Abstract
Chest imaging can represent a powerful tool for detecting the Coronavirus disease 2019 (COVID-19). Among the available technologies, the chest Computed Tomography (CT) scan is an effective approach for reliable and early detection of the disease. However, it could be difficult to rapidly identify by human inspection anomalous area in CT images belonging to the COVID-19 disease. Hence, it becomes necessary the exploitation of suitable automatic algorithms able to quick and precisely identify the disease, possibly by using few labeled input data, because large amounts of CT scans are not usually available for the COVID-19 disease. The method proposed in this paper is based on the exploitation of the compact and meaningful hidden representation provided by a Deep Denoising Convolutional Autoencoder (DDCAE). Specifically, the proposed DDCAE, trained on some target CT scans in an unsupervised way, is used to build up a robust statistical representation generating a target histogram. A suitable statistical distance measures how this target histogram is far from a companion histogram evaluated on an unknown test scan: if this distance is greater of a threshold, the test image is labeled as anomaly, i.e. the scan belongs to a patient affected by COVID-19 disease. Some experimental results and comparisons with other state-of-the-art methods show the effectiveness of the proposed approach reaching a top accuracy of 100% and similar high values for other metrics. In conclusion, by using a statistical representation of the hidden features provided by DDCAEs, the developed architecture is able to differentiate COVID-19 from normal and pneumonia scans with high reliability and at low computational cost.
Keywords: Deep denoising autoencoder, Convolutional autoencoders, Feature learning, Computed Tomography (CT), Coronavirus disease 2019, COVID-19, Pneumonia, Anomaly detection
1. Introduction
Since its inception, medical imaging has been a valid tool for making non-invasive medical diagnoses (Suetens, 2009). Among the different techniques, Computed Tomography (CT) has assumed a very important role (Hsieh, 2009). In fact, CT images represent a powerful investigation tool because they contain more detailed information than conventional X-rays. Unlike a conventional X-ray, this computerized version uses a mobile X-ray source that rotates around the patient and generate cross-sectional images of the body, called slices.
Recently, CT scans have been adopted to identify the novel coronavirus pneumonia (NCP) due to SARS-CoV-2 viral infection of the COVID-19 pandemic (Kwee & Kwee, 2020). In this regard, this method has demonstrated a high potentiality, showing a high sensitivity for detection of the disease (Adams et al., 2020, Lerum et al., 2020). Since COVID-19 is spreading rapidly all over the world, a fast and accurate screening is of primary importance for controlling the pandemic.
To this purpose, many researchers have highlighted that the COVID-19 pneumonia is different from other viral (common) pneumonia (CP) (Sharma, 2020). In this regard, some works have shown that cases of NCP tend to affect the entire lungs, unlike common diseases that are limited to small regions (Chen et al., 2020, Sharma, 2020). Pneumonia caused by the COVID-19 shows a typical hazy patch on the outer edges of the lungs. For this reason, CT scans appear to work well, as they are able to bring out three distinctive hallmarks: (i) while normal lung scans appear black, those related to COVID-19 show lighter colored spots or gray; (ii) the lung airspaces are full of fluids due to inflammation (consolidation); and, (iii) pleural effusion is present, i.e. liquid in the spaces around the lungs.
However, CT scans are not always easy to read and interpret by radiologists. In addition, in order to reduce the massive dose of radiation and avoid harmful consequences (such as tumors), it is preferable to perform scans with a low emission of radiation (Chen et al., 2020). In this case, unfortunately, the scanned images often have a degraded quality (such as blur, background noise and low contrast) that can make interpretation ambiguous and difficult to take a certain and precise diagnosis (Al-Ameen & Sulong, 2016).
In order to solve all the above problems, very recently many studies have been directed towards the use of automatic image classification techniques through Deep Learning (DL) algorithms (Shen, Wu, and Suk, 2017, Zhou et al., 2017), a branch of machine learning that uses architectures that possess many layers of processing (Goodfellow et al., 2016). The author in Sharma (2020) has recently hypothesized that machine learning techniques applied to CT scans can certainly become the first alternative screening test to the real-time reverse transcriptase-polymerase chain reaction (RT-PCR) in the near future.
Although DL techniques have been applied with great success to identify cases of the NCP (Sharma, 2020, Shen, Wu, and Suk, 2017, Zhou et al., 2017), there are nevertheless a number of challenges to be solved (Chen et al., 2020, Hammer et al., 2020). First of all, many solutions have been proposed in the literature, each of which has its own advantages and disadvantages, providing very different results: that is, there is no single solution to the problem (Sarv Ahrabi et al., 2021). Furthermore, the DL architectures have a very large number of free parameters that must be adapted by the optimization algorithm and, in order to achieve the convergence, it is necessary to have a large amount of data, which is not always possible in practice (Goodfellow et al., 2016). Also because it is not certain that having lots of scans available is enough to be of quality (Aiello et al., 2019).
This situation is further worsen by the fact that, since the CPN is a relatively recent disease, the proportion of CT scans related to COVID-19 is very limited with respect to the number of images available in the datasets freely available on the web (Chen et al., 2020). In this regard, the identification of CT scans affected by COVID-19 is a problem more similar to the anomaly detection rather than the traditional classification, since the small number of data present in each dataset (Chandola et al., 2009). It would therefore be advisable to provide an anomaly detection algorithm, light from the computational point of view and capable of identifying CT scans related to COVID-19 with high accuracy.
Motivated by these considerations, in this paper we propose an autoencoder-based approach for the detection of COVID-19 CT scans. Specifically, the denoising version of a deep convolutional autoencoder, here called DDCAE, is proposed to learn a compact and meaningful representation of the normal and common pneumonia CT scans in an unsupervised manner. Autoencoders (AEs), in fact, are unsupervised architectures trained to copy its input to the output; they possess an internal layer (the hidden features or feature vector) that is used to efficiently represent the input (Goodfellow et al., 2016). Generally, an AE is composed of two main sections: an encoder that maps the input into the hidden features, and a decoder that maps the hidden features to a reconstruction of the input. When the encoder and decoder are composed of several layers, the AE is called deep (DAE) (Goodfellow et al., 2016). In image processing, usually convolutional layers have been employed (Masci et al., 2011), particularly appropriate to extract meaningful information from images. This results in a deep convolutional AE (DCAE). In the denoising version, DDCAE, the input is stochastically corrupted, usually using a Gaussian additive noise, while the uncorrupted input is still used as the target for the optimization of the parameters (Alain and Bengio, 2014, Vincent et al., 2008).
After the proposed DDCAE has been trained on a target class (the normal or the common pneumonia), the hidden features are used to construct an average and robust statistical representation of the target class. This is accomplished by averaging all the hidden features obtained by passing the whole training set in the encoder part and then evaluating the histogram of the mean representation. This histogram is used as a reference in the inference phase. For each new test CT scan, the related hidden features are evaluate by using the trained encoder and a test histogram is obtained. At this stage, a suitable measurement of the distance between two distributions is used: if the resulting distance is below a certain threshold, the test image is classified as the same of the target class, otherwise it is considered as an anomaly and labeled as COVID-19.
Specifically, the main contributions of this paper are:
-
•
we propose an ad hoc DDCAE architecture optimized for the detection of anomaly CT scans. The proposed architecture is composed of three convolution layers in order to keep limited the computational complexity while obtaining sufficiently good and robust hidden features to obtain a high discrimination between their statistical representation;
-
•
we propose a statistical distance-based approach to label a test image either as anomaly or not. The employed distance measurements should be suitable for discriminating histograms belonging to different classes. In this paper, we compare between the Kullback–Leibler divergence, the Bhattacharyya distance, and the Euclidean one;
-
•
we perform numerical results on a well-known dataset available in the literature and compare the proposed approach to other state-of-the-art deep architectures. We expect that the proposed approach is able to obtain excellent results by keeping limited the training and inference time.
Let us remark that statistical representations of target classes and autoencoders are well known approaches for the anomaly detection problem. However, in literature these methods are used in a different way (see, for example, Chandola et al., 2009 and references therein). Specifically, the statistical representation is used to characterize the probability density or evaluating some peculiar moments of the instances by directly working on the input space. Autoencoders are typically used in anomaly detection problems by thresholding the reconstruction error (i.e., by comparing the input and the output directly). These state-of-the-art approaches have been demonstrated to be ineffective for the CT scans of COVID19 disease, as pointed out in Section 5.2. On the contrary, the approach proposed in this paper works directly on the latent space generated by the autoencoder. This means that the statistical representation is evaluated on the hidden feature vector, while the final classification is obtained by thresholding suitable-designed inter-histogram distances. To the best of the authors’ knowledge and on the basis of the broad overview of the related literature of Section 2, the proposed approach is novel and not used up to date for the classification of CT scans of infected lungs.
The rest of the paper is organized as follows. Section 2 presents the recent literature on the topic. Section 3 describes the proposed approach in terms of both used architecture and suitable distribution distances. Section 4 introduces the experimental setup, while Section 5 shows the obtained numerical results and their comparison with other state-of-the-art approaches. Finally, Section 6 concludes the paper and outlines some future works.
2. Related work
In recent years, a great attention on automatic classification of medical images has been devoted to the application of Deep Learning approaches. Although many recent works are addressed to traditional x-Ray images (Chandra et al., 2021, Ismael and Şengür, 2020, Ismael and Şengür, 2021) and use transfer learning (Vidal et al., 2021), lots of novel methods working on CT scans have been proposed. Two recent and comprehensive reviews embrace several methodologies currently used in medical screening (Ozsahin et al., 2020, Rahman et al., 2021). Obviously, automatic CT scans classification is applied for diverse diseases, as the lung nodule malignancy suspiciousness classification (Shen et al., 2017), but during the last year the main contributions focused on the COVID-19 disease. Among these works, studies are divided into two main families: those based on segmentation and those that perform the classification task directly.
Approaches based on segmentation are usually based on U-Net type architecture to identify relevant part of the CT scans and perform classification focusing the attention only on these sections (Fan et al., 2020, Saood and Hatem, 2021, Vidal et al., 2021, Yao et al., 2021). Specifically, the work in Vidal et al. (2021) is based on U-Net exploiting a multi-stage transfer learning idea to quickly develop computer aided diagnosis systems that can be used in real-time on smart-phones. Similarly, [41] performs CT scan segmentation exploiting both the U-Net and the SegNet. The authors of Fan et al. (2020), instead, propose Inf-Net, an “Infection segmentation deep Network” introduced to automatically identify infected regions from chest CT slices. Differently form these works, our proposed approach does not resort to image segmentation. Finally, [55] proposes NormNet, an approach able to recognize normal tissues and separate them from possible COVID-19 lesions. Like our approach, NormNet is trained in an unsupervised manner, however, differently from the proposed work, it uses synthetic lesions constructed by using a set of simple operations and then inserted into normal CT lung scans, while the prediction is performed by a U-Net type architecture. Moreover, the results shown in Yao et al. (2021) do not overcome a precision of 0.905 for the COVID-19 cases and hence it does not outperform our proposed idea.
The main works of the second family are based on the binary classification problem of COVID/NON-COVID images (Elmuogy et al., 2021, Mishra et al., 2020, Shah et al., 2021, Tan et al., 2021). Specifically, the work in Shah et al. (2021) is based on deep Convolutional Neural Networks (CNNs) and proposes a specific configuration called CTnet-10 while comparing the results with well-known CNN architectures, such as DenseNet-169, VGG-16, ResNet-50, InceptionV3, and VGG-19. Similarly, in Tan et al. (2021) a “super-resolution” variant of VGG-16 neural network has been introduced. The super-resolution of chest CT images has been obtained by exploiting the SRGAN neural network. The authors in Elmuogy et al. (2021) propose an automatic classification architecture based on a deep neural network called Worried Deep Neural Network (WDNN) that uses transfer learning and provide results by using different pre-trained models. Differently, the paper in Mishra et al. (2020) is based on a fusion approach. The main idea of the fusion approach is that the classification errors made by individual models may be mitigated by combining the individual predictions via a majority voting approach. The baseline models used in Mishra et al. (2020) include VGG16, InceptionV3, ResNet50, DenseNet121, and DenseNet201. Similarly, [47] proposes a voting-based approach for the screening of COVID-19 by exploiting and extending the EfficientNet neural network along with a data augmentation process and transfer learning. Differently from our approach, works in Elmuogy et al., 2021, Mishra et al., 2020, Shah et al., 2021, Silva et al., 2020 and Tan et al. (2021) are all supervised, hence they have to be trained on both the COVID and NON-COVID images.
Moreover, we point out that there are some few approaches that exploit the deep AEs in medical images (Chen et al., 2017, Li et al., 2020, Xu et al., 2016). Specifically, the work in Xu et al. (2016) proposes a Stacked Sparse Autoencoder (SSAE) plus a softmax classifier for identifying the presence or absence of nuclei in individual image patches related to the breast cancer. Authors in Chen et al. (2017) applied a deep convolutional autoencoder to pulmonary CT scans to detect lung nodules. Once again, the final detection is performed by a softmax classifier. Interestingly enough, the approach in Chen et al. (2017) has been extended for similarity measurement of lung nodules images. Topic of Li et al. (2020) is the COVID-19 diagnosis from chest CT scans exploiting a stacked autoencoder detector model. Authors propose a novel cost function to train the stacked autoencoder, regularized in a different manner for each layer, then the detection is again done by a softmax classifier. Differently from our approach, works in Chen et al., 2017, Li et al., 2020 and Xu et al. (2016) use the autoencoder to automatically construct a set of features and then use a softmax classifier on the top. We instead use the hidden features to construct a statistical representation of the input scans. Moreover, differently from Li et al. (2020) and Xu et al. (2016) that implement stacked autoencoders (trained in a layer-wise fashion), we exploit a deep autoencoder that is computationally more efficient.
In addition, there exist some approaches based on traditional Machine Learning (ML) methods. To this aim, the main goal of the contribution in Gomes et al. (2020) is to check the actual effectiveness of some low-complexity shallow supervised classifiers (namely, Support Vector Machine (SVM), Multi-Layer Perceptron (MLP) and Random Forest (RF)) to detect COVID-19 diseases by texture analysis of chest X-ray images. The goal is to properly classify COVID-19, viral pneumonia, bacterial pneumonia and healthy radiographic images by running the mentioned ML approaches on a set of features composed by the Haralick and Zernike moments. Interestingly enough, the numerical results reported in Gomes et al. (2020) support the conclusion that SVMs equipped with 2–3 degree polynomial kernels are the most performing ones and, in the carried out tests, obtain a good average accuracy, recall, precision and specificity.
Finally, the follow-up paper in Gomes et al. (2021) extends the utilization of shallow ML approaches to the classification of DNA sequences of 25 different virus classes. Specifically, the paper proposes a technique for representing DNA sequences in which each sequence is partitioned into shorter mini-sequences that partially overlap in a pseudo-convolutional fashion, in order to be represented by suitable co-occurrence matrices. These last are the extracted features utilized as input to five types of shallow supervised classifiers, namely, SVM, RF, MLP, Naïve Bayes classifier and Instance-Based-K (IBK) learner (Alpaydin, 2014). Interestingly, the multiclass classification tests carried out in Gomes et al. (2021) support the conclusion that RF classifiers are the most performing ones and they attain average accuracies around 94%.
An overview of the related work is provided in Table 1 that briefly summarizes the main approaches. Overall, on the basis of the carried out overview, we conclude that our approach is unique in: (i) applying a statistical representation of the hidden feature vector, estimated in an unsupervised manner; and, (ii) performing classification by thresholding suitable inter-histogram distances.
Table 1.
Summary of the main related work. The last column indicates whether the learning is of supervised (S) or unsupervised (U) type. See also [39] and Rahman et al. (2021) for a review on the diagnosis of COVID-19 from radiography images.
| Family | Work | Approach | Type |
|---|---|---|---|
| X-ray | [28] | Multiresolution | S |
| [29] | Deep learning | S | |
| [14] | Majority voting | S | |
| [51] | U-Net + Transfer Learning | S | |
| [19] | Shallow ML | S | |
| CT segmentation | [55] | NormNet | U |
| [41] | U-Net + SegNet | S | |
| [18] | InfNet | S | |
| CT binary | [43] | CNN (CTnet10) | S |
| classification | [50] | VGG-16 | S |
| [17] | WDNN | S | |
| [38] | Fusion + Majority voting | S | |
| [47] | Fusion + Majority voting | S | |
| Autoencoders | [54] | SSAE + Softmax | S |
| [15] | CAE + Softmax | S | |
| [36] | SSAE + Softmax | S | |
| Our approach | DDCAE + hidden features | U | |
3. Proposed approach and related deep denoising CAE architectures
The proposed approach is based on a denoising version of a Deep Convolutional Autoencoder (DCAE), here called DDCAE.
Although Autoencoders (AEs) have been introduced at the end of 80s (Bourlard & Kamp, 1988), only recently their deep versions have been exploited in practical applications (Goodfellow et al., 2016). In brief, a Deep AE (DAE) is a feed-forward neural network with hidden layers trained to (quasi) reproduce its input at the output layer. In this regard, the aim of a DAE is to learn a compact and meaningful representation (encoding, also called hidden features or feature vector) for a set of (possibly, noisy) input data, using a set of weight parameters. Then, an estimate of the input data is recovered (decoding), usually using tied weights (Goodfellow et al., 2016).
However, when used on image data, it is more convenient to resort to the convolutional version of the DAE. In the Deep Convolutional AE (DCAE) (Masci et al., 2011), each fully connected layer is replaced by a cascade of a suitable number of convolutional, pooling and normalization layers, as described in the following.
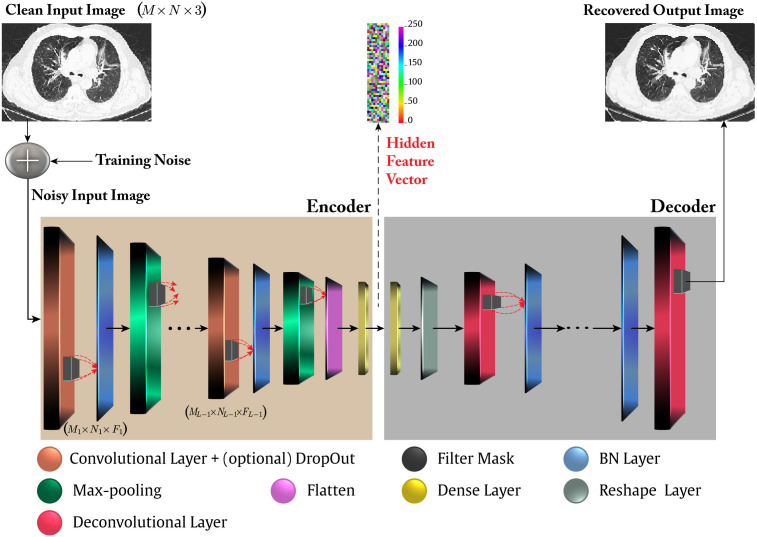
Moreover, in literature it is often used a robust variant of the AE, called Denoising AE (Alain and Bengio, 2014, Vincent et al., 2010), in which a stochastically corrupted version of the input is employed to feed the AE (usually, using a Gaussian additive noise with zero mean and variance ), while the uncorrupted input is still used as the target for the optimization of the parameters. The general idea of a DDCAE is graphically depicted in Fig. 1.
Fig. 1.
The considered DDCAE reference architecture. The input noise is present only in the DDCAE training phase while it is zeroed in the validation and test phases. Furthermore, depending on the actually considered DDCAE architecture, the innermost flattening and dense layers may be absent. Accordingly, is the depth of the Encoder, while is the number of the corresponding Convolutional+Pooling+Batch Normalization (BN) layers. Finally, the taxonomy: indicates a convolutional layer (or a filter kernel or an input image) of spatial dimension: which embraces feature maps.
Fig. 1 shows that the encoder of a DDCAE is composed of the cascade of Convolutional, Max Pooling, and Batch Normalization (BN) layers, plus and eventual final Dense layer. Not all the depicted layers are used in some specific configurations of the proposed DDCAE.
Although, the role of these layers is well-known in literature, we provide a short description of each of the involved layers to help the non expert reader.
First of all, let be the training set composed of a sequence of images of normal or CP diagnostics. Each th image is a tensor of dimension , representing the number of rows, columns, and colors.
The th convolutional layer, which is responsible for the extraction of local features, implements a set of convolutions by using filters (also called kernels) to produce a certain number of feature maps. Specifically, a kernel function is convolved with a specific region of the image of the same size of the kernel to produce an output pixel. Then the kernel is moved by a quantity , called stride, and another output pixel is produced. This operation is performed in parallel for all the filters. A stride value greater than the unity will produce an output map of reduced size with respect to its input. Let us denote with , and the corresponding number of squared 2D filters, filter size and stride coefficient, respectively, while and are the spatial size of the th 2D filter output. Mathematically, the output sample generated at the spatial position by the th spatial filter of the th convolutional layer, is provided by the following summation (Goodfellow et al., 2016):
| (1) |
for , , and , where is the set of the scalar samples of the kernel of the th 2D filter, is the set of scalar features at the input of the th channel of th layer, and is the bias term of the th filter in the th layer. All the convolutional layers, except the last of the decoder, use the ReLU activation function. The ReLU function is defined as . The last layer of the decoder uses instead a sigmoid activation in order to produce a valid pixel values inside the interval .
In the pooling layer, used for the down-sampling of the feature maps, each input map is divided into adjacent non-overlapping regions according to the size of the pooling region, and the maximum of the region is output. In this way the spatial dimension of the input is reduced after the pooling operation. Instead, the number of input feature maps is equal to the number of output feature maps in the pooling layer. Mathematically, the max pooling operation is performed as:
| (2) |
where is the th region of the th feature map and represents the th element of the th output feature map.
The batch normalization layer applies a standardization operation on the output of the layer, by forcing a zero mean and a unit variance. This operation works as a regularization, increasing the stability of the neural network and accelerating the training (Ioffe & Szegedy, 2015). This layer normalizes its output using the mean and standard deviation of the current batch of inputs , by evaluating:
| (3) |
where is a small constant used to avoid division by zero, while and are a scaling and offset parameters learned during the training phase.
Fig. 1 also shows the presence of a Flatten layer, whose role is to transform the output of last layer, which is in a tensor form, into a vector by stacking the single vectors of the output tensor one atop the others. The aim of the Flatten layer is to produce the hidden feature vector or the input to the optional final dense layer.
The dense layer is a fully-connected set of neurons where every input is connected to every output by a weight, and generally followed by a nonlinear activation function:
| (4) |
where is the activation function, usually a sigmoid or the ReLU again, and are the vectors of the inputs and the outputs, respectively, and is the matrix collecting all the weights between the th input and the th output in the last th layer.
The decoder is, in a certain sense, the mirror version of the decoder: it presents the same layers of the encoder but in a reverse order. Hence, its first layer is the dense one, if used in the encoder. At this stage we need to reshape a vector (the output of the dense layer or directly the hidden features if the encoder has not a final dense layer) into a tensor of suitable shape. This task is performed by the Reshape layer shown in Fig. 1. In the decoder of the proposed DDCAE, we need some layers implementing the deconvolution operation and the up-sampling of the image. These tasks are simply accomplished by the Deconvolutional layers in Fig. 1. Technically, we use a transposed convolutional layer, which is equivalent to first stretching the image by inserting empty rows and columns (full of zeros), and then performing a regular convolution.
3.1. The proposed deep denoising CAE (DD-CAE) architecture
In this paper we propose a specific DDCAE that will be trained on the target classes (usually the normal or common pneumonia). The proposed architecture consists in a DDCAE, formed by three convolutional layers, along with two max-pooling and two batch normalization layers, as summarized in Table 2.
Table 2.
Layers’ organization of the proposed DDCAE. The output shape refers to the tuple for the th layer.
| Layer type | Kernel size | Stride | Output Shape | Param. # |
|---|---|---|---|---|
| Conv2D | 3 × 3 | 1 | (200, 300, 256) | 7168 |
| Batch Normalization | – | – | (200, 300, 256) | 1024 |
| Conv2D | 3 × 3 | 1 | (200, 300, 128) | 295040 |
| MaxPooling | 2 × 2 | 2 | (100, 150, 128) | 0 |
| Conv2D | 3 × 3 | 1 | (100, 150, 64) | 73792 |
| Batch Normalization | – | – | (100, 150, 64) | 256 |
| MaxPooling | 2 × 2 | 2 | (50, 75, 64) | 0 |
| Total params: 377,280 | ||||
| Trainable params: 376,640 | ||||
| Non-trainable params: 640 | ||||
| Layer type | Kernel size | Stride | Output Shape | Param. # |
| Conv2D Transpose | 3 × 3 | 2 | (100, 150, 128) | 73856 |
| Batch Normalization | – | – | (100, 150, 128) | 512 |
| Conv2D Transpose | 3 × 3 | 2 | (200, 300, 256) | 295168 |
| Batch Normalization | – | – | (200, 300, 256) | 1024 |
| Conv2D Transpose | 3 × 3 | 1 | (200, 300, 3) | 6915 |
| Total params: 377,475 | ||||
| Trainable params: 376,707 | ||||
| Non-trainable params: 768 | ||||
Training of the proposed DDCAE.
The training of the proposed DDCAE is performed by minimizing the Mean Square Error (MSE) cost function that measures how similar the reconstitution is to its input . After denoting with the whole set of trainable parameters of the network, the MSE cost function can be defined as:
| (5) |
where is the size of the mini-batch, and and denote the number of rows and columns of each image, respectively.
As already introduced, since we are using the denoising version of autoencoder, in the training phase we adopt a statistically corrupted input , that is:
| (6) |
where is a tensor of the same dimension as the input image whose elements are drawn from a Gaussian distribution with zero mean and variance. In addition, since the single pixel of an image can take values inside the interval , the values of that exceed such range have been clipped.
The training of the proposed DDCAE has been performed by the Adam optimizer, a gradient-based optimization algorithm that exploits the first and second order moments to obtain a smooth and fast convergence (Kingma & Ba, 2015).
Evaluating the target statistical representation.
After the training of the DDCAE on a reference class by minimizing the reconstruction error in (5), its encoder has been used to construct a meaningful statistical representation of the image pixels.
In order to obtain a robust statistical representation, in this work we evaluate the hidden features for the entire training set by using the encoder of the trained DDCAE. Hence, a target hidden feature vector , of length , is obtained as the average of all the single hidden feature vectors. We expect that, if the number of images is sufficiently high, this target representation can be a meaningful representative of the target class.
As a statistical representation of the target feature vector , in this paper we choose the (normalized) histogram, principally due to its simplicity and efficiency in computation. A histogram is an estimate of the distribution obtained by dividing the range of values into a sequence of equally-spaced interval called bins and counting how many values fall into each interval. The histogram is then normalized to sum to unit.
The number of bins used to construct the histogram should be chosen by a trade-off between numerical stability of the used distance measurement and its discriminating capability. Although this number turned out not to be critical for the final model performance, if it is much less than the length of the hidden feature vector, we found that an excellent choice to guarantee non-empty bins is by using 50 bins, i.e. . The only observation is regarding the Euclidean distance that tends to fill the gap between the two classes if is excessively increased.
In this phase, we also evaluate the distance between the just computed target histogram and all the histograms of the single reference encoded scans, by using suitable probability dissimilarity measurements (introduced in the next subsection). Among these distances, we compute the mean and standard deviation values in order to set conveniently a suitable threshold used during the test phase to discriminate between a reference scan from an anomaly one. The idea is that the statistical distance from an anomaly scan should be greater than a reference one, hence the threshold could be set proportionally to the mean distance plus a term depending on its standard deviation. Mathematically, we set the threshold as follows:
| (7) |
where is a suitable constant. In this case, a provides good results.
Test of the proposed DDCAE.
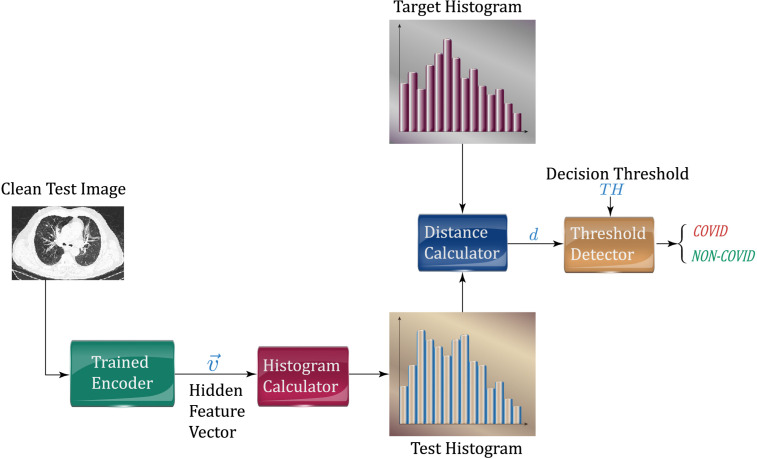
During the inference phase, each test image passes through the trained encoder and produces its latent feature vector used to evaluate a test histogram. This test histogram will be successively compared to the target one by a suitable distance measurement (the same used during the target computation). In this paper, we focus our attention on two reference classes: the normal one and the common pneumonia (CP) one. If the distance between the test and target histograms is above the set threshold , the related image will be marked as COVID (anomaly), otherwise as the reference one. The main idea is sketched in Fig. 2.
Fig. 2.
Sketch of the test phase proposed in the paper.
3.2. Unsupervised distance-based processing of the latent representation
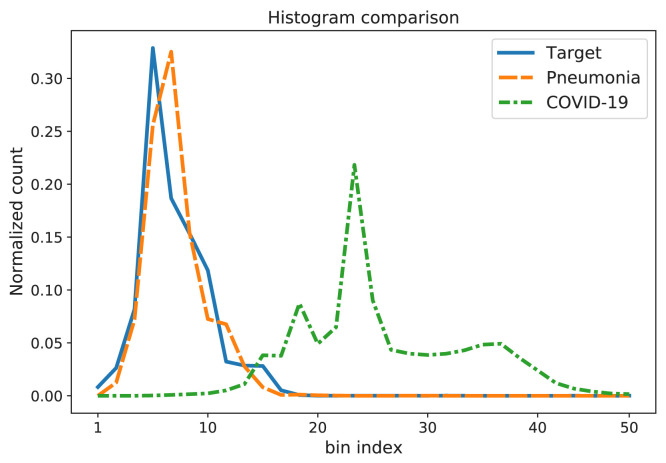
The main focus of the paper is the evaluation of the dissimilarity between the target histogram and the histogram obtained by the encoder for a test image. If the histogram of the test image is similar to the target one, we can assign it to the corresponding target class, otherwise it is labeled as a COVID-19 image. An example of target, NCP, and CP histograms are shown in Fig. 3.
Fig. 3.
Example of target, NCP, and CP histograms.
In literature, the similarity between two histograms can be evaluated by several distance measurements of the underlying distribution (Kullback, 1997). For the aims of this paper, after denoting with and the two involved histogram distributions defined over the set of interval bins , we have selected the following three distances.
-
1.Kullback–Leibler (KL) divergence:
where and are the values assumed by the and histogram in the th bin, respectively. By definition, the contribution of the th term in the summation in (8) is zero if .(8) -
2.Bhattacharyya distance:
(9) -
3.Euclidean distance:
(10)
These distances have been normalized by the number of used bins in the histogram in order to render them independent of this choice. The Bhattacharyya distance is widely used in several applications, like image processing, and, differently from the KL one has the advantages of being insensitive to the zeros of distributions. On the other hand, the Euclidean distance is very simple and smooth but it tends to treat excessively equally differences between the distributions.
In the proposed approach, if the distance (chosen between the KL, Bhattacharyya and Euclidean one) between the target and the test histograms is above the set threshold , the image under test is classified as COVID-19 (CNP), otherwise it is classified as the target class (normal or CP, depending on the used training set).
4. Experimental setup
In this section, we describe the utilized dataset for the reference classes, the employed software environment, the setting of the main parameters, and the metrics used to evaluate the performance of the proposed idea.
4.1. The utilized datasets
In response to the COVID-19 pandemic, the global open source and open access COVID-Net initiative1 made available some relevant datasets of CT scans in order to accelerate the advancement in machine learning to fight the pandemic (Gunraj et al., 2020). Recently, the COVID-Net team has released a second version of the dataset in two variants (the COVIDx CT-2 A and CT-2B datasets, respectively) (Gunraj et al., 2021). The “A” variant consists of cases with confirmed diagnoses, while the “B” variant contains all of the “A” variant and adds some cases which are assumed to be correctly diagnosed but are weakly verified. In this paper, we address our attention to the “A” variant of the dataset.2
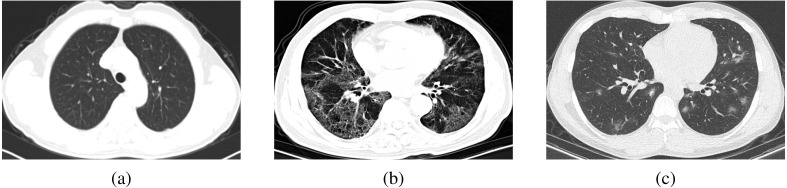
The COVIDx CT-2 A dataset has been constructed by collecting many publicly available data sources (Gunraj et al., 2021, Gunraj et al., 2020) and comprises 194,922 CT slices from 3745 patients. The dataset scans are related to three classes: novel coronavirus pneumonia due to SARS-CoV-2 viral infection (NCP), common pneumonia (CP), and normal controls. For NCP and CP CT volumes, slices marked as containing lung abnormalities were leveraged. Moreover, all the CT volumes contain the background in order to avoid model biases. An example of a representative image for each class is shown in Fig. 4. The issues pertinent to data ethics were ensured during the data collection of the dataset, according to the information reported on the related websites supporting this study.
Fig. 4.
Examples of some CT images: (a) normal, (b) common pneumonia (CP), and (c) novel coronavirus pneumonia (NCP).
In this paper, we have randomly selected 3500 and 700 images from the normal and Pneumonia classes for the training and validation of the proposed DDCAE, respectively, and 500 images from each class to test it.
Table 3.
Number of training and testing instances of each considered class. In the proposed DDCAE approach, the COVID-19 data has not been used in the training phase.
| Type | Training | Validation | Test |
|---|---|---|---|
| Normal | 3500 | 700 | 500 |
| Pneumonia | 3500 | 700 | 500 |
| COVID-19 | – | – | 500 |
4.2. Utilized simulation environment and setting of the main parameters
All the simulations described in this paper have been implemented in Python environment by using the end-to-end and open-source machine learning platform TensorFlow 2 exploiting the Keras API, with a PC having an Intel Core i7-4500U 2.4 GHz processor, 16 GB RAM, and Windows 10 operating system.
About the setting of the main parameters, Table 4 summarizes the considered default values and the meaning of each parameter. The values of these parameters have been selected by using the validation set in Table 3.
Table 4.
Main considered parameters and related default values.
| Description | Parameter | Value |
|---|---|---|
| Mini-batch size | 16 | |
| Number of epochs | 50 | |
| Learning rate | 0.001 | |
| Adam parameter | 0.9 | |
| Adam parameter | 0.999 | |
| Adam parameter | ||
| Number of bins | 50 | |
| Number of hidden features | 240,000 | |
| Parameter to set the threshold | 0.3 |
4.3. The considered performance metrics
In a binary classification problem we are interested in classifying items belonging to a positive class (P) and a negative class (N). With respect to a specific dataset, there are four basic combinations of actual data category and the assigned output category:
-
•
true positive (TP): correct positive assignments;
-
•
true negative (TN): correct negative assignments;
-
•
false positive (FP): incorrect positive assignments;
-
•
false negative (FN): incorrect negative assignments.
The set of these four quantities is usually arranged in a matrix layout, called confusion matrix (CM), which allows a simple visualization of the performance of a classification algorithm. Each column of the CM represents the instances in a predicted class while each row represents the instances in an actual class. Moreover, the combination of the previous four numbers in some powerful indicators can be a valid tool to quantitatively measure the performance of a classification algorithm (Alpaydin, 2014). Among all the possible combination, in this paper we focus our attention on the accuracy, precision, recall and F-measure metrics, whose formal definition can be found in Table 5. The accuracy is the ratio between the correct identified instances among their total number. The precision is the ratio of relevant instances among the retrieved instances, while the recall is the ratio of the total amount of relevant instances that were actually retrieved. Finally, precision and recall can be combined in a single measurements called F-measure that is mathematically defined as their harmonic mean.
We also consider the TP rate (formally coincident with the recall metric) and the FP rate. These last measures are, respectively, the ratio between the number of TP and the total positive examples and the ratio between the number of FP and the total negative examples. By plotting the TP rate on the -axis against the FP rate on the -axis in a plane, when the discrimination threshold is changed, we obtain the Receiver Operating Characteristic (ROC) curve that is a graphical representation of the performance of a binary classifier. However, the ROC curve is a two-dimensional depiction of classifier performance and often we need of a single scalar value representing the expected performance. A common method is to calculate the area under the ROC curve, abbreviated as AUC. The closer the AUC is to one, the better the classifier performance.
Table 5.
The performance metrics for the evaluation of the proposed model.
| Performance Metrics | Formula |
|---|---|
| Precision | |
| Recall | |
| F-measure | |
| Accuracy | |
| TP rate | |
| FP rate |
5. Numerical results
In this section, we show the numerical results obtained by the proposed approach. As described above, the performance of the designed DDCAE is tested by using CT scans related to two target classes: the normal images and the common pneumonia (CP) ones. Some comparisons with two state-of-the-art deep architectures will be also performed. The test set used in experiments is composed of 500 CT scans belonging to the new coronavirus pneumonia (NCP) and 500 CT scans belonging to the reference class (normal or CP). To provide a clear graphical representation of the obtained distances, the test instances have been fed to the proposed algorithm in this order: first the NCP scans (positive class) and then the reference ones (negative class).
5.1. Evaluation of the proposed approach
In the first set of experiments, we investigate the effect of the noise level on the DDCAE, i.e. we perform several test by using a different noise level in generating the tensor in (6).
In the following, experiments have been performed by using four different values of the standard deviation . Specifically, we use the set of values: . A value is meaning that the traditional deep CAE is used, i.e. the denoising idea is not implemented.
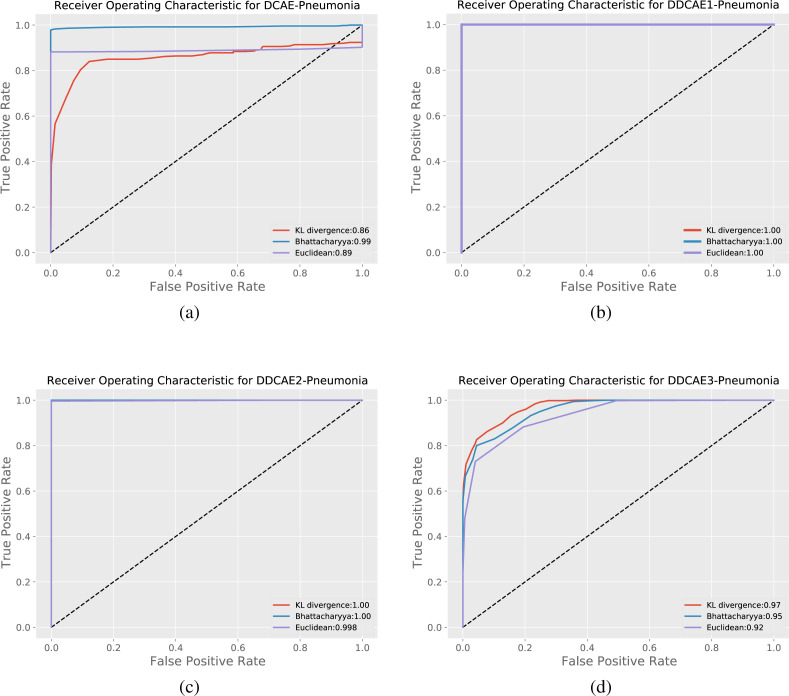
Table 6 summarizes the results obtained by the proposed DDCAE for all the tested values in terms of the Accuracy, Precision, Recall, F-measure and AUC metrics introduced in Section 4.3 and defined in Table 5, by considering the COVID-19 images as the positive class and the reference images as the negative class. Simulations have been performed for both the normal and CP reference datasets. Table 6 also provides the different metrics for all the three considered distribution distances in Section 3.2. The related ROC curves for the CP case are shown in Fig. 5. Similar curves are obtained for the normal reference scenario.
Table 6.
Results obtained by the proposed DDCAE at different noise level for the two considered scenarios.
| Normal |
Pneumonia |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Model | Accuracy | Precision | Recall | F-measure | AUC | Accuracy | Precision | Recall | F-measure | AUC | ||
| Kullback–Leibler divergence | ||||||||||||
| DCAE | 0.00 | 100.00 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 83.40 | 0.8345 | 0.8340 | 0.8342 | 0.8600 | |
| DDCAE1 | 0.01 | 100.00 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 100.00 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | |
| DDCAE2 | 0.05 | 99.60 | 0.9960 | 0.9960 | 0.9960 | 0.9990 | 100.00 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | |
| DDCAE3 | 0.10 | 92.70 | 0.9272 | 0.9270 | 0.9270 | 0.9790 | 88.33 | 0.8942 | 0.8830 | 0.8886 | 0.9710 | |
| Bhattacharyya distance | ||||||||||||
| DCAE | 0.00 | 99.60 | 0.9960 | 0.9960 | 0.9960 | 0.9990 | 98.80 | 0.9881 | 0.9880 | 0.9880 | 0.9930 | |
| DDCAE1 | 0.01 | 100.00 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 100.00 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | |
| DDCAE2 | 0.05 | 99.20 | 0.9921 | 0.9920 | 0.9920 | 0.9990 | 100.00 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | |
| DDCAE3 | 0.10 | 93.50 | 0.9352 | 0.9350 | 0.9350 | 0.9820 | 87.60 | 0.8810 | 0.8760 | 0.8785 | 0.9510 | |
| Euclidean distance | ||||||||||||
| DCAE | 0.00 | 92.00 | 0.9328 | 0.9200 | 0.9265 | 0.9070 | 91.80 | 0.9314 | 0.9180 | 0.9247 | 0.8930 | |
| DDCAE1 | 0.01 | 99.90 | 0.9990 | 0.9990 | 0.9990 | 0.9990 | 100.00 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | |
| DDCAE2 | 0.05 | 92.90 | 0.9391 | 0.9290 | 0.9340 | 0.9560 | 99.80 | 0.9980 | 0.9980 | 0.9980 | 0.9980 | |
| DDCAE3 | 0.10 | 92.90 | 0.9294 | 0.9290 | 0.9292 | 0.9710 | 74.90 | 0.8730 | 0.7490 | 0.8063 | 0.9190 | |
Fig. 5.
ROC curves of the proposed approach for the considered distance measures in the Pneumonia scenario: (a) DCAE, (b) DDCAE1, (c) DDCAE2, and (d) DDCAE3. The corresponding AUC values are reported in the legend boxes.
Table 6 demonstrates the effectiveness of the proposed idea. In fact, results in terms of all the considered metrics are generally satisfying and, interestingly enough, they reach the top result of 100% in some of the proposed settings. By a carefully examination of the rows of Table 6, we can draw some general considerations:
-
•
results obtained with reference to the normal class generally outperform those of the CP class. This is justified by the fact that scans of the NCP are more similar to the CP class rather than the normal one, hence discriminating between NCP and CP is a more complicated task with respect to the discrimination between NCP and normal;
-
•
the clean version of the deep CAE provides worsening results with respect to the denoising versions. This is justified by the fact that the input noise operates as a regularizer providing a more robust classification;
-
•
the level of the noise should be not too big. In fact, the performance obtained by using the value is lower than the corresponding ones with and ;
-
•
the proposed DDCAE with and is able to reach top performance in terms of all the considered metrics;
-
•
although all the three considered distance measurements (i.e., the KL divergence, the Bhattacharyya distance, and the Euclidean distance) provide similar results, when the performance are not at the top level, Table 6 suggests that the Bhattacharyya distance produces higher scores, while the Euclidean distance the smaller ones. This could be justified by the mathematical expressions of these distances. In fact, the Bhattacharyya distance is quite smooth and automatically takes into account for the possible zero values of a distribution, while in the KL divergence this could be a problem, even if the chosen number of bins assures the absence of zeros in the both distributions. On the other hand, the Euclidean distance, for its nature, produces not so discriminating distance;
-
•
the DDCAE with provides always the top accuracy of 100%. Just for the normal reference and Euclidean distance, the accuracy is slightly lower, i.e. 99.90%!
Motivated from these considerations, in the following tests and comparisons we use the DDCAE with and the distance are measured by the KL divergence. Moreover, since it is more challenging, we use as reference class the CP one.
In order to give a visual insights of the results in Table 6 and justify the top 100% accuracy, Fig. 6 shows the considered Kullback–Leibler divergence, Bhattacharyya distance, and Euclidean distance in the case of common pneumonia reference dataset and the DDCAE with . This figure clearly shows the effectiveness of the proposed idea. In fact, we can see that the NCP scans (the first 500 bars in Fig. 6) are much more distant with respect to the corresponding reference images (the last 500 bars). The differences between the classes is about one order of magnitude for the KL divergence and the Bhattacharyya distance, as we can see in Fig. 6, Fig. 6, while it is more limited in the case of the Euclidean distance (see Fig. 6(c)). This last case also shows a larger variance of the obtained distances with respect to the first two measures.
Fig. 6.
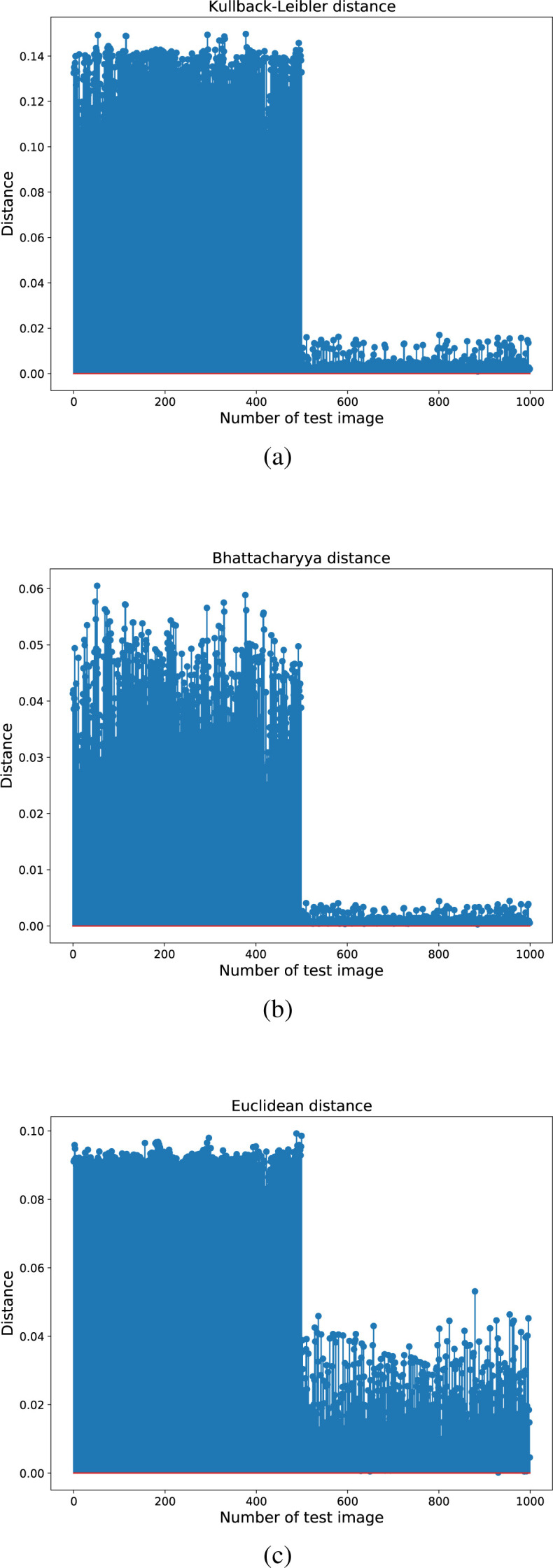
The obtained distances for the case of CP reference using the DDCAE with . The considered distances are: (a) Kullback–Leibler divergence, (b) Bhattacharyya distance, and (c) Euclidean distance.
In the following set of experiments, we compare the proposed 3-Layer DDCAE by changing the number of hidden layers. Specifically, we test two shallower architectures (with one and two layers, respectively) and a deeper one (with four hidden layers). Since the DDCAE with a single hidden layer performed very poorly, we provide results obtained by an architecture composed of a single dense layer, that is the only yellow layers in Fig. 1. The results obtained by these solutions on the CP reference dataset and employing the KL divergence to measure the histogram distances are shown in Table 7. As we can see from this table, the performance tends to decrease for both the increasing and decreasing number of hidden layers. This behavior is uniform for all the considered metrics. Similar results have also been obtained by using the normal class as reference, not shown here for space constraints. The results, shown in Table 7, justify the use of three hidden layers in the proposed DDCAE.
Table 7.
Results obtained by the proposed DDCAE at different number of layers. The metrics have been evaluated by using the KL divergence on the CP reference dataset. In italic the proposed DDCAE.
| Layers | Accuracy | Precision | Recall | F-measure | AUC |
|---|---|---|---|---|---|
| 1 | 63.90 | 0.6594 | 0.6390 | 0.6487 | 0.7250 |
| 2 | 88.20 | 0.7800 | 0.9799 | 0.8686 | 0.9210 |
| 3 | 100.00 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| 4 | 96.40 | 0.9648 | 0.9640 | 0.9644 | 0.9960 |
Remark.
We point out that we also implement the same methodology by exploiting the idea of Sparse AEs as done in Xu et al. (2016). However, the sparse AE does not yield any good results by nothing of the used metrics. Therefore, we decided to exclude it from the paper. The bad results obtained by sparse AEs are intuitively justified by the fact that our approach is based on the construction of a statistic representation by evaluating the histogram of the hidden feature vector. Since the sparse AE produces a very sparse hidden feature vector, the obtained histogram becomes not so significant.
5.2. Comparisons to state-of-the-art benchmark solutions
In this subsection, we show some comparisons with other state-of-the-art benchmark solutions. These comparisons involve both unsupervised and supervised techniques. Specifically, we employ the DDCAE itself to evaluate the anomalies. In fact, an autoencoder is trained to reconstruct its input: this is meaning that the reconstruction error in (5) should be small for test image belonging to the same class on which the architecture has been trained, otherwise the error should be bigger. Hence, the anomaly scans (i.e., the NCP images) can be obtained by thresholding the reconstruction error in (5) (Amarbayasgalan et al., 2020). We refer to this method as the first benchmark approach or BAP1.
In addition, to perform fair comparisons with the proposed approach, we also rearrange our methodology to the output space. We compute the reconstruction error of a test scan and evaluate the statistical representation of such an error by means of the histogram. Then, once again, we perform the distance measurement of this test histogram to the reference one, obtained from the average of all the error images computed from the training scans. The same three distance measurements introduced in Section 3.2 have been employed. We refer to this method as the second benchmark approach or BAP2.
Results for the BAP1 and BAP2 benchmark approaches are shown in the top part of Table 8. For space reasons, only the results related to the CP dataset as reference are shown, but similar results can be obtained by using the normal reference dataset. Regarding the BAP2 approach, the second row of Table 8 is related to the KL divergence, which provide the best results compared to the Bhattacharyya and Euclidean distances.
Table 8.
Results obtained by other state-of-the-art unsupervised and supervised approaches on CP dataset.
| Architecture | Accuracy | Precision | Recall | F-measure | AUC |
|---|---|---|---|---|---|
| Unsupervised | |||||
| BAP1 | 81.90 | 0.8513 | 0.8190 | 0.8348 | 0.8450 |
| BAP2 | 65.50 | 0.6925 | 0.6550 | 0.6732 | 0.6520 |
| Supervised | |||||
| AlexNet | 71.10 | 0.8601 | 0.7110 | 0.7785 | 0.9460 |
| GoogLeNet | 100.00 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
Results shown in the first two rows of Table 8 clearly show that the benchmark solutions are not suitable to produce high metrics. This is meaning that the hidden features, extracted by the DDCAE, are more informative than those corresponding to the reconstructed images. This, in turn, supports the proposed approach.
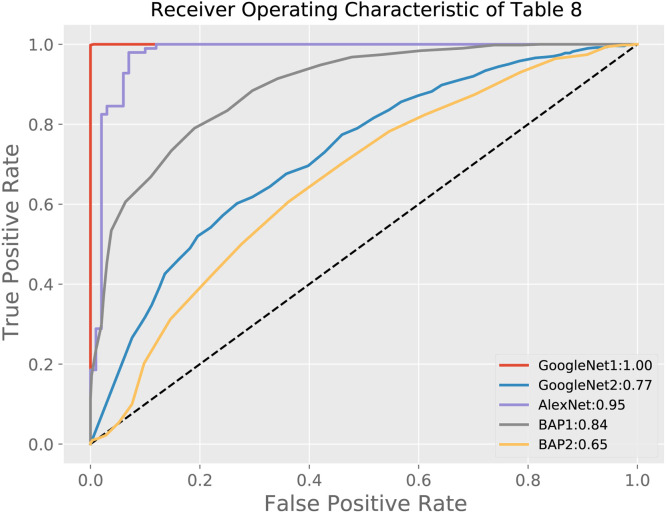
Regarding the supervised approaches, in this paper we consider some well-known feed-forward deep networks in the literature, i.e., the AlexNet (Krizhevsky et al., 2012) and the GoogLeNet (Szegedy et al., 2015). Specifically, AlexNet is composed of the cascade of five convolutional layers and three (dense) fully connected ones, while the GoogLeNet is more complicated since it is very deep and constructed by stacking three convolutional layers, nine inception modules, and two dense layers. An inception module is a particular layer obtained by concatenating several convolution operations with different filter sizes and a max pooling operation. Since these architectures are of supervised type, they have been trained by using both the CP and NCP classes in the training set, differently to our approach that has been trained only on the CP class in an unsupervised manner.
The results provided by AlexNet and GoogLeNet are shown in the bottom part of Table 8. As shown in the table, AlexNet performs worse than our approach, since it reaches only 71% accuracy, and similar other metrics. On the contrary, the performance of GoogLeNet are the same as the proposed idea, since it obtains a 100% accuracy. The ROC curves of all the considered approaches in Table 8 are shown in Fig. 7. However, we have to remark that this result is obtained with a deeper approach with a huge number of free parameters compared to the proposed approach, as shown in Table 9 that reports the number of trainable parameters and the training time (in minutes) for all the considered architectures. For the proposed approach, we have also to consider the time needed to compute the target histogram. The complexity of a single histogram computation depends on the length of the hidden feature vector plus the number of bins. Hence, the evaluation of the target histogram has an asymptotic computational complexity of . This provides an additional time of few seconds, which is negligible with respect to the training of the models. Once again, this consideration, along with the trade-off shown in Table 9, supports the proposed methodology. Moreover, since GoogLeNet is based on a supervised approach, we expect that it may work efficiently only on data belonging to the training classes, differently from our unsupervised approach, as investigated in the following subsection.
Fig. 7.
ROC curves of all the considered approaches in Table 8. The corresponding AUC values are reported in the legend. The red curve labeled “GoogleNet1” refers to results shown in the last raw of Table 8, while the blue curve labeled “GoogleNet2” refers to the experiment on the robustness of the GoogLeNet to unseen data reported in Table 11 of Section 5.4.
Table 9.
Computational complexity of the compared models. The training time (in minutes) refers to data sets composed of images of the size 300 × 200 pixels. Our proposed approach is the 3-Layer DDCAE.
| Model | Number of parameters | Training time [min] |
|---|---|---|
| Shallow Dense | 108 M | 145 |
| Shallow DCAE | 28 k | 5 |
| 2-Layer DDCAE | 426 k | 54 |
| 3-Layer DDCAE | 753 k | 96 |
| 4-Layer DDCAE | 2 M | 216 |
| AlexNet | 58 M | 223 |
| GoogLeNet | 6 M | 258 |
M stands for million of parameters, k for thousands.
5.3. Layer-wise training of DDCAEs
All the numerical results presented up to now refer to an end-to-end training approach in which each noisy input image is utilized to train end-to-end (i.e., one-shot) all the layers composing the underlying DDCAE. In order to evaluate the actual effectiveness of the considered end-to-end training approach, we have also implemented the more sophisticated layer-wise training approach described in Section 3.5 of Vincent et al. (2010). In a nutshell, according to this layer-wise approach (see, in particular, Fig. 3 of Vincent et al., 2010), a shallow denoising AE is considered at first, which is composed by only the first layer of the overall Deep AE. This shallow AE is trained on a set of noisy input training images (see Fig. 1 of Vincent et al., 2010), in order to learn its single-layer encoding function. Afterward, the learnt encoding function of the first-layer is used on a set of clean input training images, in order to train the second-layer of a two-layer denoising AE which is composed by the first two-layer of the underlying DDCAE (see the middle part of Fig. 3 of Vincent et al., 2010). So doing, the second-level encoding function is learnt. The described layer-wise procedure is replicated up to learn the encoding function of the upper-most layer of the underlying DDCAE (see the right part of Fig. 3 of Vincent et al., 2010). Finally, after finishing the layer-wise training procedure, the target histogram is numerically evaluated on a set of clean training images. The numerical results we have obtained by applying the described layer-wise procedure for training the proposed 3-Layer DDCAE of Table 2 are reported in Table 10 under the same experimental setup already considered in Section 5.1 for the (somewhat more challenging) Normal data set (see the columns labeled as “Normal” of Table 6 for the related numerical results).
Table 10.
Numerical performance of the proposed 3-Layer DDCAE of Table 2 when it is trained according to the layer-wise approach of Vincent et al. (2010) under the Normal dataset. The same simulation setup of Section 5.1 at is considered under the Normal datasets.
| Distance | Accuracy | Precision | Recall | F-measure | AUC |
|---|---|---|---|---|---|
| Kullback–Leibler | 99.61 | 0.9961 | 0.9960 | 0.9960 | 0.9992 |
| Bhattacharyya | 99.22 | 0.9923 | 0.9921 | 0.9922 | 0.9991 |
| Euclidean | 92.93 | 0.9401 | 0.9292 | 0.9346 | 0.9580 |
A comparison of the numerical results reported in Table 10 against the companion ones previously reported in Table 6 point out that the performance improvements arising from the utilization of the implemented layer-wise approach for training the considered DDCAE are, indeed, (very) marginal. Hence, since the implementation complexity of the described layer-wise procedure scales up (at least) linearly with the depth of the considered DDCAE, we conclude that, at least in the here considered application scenarios, the adopted end-to-end one-shot training procedure exhibits a more appealing performance-vs.-implementation complexity tradeoff.
5.4. Robustness of the considered approaches
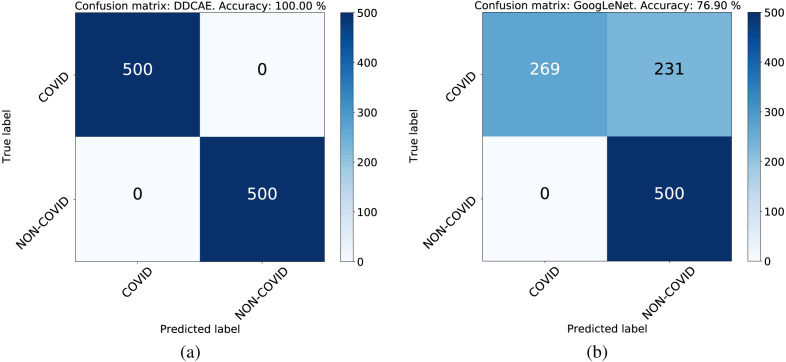
The aim of this subsection is to evaluate the robustness of the proposed DDCAE, compared to the GoogLeNet architecture, against unexpected image classes, like new variants of coronavirus and/or other classes never used in the training process.
In the following experiment, as in the previous subsection, the GoogLeNet has been trained by using both the CP and NCP classes while our DDCAE has been trained only on the CP class in an unsupervised manner. In order to check the robustness of these two considered approaches, both the architectures will be now tested on a test set composed of 500 normal scans and 500 CP scans. The obtained results are shown in Table 11, which clearly supports the effectiveness of our DDCAE. In fact, since our approach relies on an unsupervised training and a statistical distance measurement, it is able to identify anything that is different from the target class (in our case the CP class). On the other hand, we expect that the GoogLeNet can only assign previously unseen data randomly to the two output classes. This insight is confirmed by looking the confusion matrices in Fig. 8 that shows how GoogLeNet splits the unseen data with a ratio of 53.8% and 46.2% between the two classes, respectively. The ROC curve of GoogLeNet in this experiment is shown in the blue curve of Fig. 7.
Table 11.
Robustness of the proposed 3-Layer DDCAE and GoogLeNet to unseen data.
| Architecture | Accuracy | Precision | Recall | F-measure | AUC |
|---|---|---|---|---|---|
| 3-Layer DDCAE | 100.00 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| GoogLeNet | 76.90 | 1.0000 | 0.5380 | 0.6996 | 0.7690 |
Fig. 8.
The confusion matrices in the case of unseen data: (a) DDCAE and (b) GoogLeNet.
5.5. Performance comparisons with shallow classifiers working on Haralick–Zernicke input features
The recent contribution in Gomes et al. (2020) supports the conclusion that good accuracy performance in the diagnosis of COVID-19 pathologies may be obtained by Support Vector Machine (SVM), Multi-Layer Perceptron (MLP) and Random Forest (RF)-based supervised shallow classifiers (Alpaydin, 2014), working on input features that are obtained by extracting suitable Haralick and/or Zernicke moments from X-ray images. Hence, motivated by the interesting results reported in Gomes et al. (2020) on X-ray images, goal of this section is to numerically check the accuracy performance of such of kinds of simple-to-implement ML models for the binary classification of Pneumonia-vs.-COVID CT images extracted by the dataset of Table 3. Being the training of the considered shallow classifiers of supervised-type by design, the dataset of Table 3 has been augmented by including 3500 training plus 700 validation COVID-19 images picked up from the (previously described) CT-2 A dataset. Table 12 describes the main parameters and related taxonomy of the implemented shallow classifiers.
Table 12.
Main parameters of the implemented supervised shallow classifiers: SVMs with 2-degree (SVM-2D) and 3-degree (SVM-3D) polynomial, and Radial Basis Function (SVM-RBF) kernels; MLPs equipped with single hidden layers composed by 50 (MLP-50), 100 (MLP-100), and 200 (MLP-200) neurons; RFs composed by 100 (RF-100), 500 (RF-500), and 1000 (RF-1000) binary trees.
| Model | Main parameters |
|---|---|
| 2-degree polynomial kernel (SVM-2D) | |
| SVM | 3-degree polynomial kernel (SVM-3D) |
| RBF kernel (SVM-RBF) | |
| 50 neurons in the hidden layer (MLP-50) | |
| MLP | 100 neurons in the hidden layer (MLP-100) |
| 200 neurons in the hidden layer (MLP-200) | |
| RF with 100 randomly generated binary trees (RF-100) | |
| RB | RF with 500 randomly generated binary trees (RF-500) |
| RF with 1000 randomly generated binary trees (RF-1000) | |
According to Gomes et al. (2020), in the carried out tests, suitable sets of Haralick (Haralick et al., 1973) and Zernicke (Kan & Srinath, 2001) moments extracted by the (previously described) available sets of CP and NCP scans are used as input features to the shallow classifiers of Table 12. Specifically, four (256 × 256) matrices of the co-occurrences of the gray levels of row-wise, column-wise and diagonal-wise adjacent pixels are extracted from each available CT scans and, then, they are employed as Haralick input features to the classifiers of Table 12 (see the seminal paper in Haralick et al., 1973) for an in-depth presentation of the Haralick feature extraction). As detailed in Gomes et al. (2020), the corresponding Zernicke moments are obtained by computing the coefficients of the orthogonal projection of each image onto an orthogonal basis composed by a set polar functions , with each basis function labeled by a pair of non-negative integer indexes (see, for example, Eqs. (1) and (2) of Gomes et al. (2020)). In the carried out tests, the same set of index pairs reported in Table 3 of Gomes et al. (2020) is considered, so to associate 64 Zernicke moments to each processed image.
The top (resp., bottom) part of Table 13 shows the numerically evaluated results that have been obtained by running the shallow supervised binary classifiers of Table 12 when the described Haralick (resp., Haralick-plus-Zernicke) moments of the CP and NCP images of the dataset are used as input features.
Table 13.
Numerical results of the shallow models of Table 12 when they are used to classify the Haralick and Haralick-plus-Zernicke input features extracted from the CP and NCP scans of the test dataset of Table 3.
| Model | Accuracy | Precision | Recall | F-measure | AUC | Training time |
|---|---|---|---|---|---|---|
| Haralick input features | ||||||
| SVM-2D | 93.00 | 0.9386 | 0.9300 | 0.9343 | 0.9940 | 1.6 |
| SVM-3D | 90.20 | 0.9181 | 0.9200 | 0.9190 | 0.9910 | 1.8 |
| SVM-RBF | 91.20 | 0.9252 | 0.9120 | 0.9186 | 0.9700 | 1.5 |
| MLP-50 | 61.60 | 0.6160 | 0.6160 | 0.6159 | 0.6600 | 6.4 |
| MLP-100 | 70.10 | 0.8083 | 0.7010 | 0.7508 | 0.8000 | 9.3 |
| MLP-200 | 76.10 | 0.7947 | 0.7610 | 0.7775 | 0.7800 | 12.2 |
| RF-100 | 68.50 | 0.7652 | 0.6850 | 0.7229 | 0.8500 | 6.4 |
| RF-500 | 68.70 | 0.7665 | 0.7210 | 0.7431 | 0.9600 | 31.8 |
| RF-1000 | 68.90 | 0.7676 | 0.6890 | 0.7262 | 0.9620 | 63.5 |
| Haralick-plus-Zernicke input features | ||||||
| SVM-2D | 93.40 | 0.9417 | 0.9340 | 0.9378 | 0.9950 | 1.7 |
| SVM-3D | 92.70 | 0.9363 | 0.9270 | 0.9316 | 0.9930 | 2.4 |
| SVM-RBF | 91.70 | 0.9788 | 0.9170 | 0.9469 | 0.9600 | 1.6 |
| MLP-50 | 76.00 | 0.8132 | 0.7600 | 0.7857 | 0.7900 | 7.5 |
| MLP-100 | 77.40 | 0.7874 | 0.7740 | 0.7806 | 0.7200 | 10.8 |
| MLP-200 | 78.30 | 0.8123 | 0.7830 | 0.7974 | 0.7800 | 13.2 |
| RF-100 | 71.10 | 0.7852 | 0.7110 | 0.7463 | 0.9600 | 7.3 |
| RF-500 | 72.00 | 0.7903 | 0.7200 | 0.7535 | 0.9650 | 36.5 |
| RF-1000 | 72.10 | 0.7893 | 0.7210 | 0.7536 | 0.9680 | 74.0 |
Accuracy is in percentage while the training time is measured in seconds.
A comparative examination of the numerical results of Table 13 leads to two main conclusions. First, under both the tested Haralick and Haralick-plus-Zernicke input feature sets, the SVM classifier with degree-2 polynomial kernel attains the best performance metrics. In contrast, at least in the carried out tests, the implemented RF-based classifiers exhibit the worst performance. Second, a comparison of the top and bottom parts of Table 13 points out that, in all carried out tests, performance improvements are experienced by concatenating the Haralick input features to the corresponding Zernicke ones. Specifically, the experienced accuracy improvement is noticeable and around 14.5% for the (less-performing) MLP-50 classifier, while it reduces to 0.4% for the (most performing) SVM-2D model.
Finally, some insights may be acquired by comparing the performance metrics of the proposed DDCAE models of Table 6 against the corresponding ones of the shallow models of Table 13. Specifically, the comparison points out that the average accuracies of the most performing DDCAE models (namely, the proposed DDCAE1 and DDCAE2 models equipped with the KL and/or the Bhattacharyya distance of Table 6) reach 100%, while the average accuracy of the most performing tested shallow classifier (namely, the SVM-2D classifier of Table 13) remains limited up to 93%. Furthermore, since the considered shallow classifiers are supervised models, in principle, they are prone to the same robustness issues already pointed out in Section 5.4. Obviously, these pros of the proposed DDCAE models are counterbalanced by some cons arising from considerations on the corresponding implementation complexity. The training times of the shallow models provided in the last column of Table 13 show that all the compared models are faster than the corresponding ones of the proposed DDCAE models (see Table 9). This is due to the facts that these models are shallow and operate on input feature vectors of limited length, as detailed in Gomes et al. (2020). However, although these shallow models can be quickly trained, we remark that the obtained accuracies of Table 13 are not suitable for a reliable system of automatic diagnosis. Hence, all in all, we conclude that the proposed DDCAE models may represent an appealing solution in application scenarios in which very high accuracy (we say, accuracy over than 95%) and robustness against unseen input features are the main quality-of-service requirements. In fact, although the training time of the proposed DDCAE is higher than the corresponding ones of the tested shallow methods, the resulting test accuracy-vs.-training time tradeoffs compare favorably with respect to competing deep methods with similar test accuracies.
5.6. Performance sensitivity on the size of the training dataset
The aim of this last subsection is to evaluate the sensitivity of the proposed DDCAE and related compared approaches to the size of the training set. Specifically, the COVIDx CT-2 A dataset described in Section 4.1 contains about 194,922 scans partitioned in Normal, CP, and NCP classes. The available images have already split in training, validation, and test sets. The training and validation sets are composed of a variable number of images, and they provide at least 25,000 and 7000 scans for each class, respectively. For uniformity between all the considered classes and hardware constraints, in the carried out tests, we train again the proposed architecture and the baseline Deep ones (i.e., AlexNet and GoogLeNet) on the dataset by selecting 25,600 training scans and 6400 validation images. Since our proposed approaches are unsupervised, they have been trained only on the reference class, while the compared supervised deep architectures have been trained on both the selected reference class and the COVID-19 one.
Results in terms of the considered metrics for the proposed DDCAEs remain more or less the same and present numerical values similar to those presented in Table 6. Hence, we do not explicitly report these results in a tabular form. Once again, the proposed DDCAE1 and DDCAE2 models using the KL and/or the Bhattacharyya distance attain top accuracies of 100% and similar metrics.
Results of AlexNet, in this case, show an enhanced performance since now AlexNet reaches an accuracy of 100%, and a precision, recall, F-measure, and AUC of 1.000. Performance indexes of GoogLeNet remain unchanged and are the same as in the last line of Table 8. The performance improvement experienced by AlexNet is due to its huge number of free parameters (see Table 9): using the partial dataset as in Table 3 is not sufficient for avoiding underfitting phenomena in the training phase of AlexNet.
Motivated by these considerations, we stress the effectiveness of the proposed approach. In fact, the proposed 3-Layer DDCAE is able to reach the 100% of accuracy, like also AlexNet and GoogLeNet, but with a smaller number of parameters. This means, in turn, that it can be trained quickly by using limited datasets. These aspects surely represent a great added value to tools for the automatic clinical diagnosis.
6. Conclusion
In this paper, we propose an unsupervised approach to detect the new coronavirus pneumonia from CT scans. Since the number of these images is usually limited, we train a deep denoising convolutional autoencoder (DDCAE) on some target classes (normal and common pneumonia) and construct a robust statistical representation by evaluating the histogram of the hidden features averaged over all the training scans. A suitable distribution distance is then used to compute how far this target histogram is from the corresponding histogram evaluated for an unknown test scan: if this distance is above a threshold the test image is classified as anomaly, i.e. affected by the COVID-19 disease, otherwise it is classified the same as the target class. Some numerical results evaluated on an open-source dataset, known in literature, demonstrate the effectiveness of the proposed idea, since it is able to obtain the top 100% of the considered metrics (accuracy, precision, recall, F-measure and AUC) with a limited computational complexity, outperforming the corresponding state-of-the-art approaches.
In future works, we aim at extending our methodology towards different types of medical images, other than CT, and/or different diseases. We expect, in fact, that the automatic screening of pathological images can take a great advantage by the simplicity of our methodology, in both of the resulting accuracy and prediction time. A second line of future research can be addressed towards to use of Generative Adversarial Networks (GANs) for generating additional examples in the case of new variants of COVID-19, in order to be fast in the automatic discrimination of these scans without awaiting the construction of sufficiently copious dataset. Finally, a third research hint can be focused on the implementation of the proposed methodology in a distributed Cloud/Fog networked technological platforms (Baccarelli et al., 2021, Baccarelli et al., 2017), in order to produce in fast and reliable clinical responses by exploiting the low-delay and (possibly, adaptive Baccarelli & Cusani, 1996 and/or smart-antenna empowered Baccarelli and Biagi, 2003, Baccarelli et al., 2007) capability of virtualized Fog computing infrastructures in wireless-oriented application environments.
CRediT authorship contribution statement
Michele Scarpiniti: Writing – original draft, Conceptualization, Methodology, Investigation, Software. Sima Sarv Ahrabi: Data curation, Software, Validation, Visualization. Enzo Baccarelli: Methodology, Supervision, Funding acquisition. Lorenzo Piazzo: Data curation, Investigation. Alireza Momenzadeh: Reviewing and editing, Visualization.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This work has been supported by the projects: “SoFT: Fog of Social IoT” funded by Sapienza University of Rome Bando 2018 and 2019, Italy; “End-to-End Learning for 3D Acoustic Scene Analysis (ELeSA)” funded by Sapienza University of Rome Bando Acquisizione di medie e grandi attrezzature scientifiche 2018, Italy; and, “DeepFog — Optimized distributed implementation of Deep Learning models over networked multitier Fog platforms for IoT stream applications” funded by Sapienza University of Rome Bando 2020, Italy .
Footnotes
It can be downloaded from: https://www.kaggle.com/hgunraj/covidxct.
References
- Adams H.J.A., Kwee T.C., Yakar D., Hope M.D., Kwee R.M. Chest CT imaging signature of coronavirus disease 2019 infection: In pursuit of the scientific evidence. Chest Infections. 2020;158:1885–1895. doi: 10.1016/j.chest.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiello M., Cavaliere C., D’Albore A., Salvatore M. The challenges of diagnostic imaging in the era of big data. Journal of Clinical Medicine. 2019;8:316. doi: 10.3390/jcm8030316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Ameen Z., Sulong G. Prevalent degradations and processing challenges of computed tomography medical images: A compendious analysis. International Journal of Grid and Distributed Computing. 2016;9:107–118. doi: 10.14257/ijgdc.2016.9.10.10. [DOI] [Google Scholar]
- Alain G., Bengio Y. What regularized auto-encoders learn from the datagenerating distribution. Journal of Machine Learning Research. 2014;15:3743–3773. [Google Scholar]
- Alpaydin E. 3rd ed. Mit Press; Cambridge, MA: 2014. Introduction to machine learning. [Google Scholar]
- Amarbayasgalan T., Pham V.H., Theera-Umpon N., Ryu K.H. Unsupervised anomaly detection approach for time-series in multi-domains using deep reconstruction error. Simmetry. 2020;12:1251. doi: 10.3390/sym12081251. [DOI] [Google Scholar]
- Baccarelli E., Biagi M. In: IFIP international conference on personal wireless communications. Conti M., Giordano S., Gregori E., Olariu S., editors. Vol. 2775. 2003. Optimized power allocation and signal shaping for interference-limited multi-antenna “Ad Hoc” networks; pp. 138–152. (Lecture notes in computer science). [DOI] [Google Scholar]
- Baccarelli E., Biagi M., Pelizzoni C., Cordeschi N. Optimized power-allocation for multiantenna systems impaired by multiple access interference and imperfect channel estimation. IEEE Transactions on Vehicular Technology. 2007;56:3089–3105. doi: 10.1109/TVT.2007.900514. [DOI] [Google Scholar]
- Baccarelli E., Cusani R. Recursive Kalman-type optimal estimation and detection of hidden Markov chains. Signal Processing. 1996;51:55–64. doi: 10.1016/0165-1684(96)00030-8. [DOI] [Google Scholar]
- Baccarelli E., Scarpiniti M., Momenzadeh A., Sarv Ahrabi S. Learning-in-the-fog (LiFo): Deep learning meets fog computing for the minimum-energy distributed early-exit of inference in delay-critical IoT realms. IEEE Access. 2021;9:2571–25757. doi: 10.1109/ACCESS.2021.3058021. [DOI] [Google Scholar]
- Baccarelli E., Vinueza Naranjo P.G., Shojafar M., Scarpiniti M. Q*: Energy and delay-efficient dynamic queue management in TCP/IP virtualized data centers. Computer Communications. 2017;102:89–106. doi: 10.1016/j.comcom.2016.12.010. [DOI] [Google Scholar]
- Bourlard H., Kamp Y. Auto-association by multilayer perceptrons andsingular value decomposition. Biological Cybernetics. 1988;59:291–294. doi: 10.1007/BF00332918. [DOI] [PubMed] [Google Scholar]
- Chandola V., Banerjee A., Kumar V. Anomaly detection: A survey. ACM Computing Surveys. 2009;41:15. doi: 10.1145/1541880.1541882. [DOI] [Google Scholar]
- Chandra T.B., Verma K., Singh B.K., Jain D., Netam S.S. Coronavirus disease (COVID-19) detection in chest X-ray images using majority voting based classifier ensemble. Expert Systems with Applications. 2021;165 doi: 10.1016/j.eswa.2020.113909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S.-G., Chen J.-Y., Yang Y.-P., Chien C.-S., Wang M.-L., Lin L.-T. Use of radiographic features in COVID-19 diagnosis: Challenges and perspectives. Journal of the Chinese Medical Association. 2020;83:644–647. doi: 10.1097/JCMA.0000000000000336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Shi X., Zhang Y., Wu D., Guizani M. Deep features learning for medical image analysis with convolutional autoencoder neural network. IEEE Transactions on Big Data. 2017:1–10. doi: 10.1109/TBDATA.2017.2717439. [DOI] [Google Scholar]
- Elmuogy S., Hikal N.A., Hassan E. An efficient technique for CT scan images classification of COVID-19. Journal of Intelligent & Fuzzy Systems. 2021;40:5225–5238. doi: 10.3233/JIFS-201985. [DOI] [Google Scholar]
- Fan D.-P., Zhou T., Ji G.-P., Zhou Y., Chen G., Fu H. Inf-Net: Automatic COVID-19 lung infection segmentation from CT images. IEEE Transactions on Medical Imaging. 2020;39:2626–2637. doi: 10.1109/TMI.2020.2996645. [DOI] [PubMed] [Google Scholar]
- Gomes J.C., de Barbosa V.A., Santana M.A., Bandeira J., Valença M.J.S., de Souza R.E. IKONOS: An intelligent tool to support diagnosis of COVID-19 by texture analysis of X-ray images. Research on Biomedical Engineering. 2020 doi: 10.1007/s42600-020-00091-7. [DOI] [Google Scholar]
- Gomes J.C., Masood A.I., de S. Silva L.H., da Cruz Ferreira J.R.B., Freire Junior A.A.F., dos Santos Rocha A.L. COVID-19 diagnosis by combining RT-PCR and pseudo-convolutional machines to characterize virus sequences. Scientific Reports. 2021;11 doi: 10.1038/s41598-021-90766-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow I., Bengio Y., Courville A. MIT Press; Cambridge, MA: 2016. Deep learning. [Google Scholar]
- Gunraj H., Sabri A., Koff D., Wong A. 2021. COVID-Net CT-2: Enhanced deep neural networks for detection of COVID-19 from chest CT images through bigger, more diverse learning. arXiv:2101.07433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunraj H., Wang L., Wong A. COVIDNet-CT: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest CT images. Frontiers in Medicine. 2020;7 doi: 10.3389/fmed.2020.608525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer M.M., Raptis C.A., Henry T.S., Shah A., Bhalla S., Hope M.D. Challenges in the interpretation and application of typical imaging features of COVID-19. The LANCET Respiratory Medicine. 2020;8:534–536. doi: 10.1016/S2213-2600(20)30233-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haralick R.M., Shanmungam S., Dinstein I. Textural features for images classification. IEEE Transactions on Systems, Man, and Cybernetics. 1973;SMC-3:610–621. doi: 10.1109/TSMC.1973.4309314. [DOI] [Google Scholar]
- Hsieh J. 2nd ed. John Wiley & Sons; 2009. Computed tomography: principles, design, artifacts, and recent advances. [Google Scholar]
- Ioffe S., Szegedy C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. Lille, France; 2015. pp. 448–456. [Google Scholar]
- Ismael A.M., Şengür A. The investigation of multiresolution approaches for chest X-ray image based COVID-19 detection. Health Information Science and Systems. 2020;8:29. doi: 10.1007/s13755-020-00116-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismael A.M., Şengür A. Deep learning approaches for COVID-19 detection based on chest X-ray images. Expert Systems with Applications. 2021;164 doi: 10.1016/j.eswa.2020.114054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan, C., & Srinath, M. D. (2001). Combined features of cubic B-spline wavelet moments and Zernicke moments for invariant character recognition. In Proceedings international conference on information technology: coding and computing (pp. 511–515). Las Vegas, NV, USA. 10.1109/ITCC.2001.918848. [DOI]
- Kingma, D. P., & Ba, J. (2015). Adam: A method for stochastic optimization. In 3rd international conference for learning representations (pp. 1–15). San Diego, USA. URL: https://arxiv.org/abs/1412.6980.
- Krizhevsky A., Sutskever I., Hinton G.E. 25th international conference on neural information processing systems. 2012. ImageNet classification with deep convolutional neural networks; pp. 1097–1105. [DOI] [Google Scholar]
- Kullback S. Dover Pubns; Mineola, N.Y., USA: 1997. Information theory and statistics. [Google Scholar]
- Kwee T.C., Kwee R.M. Chest CT in COVID-19: What the radiologist needs to know. RadioGraphics. 2020;40:1848–1865. doi: 10.1148/rg.2020200159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerum T.V., Aaløkken T.M., Brønstad E., Aarli B., Ikdahl E., Lund K.M.A. Dyspnoea, lung function and CT findings three months after hospital admission for COVID-19. European Respiratory Journal. 2020;57 doi: 10.1183/13993003.03448-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Fu Z., Xu J. Stacked-autoencoder-based model for COVID-19 diagnosis on CT images. Applied Intelligence: The International Journal of Artificial Intelligence, Neural Networks, and Complex Problem-Solving Technologies. 2020 doi: 10.1007/s10489-020-02002-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masci J., Meier U., Cireşan D., Schmidhuber J. Proceedings of the 21st international conference on artificial neural networks. Vol. 6791. Springer; 2011. Stacked convolutional auto-encoders for hierarchical feature extraction; pp. 52–59. (Lecture notes in computer science). [DOI] [Google Scholar]
- Mishra A.K., Das S.K., Roy P., Bandyopadhyay S. Identifying COVID19 from chest CT images: A deep convolutional neural networks based approach. Journal of Healthcare Engineering. 2020;2020 doi: 10.1155/2020/8843664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsahin I., Sekeroglu B., Musa M.S., Mustapha M.T., Ozsahin D.U. Review on diagnosis of COVID-19 from chest CT images using artificial intelligence. Computational and Mathematical Methods in Medicine. 2020;2020 doi: 10.1155/2020/9756518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman S., Sarker S., Miraj M.A.A., Nihal R.A., Haque A.K.M.N., Noman A.A. Deep learning–driven automated detection of COVID-19 from radiography images: A comparative analysis. Cognitive Computation. 2021 doi: 10.1007/s12559-020-09779-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saood A., Hatem I. COVID-19 lung CT image segmentation using deep learning methods: U-Net versus SegNet. BMC Medical Imaging. 2021;21:19. doi: 10.1186/s12880-020-00529-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarv Ahrabi S., Scarpiniti M., Baccarelli E., Momenzadeh A. An accuracy vs. complexity comparison of deep learning architectures for the detection of COVID-19 disease. Computation. 2021;9:3. doi: 10.3390/computation9010003. [DOI] [Google Scholar]
- Shah V., Keniya R., Shridharani A., Punjabi M., Shah J., Mehendale N. Diagnosis of COVID-19 using CT scan images and deep learning techniques. Emergency Radiology. 2021:1–9. doi: 10.1007/s10140-020-01886-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S. Drawing insights from COVID-19-infected patients using CT scan images and machine learning techniques: A study on 200 patients. Environmental Science and Pollution Research. 2020;27:37155–37163. doi: 10.1007/s11356-020-10133-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen D., Wu G., Suk H. Deep learning in medical image analysis. Annual Review of Biomedical Engineering. 2017;19:221–248. doi: 10.1146/annurev-bioeng-071516-044442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W., Zhou M., Yang F., Yu D., Dong D., Yang C. Multi-crop convolutional neural networks forl ung nodule malignancy suspiciousness classification. Pattern Recognition. 2017;61:663–673. doi: 10.1016/j.patcog.2016.05.029. [DOI] [Google Scholar]
- Silva P., Luz E., Silva G., Moreira G., Silva R., Lucio D. COVID-19 detection in CT images with deep learning: A voting-based scheme and cross-datasets analysis. Informatics in Medicine Unlocked. 2020;20 doi: 10.1016/j.imu.2020.100427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetens P. 2nd ed. Cambridge University Press; Cambridge, UK: 2009. Fundamentals of medical imaging. [DOI] [Google Scholar]
- Szegedy, C., Liu, W., Jia, Y., Sermanet, P., Reed, S., & Anguelov, D. (2015). Going deeper with convolutions. In 2015 IEEE conference on computer vision and pattern recognition. Boston, MA, USA. 10.1109/CVPR.2015.7298594. [DOI]
- Tan W., Liu P., Li X., Liu Y., Zhou Q., Chen C. Classification of COVID-19 pneumonia from chest CT images based on reconstructed super-resolution images and VGG neural network. Health Information Science and Systems. 2021;9:10. doi: 10.1007/s13755-021-00140-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal P.L., de Moura J., Novo J., Ortega M. Multi-stage transfer learning for lung segmentation using portable X-ray devices for patients with COVID-19. Expert Systems with Applications. 2021;173 doi: 10.1016/j.eswa.2021.114677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent P., Larochelle H., Bengio Y., Manzagol P.-A. Proceedings of the 25th international conference on machine learning. 2008. Extracting and composing robust features with denoising autoencoders; pp. 1096–1103. [DOI] [Google Scholar]
- Vincent P., Larochelle H., Lajoie I., Bengio Y., Manzagol P.A. Stacked denoising autoencoders: Learning useful representationsina deep network with a local denoising criterion. Journal of Machine Learning Research. 2010;11:3371–3408. [Google Scholar]
- Xu J., Xiang L., Liu Q., Gilmore H., Wu J., Tang J. Stacked sparse autoencoder (SSAE) for nuclei detection on breast cancer histopathology images. IEEE Transactions on Medical Imaging. 2016;35:119–130. doi: 10.1109/TMI.2015.2458702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Q., Xiao L., Liu P., Zhou S.K. Label-free segmentation of COVID-19 lesions in lung CT. IEEE Transactions on Medical Imaging. 2021 doi: 10.1109/TMI.2021.3066161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Academic Press; 2017. Deep learning for medical image analysis. [Google Scholar]