Abstract
A specific serotype, O3:K6, of Vibrio parahaemolyticus has recently been causing epidemics of gastroenteritis in Southeast Asia, Japan, and North America. To examine whether the new O3:K6 strains possess characteristics that may exacerbate outbreaks, we compared V. parahaemolyticus O3:K6 strains with non-O3:K6 strains using strains isolated from individuals with traveler's diarrhea at Kansai Airport Quarantine Station, Osaka, Japan. All 24 O3:K6 strains possessed a common plasmid, pO3K6 (DNA size, 8,782 bp, with 10 open reading frames [ORFs]). The gene organization of pO3K6 was similar to that of Vf33, a filamentous phage previously described in V. parahaemolyticus. We isolated a phage (phage f237) from the culture supernatant of V. parahaemolyticus O3:K6 strain KXV237, which formed a turbid plaque on an indicator strain. The genome of f237 was single-stranded DNA, and the double-stranded DNA obtained by treatment of the genome with DNA polymerase was identical to that of pO3K6 when analyzed by agarose gel electrophoresis after HindIII digestion. Furthermore, the N-terminal amino acid sequence of the f237 major coat protein was found in ORF4 of pO3K6. Our results showed that pO3K6 is a replicative form of f237. Among the ORFs found in the f237 genome, the sequence of ORF8 had no significant homology to those of any proteins in databases. ORF8 was located on a region corresponding to the distinctive region of Vf33, and its G+C content was apparently lower than that of the remaining DNA sequence of f237. By colony hybridization, ORF8 was detected only in O3:K6 strains isolated since 1996 and was not found in O3:K6 strains isolated before 1996 and clinical V. parahaemolyticus strains other than those of serotype O3:K6. Thus, this study shows that f237 is exclusively associated with recent V. parahaemolyticus O3:K6 strains. The ORF8 gene can be a useful genetic marker for the identification of the recently widespread O3:K6 strains of V. parahaemolyticus.
Vibrio parahaemolyticus is a gram-negative bacterium that causes seafood-related gastroenteritis and traveler's diarrhea (8). The thermostable direct hemolysin (TDH) produced by V. parahaemolyticus has been found to be closely associated with the enteropathogenicity of the organism (8). In late 1980s, cases of gastroenteritis due to V. parahaemolyticus strains that showed hemolytic activity but that did not produce TDH were found (9). The hemolysin produced by the strains, which was immunologically related to but not identical to TDH, was named TDH-related hemolysin (TRH). Currently, both TDH and TRH are regarded as important virulence factors for V. parahaemolyticus (8).
Since 1996, gastroenteritis caused by a specific serotype, O3:K6, of V. parahaemolyticus has been widespread in Southeast Asia, East Asia including Japan, and North America (1, 3, 17). Such an epidemic specifically caused by a particular serotype of V. parahaemolyticus has not been reported previously. The V. parahaemolyticus O3:K6 strains isolated since 1996 produce TDH but not TRH, while the O3:K6 strains isolated before 1996 produce only TRH (9, 17). The new O3:K6 strains isolated since 1996 were reported to be derived from a single clone (17). It has remained unclear, however, why the new V. parahaemolyticus O3:K6 strains rather than other strains are implicated so strongly in epidemics.
To discover which bacterial factors of the new O3:K6 strains may be implicated in an epidemic that they cause, we compared the characteristics of new V. parahaemolyticus O3:K6 strains to those of other clinical V. parahaemolyticus isolates. Our results show that a specific filamentous phage is associated with the new V. parahaemolyticus O3:K6 strains.
MATERIALS AND METHODS
Bacterial strains and media.
V. parahaemolyticus strains (99 in total) were isolated from patients with traveler's diarrhea arrived at Kansai Airport Quarantine Station, Osaka, Japan, from Southeast Asian countries (mainly Thailand, Singapore, and the Philippines) between January and August 1996. Fecal sample collection and isolation of V. parahaemolyticus from the samples were carried out at the time when the patients arrived at the airport. Among the isolates, 24 strains were of the O3:K6 serotype; during the sampling period these were isolated only after April. After evaluation with the TDH Detection Kit by reversed passive latex agglutination (Denka Seiken, Tokyo, Japan), all of the O3:K6 isolates were found to produce TDH. The isolated strains were stored at −80°C in Luria-Bertani (LB) broth (1% tryptone [Difco Laboratories, Detroit, Mich.], 0.5% yeast extract [Difco], 0.5% NaCl) supplemented with 15% glycerol until use. One of the isolated V. parahaemolyticus O3:K6 strains, KXV237, was used for phage preparation, and V. parahaemolyticus strain KXV191 (serotype O4:K12) was used as an indicator strain to detect the phage. To isolate plasmids, all the strains were grown at 37°C with shaking for 8 h in LB broth supplemented with 3% NaCl. Heart infusion broth (Difco) supplemented with 3% NaCl (3% NaCl–HIB) was used for phage isolation.
DNA techniques.
General DNA techniques such as treatment of DNA with nucleases (DNase, RNase, and mung bean nuclease) were performed as described previously (20). These nucleases were purchased from Takara Shuzo (Kyoto, Japan). Plasmid DNA was isolated by the method of Birnboim and Doly (2). Southern hybridization techniques were carried out as described previously (11). The hybridization temperature was 42°C.
Preparation of DNA templates for sequencing.
We sonicated 10 μg of the plasmid DNA with a sonic disrupter (Tomy Seiko, Co., Ltd., Tokyo, Japan) at 3.6 W for 30 s. Then, the sonicated DNA was blunt ended with a DNA blunting kit (Takara Shuzo) and was fractionated by polyacrylamide gel electrophoresis on a 6% polyacrylamide gel. A gel slice containing DNA was eluted by immersing the gel in MG elution buffer (500 mM ammonium acetate, 10 mM magnesium acetate, 1 mM EDTA, 0.1% [wt/vol] sodium dodecyl sulfate [SDS]) overnight at 37°C. The recovered DNA was ligated to the SmaI site of vector M13mp18 DNA (26), which had been pretreated with bacterial alkaline phosphatase, and was introduced into competent Escherichia coli DH11S cells (13, 18) with an electroporator. These cells were plated with LB soft agar containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; 100 μg/ml; Wako Pure Chemical Industries, Ltd., Osaka, Japan), isopropyl-β-d-thiogalactopyranoside (1 mM), and indicator cells to screen the transfectants. The isolated plaques were inoculated into 100-fold-diluted suspensions of overnight cell cultures (incubated at 37°C with shaking for 12 h) and grown for 3 h at 37°C. These cultures were used as templates for PCR. The DNA inserts in the phage clones were amplified by PCR with a TaKaRa EX Taq kit (Takara Shuzo) with two primers flanking the cloning site: primer 1, 5′-TCCGGCTCGTATGTTGTGTGGA-3′; primer 2, 5′-GTGCTGCAAGGCGATTAAGTTGG-3′. The PCR products were treated with a PCR product presequencing kit containing exonuclease I and shrimp alkaline phosphatase (Amersham Japan, Ltd., Tokyo, Japan).
DNA sequencing, sequence assembly, and data analysis.
The sequencing reactions were performed by using a Dye Primer Cycle Sequencing FS Ready Reaction Kit and a thermal cycler (GeneAmp PCR System 9600; Perkin-Elmer Japan Co., Ltd., Tokyo, Japan). DNA sequences were analyzed with an ABI Prism 373XL DNA sequencer (Perkin-Elmer). Sequences were assembled by using Factura, version 2.0, feature identification software and Autoassembler DNA sequence assembly software, version 2.0 (both were from Perkin-Elmer). To complete the sequencing of plasmid pO3K6, 112 shotgun clones were sequenced by using −21M13 and/or M13Rev primers (Perkin-Elmer). The BLAST program was used to analyze and search for homologous genes from databases, and the DNASIS program (Hitachi Software Engineering, Yokohama, Japan) was used to determine the G+C contents and to search for motifs.
Phage isolation and assay for lytic activity.
V. parahaemolyticus KXV237 was inoculated into 3% NaCl–HIB and incubated at 37°C for 15 h with shaking. The supernatant was filtered through a 0.22-μm-pore-size membrane (PES membrane; Iwaki Glass, Chiba, Japan) and was salted out with 40% saturated ammonium sulfate. Then, the precipitate was dialyzed to SM buffer (0.1 M NaCl, 0.01% gelatine, 8 mM MgSO4 · 7H2O, 50 mM Tris-HCl [pH 7.5]). After this, the solution was fractionated by CsCl step-gradient (d = 1.5, 1.3, and 1.15 g/cm3) centrifugation (77,000 × g, 2 h). The fractions that showed lytic activity when the plaque formation test was performed were collected as a phage solution and were dialyzed against SM buffer. We carried out the plaque formation test as follows: a 0.2-ml culture of indicator strain KXV191 that had been incubated at 37°C for 6 h in LB broth was mixed well with 10 ml of soft agar (1% tryptone, 0.5% yeast extract, 3% NaCl, 0.6% agar) at 45°C and spread onto LB agar (LB broth with 1.5% agar) containing 10 mM MgCl2. Ten microliters of phage solution was then spotted onto the plate. Plaque formation was observed after incubation at 37°C for 6 h. To isolate the phage DNA we precipitated the phage solution with 4% polyethylene glycol 6000 and 0.5 M NaCl and allowed the mixture to stand at room temperature for 30 to 60 min. The precipitate was separated by centrifugation and was then subjected to phenol-chloroform extraction and ethanol precipitation as described previously (20).
dsDNA synthesis.
Phage f237 genomic DNA was treated with T4 polymerase to obtain double-stranded DNA (dsDNA). The primers used to synthesize the dsDNA were primer 8S (5′-GTTCGCATACAGTTGAGG-3′), which was derived from the sense strand of ORF8, or primer 8A (5′-AAGTACAGCAGGAGTGAG-3′), which was derived from the antisense strand of ORF8.
Electron microscopy.
The purified phage was negatively stained with 4% (wt/vol) uranyl acetate and was observed with a Hitachi H-7100 microscope. Electron microscopic analysis of the phage was performed on a carbon-coated Formvar grid.
SDS-PAGE and amino acid sequencing.
SDS-polyacrylamide gel electrophoresis (PAGE) was performed as described by Okajima et al. (16). The N-terminal amino acid sequence was analyzed by automated Edman degradation on an Applied Biosystems (Foster City, Calif.) 473A protein sequencer.
Colony hybridization.
To determine the distributions of ORF7 and ORF8 in V. parahaemolyticus strains, colony hybridization was carried out as described previously (11). The hybridization temperature was 42°C. Probes for ORF7 and ORF8 were prepared by PCR with oligonucleotide primers that were synthesized on the basis of the sequences of the open reading frames (ORFs). Each probe was labeled by a random primer-extension method with a digoxigenin DNA labeling kit (Boehringer Mannheim GmbH, Mannheim, Germany).
Nucleotide sequence accession number.
The nucleotide sequence data reported in this work will appear in the EMBL, Genbank, and DDBJ Sequence Databases under accession no. AP000581.
RESULTS
Plasmids possessed by clinical V. parahaemolyticus isolates.
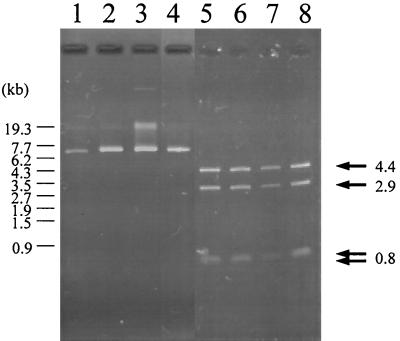
We examined 99 strains of V. parahaemolyticus that were isolated from patients with diarrhea at Kansai Airport Quarantine Station in 1996 for the possession of plasmids. All 24 V. parahaemolyticus O3:K6 strains possessed a plasmid (we named it plasmid pO3K6), which was detected slightly ahead of a 7.7-kb size marker after agarose gel electrophoresis (Fig. 1, lanes 1 to 4). HindIII digestion of the purified plasmid separated it into four fragments (4.4- and 2.9-kb fragments and two fragments of 0.8 kb) (Fig. 1, lanes 5 to 8). In contrast, this plasmid was not detected in any of the 75 remaining non-O3:K6 V. parahaemolyticus isolates (data not shown). These results suggest that all of the new V. parahaemolyticus O3:K6 strains possess a common plasmid, pO3K6.
FIG. 1.
Plasmids of V. parahaemolyticus O3:K6. Lanes: 1 and 5, KXV226; 2 and 6, KXV237; 3 and 7, KXV260; 4 and 8, KXV278. Lanes: 1 to 4, undigested plasmids; 5 to 8, plasmids digested with HindIII. The arrows indicate the HindIII digestion products (4.4 kb, 2.9 kb, 0.8 kb) for each plasmid. Electrophoresis carried out under optimal conditions separated the 0.8-kb band into two, indicating that two 0.8-kb fragments were present (data not shown). The numbers at the left of the panel show the size of the StyI-digested bacteriophage λ marker. The bands at about 19 kb found in lanes 1 to 4 indicate the changed conformation of the plasmids. KXV260 has another plasmid in addition to pO3K6.
Nucleotide sequence of pO3K6.
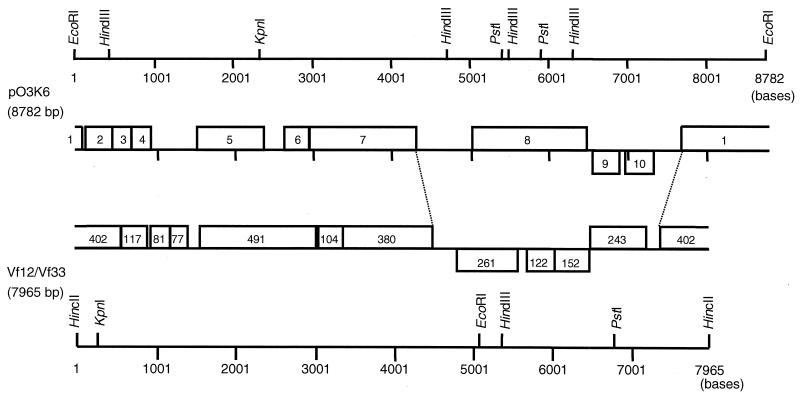
Plasmid pO3K6 was purified from a V. parahaemolyticus O3:K6 strain, KXV237, and the nucleotide sequence was determined. We found that pO3K6 had a total size of 8,782 bp and had 10 ORFs (Fig. 2): 8 ORFs were found on one strand, and ORF9 and ORF10 were found on the other.
FIG. 2.
Restriction map and ORFs of plasmid pO3K6 from KXV237. The restriction maps and ORFs of pO3K6 and the Vf33 and Vf12 genomes are shown. The restriction map of pO3K6 is a linear representation, with the site of EcoRI as point zero and with numbering in the 5′-to-3′ direction. The ORFs of pO3K6 and Vf33 are represented as blocks. The numbers in the blocks of pO3K6 denote ORF numbers, but numbers in the Vf33 and Vf12 sequences indicate the number of deduced amino acid residues. The blocks shown above the line are ORFs found in the 5′-to-3′ direction, while the blocks below are ORFs on the opposite strand. The source of the genetic map of Vf33 and Vf12 was Chang et al. (4).
Table 1 shows the results of a homology search for ORFs that resemble those on pO3K6. Some ORFs on pO3K6 were similar to the ORFs of filamentous phages. The deduced amino acid sequence of ORF1 showed close similarity to the sequence of Vpf402 of Vf33, a filamentous phage of V. parahaemolyticus (4, 21). In addition, ORF1 was similar to ORF166 and ORF208 of fs1, a filamentous phage of Vibrio cholerae O139 (10, 14), and to RstA1 or RstA2 of the V. cholerae CTX phage (24, 25). RstA1 and RstA2 are considered necessary for phage replication and integration into the host chromosome. The deduced amino acid sequence of ORF2 was homologous to the sequence of Vpf117 of Vf33 and that of ORF112 of fs1. Vpf117 is homologous with RstB of the CTX phage, which is necessary for integration of the phage with the chromosome of a host bacterium, V. cholerae. The protein encoded by ORF4 was homologous with gene 8, a major coat protein of the Pseudomonas aeruginosa Pf1 phage (7). The amino acid sequence of ORF5 was homologous with those of Vpf491 of Vf33 and ORF193 of fs1. Both Vpf491 and ORF193 are thought to be structural proteins of each phage. The product of ORF7 showed homology with the products of ORF424 of Pf1 and the zonula occludens toxin (Zot) of CTX phage of V. cholerae; ORF424 of Pf1 is considered indispensable for phage growth (7). Although ORF9 and ORF10 did not reveal significant homologies with any protein, some of the nucleotide sequences of ORF9 were similar to those of Vpf122, and some of the nucleotide sequences of ORF10 were similar to those of Vpf152 of Vf33. The overall gene organization of pO3K6 was similar to that of Vf33 (Fig. 2).
TABLE 1.
Peptides homologous to ORFs of pO3K6
| ORF | No. of amino acids | % G+C content | Homologous peptide(s) | Function of homologous peptide | Ratioa of identity (%) | Ratioa of similarity (%) |
|---|---|---|---|---|---|---|
| 1 | 402 | 48.3 | Vpf402b | 400/402 (99) | 400/402 (99) | |
| ORF166 and ORF208c | 144/370 (39) | 196/370 (53) | ||||
| RstA1 or RstA2d | Phage replication | 133/372 (35) | 182/372 (48) | |||
| 2 | 119 | 46.8 | Vpf117b | 111/115 (96) | 114/115 (99) | |
| ORF112c | 42/94 (44) | 58/94 (61) | ||||
| 3 | 76 | 46.5 | ||||
| 4 | 81 | 50.5 | Gene 8e | Major coat protein | 16/54 (29) | 29/54 (53) |
| 5 | 346 | 49.3 | Vpf491b | 137/305 (44) | 178/305 (57) | |
| ORF193c | 33/117 (28) | 58/117 (49) | ||||
| 6 | 114 | 44.9 | ||||
| 7 | 461 | 48.7 | Zotd | Affects intercellular tight junctions | 80/289 (27) | 123/289 (41) |
| ORF424e | Phage growth | 82/288 (28) | 132/288 (45) | |||
| 8 | 497 | 36.4 | ||||
| 9 | 121 | 37.4 | ||||
| 10 | 122 | 40.9 |
We speculate that pO3K6 is a replicative form (RF) of a filamentous phage because the overall gene organization and some of the ORFs of pO3K6 resemble those of Vf33.
Phage extraction from the supernatant of KXV237 culture.
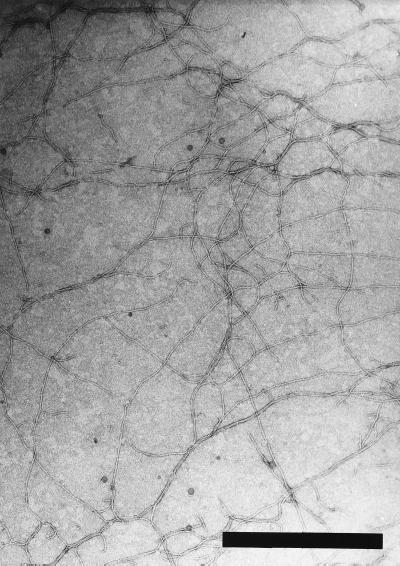
In light of this strong indication that pO3K6 was likely to be a RF of a filamentous phage, we attempted to isolate the phage from a V. parahaemolyticus O3:K6 strain. The culture supernatant from KXV237 formed a turbid plaque on a lawn of indicator cells (strain KXV191 of V. parahaemolyticus O4:K12) (data not shown). A phage was purified from the culture supernatant as described in Materials and Methods. This purified sample was negatively stained and was observed with an electron microscope. Flexible filaments, approximately 8 nm wide and usually appearing to be about 2.5 μm long, were observed in the sample (Fig. 3). The solution was considered to be a purified phage and was named f237.
FIG. 3.
Electron microscopic observation of f237 phage. Bar, 0.6 μm.
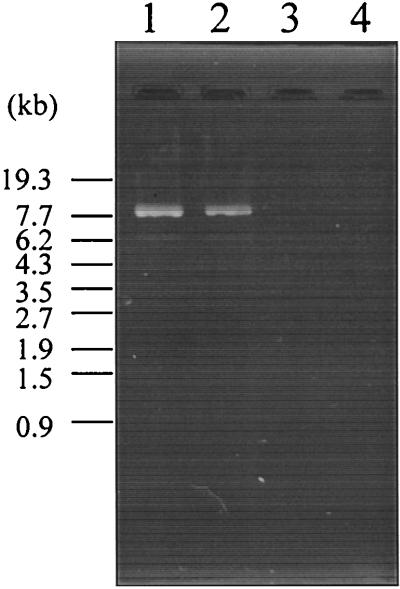
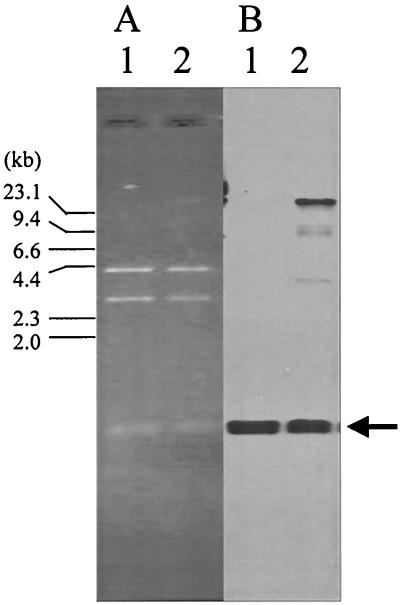
Nucleic acids extracted from the purified phage by phenol-chloroform extraction were detected at about a 7.7-kb size marker after agarose gel electrophoresis. It was resistant to RNase I but was sensitive to DNase I and mung bean nuclease, suggesting that it is single-stranded DNA (ssDNA) (Fig. 4). To confirm that the ssDNA was derived from pO3K6, the f237 genomic DNA was treated with T4 polymerase to obtain dsDNA. The HindIII digestion products of the resultant dsDNA (see below) were identical to those of pO3K6 (Fig. 5A). The dsDNA also hybridized with a probe for pO3K6 (Fig. 5B). Furthermore, the plasmid was isolated from f237-infected KXV191 cells by a rapid alkaline extraction procedure described by Birnboim and Doly (2). The HindIII digestion products of the plasmid separated in the same way as those of pO3K6 after agarose gel electrophoresis (data not shown). These results suggest that pO3K6 is the RF of phage f237.
FIG. 4.
Agarose gel electrophoresis of the genome of f237 phage. Lanes: 1, untreated f237 genome; 2, f237 genome treated with RNase I; 3, f237 genome treated with DNase I; 4, f237 genome treated with mung bean nuclease. The numbers at the left of the panel show the size of the StyI-digested bacteriophage λ marker.
FIG. 5.
(A) Agarose gel electrophoresis of pO3K6 and dsDNA derived from f237 genome. (B) Southern hybridization with the probe for ORF8. Lanes: 1, HindIII digests of pO3K6; 2, HindIII digests of dsDNA obtained by DNA polymerase treatment of f237 genome. Primer 8A was used for DNA synthesis. The arrow indicates the fragments detected by the probe for ORF8. The larger DNA fragment observed in panel B, lane 2, is an undigested form of open circular DNA that was synthesized by T4 polymerase treatment of the f237 genome. The numbers at the left of the panel show the size of the HindIII-digested bacteriophage λ marker.
When the f237 genome was treated with DNA polymerase to make dsDNA, the dsDNA was obtained only when primer 8A (designed from the antisense sequence of ORF8) was used and was not obtained when the 8S primer, from the sense sequence of ORF8, was used (data not shown). Thus, we concluded that the ssDNA possessed by the f237 phage is the strand which encodes ORF1 to ORF8, but not ORF9 and ORF10 (Fig. 2).
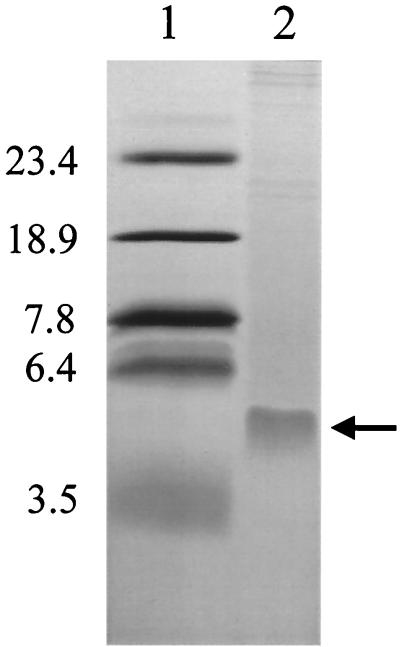
SDS-PAGE of purified f237 revealed a major protein band of approximately 5 kDa (Fig. 6). The N-terminal amino acid sequence of the band was analyzed and was revealed to be EVDITGAINS, and this amino acid sequence was found in ORF4 of pO3K6. The deduced molecular mass of the mature protein after cleavage of the putative signal sequence of the ORF4 product is estimated to be about 5.2 kDa. These results suggest that the ORF4 product is a major coat protein of f237.
FIG. 6.
SDS-PAGE of f237 phage. Lanes: 1, polypeptide SDS-PAGE molecular size standards (in kilodaltons) (Bio-Rad Laboratories, Hercules, Calif.); 2, purified f237 phage. The arrow indicates a major protein band.
Distribution of ORF7 and ORF8.
We further compared the ORFs in the f237 genome with those in the Vf33 genome (4). ORF3 corresponded in size and location to Vpf81. Vpf81 is similar to the TraK protein of the conjugative plasmid IncP-Beta RP4 of Escherichia coli, which is an ssDNA-binding protein with a transfer origin (28). ORF6 matched Vpf104 in size and location; Vpf104 is similar to the accessory cholera enterotoxin (Ace), which is capable of altering cellular ion fluxes, of the CTX phage (22, 25). From its size and location, we speculate that ORF9 corresponds to Vpf122 and that ORF10 corresponds to Vpf152. We could not, however, find an ORF in the genome of Vf33 that showed any correspondence with ORF8. Nor did we find any significant homology between ORF8 and any other genes recorded in the databases (Fig. 2; Table 1). The G+C contents of ORF8, ORF9, and ORF10 were apparently lower than those of the remaining regions of the f237 genome (Fig. 2). ORF8, ORF9, and ORF10 were located in a region that corresponded to the distinctive region of Vf33 (4). These results suggest that, when compared with the gene organization of Vf33, ORF8 is an ORF peculiar to the f237 genome.
To analyze whether ORF8 is peculiar to O3:K6 strains, we tested the 99 strains of V. parahaemolyticus isolates for the possession of ORF8 using colony hybridization with the probe for ORF8. As a control, we also tested for the possession of ORF7. We found ORF7 in all of the O3:K6 strains tested and four other non-O3:K6 strains, but ORF8 was detected only in the 24 O3:K6 strains tested. These results suggest that ORF8 is exclusively associated with O3:K6 strains.
DISCUSSION
In this study, we examined 99 strains of V. parahaemolyticus isolated from patients with diarrhea at the Kansai Airport Quarantine Station in 1996 for the possession of plasmids. All 24 strains of V. parahaemolyticus O3:K6 tested had in common a plasmid, pO3K6, which is an RF of phage f237. Our results suggest that f237 is specifically associated with the recently clinically widespread organism V. parahaemolyticus O3:K6.
Nakasone et al. (15) have reported the presence of a filamentous phage (lvpf5) from a V. parahaemolyticus O3:K6 strain (LVP5) isolated in Laos. A comparison of the restriction maps of the RF and the N-terminal amino acid sequences of the major coat protein show that f237 is clearly different from lvpf5 (15). In that study the investigators isolated lvpf5 from only a single V. parahaemolyticus O3:K6 strain (LVP5), so it is still unclear whether lvpf5 is exclusively associated with V. parahaemolyticus O3:K6 strains.
We found that the overall gene organization of f237 is similar to that of Vf33, a filamentous phage isolated from V. parahaemolyticus (4, 21), while the restriction map and several of the ORFs of the f237 genome were clearly different from those of Vf33. A gene organization similar to those of f237 and Vf33 is also found in the genomes of ssDNA phages, such as the CTX phage of V. cholerae (25) and the M13 phage of E. coli (23). Thus, f237 might be phylogenetically related to these phages.
The amino acid sequences of ORF1, ORF2, ORF4, ORF5, and ORF7 in f237 were homologous with the ORFs of both the V. cholerae fs1 phage (10) and those of the P. aeruginosa Pf1 phage (7). Although in these two cases the gene organization was not similar to that of f237, there could be a phylogenetic relationship between f237 and these two phages, albeit more distant than the putative relationship with Vf33, the CTX phage, and the M13 phage. It is also possible that genetic exchange by horizontal gene transfer, as has been reported for dsDNA phages (6), might have occurred between f237 and these phages.
Here we report that ORF8, a specific marker of f237, has been common and specific among the O3:K6 strains since 1996. For further corroboration we used colony hybridization to test for the presence of ORF8 in more recent V. parahaemolyticus O3:K6 isolates from 1998 outbreaks in Aomori and Shiga in Japan and found that ORF8 was common to all O3:K6 strains tested (data not shown). We also found that ORF8 was not associated with O3:K6 isolates that produced TRH and that were collected before 1996 (data not shown). These results confirm that f237 is specifically associated with recent V. parahaemolyticus O3:K6 strains isolated since 1996.
The G+C contents of ORF8, ORF9, and ORF10 were lower than those of the remaining regions of the f237 genome (Table 1). These three ORFs are located on a region that corresponds to the distinctive region of Vf33 (4). In size and location, ORF9 is similar to Vpf122 and ORF10 is similar to Vpf152 of Vf33, but ORF8 does not match the size or location of any ORF on the Vf33 genome. Thus, ORF8 seems to be an ORF peculiar to the f237 genome.
One possible explanation for the epidemic potency of the recent V. parahaemolyticus O3:K6 strains is that the strains have recently acquired ORF8 by f237 infection, which resulted in an as yet unknown phenotypic alteration that has given them epidemic potency; as yet there is no evidence for this, and the suggestion remains purely speculative. The deduced amino acid sequence of ORF8 shows an RGD sequence in the N-terminal region, a leucine zipper in the C-terminal region, and a hydrophobic stretch between the two. This hydrophobic stretch could be a transmembrane domain. A similar combination of motifs in a single protein is found in the Drosophila plx gene, which encodes a novel adhesion molecule (27). If ORF8 encodes a protein that has an adhesive function similar to that of the protein encoded by plx, the recent V. parahaemolyticus O3:K6 strains that possess ORF8 could be more adhesive, possibly to host intestine cells or to the surface of marine plankton, and this adhesiveness could account for the epidemic potency of V. parahaemolyticus O3:K6 strains. We are examining the function of the ORF8 product.
In conclusion, we isolated a filamentous phage, f237, which is associated with recent V. parahaemolyticus O3:K6 strains. We propose that the ORF8 gene is a useful genetic marker for characterization of the recently widespread O3:K6 strains of V. parahaemolyticus in clinical laboratories, because ORF8 in f237 is a unique ORF which is not found in other filamentous phages. Although the function of ORF8 must still be clarified, the product of ORF8 could be a novel virulence factor of V. parahaemolyticus.
ACKNOWLEDGMENTS
This work was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan, a Grant for International Health Cooperation Research from the Ministry of Health and Welfare of Japan, and the “Research for the Future” Program of the Japan Society for the Promotion of Sciences (grant no. JSPS-RFTF 97L00704).
REFERENCES
- 1.Bag P K, Nandi S, Bhadra R K, Ramamurthy T, Bhattacharya S K, Nishibuchi M, Hamabata T, Yamasaki S, Takeda Y, Nair G B. Clonal diversity among recently emerged strains of Vibrio parahaemolyticus O3:K6 associated with pandemic spread. J Clin Microbiol. 1999;37:2354–2357. doi: 10.1128/jcm.37.7.2354-2357.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. Outbreak of Vibrio parahaemolyticus infection associated with eating raw oysters and clams harvested from Long Island Sound—Connecticut, New Jersey, and New York, 1998. Morbid Mortal Weekly Rep. 1999;48:48–51. [PubMed] [Google Scholar]
- 4.Chang B, Taniguchi H, Miyamoto H, Yoshida S. Filamentous bacteriophages of Vibrio parahaemolyticus as a possible clue to genetic transmission. J Bacteriol. 1998;180:5094–5101. doi: 10.1128/jb.180.19.5094-5101.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fasano A, Baudry B, Pumplin D W, Wasserman S S, Tall B D, Ketley J M, Kaper J B. Vibrio cholerae produces a second enterotoxin, which affects intestinal tight junction. Proc Natl Acad Sci USA. 1991;88:5242–5246. doi: 10.1073/pnas.88.12.5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hendrix R W, Smith M C M, Burns R N, Ford M E, Hatfull G F. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc Natl Acad Sci USA. 1999;96:2192–2197. doi: 10.1073/pnas.96.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hill D F, Short N J, Perham R N, Petersen G B. The DNA sequence of the filamentous bacteriophage Pf1. J Mol Biol. 1991;218:349–364. doi: 10.1016/0022-2836(91)90717-k. [DOI] [PubMed] [Google Scholar]
- 8.Honda T, Iida T. The pathogenicity of Vibrio parahaemolyticus and the role of the thermostable direct haemolysin and related haemolysins. Rev Med Microbiol. 1993;4:106–113. [Google Scholar]
- 9.Honda T, Ni Y, Miwatani T. Purification and characterization of a haemolysin produced by a clinical isolate of Kanagawa phenomenon-negative Vibrio parahaemolyticus and related to be the thermostable direct haemolysin. Infect Immun. 1988;56:961–965. doi: 10.1128/iai.56.4.961-965.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Honma Y, Ikema M, Toma C, Ehara M, Iwanaga M. Molecular analysis of a filamentous phage (fs1) of Vibrio cholerae O139. Biochim Biophys Acta. 1997;1362:109–115. doi: 10.1016/s0925-4439(97)00055-0. [DOI] [PubMed] [Google Scholar]
- 11.Iida T, Suthienkul O, Park K-S, Tang G-Q, Yamamoto R K, Ishibashi M, Yamamoto K, Honda T. Evidence for genetic linkage between the ure and trh genes in Vibrio parahaemolyticus. J Med Microbiol. 1997;46:639–645. doi: 10.1099/00222615-46-8-639. [DOI] [PubMed] [Google Scholar]
- 12.Landshulz W H, Johnson P F, McKnight S L. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988;240:1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- 13.Lin J J, Smith M, Jessee J, Bloom F. DH11S: an Escherichia coli strain for preparation of single-stranded DNA from phagemid vectors. BioTechniques. 1992;12:718–721. [PubMed] [Google Scholar]
- 14.Nakasone N, Honma Y, Toma C, Yamashiro T, Iwanaga M. Filamentous phage fs1 of Vibrio cholerae O139. Microbiol Immunol. 1998;42:237–239. doi: 10.1111/j.1348-0421.1998.tb02277.x. [DOI] [PubMed] [Google Scholar]
- 15.Nakasone N, Ikema M, Higa N, Yamashiro T, Iwanaga M. Filamentous phage of Vibrio parahaemolyticus O3:K6 isolated in Laos. Microbiol Immunol. 1999;43:385–388. doi: 10.1111/j.1348-0421.1999.tb02420.x. [DOI] [PubMed] [Google Scholar]
- 16.Okajima T, Tanabe T, Yasuda T. Nonurea sodium dodecyl sulfate-polyacrylamide gel electrophoresis with high-molarity buffers for the separation of proteins and peptides. Anal Biochem. 1993;211:293–300. doi: 10.1006/abio.1993.1272. [DOI] [PubMed] [Google Scholar]
- 17.Okuda T, Ishibashi M, Hayakawa E, Nishino T, Takeda Y, Mukhopadhyay A K, Garg S, Bhattacharya S K, Nair G B, Nishibuchi M. Emergence of a unique O3:K6 clone of Vibrio parahaemolyticus in Calcutta, India, and isolation of strains from the same clonal group from Southeast Asian travelers arriving in Japan. J Clin Microbiol. 1997;35:3150–3155. doi: 10.1128/jcm.35.12.3150-3155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raleigh E A, Murray N E, Revel H, Blumenthal R M, Westaway D, Reith A D, Rigby P W, Elhai J, Hanahan D. McrA and McrB restriction phenotypes of some E. coli strains and implications for gene cloning. Nucleic Acids Res. 1988;16:1563–1575. doi: 10.1093/nar/16.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruoslahti E. RGD and other recognition sequences for integrins. Annu Rev Cell Dev Biol. 1996;12:697–715. doi: 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 21.Taniguchi H, Sato K, Ogawa M, Udou T, Mizuguchi Y. Isolation and characterization of a filamentous phage, Vf33, specific for Vibrio parahaemolyticus. Microbiol Immunol. 1984;28:327–337. doi: 10.1111/j.1348-0421.1984.tb00684.x. [DOI] [PubMed] [Google Scholar]
- 22.Trucksis M, Galen J E, Michalski J, Fasano A, Kaper J B. Accessory cholera enterotoxin (Ace), the third toxin of a Vibrio cholerae virulence cassette. Proc Natl Acad Sci USA. 1993;90:5267–5271. doi: 10.1073/pnas.90.11.5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Wezenbeek P M, Hulsebos T J, Schoenmakers J G. Nucleotide sequence of the filamentous bacteriophage M13 DNA genome: comparison with phage fd. Gene. 1980;11:129–148. doi: 10.1016/0378-1119(80)90093-1. [DOI] [PubMed] [Google Scholar]
- 24.Waldor M K, Rubin E J, Pearson G D N, Kimsey H, Mekalanos J J. Regulation, replication, and integration functions of the Vibrio cholerae CTXφ are encoded by region RS2. Mol Microbiol. 1997;24:917–926. doi: 10.1046/j.1365-2958.1997.3911758.x. [DOI] [PubMed] [Google Scholar]
- 25.Waldor M K, Mekalanos J J. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 26.Yanish-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 27.Zhang S-D, Kassis J, Olde B, Mellerick D M, Odenwald W F. Pollux, a novel Drosophila adhesion molecule, belongs to a family of proteins expressed in plants, yeast, nematodes, and man. Genes Dev. 1996;10:1108–1119. doi: 10.1101/gad.10.9.1108. [DOI] [PubMed] [Google Scholar]
- 28.Ziegelin G, Pansegrau W, Strack B, Balzer D, Kroeger M, Kruft U, Lanke E. Nucleotide sequence and organization of genes flanking the transfer origin of promiscuous plasmid RP4. DNA Sequence. 1991;1:303–327. doi: 10.3109/10425179109020786. [DOI] [PubMed] [Google Scholar]