Abstract
We have evaluated the new Digene Hybrid Capture II HBV DNA Test (HCII HBV), which is a 96-well microtiter plate-based signal amplification assay. This test uses hybrid capture technology that specifically detects RNA-DNA hybrids. HCII HBV is able to quantify hepatitis B virus (HBV) DNA at between 1.4 × 105 and 1.7 × 109 HBV copies per ml in a standard format. By using a modified sample preparation method, which allows the input of 30-fold more serum for an ultrasensitive format, the sensitivity of the assay can be increased reproducibly to approximately 8,000 copies of HBV per ml. By using a combination of these two formats, the assay can quantify over a total range of 6 logs. In our multicenter evaluation study, the mean laboratory-to-laboratory coefficients of variation were 22, 7, and 12% at the three sites, respectively, with a combined specificity of 98.4%. The linearities of both the standard test and the ultrasensitive test were excellent, with Spearman correlation coefficients of 0.997 and 0.999, respectively. Furthermore, the intra-assay reproducibility for the standard assay gave coefficients of variation of from 13 to 33, 9 to 21, and 3 to 8% at the three sites, respectively. HCII HBV was shown to be genotype independent when the EUROHEP standards for genotypes A and D were used. This assay allows the accurate measurement of HBV DNA levels in serum and can be clinically used for the monitoring of responses to antiviral agents for patients chronically infected with HBV.
It has been estimated that worldwide approximately 300 million individuals are chronic carriers of hepatitis B virus (HBV). The measurement of HBV DNA levels in serum has become an important tool for the identification of individuals with high levels of viral replication who might benefit from antiviral therapy, monitoring of patients on therapy, and prediction of whether antiviral therapy will be successful. With the introduction of new antiviral agents like lamivudine [(−)-2′,3′-dideoxy-3′-thiacytidine], close monitoring of patients has become increasingly important due to the occurrence of antiviral agent-resistant strains or the presence of flares after withdrawal from antiviral therapy (8, 16).
Several molecular approaches, either commercially available or homemade tests, have been used in the last few years to quantify serum HBV DNA levels more accurately. Unfortunately, these assays generate highly divergent results due to a lack of standardization and differences in the dynamic ranges of the assays (4, 10, 12, 13, 19, 23).
Here, we describe the results obtained with a second-generation microplate assay, the Digene Hybrid Capture II HBV DNA Test (HCII HBV), which is a signal amplification-based assay. By using a high-speed centrifugation step, the assay's analytical sensitivity can be increased 30-fold, giving a lower detection limit of approximately 8,000 HBV copies/ml. We evaluated the sensitivity, specificity, linear range, reproducibility, and precision of the test at three different laboratories. In addition, by using the EUROHEP HBV standards of HBV genotypes ad and ay, quantification was shown to be genotype independent. HCII HBV was also compared to the previous generation tube-based assay, the Hybrid Capture System (HCS) HBV DNA assay.
MATERIALS AND METHODS
Clinical samples.
Specimens used for the multicenter evaluation were well-characterized samples obtained from a commercial manufacturer (Boston Biomedica Incorporated, West Bridgewater, Mass.). These samples were representative of clinical specimens. The samples and dilutions for the multicenter evaluation were prepared centrally (Digene Corporation, Gaithersburg, Md.). The samples were serially diluted twofold in human HBV-negative serum, including negativity for hepatitis B surface antigen, (HBsAg), anti-hepatitis C virus antibodies, anti-human immunodeficiency virus type 1 and 2 antibodies, anti-hepatitis B core antigen antibodies, and anti-HBsAg antibodies (Boston Biomedica Incorporated). Selection of samples for the multicenter study was based on the presence HBsAg and/or hepatitis B e antigen positivity. All samples were tested at Digene Corporation prior to distribution to the centers. The values obtained were depicted as expected number of HBV copies per milliliter.
An additional 89 clinical samples for the correlation between HCII HBV and the HCS tube-based assay were obtained from the Virology Department of the University Hospital Rotterdam. With the exception of the EUROHEP standards, which are plasma samples, only serum samples were used. Samples that required dilution, including the EUROHEP standards, were diluted in known HBV DNA-negative serum. All aliquots were stored frozen at −20°C or lower within 2 h of collection.
EUROHEP standards.
For an evaluation of the genotype detection characteristics of the assay, an international reference plasma preparation was obtained from the EUROHEP Pathobiology Group (W. H. Gerlich, University of Giessen, Giessen, Germany). These standards contained well-characterized levels of HBV DNA of serotypes ad (genotype A) and ay (genotype D). On the basis of extensive testing, these samples contain 2.7 × 109 and 2.6 × 109 HBV molecules per ml, respectively (7). These two EUROHEP reference panels have already been used for standardization of the test kits and in quality control trials (19). The plasma with genotype A will probably be the basis of the World Health Organization reference sample.
HCS-based tube assay.
The HCS tube-based assays were performed according to the instructions of the manufacturer. Briefly, 50-μl serum samples, controls, and standards or calibrators with HBV DNA at concentrations ranging from 5 to 2,000 pg/ml (equivalent to 1.42 × 106 to 5.6 × 108 HBV copies per ml) were each prepared in separate 2-ml reaction tubes, together with 25 μl of sample diluent and 25 μl of sample preparation reagent. The tubes were incubated for 20 min at 65°C for lysis of the viral particles. The sample preparation reagent contained proteinase K. After incubation, 50 μl of a sodium hydroxide solution (the DNA denaturation reagent) was added for 30 min at 65°C. Once the probe mixture was prepared, 50 μl of the probe mixture was added to the denatured HBV DNA, followed by incubation for another 60 min at 65°C. The probe contains full-length HBV RNA of two different genotypes (genotypes A and D). During this hybridization step, RNA-DNA hybrids were formed. These were subsequently detected by transferring the solution into a capture tube coated with anti-RNA-DNA antibodies. RNA-DNA hybrid capture was performed at room temperature on a rotary shaker. The immobilized hybrid was reacted with an anti-RNA-DNA hybrid antibody conjugated to alkaline phosphatase and was detected with the chemiluminescent substrate LumiPhos 530. The concentration of the HBV DNA was subsequently calculated from the external standard-calibration curve with the manufacturer's software. Processing of 60 samples required approximately 6 h.
HCII HBV.
HCII HBV was performed according to the manufacturer's instruction. Briefly, 30-μl serum samples, controls, and standards or calibrators with HBV DNA at concentrations ranging from 0.5 to 6,000 pg/ml (equivalent to 1.42 × 105 to 1.7 × 109 HBV copies per ml) were incubated with 30 μl of sodium hydroxide solution (denaturation reagent) for 30 min at 65°C in a 96-well microplate. No extra sample preparation step was needed. After preparation of the probe mixture, 30 μl of RNA probe was added to each well and the plate was incubated for 1 h at 65°C. The RNA probes were identical to those used in the first-generation HCS tube-based assay. To capture the RNA-DNA hybrids, 75 μl of each solution in the microplate was transferred to the corresponding well of the anti-RNA-DNA hybrid antibody-coated capture microplate, and the plate was subsequently shaken at room temperature for 1 h. The hybrid was reacted with an anti-hybrid antibody conjugated to alkaline phosphatase and detected with the chemiluminescent substrate CDP-star with Emerald II. The testing process for 96 samples (including controls) required approximately 3.5 h.
In the ultrasensitive format of the assay, 1 ml of serum sample and controls along with 50 μl of precipitation buffer was centrifuged at 33,000 × g at 4°C in a Hereaus Stratos Biofuge or Jouan high-speed tabletop centrifuge for 110 min. This procedure yields a 30-fold increase in sensitivity and enhances the lower detection limit of the assay to approximately 8,000 HBV copies/ml.
Statistics.
x-y scatter diagrams and the correlation coefficients (r2) or Spearman correlation (r) as well as linear regression analysis were prepared or calculated by using the statistical functions of SPSS, version 8.0, software. In order to compare to what extent the data obtained with the first- and second-generation assays were in agreement, the data were also analyzed as described by Bland and Altman (2), which is based on a comparison of the differences between measurements for the same sample by plotting the differences against the average.
RESULTS
Correlation between HCII HBV and HCS tube-based assay.
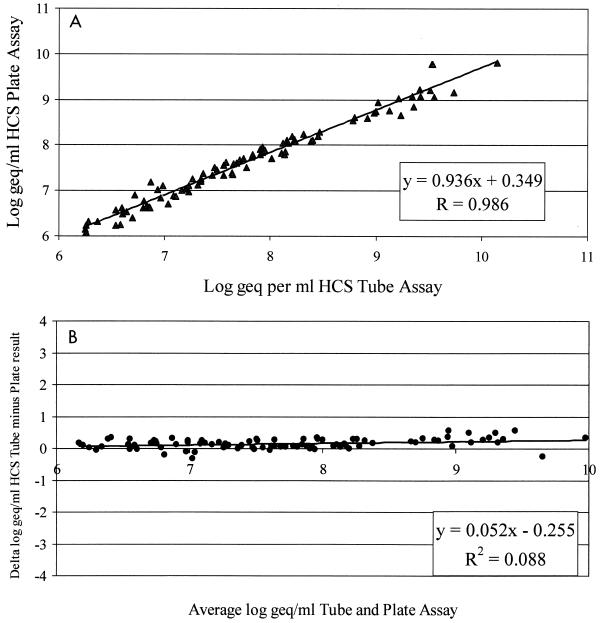
The correlation between the HCII HBV and the HCS tube-based assay was determined with 89 positive clinical samples with loads of more than 1.5 × 106 copies of HBV DNA per ml prepared by both methods (Fig. 1A). If the samples contained HBV DNA at levels above the dynamic range of the assays, they were diluted in HBV DNA-negative serum. The slope, intercept, and Spearman correlation coefficient (r = 0.986) of the log-transformed calculated DNA concentrations were determined by linear regression analysis.
FIG. 1.
(A) Correlation of HCS tube-based assay with HCII HBV. The calculated log10 DNA concentrations were calculated for a set of 89 randomly selected clinical samples. Linear regression analysis showed a Spearman correlation of 0.986. (B) Comparison of HCS tube-based assay and HCII HBV as described by Bland and Altman (2). Only data for samples that were positive and above the detection level of the HCS tube-based assay are included. geq, genome equivalents.
The correlation between the two assays was also calculated by the method of Bland Altman (2) by plotting the differences against the average (Fig. 1B). The correlation coefficient (r2) was 0.088, and the high P value (P = 0.296) indicates that the differences observed were independent from the average. However, the viral loads obtained by the HCS tube-based assay were on average 0.25 log higher than those obtained by HCII HBV.
Linear range of HCII HBV.
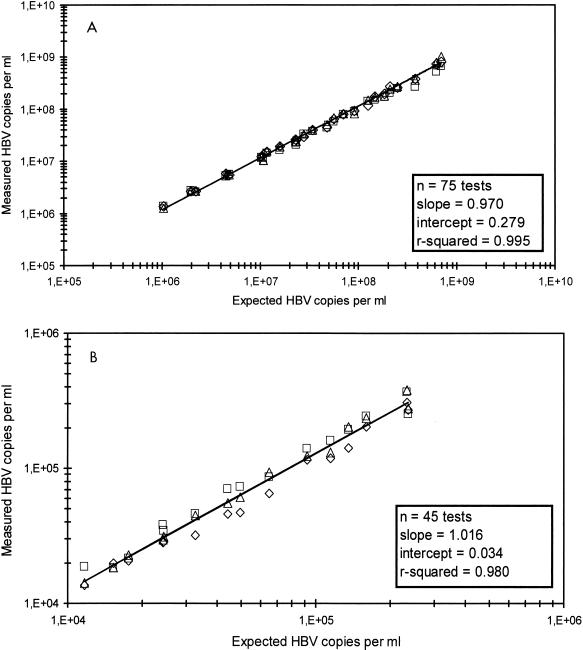
The linearities of the standard and ultrasensitive formats were determined. For the standard format of the assessment test, five serum samples which were serially twofold diluted were used. The quantities in the dilutions ranged from 1.4 × 106 to 8.5 × 108 HBV DNA copies per ml (Fig. 2A). For the ultrasensitive format of the assessment test, three serially twofold diluted serum samples were used. The quantities in the dilutions ranged from 1.5 × 104 to 3.5 × 105 HBV copies per ml (Fig. 2B). The calculated log10 concentration for each replicate was compared to the expected log10 input concentration. Regression analysis showed that the linear range of concentrations of the ultrasensitive assay format and the standard assay format were close to the expected concentrations because the slopes approached 1.0 (1.016 and 0.97, respectively) and the intercepts approached zero (0.034 and 0.279, respectively).
FIG. 2.
(A) Linear range of HCII HBV determined in the standard format. Five twofold serial dilutions were made from five different patients. Each sample was analyzed three times at the three different sites. The data for each site are marked with a different symbol. Linear regression was performed for input DNA concentrations. (B) Linear range of HCII HBV determined in the ultrasensitive format. Five twofold serial dilutions were made from three different patients. Each sample was analyzed three times at the three different sites. The data for each site are marked with a different symbol. Linear regression was performed for input DNA concentrations.
Specificity.
The analytical specificity of the assay in the standard format was determined by testing 15 HBsAg-negative specimens at the three different sites multiple times, giving a total of 63 datum points. One sample gave a value above the mean for calibrator 2 that resulted in a false-positive result. Repeat testing of this sample identified it as negative, indicating that the false-positive result was more likely due to a technical error. This resulted in an overall specificity of 98.4%. The specificity of the assay in the ultrasensitive format was not determined in this multicenter evaluation. Testing at one laboratory resulted in an overall specificity of 99%.
Interlaboratory variation.
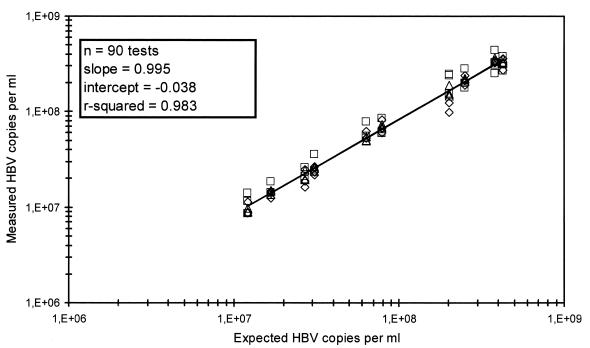
To determine the variability of the method when used by the different laboratories, 10 samples were tested at the three different evaluation sites on 3 different days by the same operator at each site. The mean percent coefficients of variation (CVs) for the three sites were 22, 7, and 12%, respectively, and the overall mean CV for the combined results was less than 14% (Fig. 3).
FIG. 3.
Combined results of interlaboratory variation for quantitation of HBV DNA in 10 samples with viral loads of between 8.7 × 106 and 4.4 × 108 HBV DNA copies per ml. Each sample was tested singly on 3 consecutive days at each of the three sites. The data from the individual sites are depicted with different symbols.
Intralaboratory variation.
To determine the variability of the method within a given laboratory, three samples were tested at each laboratory by one assay by using six replicates per sample. The CVs for the three sites ranged from 13 to 33, 9 to 21, and 3 to 8%, respectively (Table 1). The overall mean CV for the combined sites was less than 13%.
TABLE 1.
Intratest reproducibility of HCII HBVa
| Sample no. | Site 1
|
Site 2
|
Site 3
|
|||
|---|---|---|---|---|---|---|
| Avga no. of HBV DNA copies/ml | % CV | Avg no. of HBV DNA copies/ml | % CV | Avg no. of HBV DNA copies/ml | % CV | |
| 1 | 1.8 × 107 | 33 | 1.4 × 107 | 9 | 1.5 × 107 | 4 |
| 2 | 1.6 × 108 | 16 | 8.1 × 107 | 21 | 1.5 × 108 | 3 |
| 3 | 3.6 × 108 | 13 | 3.7 × 108 | 8 | 3.7 × 108 | 8 |
The average of six replicates for each HBV-positive sample was determined for the standard test, and the CV was calculated.
Correlation between concentrations by HCII HBV and concentrations in EUROHEP standards.
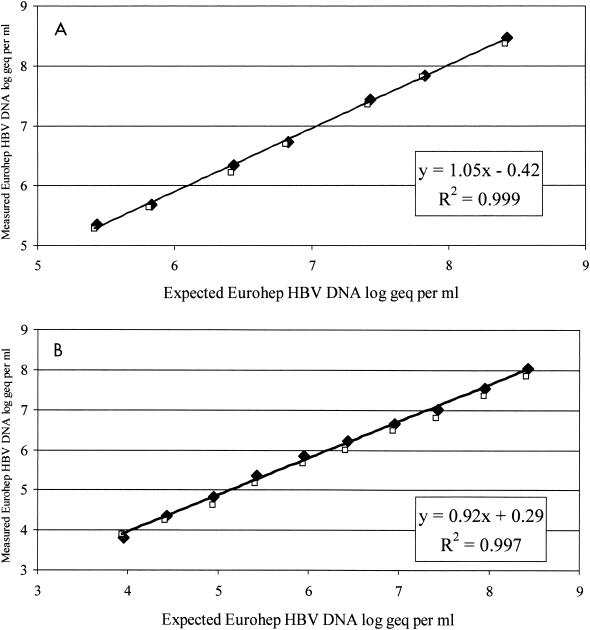
In order to determine whether the microplate assay was HBV genotype independent, serial dilutions from the EUROHEP standards of genotypes A and D were made and analyzed in the standard format of HCII HBV. The differences between the expected HBV DNA concentration and the measured HBV DNA concentration were 0.05 log10 for genotype A and 0.11 log10 for genotype D, indicating that the difference between the two EUROHEP standards was well within the intra-assay variability. From this analysis it could furthermore be concluded that the detection limit of the standard assay was 190,000 copies of HBV DNA (Fig. 4A). The detection limit of the assay in the ultrasensitive format was determined by using half-log serial dilutions of the EUROHEP standards and was shown to be approximately 8,000 copies of HBV DNA per ml for both genotypes A and D (Fig. 4B).
FIG. 4.
(A) Genotype-specific linear range of HCII HBV determined in the standard format. Serial dilutions were made from EUROHEP standards of genotypes A (⧫) and D (□). Each sample was analyzed at least three times. Linear regression was performed for input DNA concentrations. (B) Genotype-specific linear range of HCII HBV determined in the ultrasensitive format. Serial dilutions were made from EUROHEP standards of genotypes A (⧫) and D (□). Each sample was analyzed at least three times. Linear regression was performed for input DNA concentrations, geq, genome equivalents.
DISCUSSION
In the present paper, we have demonstrated that the analytical sensitivity of HCII HBV was approximately 10-fold higher than that of the HCS tube-based assay, with a detection limit of 200,000 copies of HBV DNA per ml. A simple high-speed centrifugation step further increased the sensitivity of HCII HBV down to 8,000 copies per ml. This step did not interfere with the binding of the RNA probe to the HBV DNA or the capture of the RNA-DNA hybrid to the 96-well capture plate and led to a more than 300-fold higher sensitivity than that obtained with the HCS tube-based assay. The detection limit thus obtained is more in the range of the detection limits of standard target amplification assays, like PCR. A great practical advantage of these two formats is that within a single plate the HBV DNA loads in samples with HBV DNA load differences of more than 6 logs can be simultaneously determined. Current commercially available PCR-based techniques can detect as few as 1,000 copies per ml if the EUROHEP panel is used as a standard, which is a narrower range of detection (11). This indicates that with the new technology, the differences between standard hybridization assays, also called signal amplification assays and target amplification assays, like PCR or nucleic acid sequence-based amplification (NASBA), are greatly reduced.
Minor differences have been observed between the HCS tube-based assay and HCII HBV. The results obtained by HCII HBV were, on average, 0.25 log lower than those obtained by the HCS tube-based Assay. This difference might be obtained due to a slightly modified sample preparation step. The HCS tube-based assay uses a proteinase K digestion step, whereas HCII HBV does not. Since in HBV DNA the polymerase protein is covalently attached to the DNA, a proteinase K digestion step in the sample preparation procedure of the HCS tube-based assay might explain the observed difference (7).
The lack of standardization has been a problem for nucleic acid detection assays, although in theory, molecular biology-based assays are ideal for standardization purposes (3, 6). With standard hybridization-based assays, sensitivity is generally the biggest problem. Although the basic PCR assay can be exquisitely sensitive, the first international quality control program for HBV has shown great differences in sensitivity and specificity among results from different laboratories (19). This phenomenon is not specific for HBV DNA detection assays but applies to target amplification-based assays in general (6, 22). The lack of standardization has contributed to these differences in sensitivity. From the data obtained with the two EUROHEP reference samples for genotypes A and D (7), it has also become clear that HCII HBV is not genotype dependent. This is in contrast to the Abbott Genostics assay, which is widely used in clinical studies worldwide. By that assay genotype differences can lead to a 35-fold differences in output signals (23). Furthermore, the EUROHEP standard could be used to determine the sensitivity of the assay.
HCII HBV proved to be reproducible, with an overall CV of less then 13% and a maximum CV of 33%. This indicates that the results of tests with a single sample will typically vary by a factor 2. When the three different laboratories tested the same samples in a test of linearity for both the standard and the ultrasensitive formats, the results that were obtained did not differ by more than a factor of 2. This implies that results from different laboratories may be compared if the results are within these limits. Additional sample volume in the preparation step, as has also been shown for signal and target amplification-based assays targeted at human immunodeficiency virus type 1 (5, 20) or hepatitis C virus (14), will probably further increase the sensitivity of the test.
Determination of the serum HBV DNA level has been shown to be useful for monitoring of the effect of antiviral treatment and for patient management. Furthermore, the emergence of resistance to antiviral drugs can be determined by the detection of increases in viral loads during treatment (9, 15, 21). Typically, signal amplification-based assays are less sensitive, which may not allow the detection of early elevations in viral loads until they reach 106 HBV DNA copies per ml (23). Standard antiviral treatment with nucleoside analogues will reduce HBV DNA levels within a few weeks to a level not detectable by these assays (9). HCII HBV increases the sensitivity approximately 300-fold. Limited information is available on what levels will be reached during antiviral treatment or what HBV DNA levels are present in asymptomatic carriers. Only Niitsuma et al. (17) found that an HBV DNA level of approximately 10,000 HBV DNA copies per ml should be reached after successful antiviral treatment. Below this level, no hepatitis occurred in their study group. This is a detection level just above the cutoff of the ultrasensitive format of HCII HBV.
In summary, the performance characteristics of HCII HBV indicate that this assay is a reliable tool for the accurate measurement of HBV DNA levels in serum and therefore could be used to monitor the effects of antiviral therapy. In its ultrasensitive format, the assay has a sensitivity which is just above the range of the currently available PCR-based assays (1, 11, 18).
REFERENCES
- 1.Abe A, Inoue K, Tanaka T, Kato J, Kajiyama N, Kawaguchi R, Tanaka S, Yoshiba M, Kohara M. Quantitation of hepatitis B virus genomic DNA by real-time detection PCR. J Clin Microbiol. 1999;37:2899–2903. doi: 10.1128/jcm.37.9.2899-2903.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bland J M, Altman D G. Comparing methods of measurement: why plotting difference against standard method is misleading. Lancet. 1995;346:1085–1087. doi: 10.1016/s0140-6736(95)91748-9. [DOI] [PubMed] [Google Scholar]
- 3.Bresters D, Cuypers H T, Reesink H W, Mauser-Bunschoten E P, van den Berg H M, Schaasberg W P, Wilber J C, Urdea M S, Neuwald P, Lelie P N. Comparison of quantitative cDNA-PCR with the branched DNA hybridization assay for monitoring plasma hepatitis C virus RNA levels in haemophilia patients participating in a controlled interferon trial. J Med Virol. 1994;43:262–268. doi: 10.1002/jmv.1890430313. [DOI] [PubMed] [Google Scholar]
- 4.Butterworth L A, Prior S L, Buda P J, Faoagali J L, Cooksley W G. Comparison of four methods for quantitative measurement of hepatitis B viral DNA. J Hepatol. 1996;24:686–691. doi: 10.1016/s0168-8278(96)80264-9. [DOI] [PubMed] [Google Scholar]
- 5.Collins M L, Irvine B, Tyner D, Fine E, Zayati C, Chang C, Horn T, Ahle D, Detmer J, Shen L P, Kolberg J, Bushnell S, Urdea M S, Ho D D. A branched DNA signal amplification assay for quantification of nucleic acid targets below 100 molecules/ml. Nucleic Acids Res. 1997;25:2979–2984. doi: 10.1093/nar/25.15.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Damen M, Cuypers H T, Zaaijer H L, Reesink H W, Schaasberg W P, Gerlich W H, Niesters H G, Lelie P N. International collaborative study on the second EUROHEP HCV-RNA reference panel. J Virol Methods. 1996;58:175–185. doi: 10.1016/0166-0934(96)02011-3. [DOI] [PubMed] [Google Scholar]
- 7.Heermann K H, Gerlich W H, Chudy M, Schaefer S, Thomssen R The Eurohep Pathobiology Group. Quantitative detection of hepatitis B virus DNA in two international reference plasma preparations. J Clin Microbiol. 1999;37:68–73. doi: 10.1128/jcm.37.1.68-73.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Honkoop P, de Man R A, Heijtink R A, Schalm S W. Hepatitis B reactivation after lamivudine. Lancet. 1995;346:1156–1157. doi: 10.1016/s0140-6736(95)91829-9. [DOI] [PubMed] [Google Scholar]
- 9.Honkoop P, Niesters H G, de Man R A, Osterhaus A D, Schalm S W. Lamivudine resistance in immunocompetent chronic hepatitis B. Incidence and patterns. J Hepatol. 1997;26:1393–1395. doi: 10.1016/s0168-8278(97)80476-x. [DOI] [PubMed] [Google Scholar]
- 10.Janssen H L, Schoenmaker-Weber Y A, Kruining H, Schalm S W, Heijtink R A. Quantitative assessment of hepatitis B virus DNA in chronic hepatitis B: comparison of two solution hybridization assays. J Med Virol. 1993;40:307–312. doi: 10.1002/jmv.1890400408. [DOI] [PubMed] [Google Scholar]
- 11.Kessler H H, Pierer K, Dragon E, Lackner H, Santner B, Stunzner D, Stelzl E, Waitzl B, Marth E. Evaluation of a new assay for HBV DNA quantitation in patients with chronic hepatitis B. Clin Diagn Virol. 1998;9:37–43. doi: 10.1016/s0928-0197(97)10008-3. [DOI] [PubMed] [Google Scholar]
- 12.Krajden M, Minor J, Cork L, Comanor L. Multi-measurement method comparison of three commercial hepatitis B virus DNA quantification assays. J Viral Hepatol. 1998;5:415–422. doi: 10.1046/j.1365-2893.1998.00129.x. [DOI] [PubMed] [Google Scholar]
- 13.Lai V C H, Guan R, Wood M L, Lo S K, Yuen M F, Lai C L. Nucleic acid-based cross-linking assay for detection and quantification of hepatitis B virus DNA. J Clin Microbiol. 1999;37:161–164. doi: 10.1128/jcm.37.1.161-164.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McHutchison J G, Blatt L M, Ponnudurai R, Goodarzi K, Russell J, Conrad A. Ultracentrifugation and concentration of a large volume of serum for HCV RNA during treatment may predict sustained and relapse response in chronic HCV infection. J Med Virol. 1999;57:351–355. [PubMed] [Google Scholar]
- 15.Melegari M, Scaglioni P P, Wands J R. Hepatitis B virus mutants associated with 3TC and famciclovir administration are replication defective. Hepatology. 1998;27:628–633. doi: 10.1002/hep.510270243. [DOI] [PubMed] [Google Scholar]
- 16.Niesters H G, Honkoop P, Haagsma E B, de Man R A, Schalm S W, Osterhaus A D. Identification of more than one mutation in the hepatitis B virus polymerase gene arising during prolonged lamivudine treatment. J Infect Dis. 1998;177:1382–1385. doi: 10.1086/517819. [DOI] [PubMed] [Google Scholar]
- 17.Niitsuma H, Ishii M, Miura M, Kobayashi K, Toyota T. Low level hepatitis B viremia detected by polymerase chain reaction accompanies the absence of HBe antigenemia and hepatitis in hepatitis B virus carriers. Am J Gastroenterol. 1997;92:119–123. [PubMed] [Google Scholar]
- 18.Noborg U, Gusdal A, Pisa E K, Hedrum A, Lindh M. Automated quantitative analysis of hepatitis B virus DNA by using the Cobas Amplicor HBV Monitor test. J Clin Microbiol. 1999;37:2793–2797. doi: 10.1128/jcm.37.9.2793-2797.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quint W G V, Heijtink R A, Schirm J, Gerlich W H, Niesters H G M. Reliability of methods for hepatitis B virus DNA detection. J Clin Microbiol. 1995;33:225–228. doi: 10.1128/jcm.33.1.225-228.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun R, Ku J, Jayakar H, Kuo J C, Brambilla D, Herman S, Rosenstraus M, Spadoro J. Ultrasensitive reverse transcription-PCR assay for quantitation of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 1998;36:2964–2969. doi: 10.1128/jcm.36.10.2964-2969.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolters L M, Niesters H G M, de Man R A, Schalm S W. Antiviral treatment for human immunodeficiency virus patients co-infected with hepatitis B virus: combined effect for both infections, an obtainable goal? Antivir Res. 1999;42:71–76. doi: 10.1016/s0166-3542(99)00016-9. [DOI] [PubMed] [Google Scholar]
- 22.Zaaijer H L, Cuypers H T, Reesink H W, Winkel I N, Gerken G, Lelie P N. Reliability of polymerase chain reaction for detection of hepatitis C virus. Lancet. 1993;341:722–724. doi: 10.1016/0140-6736(93)90488-3. [DOI] [PubMed] [Google Scholar]
- 23.Zaaijer H L, ter Borg F, Cuypers H T, Hermus M C, Lelie P N. Comparison of methods for detection of hepatitis B virus DNA. J Clin Microbiol. 1994;32:2088–2091. doi: 10.1128/jcm.32.9.2088-2091.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]