.Mao S, Lu Z, Zheng S, Zhang H, Zhang G, Wang F, Huang J, Lei Y, Wang X, Liu C, et al. Exosomal miR-141 promotes tumor angiogenesis via KLF12 in small cell lung cancer. J Exp Clin Cancer Res. 2020;39(193) doi: 10.1186/s13046-020-01680-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen X, Lou N, Anming R, Qiu B, Yun Y, Wang X, Du Q, Ruan H, Han W, Wei H, et al. miR-224/miR-141 ratio as a novel diagnostic biomarker in renal cell carcinoma. Oncol Lett. 2018;16:1666–1674. doi: 10.3892/ol.2018.8874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moriya Y, Nohata N, Kinoshita T, Mutallip M, Okamoto T, Yoshida S, Suzuki M, Yoshino I, Seki N. Tumor suppressive microRNA-133a regulates novel molecular networks in lung squamous cell carcinoma. J Hum Genet. 2012;57:38–45. doi: 10.1038/jhg.2011.126. [DOI] [PubMed] [Google Scholar]
- 50.Janowski BA, Younger ST, Hardy DB, Ram R, Huffman KE, Corey DR. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat Chem Biol. 2007;3:166–173. doi: 10.1038/nchembio860. [DOI] [PubMed] [Google Scholar]
- 51.Khraiwesh B, Arif MA, Seumel GI, Ossowski S, Weigel D, Reski R, Frank W. Transcriptional control of gene expression by microRNAs. Cell. 2010;140:111–122. doi: 10.1016/j.cell.2009.12.023. [DOI] [PubMed] [Google Scholar]
- 52.Liu Y, Ding Y, Huang J, Wang S, Ni W, Guan J, Li Q, Zhang Y, Ding Y, Chen B, Chen L. miR-141 suppresses the migration and invasion of HCC cells by targeting Tiam1. PLoS One. 2014;9(e88393) doi: 10.1371/journal.pone.0088393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ahn JH, Choi YS, Choi JH. Leptin promotes human endometriotic cell migration and invasion by up-regulating MMP-2 through the JAK2/STAT3 signaling pathway. Mol Hum Reprod. 2015;21:792–802. doi: 10.1093/molehr/gav039. [DOI] [PubMed] [Google Scholar]
- 54.Liu F, Zhang T, Zou S, Jiang B, Hua D. B7-H3 promotes cell migration and invasion through the Jak2/Stat3/MMP9 signaling pathway in colorectal cancer. Mol Med Rep. 2015;12:5455–5460. doi: 10.3892/mmr.2015.4050. [DOI] [PubMed] [Google Scholar]
- 55.Wang Y, Jing Y, Ding L, Zhang X, Song Y, Chen S, Zhao X, Huang X, Pu Y, Wang Z, et al. Epiregulin reprograms cancer-associated fibroblasts and facilitates oral squamous cell carcinoma invasion via JAK2-STAT3 pathway. J Exp Clin Cancer Res. 2019;38(274) doi: 10.1186/s13046-019-1277-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu D, Ma Y, Feng C, Ma Z, Guo J, Chen H, He T, Guo J, Sun X, Qin Q, et al. PBX1 increases the radiosensitivity of oesophageal squamous cancer by targeting of STAT3. Pathol Oncol Res. 2020;26:2161–2168. doi: 10.1007/s12253-020-00803-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.