Abstract
Background:
Food protein-induced enterocolitis syndrome (FPIES) is a non-IgE-mediated food allergy characterized by profuse vomiting within hours of ingestion of the causative food. We have previously reported that FPIES is associated with systemic innate immune activation in the absence of a detectable antigen-specific antibody or T cell response. The mechanism of specific food recognition by the immune system remains unclear.
Objective:
To identify immune mechanisms underlying FPIES reactions by proteomic and flow cytometric analysis of peripheral blood.
Methods:
Children with a history of FPIES underwent a supervised oral food challenge. Blood samples were taken at baseline, upon symptom onset, and 4 h post-symptoms. We analyzed samples from 23 children (11 reactors, 12 outgrown). 184 protein markers were analyzed by proximity ligation assay and verified by multiplex immunoassay. Analysis of cell subset activation was performed by mass cytometry and spectral cytometry.
Results:
Symptomatic FPIES challenges were associated with significant elevation of cytokines and chemokines including IL-17 family markers (IL-17A, IL-22, IL-17C, CCL20), T cell activation (IL-2), and innate inflammatory markers (IL-8, Oncostatin M, LIF, TNFα, IL-10, IL-6). The mucosal damage marker REG1A was also significantly increased. These biomarkers were not increased in asymptomatic challenges or IgE-mediated allergy. Phospho-STAT3 was significantly elevated in myeloid and T cells post-challenge in individuals with symptoms. Mass cytometry indicated preferential activation of non-conventional T cell populations, including γδ T cells and CD3+CD4-CD8-CD161+ cells, however the potential sources of IL-17 in PBMCs were primarily CD4+ Th17 cells.
Conclusions:
These results demonstrate a unique IL-17 signature and activation of innate lymphocytes in FPIES.
Keywords: Food protein-induced enterocolitis syndrome (FPIES), oral food challenge, proteomics, Th17, STAT3, inflammation, innate immunity, mucosal barrier
Capsule Summary:
Symptomatic FPIES challenges resulted in a systemic release of markers associated with an innate IL-17 response and mucosal barrier damage.
Introduction
Food protein-induced enterocolitis syndrome (FPIES) is a non-IgE-mediated food allergy that commonly presents in infancy 1. Although typically outgrown in the first years of life, symptoms can persist into adolescence and adulthood. An adult-onset form of FPIES to shellfish has also been described 2. Cow’s milk is the most common trigger of FPIES in infancy, however a wide range of foods can trigger reactions, and these foods show limited overlap with foods that induce IgE-mediated food allergy 3–5. FPIES reactions involve profuse, repetitive vomiting within 1–4 hours of food ingestion, and patients with severe reactions may experience shock-like symptoms including lethargy and pallor.
The mechanisms responsible for immune recognition of FPIES food triggers remain unclear. An increase in circulating neutrophils 4 hours post-challenge is part of the criteria for defining positive FPIES oral food challenges (OFC)1. We have also found that acute FPIES reactions are associated with a broad systemic innate activation, including neutrophils, monocytes, eosinophils, and pan-lymphocyte activation observed 4 h after symptom onset 6. This was supported by work from Mehr et al describing changes in innate inflammatory gene expression in whole blood after an FPIES OFC 7. There is no detectable food-specific antibody response in acute FPIES, and there is a lack of direct evidence for a pathogenic antigen-specific T cell response 6, 8, 9. To obtain a better understanding of immune mechanisms associated with symptoms in FPIES, we measured a broad panel of biomarkers in serum of patients undergoing a supervised OFC and examined these markers at baseline, at symptom onset, and 4 h post-symptoms.
Results/Discussion
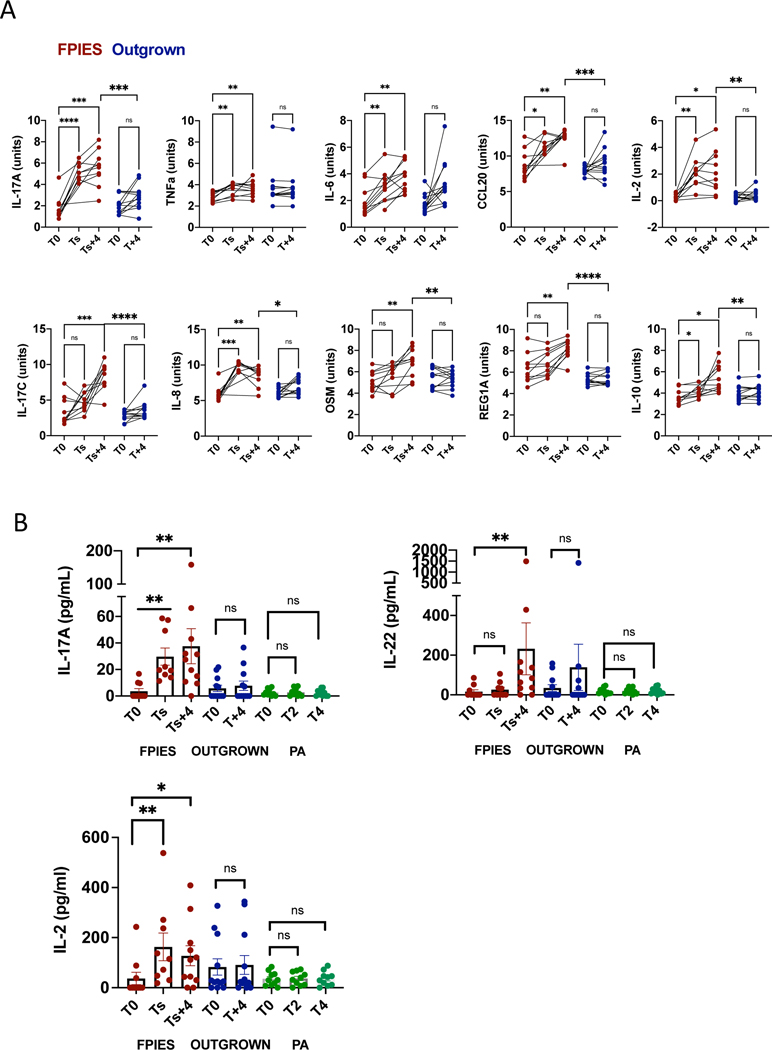
Patients with a history of FPIES were challenged as part of their clinical care to determine if reactivity to foods had been outgrown. Since 07/2014, biospecimens from 68 OFCs had been collected. Pre/post serum samples were available for 11 positive reactions, and specimens from 12 asymptomatic challenges from those with outgrown FPIES were selected as controls. Additional patient information is shown in Table E1. Participants ranged in age from 1.5 to 16 years. Serum samples were obtained at baseline (T0), at time of symptoms (Ts, approximately 2 h after challenge), and 4 h after symptom onset in those with reactions (Ts+4). In those with no symptoms, a second serum sample was obtained prior to discharge, approximately 4 h after challenge (T+4). We used two panels to measure a total of 184 biomarkers by proximity ligation assay (Olink). Results were analyzed first by comparing post-challenge values to baseline in those with active FPIES. At 4h post-symptoms, 14 biomarkers were significantly different (q value <0.05) compared to baseline (Table 1, Figure 1A). Three of the top four differentially expressed analytes were in the Th17 pathway (IL-17A, IL-17C, CCL20). IL-23 was below the level of detection. The most differentially expressed protein (by q value) was REG1A, which is expressed in the pancreas and intestine and regulates mucosal barrier function downstream of the Th17 cytokine IL-22. Consistent with previous reports 10, IL-2 was also increased after a FPIES reaction. Chemokines produced by intestinal epithelial cells (CCL25, CXCL9, IL-8) were increased, as were a number of innate cytokines known to be produced by monocytes (IL-6, IL-10, OSM, LIF, TNF). At symptom onset, IL-17A, IL-8, and IL-2 were significantly elevated (Table 1, Figure 1A). In samples obtained post-challenge from asymptomatic challenges (outgrown FPIES), no biomarkers were significantly elevated compared to baseline (Table 1, Figure 1A).
Table 1:
Differentially abundant plasma biomarkers
| Ts+4 vs T0 | Ts vs T0 | T4 vs T0 (Outgrown) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| P value | Difference | q value | P value | Difference | q value | P value | Difference | q value | |
| REG1A | 0.00000 | 1.649 | 0.000 | 0.0479 | 0.4157 | >0.99 | 0.945 | 0.014 | >0.99 |
| IL-17A | 0.00000 | 3.879 | 0.000 | 0.00008 | 3.042 | 0.004 | 0.0623 | 0.814 | >0.99 |
| CCL20 | 0.00004 | 3.888 | 0.002 | 0.01238 | 2.399 | 0.145 | 0.1227 | 0.960 | >0.99 |
| IL-17C | 0.00006 | 4.648 | 0.002 | 0.21497 | 1.028 | 0.963 | 0.1153 | 0.682 | >0.99 |
| IL8 | 0.00017 | 2.809 | 0.004 | 0.00001 | 3.498 | 0.001 | 0.0220 | 0.913 | 0.839 |
| CXCL9 | 0.00193 | 1.450 | 0.021 | 0.53305 | 0.172 | 0.963 | 0.8391 | −0.059 | >0.99 |
| IL10 | 0.00152 | 1.664 | 0.021 | 0.18411 | 0.376 | 0.950 | 0.5412 | 0.175 | >0.99 |
| IL2 | 0.00183 | 2.271 | 0.021 | 0.00064 | 1.722 | 0.020 | 0.3822 | 0.126 | >0.99 |
| IL6 | 0.00161 | 1.779 | 0.021 | 0.03533 | 1.198 | 0.289 | 0.0063 | 1.588 | 0.611 |
| OSM | 0.00223 | 1.769 | 0.021 | 0.52136 | 0.331 | 0.963 | 0.8642 | −0.062 | >0.99 |
| LIF | 0.00295 | 0.614 | 0.025 | 0.01899 | 0.191 | 0.177 | 0.1598 | 0.394 | >0.99 |
| CCL25 | 0.00672 | 0.740 | 0.048 | 0.26773 | 0.254 | 0.963 | 0.9124 | −0.023 | >0.99 |
| TNF | 0.00673 | 0.767 | 0.048 | 0.00721 | 0.729 | 0.120 | 0.9850 | 0.014 | >0.99 |
| HGF | 0.00761 | 0.591 | 0.050 | 0.44960 | 0.134 | 0.963 | 0.2414 | −0.131 | >0.99 |
| CCL4 | 0.00982 | 1.152 | 0.056 | 0.00773 | 1.299 | 0.120 | 0.7342 | 0.094 | >0.99 |
| TGF-a | 0.00981 | 1.359 | 0.056 | 0.16102 | 0.450 | 0.950 | 0.9260 | −0.023 | >0.99 |
| CCL3 | 0.02001 | 0.923 | 0.108 | 0.01488 | 1.028 | 0.155 | 0.5454 | 0.150 | >0.99 |
| CXCL1 | 0.03006 | 0.729 | 0.152 | 0.01185 | 0.781 | 0.145 | 0.6001 | −0.161 | >0.99 |
| MCP-1 | 0.04146 | 1.061 | 0.198 | 0.00479 | 1.358 | 0.112 | 0.2082 | 0.382 | >0.99 |
Difference = normalized protein units (Log2 scale). Significant values bolded. T0 = baseline, Ts = Time of symptoms, Ts+4 = 4 hours after symptoms. In asymptomatic (outgrown) challenges, a single post-challenge sample was obtained 4 hours after challenge (T4).
Figure 1: Biomarkers in serum during FPIES challenges.
A.Biomarkers measured by Olink were measured in blood taken at baseline (T0), time of symptom onset (Ts), and 4 h after symptoms (Ts + 4). In those who tolerated the challenge (Outgrown, in blue), blood was obtained at baseline (T0) and 4 h after challenge (T+4). Data are expressed as normalized protein units (Log2 scale). Within group analysis by paired T test, between group analysis by T test. *p<0.05, **<0.01, ***<0.001, ****<0.0001. Each data point is one individual. B. Multiplex immunoassay detection of IL-17, IL-22, and IL-2. For additional disease controls, serum samples from 10 peanut allergic (PA) children who experienced vomiting during peanut challenges were measured. Within group analysis by Mann-Whitney U test.
To verify key changes using a quantitative assay, we measured serum cytokines by multiplex immunoassay. As shown in Figure 1B, IL-17, IL-22, and IL-2 were significantly elevated during symptomatic but not asymptomatic FPIES challenges. As a disease specificity control, we used 10 samples from children undergoing a double-blind placebo-controlled peanut challenge who experienced vomiting during their peanut challenge. Cytokines associated with FPIES challenge were unchanged during IgE-mediated reactions to peanut (Figure 1B). IL-17, IL-22, and IL-2 values were highly and significantly correlated with each other (Rs=0.84, p = 3×10−9, (IL-17/IL-22), Rs=0.77, p=1 × 10−11 (IL-17/IL-2).
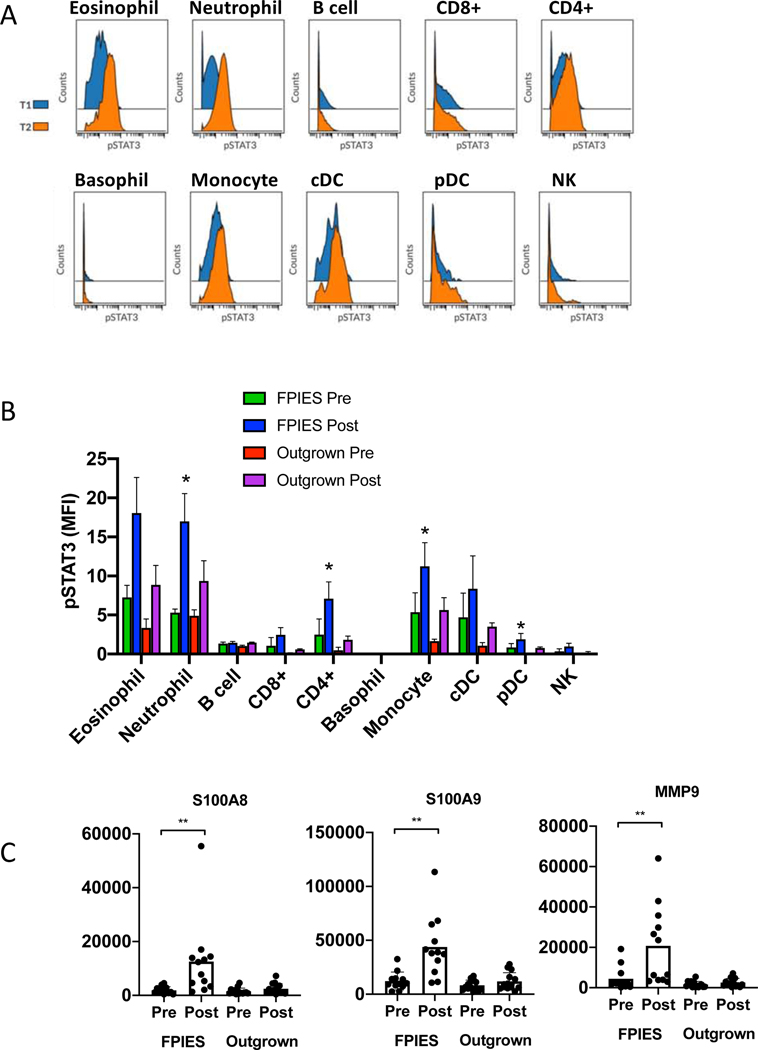
To assess the impact of elevated cytokines on peripheral immune cells, we examined cell subsets in whole blood obtained before and after OFC by mass cytometry. We used surface markers and detection by intracellular phospho-proteins (p38, ERK1/2, STAT1, STAT3, STAT6). Markers are shown in Table E2. STAT3 was significantly elevated post-challenge in individuals with reactions but not individuals who tolerated the challenge (Figure 2A,B). Signaling was observed in myeloid cells (eosinophils, neutrophils, monocytes), pDCs, and CD4+ T cells. Gating of cell subsets is shown in Figure E1. STAT3 is downstream of signaling through IL-6, IL-10, LIF, and OSM, all elevated in FPIES reactions. This suggests that the systemic innate immune activation observed in FPIES reactions, including pan-T cell activation, is likely mediated by cytokines. To determine if IL-17 had systemic effects during FPIES reactions, we examined target genes downstream of IL-17 from our previously published dataset of RNA sequencing of whole blood pre/post challenge 6. We observed significant upregulation of the S100A8 and S100A9 genes, and MMP9, all known IL-17 target genes (Figure 2C).
Figure 2: Functional impact of cytokine release.
A: Representative histograms showing p-STAT3 in identified subsets at baseline (blue) and 4 h after symptoms (orange) as measured by mass cytometry. B: summary data of median intensity of STAT3 in cell subsets before and after challenge, in those with active (n=4) or outgrown (n=3) FPIES. C: Expression of IL-17A target genes measured by RNA sequencing of whole blood obtained before or after FPIES challenge. Each dot indicates one patient. *p<0.05 by paired analysis.
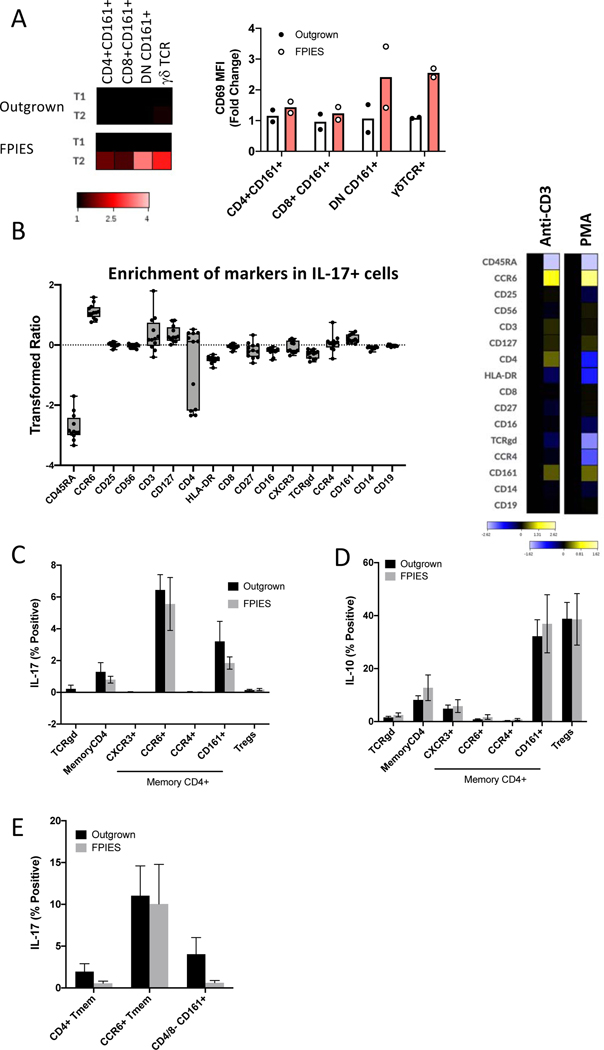
The next question was the potential source of IL-17. We had previously not been able to detect increased food antigen-responsive T cells in peripheral blood 6, consistent with other findings 9. In addition to CD4+ Th17 cells, mucosal associated invariant T cells 11, γδ T cells12, and ILC3s 13 have been described as sources of IL-17. We re-analyzed previously published mass cytometry data where we had used activation markers to identify cells activated during challenge 6. We gated on CD4+CD45RA-CD161+ cells to enrich for Th17 cells, CD3+CD4-CD45RA-CD161+ cells to enrich for invariant mucosal T cells, and γδ T cells. Using the activation marker CD69, we observed that the greatest degree of activation post-challenge was observed in γδ T cells and CD161+CD4- T cells (Figure 3A). However, activation does not necessarily equate with cytokine production.
Figure 3: Source of IL-17A in peripheral blood after stimulation.
A. T cell subset activation in peripheral blood as defined by CD69 upregulation. B. To assess potential sources of IL-17, PBMCs from active or outgrown FPIES were stimulated with anti-CD3/CD28 or PMA for 6 h. IL-17+ live singlets were gated, and expression of cell surface markers compared to all live singlets. Left: Quantification of enrichment of each marker across 8 samples. Right: Representative heat map of the transformed ratio of expression of markers under CD3/28 or PMA stimulation conditions comparing IL-17+ cells to all live singlets. C-E: T cell subsets were gated manually and IL-17 (C) and IL-10 (D) expression quantified after stimulation with CD3/CD28 (C,D) or PMA (E). (n=4/group)
We were not able to examine IL-17 production in vivo from any of our whole blood mass cytometry samples as the samples were fixed prior to storage. To assess the potential of different cell subsets for IL-17 production in active and outgrown FPIES subjects, we stimulated PBMCs with PMA or anti-CD3/CD28 prior to staining with cell subset markers and intracellular cytokines (Table E3). We first gated on all IL-17+ cells after stimulation (first gating on live singlets) and examined cell subset markers (Figure 3B). We compared marker expression in IL-17+ cells versus all live cells. This approach was used to determine the dominant features of IL-17-positive cells. There was enrichment of CD4, CCR6, and CD161 in the IL-17+ population compared to all cells after stimulation with anti-CD3/CD28, but no enrichment of CD8, γδ TCR or CD56. A similar pattern was observed after PMA stimulation (with the exception of CD4, which was downregulated by PMA). These markers are consistent with conventional Th17 cells.
We next gated known cell populations to examine IL-17 expression (gating shown in Figure E2). IL-17 was primarily restricted to the memory CD4+CD45RA-CCR6+ or CD4+CD45RA-CD161+ compartment after stimulation with anti-CD3/CD28 or PMA, consistent with conventional Th17 cells (Figure 3C). The finding of correlated release of IL-2, IL-17 and IL-22 in serum are also consistent with Th17 cells as the source of IL-17. There was no significant difference in the quantity of IL-17 expressed by T cell subsets from active or outgrown FPIES subjects after stimulation with anti-CD3/28 (Figure 3C) or PMA (Figure 3E). We enriched for ILCs by gating on lineage negative (CD3-CD19-CD14-CD16-CD123-) CD127+ CD25+ cells. Although we did not have ILC3-specific markers, there was no IL-17 expression in this compartment after PMA stimulation (data not shown). IL-10 and TNFα, produced by Tregs and all T cell subsets, respectively, were not different between active and outgrown FPIES (Figure 3D and not shown). IL-17/TNFα co-expression was also not different between FPIES and outgrown samples when stimulated with polyclonal stimulation (not shown). We do not find evidence for an aberrant source or quantity of IL-17 produced after polyclonal stimulation. Thus, the exaggerated type 17/inflammatory response seen during FPIES reactions is elicited with specificity (to food exposure in vivo) rather than hyperresponsiveness to polyclonal or innate stimuli.
We and others have tried without success to identify a pathogenic antigen-specific T cell response to foods in FPIES using ex vivo stimulation of peripheral blood 6, 9. This approach is successful at detecting responsive Th2 cells in IgE-mediated food allergy 14, 15. Others have observed a Th2 response together with TNFα and IL-6 in FPIES 8, 16, but these results have failed to provide distinguishing characteristics of the T cells compared to IgE-mediated food allergy or atopic controls. We speculate that the pathogenic T cell response to foods is restricted to tissue resident memory cells that are not detectable in peripheral circulation. In celiac disease, another non-IgE-mediated food-triggered disease, antigen-specific T cells are observed in circulation after 6–14 days of gluten challenge 17, 18. Tissue-resident human CD4+ T cells in the gastrointestinal tract persist over a long period of time, are polyfunctional cytokine producers including IL-17, and can also produce perforin and granzyme capable of mediating gastrointestinal mucosal damage 19, 20. In the absence of access to gastrointestinal tissue in FPIES (endoscopy and biopsy are not required for diagnosis or management), a prolonged low-dose exposure to the food may be necessary to dislodge T cells from the tissue or expand them such that they can be observed ex vivo.
In summary, we found that FPIES reactions were associated with a release of pro-inflammatory cytokines including IL-17 family cytokines. Functional enrichment analysis (KEGG pathways) of the upregulated markers identified IL-17 signaling (p=7.7 × 10−13) and rheumatoid arthritis (p=5.8 × 10−13), indicating shared inflammatory pathways with IL-17-mediated autoimmune disease. The release of cytokines was associated with signaling in myeloid cells and CD4+ T cells in peripheral blood and changes in gene expression in peripheral blood. We speculate that pathogenic T cells specifically activated by foods are restricted to the intestinal mucosa, hindering detection and phenotypic analysis of these cells in peripheral blood.
Supplementary Material
Key Messages:
Acute FPIES reactions are associated with an elevated IL-17 pathway response detectable at symptom onset and persisting through symptom resolution.
STAT3 activation demonstrated systemic cytokine activity, and IL-17 target genes were found to be upregulated by mRNA expression in whole blood.
FPIES reactions were associated with activation of CD3+CD4-CD8- CD161+ and γδ+ T lymphocytes, however potential sources of IL-17 include Th17 cells.
Acknowledgements
We thank the team of the Human Immune Monitoring Core (Manishkumar Patel, Hui Xie, Geoffrey Kelly, John Leech, Adeeb Rahman, Seunghee Kim-Schulze) for Olink and mass cytometry analysis, and we thank Bryan Fernandez (Biomedical Sciences Program, Center for Excellence in Youth Education) for his assistance with the FPIES research program.
The work was funded in part by the Katy and Kyle Miller Family Fund and NIH grants AI136053 and AI151707.
MGB: Dr. Baker was supported in part by the Louis and Rachel Rudin Foundation
JAB: Research support: NIH-NIAID, Genentech, Aimmune, DBV Technologies, FARE, Astellas; Consulting: Before Brands, AllerGenis, Novartis; Medical Advisory Board: International FPIES Association
ANW: Anna Nowak-Wegrzyn received research support from National Institute of Allergy and Infectious Diseases, DBV Technologies, Astellas Pharma, Danone and Nestle, and consultancy fees from Regeneron and Gerber Institute; she serves as the Deputy Editor for the Annals of Allergy, Asthma and Immunology and the Chair of the Medical Advisory Board of the International FPIES Association.
Abbreviations:
- FPIES
Food protein-induced enterocolitis syndrome
- IgE
immunoglobulin E
- LIF
leukemia inhibitory factor
- OFC
Oral food challenge
- OSM
oncostatin M
- REG1A
regenerating family member 1 alpha
Footnotes
Disclosures:
MCB, DLO, CA: no conflicts of interest to disclose
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- 1.Nowak-Wegrzyn A, Chehade M, Groetch ME, Spergel JM, Wood RA, Allen K, et al. International consensus guidelines for the diagnosis and management of food protein-induced enterocolitis syndrome: Executive summary-Workgroup Report of the Adverse Reactions to Foods Committee, American Academy of Allergy, Asthma & Immunology. J Allergy Clin Immunol 2017; 139:1111–26 e4. [DOI] [PubMed] [Google Scholar]
- 2.Du YJ, Nowak-Wegrzyn A, Vadas P. FPIES in adults. Ann Allergy Asthma Immunol 2018. [DOI] [PubMed] [Google Scholar]
- 3.Caubet JC, Ford LS, Sickles L, Jarvinen KM, Sicherer SH, Sampson HA, et al. Clinical features and resolution of food protein-induced enterocolitis syndrome: 10-year experience. J Allergy Clin Immunol 2014; 134:382–9. [DOI] [PubMed] [Google Scholar]
- 4.Mehr S, Frith K, Barnes EH, Campbell DE. Food protein-induced enterocolitis syndrome in Australia: A population-based study, 2012–2014. J Allergy Clin Immunol 2017; 140:1323–30. [DOI] [PubMed] [Google Scholar]
- 5.Ruffner MA, Ruymann K, Barni S, Cianferoni A, Brown-Whitehorn T, Spergel JM. Food protein-induced enterocolitis syndrome: insights from review of a large referral population. J Allergy Clin Immunol Pract 2013; 1:343–9. [DOI] [PubMed] [Google Scholar]
- 6.Goswami R, Blazquez AB, Kosoy R, Rahman A, Nowak-Wegrzyn A, Berin MC. Systemic innate immune activation in food protein-induced enterocolitis syndrome. J Allergy Clin Immunol 2017; 139:1885–96 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mehr S, Lee E, Hsu P, Anderson D, de Jong E, Bosco A, et al. Innate immune activation occurs in acute food protein-induced enterocolitis syndrome reactions. J Allergy Clin Immunol 2019; 144:600–2 e2. [DOI] [PubMed] [Google Scholar]
- 8.Caubet JC, Bencharitiwong R, Ross A, Sampson HA, Berin MC, Nowak-Wegrzyn A. Humoral and cellular responses to casein in patients with food protein-induced enterocolitis to cow’s milk. J Allergy Clin Immunol 2016; 139:572–83. [DOI] [PubMed] [Google Scholar]
- 9.Adel-Patient K, Lezmi G, Castelli FA, Blanc S, Bernard H, Soulaines P, et al. Deep analysis of immune response and metabolic signature in children with food protein induced enterocolitis to cow’s milk. Clin Transl Allergy 2018; 8:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kimura M, Ito Y, Shimomura M, Morishita H, Meguro T, Adachi Y, et al. Cytokine profile after oral food challenge in infants with food protein-induced enterocolitis syndrome. Allergol Int 2017; 66:452–7. [DOI] [PubMed] [Google Scholar]
- 11.Dusseaux M, Martin E, Serriari N, Peguillet I, Premel V, Louis D, et al. Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17-secreting T cells. Blood 2011; 117:1250–9. [DOI] [PubMed] [Google Scholar]
- 12.Lockhart E, Green AM, Flynn JL. IL-17 production is dominated by gammadelta T cells rather than CD4 T cells during Mycobacterium tuberculosis infection. J Immunol 2006; 177:4662–9. [DOI] [PubMed] [Google Scholar]
- 13.Montaldo E, Juelke K, Romagnani C. Group 3 innate lymphoid cells (ILC3s): Origin, differentiation, and plasticity in humans and mice. Eur J Immunol 2015; 45:2171–82. [DOI] [PubMed] [Google Scholar]
- 14.Berin MC, Grishin A, Masilamani M, Leung DYM, Sicherer SH, Jones SM, et al. Egg-specific IgE and basophil activation but not egg-specific T-cell counts correlate with phenotypes of clinical egg allergy. J Allergy Clin Immunol 2018; 142:149–58 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiang D, Chen X, Jones SM, Wood RA, Sicherer SH, Burks AW, et al. Single-cell profiling of peanut-responsive T cells in patients with peanut allergy reveals heterogeneous effector TH2 subsets. J Allergy Clin Immunol 2018; 141:2107–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morita H, Nomura I, Orihara K, Yoshida K, Akasawa A, Tachimoto H, et al. Antigen-specific T-cell responses in patients with non-IgE-mediated gastrointestinal food allergy are predominantly skewed to T(H)2. J Allergy Clin Immunol 2013; 131:590–2 e1–6. [DOI] [PubMed] [Google Scholar]
- 17.Han A, Newell EW, Glanville J, Fernandez-Becker N, Khosla C, Chien YH, et al. Dietary gluten triggers concomitant activation of CD4+ and CD8+ alphabeta T cells and gammadelta T cells in celiac disease. Proc Natl Acad Sci U S A 2013; 110:13073–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sarna VK, Skodje GI, Reims HM, Risnes LF, Dahal-Koirala S, Sollid LM, et al. HLA-DQ:gluten tetramer test in blood gives better detection of coeliac patients than biopsy after 14-day gluten challenge. Gut 2018; 67:1606–13. [DOI] [PubMed] [Google Scholar]
- 19.Bartolome-Casado R, Landsverk OJB, Chauhan SK, Saetre F, Hagen KT, Yaqub S, et al. CD4(+) T cells persist for years in the human small intestine and display a TH1 cytokine profile. Mucosal Immunol 2020. [DOI] [PubMed] [Google Scholar]
- 20.Hegazy AN, West NR, Stubbington MJT, Wendt E, Suijker KIM, Datsi A, et al. Circulating and Tissue-Resident CD4(+) T Cells With Reactivity to Intestinal Microbiota Are Abundant in Healthy Individuals and Function Is Altered During Inflammation. Gastroenterology 2017; 153:1320–37 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.