Abstract
Bacterial cancer therapy (BCT) shows great promise for treatment of solid tumors, yet basic mechanisms of bacterial-induced tumor suppression remain undefined. Attenuated strains of Salmonella enterica serovar Typhimurium (STm) have commonly been used in mouse models of BCT in xenograft and orthotopic transplant cancer models. We aimed to better understand the tumor epithelium–targeted mechanisms of BCT by using autochthonous mouse models of intestinal cancer and tumor organoid cultures to assess the effectiveness and consequences of oral treatment with aromatase A–deficient STm (STmΔaroA). STmΔaroA delivered by oral gavage significantly reduced tumor burden and tumor load in both a colitis-associated colorectal cancer (CAC) model and in a spontaneous Apcmin/+ intestinal cancer model. STmΔaroA colonization of tumors caused alterations in transcription of mRNAs associated with tumor stemness, epithelial-mesenchymal transition, and cell cycle. Metabolomic analysis of tumors demonstrated alteration in the metabolic environment of STmΔaroA-treated tumors, suggesting that STmΔaroA imposes metabolic competition on the tumor. Use of tumor organoid cultures in vitro recapitulated effects seen on tumor stemness, mesenchymal markers, and altered metabolome. Furthermore, live STmΔaroA was required, demonstrating active mechanisms including metabolite usage. We have demonstrated that oral BCT is efficacious in autochthonous intestinal cancer models, that BCT imposes metabolic competition, and that BCT has direct effects on the tumor epithelium affecting tumor stem cells.
Keywords: Gastroenterology
Keywords: Bacterial infections, Colorectal cancer
Introduction
The use of bacteria as cancer therapeutics (bacterial cancer therapy [BCT]) dates back to the late 1800s, when the field of BCT was initiated by William Coley (1). Prior observations of spontaneous tumor regression following infection of patients’ tumors had led Coley to treat cancer patients with intratumoral injections of bacterial preparations (1). Despite this early work on BCT, there is only 1 currently in the clinic — Bacillus Calmette-Guérin vaccine (BCG) therapy for superficial bladder cancer (2).
The advancement of molecular genetics has enabled bacteria to be effectively attenuated to remove adverse effects and has been engineered to deliver different payloads. Therefore, there has been a resurgence in interest in BCT in the past 20 years, with many studies showing efficacy of attenuated bacterial treatments in xenograft and orthotopic transplant tumor models, with Salmonella enterica serovar Typhimurium (STm) being by far the most studied (3–9). Attenuated STm are extremely tumor tropic; a therapy that can give such tumor-tissue selectivity is very desirable and enables further engineering to deliver drugs, immune adjuvants, or other antitumor agents (3). Despite this interest, very little is understood about the underlying mechanisms of BCT-mediated tumor suppression, which is hampering its practical application.
Given that BCG is delivered directly to the bladder epithelium, and that direct interaction is necessary for direct cytotoxic effects (2, 10), we hypothesized that other BCT can exert direct effects on tumor cells. To date, there have been limited BCT studies using autochthonous models of cancer. Studies on BCT thus far have utilized xenograft or orthotopic transplant models of cancer, often in immunocompromised mice, and these may not fully model complex disease in patients and therefore may not be entirely predictive of efficacy or mechanism of BCT. One study using an autochthonous model of prostate cancer showed limited efficacy of a heavily attenuated STm strain (CRC2631) when injected i.p. on a weekly basis (11).
Since intestinal innate immune pathways protect from both STm infection and tumorigenesis (12, 13), we aimed to determine if STm treatment could effectively treat autochthonous tumors of the intestine. We reasoned that intestinal cancer would be an appropriate target for BCT using STm, as the natural route of infection is via the intestine and, therefore, would likely enable better invasion and interaction between the bacteria and the tumor, which would overcome issues of poor dissemination of orally administered STm to tumors at nongastrointestinal sites (14). Treating colorectal cancer (CRC) patients by oral delivery of attenuated STm is feasible since oral vaccines for S. typhi are widely used and tolerated (15). Oral delivery of STm may also avoid problems of tumor homing and toxicity that have been observed when delivering STm i.v. to patients (16).
We utilized STm deficient for aromatase A (STmΔaroA) (UF020; ref. 17) to assess whether BCT could be effective for treating mouse models of colorectal cancer. Aromatase A–deficient STm are auxotrophic for aromatic amino acids (AAs). The tumor microenvironment is often enriched with AAs (18, 19), which may aid its specific colonization of tumor tissue compared with normal intestine where they are not freely available. STmΔaroA is commonly used as a vaccine strain (20) and has also previously been used successfully as a BCT in tumor transplant models (14, 21–23). Using a model of colitis-associated colorectal cancer (CAC) and a spontaneous model of intestinal cancer, Apcmin/+ mice, we show that oral delivery of an attenuated STm potently reduces tumor burden. Transcriptomic and metabolomic analyses, coupled with use of tumor organoids in vitro, demonstrated restoration of epithelial markers by STm, including reduced tumor stem markers, and found that STm impose metabolic competition, which is likely central to antitumor effects.
Results
Orally administered STmΔaroA reduces gastrointestinal tumor burden.
We first determined whether orally administered STmΔaroA would effectively colonize intestinal polyps in the Apcmin/+ mouse model. These mice carry a mutation in the adenomatous polyposis coli gene (Apc), which results in multiple intestinal neoplasia (min), serving as a model of human familial adenomatous polyposis (FAP). In mice, the Apc mutation results largely in small intestinal (SI) neoplasia (100% penetrance) and not colonic neoplasia (approximately 50% penetrance with few tumors). We treated Apcmin/+ or littermate Apc+/+ mice with oral gavage of 5 × 109 CFU STmΔaroA and assessed bacterial burden in a range of tissues at various time points after administration. Indeed, STmΔaroA colonized polyps in the ileum within 4 hours of treatment, followed by a peak in number at 24 hours and a contraction by 1 week after administration. Lower levels could still be observed 2 weeks after administration (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.139900DS1). In contrast, there were much lower CFUs in the normal SI tissue, though showing a similar trajectory over time, and WT non–tumor-bearing mice showed even lower burden in the normal SI (Supplemental Figure 1). This is likely reflected in the fact that Apcmin/+ mice have extensive polyps and aberrant crypts throughout the SI. Mesenteric lymph nodes showed a gradual increase in STmΔaroA CFUs over 2 weeks, with slightly higher levels in tumor-bearing mice than in non–tumor-bearing mice, though these levels were far less than seen within tumors (Supplemental Figure 1). Peyer’s patches showed initial colonization at 24 hours, which decreased over time, comparable in tumor-bearing mice and non–tumor-bearing mice (Supplemental Figure 1). Analysis of spleen CFUs showed some low-level colonization in few mice (1 from each genotype) 2 weeks after administration (Supplemental Figure 1). Finally, analysis of ileal content and feces showed a surprisingly low number of CFUs. Tumor-bearing mice had greater levels in the ileal content 24 hours after administration. CFUs recovered from the feces demonstrated a delayed peak (at 72 hours compared with 24 hours) in non–tumor-bearing mice. Overall, this analysis showed that, as per previous publications (4–9), attenuated STm preferentially colonize tumor tissue over normal tissues and that, within intestinal polyps, colonization decreases by 2 weeks. We therefore proceeded to assess the efficacy of STmΔaroA treatment in 2 models of intestinal cancer by giving weekly oral dosing.
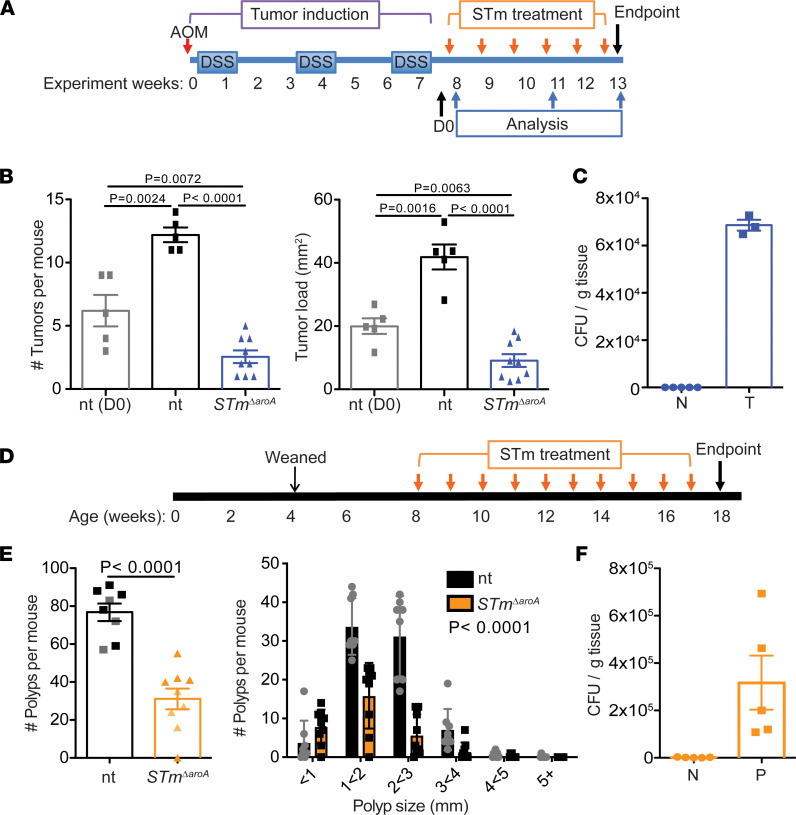
We induced colon tumors in C57B6/J mice using a well-described model of CAC, which has 100% penetrance (13, 24) (Figure 1A). After tumor induction, mice were split into treatment groups, ensuring equivalent colitis severity between groups. Supplemental Figure 2 shows weight loss during the azoxymethane/dextran sodium sulphate (AOM/DSS) protocol. Following recovery from the final dose of DSS (1 to 2 weeks), mice were given 5 × 109 CFU STmΔaroA, or vehicle control (PBS), by oral gavage once per week for 6 weeks. From the start of treatment (denoted D0), mice had well-developed colonic tumors (Figure 1B). Tumor burden and tumor load was significantly decreased in STmΔaroA-treated mice, compared with both D0 and 6-week control-treated mice (Figure 1B). This indicates that STmΔaroA treatment by oral delivery could reduce existing tumor burden and prevent further tumor development or growth. We measured STmΔaroA CFUs in tumors at the end of the protocol and could confirm colonization in the colon tumor but not normal tissue (Figure 1C).
Figure 1. Oral delivery of attenuated STm reduces intestinal tumor burden.
(A) Schematic of AOM/DSS-induced CAC model and STmΔaroA treatment. (B) Tumor burden (number of tumors/mouse) and tumor load (cumulative tumor size per mouse, mm2) in nontreated (nt) and STmΔaroA-treated mice. n = 5 for D0 and nt groups; n = 9 for STmΔaroA-treated mice. Representative of 4 independent experiments. Female mice were used in this experiment. (C) CFU of STmΔaroA in normal (N) and tumor (T) tissue from STmΔaroA-treated mice in the CAC model. (D) Schematic of Apcmin/+ mouse STmΔaroA treatment. (E) Polyp burden and polyp size per mouse in nontreated (nt) and STmΔaroA-treated mice. Data pooled from 2 independent experiments using both male and female mice, nt n = 8 (4F, 4M), STmΔaroA-treated n = 9 (5F, 4M). Lighter shaded mice in NT and STm indicate mice used for RNA analysis in Figure 4B. (F) CFU of STmΔaroA in normal (N) and polyp (P) tissue from STmΔaroA-treated mice in the Apcmin/+ model; data are shown as mean ± SD. One-way ANOVA (B) or 2-tailed t test (E) were used; data are shown as mean ± SD.
Next, we tested STmΔaroA treatment in Apcmin/+ mice. We treated Apcmin/+ mice with 5 × 109 CFU STmΔaroA by oral gavage once per week for 10 weeks, from 8 weeks of age (Figure 1D). At this age, the SI had already developed a large number of polyps and they continued to grow in size, with mice at 18 weeks showing large well-developed polyps throughout the SI tract. Treatment of Apcmin/+ mice with STmΔaroA substantially reduced both the polyp burden and size (Figure 1E). Colonization of SI polyps by STmΔaroA was confirmed at the end of the treatment, with no colonies observed in the normal surrounding tissue (Figure 1F).
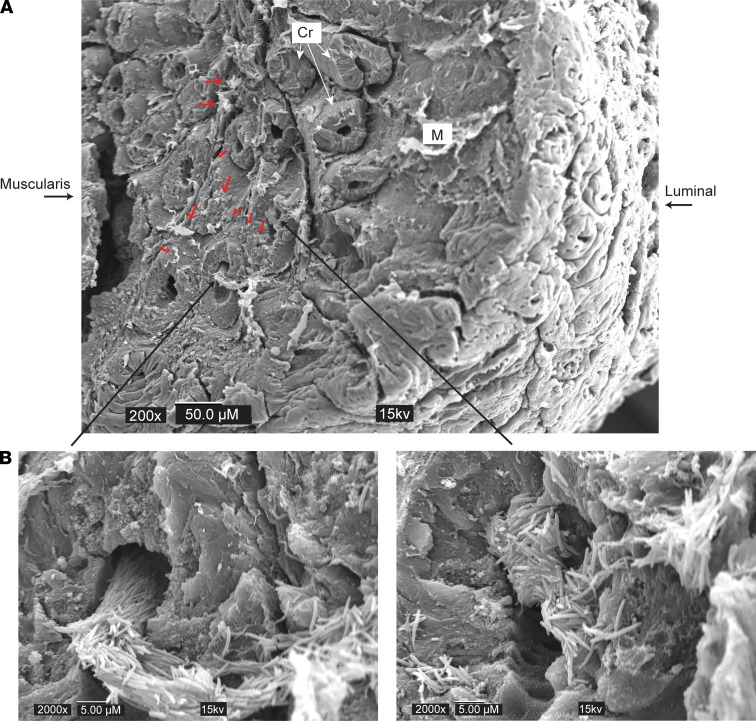
We next employed scanning electron microscopy (SEM) to view bacterial colonization in greater detail. Colonic tumors were analyzed 24 hours after administration, which showed the greatest colonization of STmΔaroA. Exceptionally large colonies of STmΔaroA were found within the tumor mass just 24 hours after administration (Figure 2, see insets). These were reminiscent of previous observations by Crull et al., in which they found large extracellular colonies of STm in CT26 tumors 2 days after administration (25). The large size of the bundles suggested that they were rapidly dividing within the tumor extracellular spaces. This is consistent with the CFUs observed at this time point (Supplemental Figure 1) and suggests that initial seeding of the tumor results in a dramatic proliferation of the bacteria, which then recedes. We could also find instances of single or multiple bacteria (Figure 2, red arrows). No bacteria could be observed in nontreated mice (Supplemental Figure 3, A–D), strongly implying that normal microbiota are not penetrating tumor tissue to form mass colonies as observed with the STmΔaroA. It is likely that small amounts of microbiota do invade via the disrupted barrier as previously described (26); however, this would be difficult to detect with SEM. IF staining detecting mCherry-expressing STmΔaroA further supports the SEM data showing large aggregates of STmΔaroA commonly occurring, with some punctate staining indicating individual bacterium (Supplemental Figure 4). Supplemental Figure 5 shows the histological appearance of colon after CAC induction in nontreated and STmΔaroA-treated mice, with boxes indicating the type of region imaged in the IF staining of STmΔaroA in Supplemental Figure 4.
Figure 2. Scanning electron microscopy of STmΔaroA-treated tumors.
Mice bearing CAC colon tumors were given STmΔaroA or control vehicle by oral gavage and tissues were taken 24 hours later. Whole sections of colon with tumors were prepared for SEM by glutaraldehyde fixation, dehydration, and freeze drying. Tumors were cut on the sagittal plane and mounted for platinum coating and SEM imaging. (A) Top image shows lower magnification view of a tumor area. Scale bar: 50 μm. Luminal side indicates the top of the tumor that was facing the intestinal lumen, and muscularis side indicates the inner side of tumor reaching the lamina propria and muscularis mucosa. Small red arrows indicate small STmΔaroA colonies or individual bacteria. (B) Large black arrows indicate areas shown in higher magnification. Scale bar: 5 μm. Cr, Crypt; M, Mucous.
STmΔaroA treatment does not alter the colonic microbiota.
Infection with WT STm induces changes in the microbiota, which lead to and support an inflammatory environment within the intestine that favors Salmonella growth (27). In addition, different microbiomes have been associated with better outcome in cancer and cancer therapy with checkpoint blockades (28, 29). We therefore assessed whether oral administration of STmΔaroA altered the microbiota composition. Colonic content was taken from AOM/DSS-induced mice following 6 weeks of treatment with STmΔaroA (as per Figure 1A) and subjected to 16s rRNA-Seq. The observed total number of operational taxonomic units (OTUs) was not different between nontreated and STmΔaroA-treated mice (Supplemental Figure 6A). Analysis of the α diversity using multiple statistical models (Chao1 and the Shannon and Simpson’s Diversity Index) also showed that there were no differences between the abundance or evenness of microbial species present in nontreated and STmΔaroA-treated mice (Supplemental Figure 6B). Analysis by weighted UniFrac for β-diversity also showed no differences in the quantitative abundance of species between groups (analysis of similarities [ANOSIM] test; r = 0.214, P = 0.068) (Supplemental Figure 6C). This would suggest that, unlike infection with WT Salmonella, STmΔaroA infection does not elicit changes in the microbiome at the time point tested, which would be consistent with the very low levels of infection in normal tissue (Supplemental Figure 1). There remains the possibility that the microbiota is altered during initial exposure to STmΔaroA when its abundance in the gut lumen is higher. However, Supplemental Figure 1 shows that STmΔaroA is rapidly cleared from the feces.
To further test whether the microbiota is involved in the efficacy of BCT, we induced colorectal tumors in germ-free (GF) mice using AOM and DSS and then treated by oral gavage with STmΔaroA. GF mice are incredibly sensitive to DSS treatment due to reduced barrier function and altered mucosal immunity (30); therefore, even with low dose DSS, weight loss was extreme and many mice reached the ethical end point. The remaining GF mice (4) were treated either with PBS or STmΔaroA (1 × 107 CFU) by oral gavage. GF mice showed susceptibility to the attenuated STmΔaroA strain and displayed rapid weight loss, which was then maintained (Supplemental Figure 6D). Mice therefore only received 1 dose of STmΔaroA and were sacrificed 11 days after the treatment, and tumor burden was analyzed. Given the caveat of there being 2 mice per group, there was a clear abolition of tumors in the STmΔaroA-treated GF mice (Supplemental Figure 6E). These mice did have areas of hyperplasia, which were increased compared with NT mice and may represent the former tumor areas (Supplemental Figure 6E). Because mice showed signs of systemic infection (weight loss), we checked the CFU in the spleens and indeed found dissemination of STmΔaroA (Supplemental Figure 6F). These data show that the presence of microbiota may, to a degree, impede STmΔaroA persistence, likely through competition for space within the intestine. However, GF mice are susceptible to bacterial dissemination, demonstrating the necessity of the microbiota to instruct barrier function. Altogether, these data imply that the presence of the gut microbiota can control the outgrowth of STmΔaroA, but there are no appreciable alterations in the gut microbiota that might explain the treatment outcome.
STmΔaroA alters the transcriptional landscape of tumors.
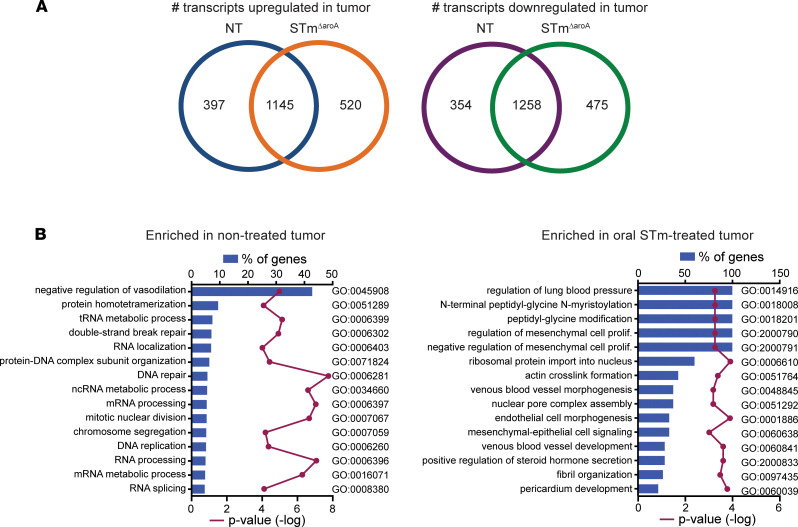
Next, to gain an understanding of the differences between nontreated and STmΔaroA-treated tumors, we performed RNA-Seq on RNA isolated from whole tumor (T) or adjacent normal tissue (N) dissected from AOM/DSS-induced CAC-bearing mice after 4 weeks treatment. Tumor burden and size for this cohort of mice are shown in Supplemental Figure 7A. Mice treated for 4 weeks with STmΔaroA had a trend toward significantly reduced tumor burden and size. Tumors used for RNA isolation was similar between groups (Supplemental Figure 7A). First, we identified the transcripts that were differentially regulated between N and T tissue in the nontreated and STmΔaroA-treated groups. Figure 3A shows the number of overlapping and unique genes for each treatment. It is interesting to note that approximately one quarter of genes either up- or downregulated in STmΔaroA-treated tumor tissue are unique to STm treatment. These differentially expressed genes (DEGs) were then analyzed by gene ontology (GO) analysis using DAVID (31, 32), revealing terms enriched in either the nontreated tumors or in the treated tumors, which intriguingly were vastly different (Figure 3B). As expected, nontreated tumors exhibited enrichment of mRNAs involved in cell cycle processes, mitosis, cell division, DNA repair, and more, whereas STmΔaroA-treated tumors displayed enrichment of mRNAs for processes involving regulation of mesenchymal cell proliferation and mesenchymal-epithelial cell signaling, as well as regulation of blood vessel development (Figure 3B and Supplemental Figure 8). Several genes involved in DNA repair, DNA damage response, RNA synthesis, and epithelial-mesenchymal transition were significantly reduced following STmΔaroA treatment (Supplemental Figure 8), suggesting major changes in cell proliferation rates.
Figure 3. STmΔaroA treatment alters the transcriptional landscape of tumors.
Normal and tumor tissue were dissected from CAC-bearing mice after 4 weeks of STmΔaroA or control treatment. RNA was isolated and used for RNA-Seq analysis. (A) Number of transcripts upregulated or downregulated in tumor compared with normal tissue, with overlapping and unique transcripts depicted. (B) Differentially expressed genes (DEGs) in tumors compared with normal tissue for each treatment identified in A were compared by GO analysis. Data represents the percentage of genes of a given pathway that are enriched in either nontreated or treated tumors, with –log P value.
There was no signature of inflammatory processes picked up in the RNA-Seq by GO analysis. We checked classically proinflammatory cytokines by transcript and found an increase in Il1b mRNA, a trend toward increased Il6, but no differences in Il17, Tnfa,and Ifny mRNAs (Supplemental Figure 9A). Analysis of tissue homogenates by Luminex cytokine array found increased levels of IL-1β and TNF-α in tumor tissue compared with normal tissue, but no differences were found between treated and nontreated groups (Supplemental Figure 9B). Other cytokines on the array (including IFN-γ, IL-2, and IL-10) were not detected. This is consistent with another report showing that an auxotrophic STm mutant does not induce inflammation in the mucosa but still induces protective immunity with mucosal invasion–associated virulence factors driving immunogenicity (33).
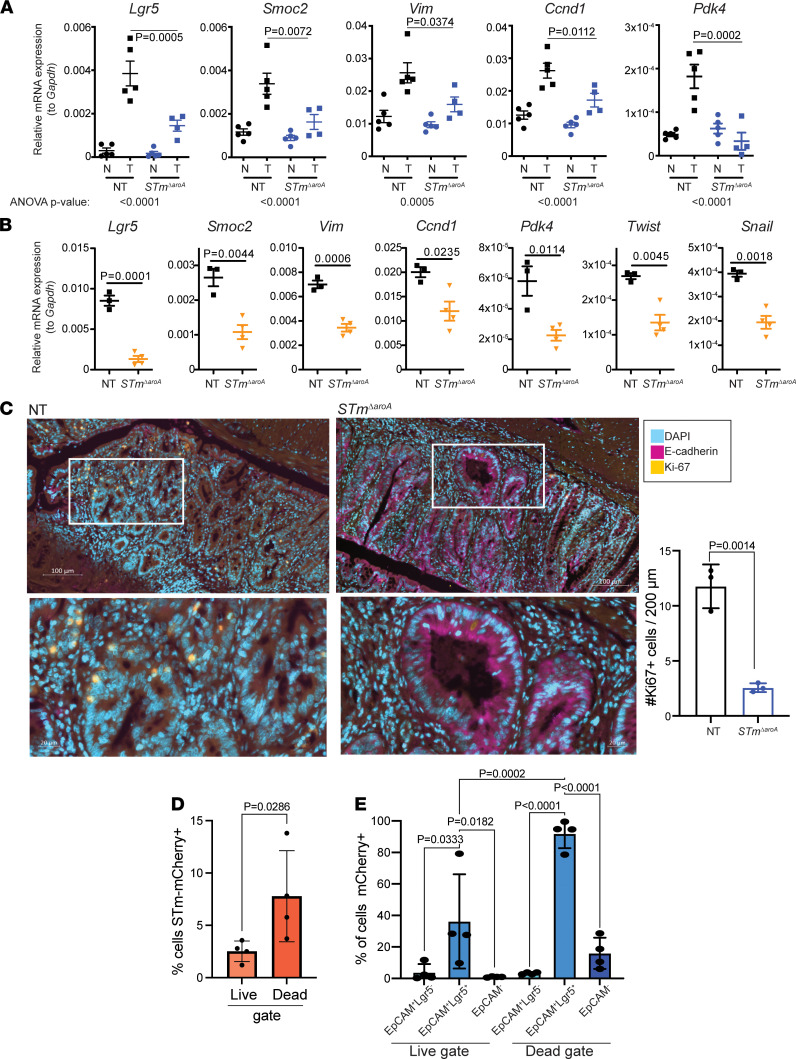
Next, we homed in on stem cell, EMT, and metabolism-related genes, and we confirmed a selection of targets by quantitative PCR (qPCR) in independent experiments where mice were treated for 6 weeks. As previously reported, transcripts for epithelial stem cells, proliferation, or epithelial-to-mesenchymal transition–related processes — including Lgr5 (leucine-rich repeat-containing G-protein coupled receptor), Smoc2 (SPARC-related modular calcium binding 2), Vim (Vimentin), Ccnd1 (Cyclin D1), and Pdk4 (pyruvate dehydrogenase kinase 4) (34–40) — were increased in tumor tissue when compared with normal tissue (Figure 4A). Strikingly, these transcripts were largely decreased following STmΔaroA treatment (Figure 4A). We confirmed these mRNA changes in the Apcmin/+ model, comparing tumor tissue from nontreated and STmΔaroA treatment. In line with results from the CAC model, STmΔaroA treatment altered the transcriptional levels of the above-mentioned genes and additional EMT-related genes Twist and Snail (Figure 4B). We also analyzed gene expression in normal, tumor (control-treated) or hyperplasia (STmΔaroA-treated) colon tissue from GF mice (from Supplemental Figure 8B) by qPCR. Tumors from GF mice showed similar upregulation of stem cell–associated, mesenchymal, proliferation, and metabolic genes as observed in specific pathogen–free (SPF) tumor-bearing mice, and the hyperplasic tissue taken from the STmΔaroA-treated GF mice looked more similar to normal tissue than to tumors from nontreated GF mice (Supplemental Figure 8B).
Figure 4. Altered tumor phenotype in STmΔaroA-treated mice.
(A) Quantitative PCR confirmation of genes identified (or pathway related) by RNA-Seq in CAC tumor–bearing mice after 6 weeks of treatment. Nontreated, NT; Salmonella treated, STmΔaroA; normal tissue, N; tumor tissue, T. Size of tumors used to isolate RNA are shown in Supplemental Figure 7. Data are representative of 3 independent experiments. One-way ANOVA with Turkey’s multiple-comparison test was conducted. ANOVA P values are indicated below the graphs, and an individual post hoc test comparing T from each treatment is shown on the graphs. (B) Analysis of indicated transcripts in Apcmin/+ tumor tissue after 10 weeks of treatment. Data come from 3 (NT) or 4 (STm) mice shown in Figure 1E. Similarly sized polyps were obtained from each group. Data are representative of 2 independent experiments. Unpaired 2-tailed t tests were used. (C) Representative immunofluorescence of E-cadherin (purple) and Ki67 (yellow) counterstained with DAPI (blue) in NT and STmΔaroA-treated (6 weeks) CAC mice. Scale bar: 100 μm. Lower images are magnification of upper images. Scale bar: 20 μm. For orientation reference, Supplemental Figure 5 shows the type of area (not taken from exact mouse/tumor) imaged here. Quantification of the number of Ki67+ cells within 200 μm field of view (FOV) shown to the right. Ten FOV from 2–3 tumors per mouse. Each dot represents the average number for each mouse. (D) Lgr5-GFP reporter mice were induced with CAC, as per Figure 1A. Mice were then gavaged with mCherry-expressing STmΔaroA, and tumors were collected for flow cytometry analysis 24 hours later. Cells were stained for live/dead marker and EpCAM (CD326); Lgr5-GFP and mCherry were expressed via reporters. Two-tailed Student’s t test. (E) From D, cells were first gated based on EpCAM and Lgr5 expression (as indicated) and the percentage of mCherry+ in each population is shown. One-way ANOVA with multiple-comparison post hoc test. Each point represents pooled tumors from 1 mouse. All data are shown as mean ± SD.
Loss of E-cadherin protein expression is an important feature of epithelial-derived tumor progression. Cdh (encoding E-cadherin) was consistently decreased at the mRNA level in tumors and showed a trend toward increasing in STmΔaroA-treated tumors (not significant in all experiments; data not shown). Since translation and protein localization of E-cadherin is important for its function (41), we checked E-cadherin protein expression by IHC staining of sections taken from CAC tumor–bearing mice. Nontreated tumor sections showed very little E-cadherin protein (Figure 4C). In contrast, tumors from STmΔaroA-treated mice showed significantly higher levels of E-cadherin within tumor areas (Figure 4C). Thus, it appears that STmΔaroA treatment diminishes tumors, reducing tumor stemness markers and restoring epithelial identity. As we had observed enrichment of proliferation-related genes in NT tumors compared with tumors from STm-treated mice, and decreased tumor size, we assessed proliferation within tumors by Ki67 staining at 6 weeks after treatment. There was an increase in Ki67+ cells in NT tumors compared with STmΔaroA-treated tumor sections (Figure 4C), which is consistent with the transcriptomic and macroscopic changes in the tumor size. This likely reflects the change in tumor size at this time point. STmΔaroA infection of tumor cells was assessed by flow cytometry, and we found that mCherry+ cells were more likely to be within the dead gate (Figure 4D). This aligns with previous findings in the literature that intracellular infection can lead to cell death by a range of cell-death pathways (12, 42–44) and implies discreet induction of cell death where bacteria do invade intracellularly. More intriguingly, by use of an Lgr5-GFP reporter mouse, we observed that a higher proportion of Lgr5+ cells was infected compared with other epithelial (EpCAM+Lgr 5–) or nonepithelial cells, with up to 90% of Lgr5+ cells being mCherry+ in the dead gate and around 30% in the live gate, compared with other cell types, which showed around 1%–5% of cells infected (Figure 4E). It has previously been reported that Salmonella and other intracellular pathogens preferentially invade mitotic and dividing cells; thus, Lgr5+ stem cells may be more prone to infection (45, 46). This raises the interesting possibility that STm could be used as a tool to directly affect cancer stem cells. However, it is unlikely that STm will reach every tumor stem cell necessary (see STm form distinct extracellular colonies) to eradicate the tumor by that mechanism alone, and many other cells types play a role in tumor progression. Taken together, these data show that STm can potently modulate the transcriptional landscape of tumors, and reduction in stem cell–associated transcripts is supported by the flow cytometry analysis showing an accumulated of infected Lgr5+ cells within the dead fraction.
STmΔaroA alters the metabolic environment of tumors.
Previous studies have demonstrated that BCT can affect tumor growth by utilizing excess nutrients, such as ethanolamine (47), or are attracted to tumors due to high levels of metabolites such as ribose or leucine (48). Our observation of large intratumoral, extracellular STmΔaroA colonies led us to question whether the tumor metabolome would be altered following treatment. From 4 hours to 24 hours after infection, there is a large increase in CFUs, and along with the appearance of the microbes in the SEM analysis (Figure 2) we hypothesized that bacteria would be rapidly dividing and, therefore, competing for essential metabolites within the tumor environment. Tumor and normal tissue from nontreated or STmΔaroA-treated CAC–tumor-bearing mice after 6 weeks or 24 hour of treatment were analyzed by gas chromatography–mass spectrometry (GC-MS) analysis for polar metabolites. Unit variance–scaled (UV-scaled) GC-MS data were analyzed, and orthogonal partial least squares–discriminant analysis (OPLS-DA) plots revealed a separation between nontreated and treated tumors after 6 weeks — but more importantly within 24 hours (Figure 5, A and B) (6 weeks treatment in vivo, R2 = 0.99; Q2 = 0.52; 24 hours treatment in vivo, R2 = 0.99; Q2 = 0.67). It is possible that the alteration in the metabolome status at 6 weeks could be due to the reduced tumor burden. However, we selected remaining larger tumors for analysis, which did not differ overall in size (Supplemental Figure 7B), though this does not rule out altered tumor characteristics. Importantly, at 24 hours, there is no difference in tumor burden between treatment groups (Supplemental Figure 7C); however, we observed dramatic changes in the metabolome, which is concurrent with the large increase in bacterial CFU, illustrating a direct impact of STmΔaroA on the tumor metabolic environment early after invasion. This precedes reduction in tumor size and likely aids in driving the reduction in tumor burden.
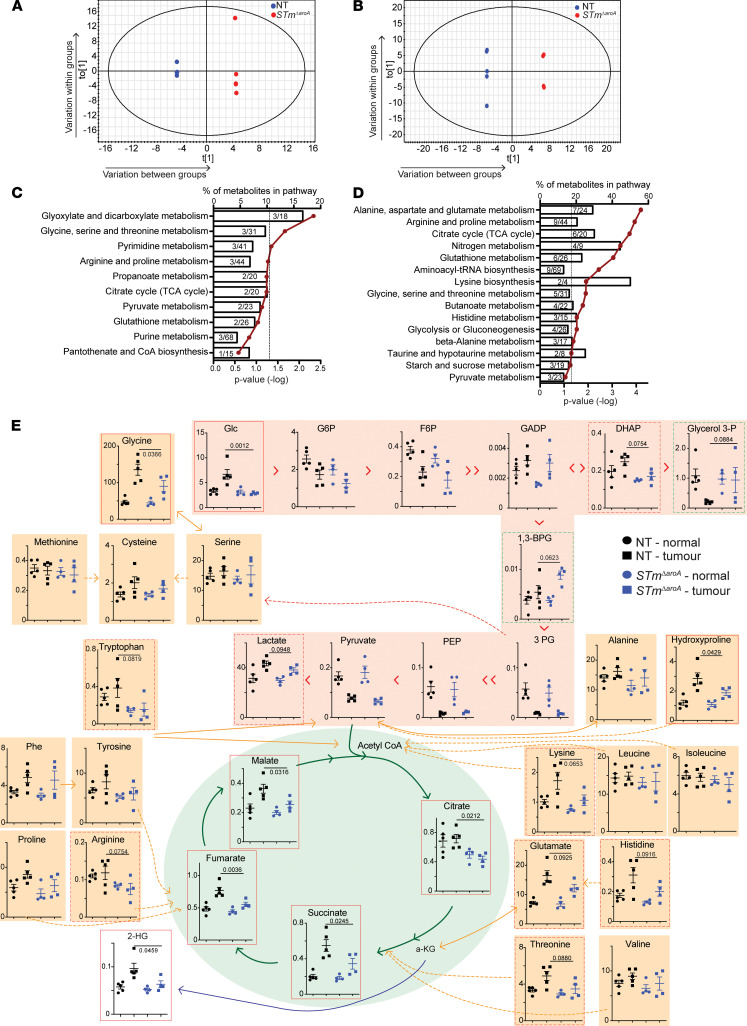
Figure 5. STmΔaroA treatment alters the metabolic environment of CAC tumors.
Tumor metabolites of CAC-induced colon tumors were assessed by GC-MS. (A and B) OPLS analysis of metabolites comparing nontreated (NT) and STmΔaroA-treated tumors after 6 weeks (A) and 24 hours (B) of treatment. The size of tumors used for analysis is shown in Supplemental Figure 7, B and C. All metabolites significantly different between STmΔaroA-treated and nontreated tumors (VIP score > 1) were submitted to pathway analysis (MetaboAnalyst). (C and D) Pathway analysis for 6 weeks of STmΔaroA treatment (C) and 24 hours treatment (D), represented as the percentage of metabolites in a pathway that were altered, against P value (–log); hypergeometric test used. (E) Metabolites detected from glycolysis (pink shading) and TCA cycle (green shading), and amino acids (orange shading), with interrelationships depicted (24 hours after treatment). The x axis shows nmol/g. One-way ANOVA was performed with Bonferroni multiple-comparison test; P values shown are the multiple-comparison statistic. Data are shown as mean ± SD. Both 6-week and 24-hour analyses were performed on 2 independent experiments, with similar changes observed in both sets.
We performed pathway analysis on metabolites with a variable importance on the projection (VIP) score greater than 1 using MetaboAnalyst 3.0 (49, 50) (Supplemental Tables 3 and 4 show the complete list). Common pathways affected by STmΔaroA treatment at both time points (6 weeks and 24 hours) included glycine, serine, and threonine metabolism; arginine and proline metabolism; and citric acid cycle, among others (Figure 5, C and D). As previously described (19), many metabolites — and particularly amino acids and tricarboxylic acid (TCA) cycle intermediates — are increased in tumor tissue compared with normal tissue (Figure 5E and Supplemental Figures 10 and 11). This likely reflects the increased energy and anabolic requirements of tumors. Strikingly, many metabolites were decreased following STmΔaroA treatment. Figure 5E shows metabolites detected in glycolysis and the TCA cycle, as well as amino acids 24 hours after treatment. Perhaps not surprisingly, glucose was significantly reduced in STmΔaroA-treated tumors (Figure 5E). Other glycolysis intermediates were only mildly affected and glycerol-3-phosphate, and 1,3 bisphosphoglycerate (1,3-BPG) trended to increase in STmΔaroA-treated tumors. Several TCA cycle intermediates (citrate, succinate, fumarate, and malate) were reduced following treatment (Figure 5E). Furthermore, several amino acids, which can feed different parts of the TCA cycle were reduced. This included glutamate (glutamine was not detected) (Figure 5E), which is an important fuel for glutaminolysis, upon which tumors can be quite dependent (51).
Other important oncometabolites were also affected. The polyamine synthesis pathway appears affected at both 24 hours and 6 weeks after treatment. Ornithine was reduced in STmΔaroA-treated tumors 24 hours and 6 weeks after treatment, while putrescine was significantly affected after 6 weeks of treatment and spermidine after 24 hours (Supplemental Figures 10 and 11). 2-Hydroxyglutarate (2-HG) can accumulate in tumors due to mutant or overactive isocitrate dehydrogenase 1/2 (IDH2) activity, converting α-ketoglutarate (α-KG) to 2-HG, which can inhibit α-KG–dependent dioxygenases, leading to increased histone and DNA methylation (51). We saw increased 2-HG in CAC tumors, and this was decreased after 24 hours of treatment (Figure 5E). Thus, several important fuel sources, metabolic intermediates, and oncometabolites are decreased following STmΔaroA treatment, presumably through metabolic competition between the bacteria and tumor cells.
STmΔaroA directly affects tumor epithelium.
Our initial hypothesis was that BCT would have a direct effect on tumor epithelium. The effects that we have described so far on tumor stem cell markers, namely an increase in epithelial identity and a change in the tumor metabolome in STmΔaroA-treated tumors, suggests an impact of STmΔaroA treatment on the tumor environment and on tumor cells. To directly test this hypothesis, we utilized tumor 3D organoid cultures. We generated tumor organoid lines from CAC-induced colorectal tumors and from Apcmin/+ SI and colonic polyps/tumors. These grow independently of exogenous Wnt pathway agonists (R-spondin and noggin), and the only supplement provided in the culture was EGF (52). Representative images of organoid appearance are shown in Supplemental Figure 12, A–C. Tumor organoids were infected with STmΔaroA by inoculating the culture medium (1 × 108 CFU). STmΔaroA were able to invade the Matrigel and infect the organoids (Figure 6A and Supplemental Figure 12B). After 2 hours of infection, the culture medium was washed off, and fresh medium containing gentamycin was added, so only bacteria that had infected organoids could grow, preventing any effects purely from bacterial overgrowth. Organoids were then collected for analysis 24 hours after the initial infection. CFU analysis was performed to determine bacterial burden (Figure 6B). We detected approximately 1 × 105 CFU per well (Supplemental Figure 12D), which contains around 1 × 106 cells within the organoid structures. Importantly, treatment of organoids with STmΔaroA could recapitulate effects on gene expression seen in vivo, with a substantial reduction in transcripts for Lgr5, Smoc2, and Vim in both CAC-derived and Apcmin/+-derived tumor organoids, as well as Pdk4 in Apcmin/+ organoids (expression was very low in CAC organoids) (Figure 6, C and D). As seen with the RNA-Seq data set (Figure 3), transcripts were not only decreasing after STmΔaroA treatment, but they showed dynamic changes. For example, an innate immune protein known to respond to bacterial infection, lipocalin-2 (Lcn2) (53), shows robust induction following organoid infection (Figure 6C). This confirms that the reduction in specific transcripts — for example, affecting stem markers — is not a global transcriptional repression. Of note, mRNA quality and amount was consistently similar between treatment groups, and Ct values for housekeeping genes were also the same between groups, showing that decreases in certain transcripts are not due to dying cells
Figure 6. In vitro treatment of tumor-derived organoids with STmΔaroA.
Tumor organoids derived from CAC-induced tumors and Apcmin/+ tumors were established and infected with GFP-expressing STmΔaroA for 2 hours. Infection was washed off, and then, organoids were cultured with gentamycin for a further 24 hours. (A) Merged bright-field and fluorescence microscope image of organoids within matrigel after 24 hours of infection shows association of STmΔaroA with tumor organoids. Scale bar: 50 μm. (B) CFU of STmΔaroA per well after 24 hours of infection. (C and D) qPCR analysis of the indicated transcripts in CAC-derived (C) and Apcmin/+ tumor (D) organoids. Representative of > 3 independent experiments with 4 independently derived organoid lines, with between 3 and 5 technical replicates per experiment. One replicate is 1 well of a 24-well plate culture with a 50 μL Matrigel dome. (E and F) Tumor organoid metabolites were assessed by GC-MS, OPLS-DA analysis, and pathway analysis of metabolites with a VIP score > 1. (G) CAC-derived tumor organoids were cultured with live or heat-killed (dead) STmΔaroA or with supernatant (SN) of STmΔaroA grown in organoid culture medium and the indicated mRNAs analyzed by qPCR. (H) Analysis of succinate levels in tumor organoids treated as described in G. Individual 2-tailed Student’s t tests (C and D) or Kruskal-Wallis tests (G and H) were performed. Data are shown as mean ± SD.
Next, we tested whether STmΔaroA treatment in vitro would have an effect on the cellular metabolome of the organoids. As with the in vivo findings, the organoid metabolome demonstrated separation of nontreated and treated organoids by OPLS analysis (Figure 6E). Taking all metabolites with a VIP score > 1 (Supplemental Table 5) and analyzing by MetaboAnalyst revealed similarly affected metabolic pathways following in vitro STmΔaroA treatment as for in vivo treatment, with amino acid metabolism pathways, TCA cycle, and glycolysis being altered (Figure 6F and Supplemental Figure 13). These data suggest that bacterial colonization imposes direct metabolic competition, leading to an altered cellular metabolome. These results provide evidence that STmΔaroA treatment can directly affect the tumor cells, independently of effects involving other systems/cell types, such as the immune system.
To further dissect whether live bacteria are required to mediate the observed effects of STmΔaroA on tumor organoids, or whether the presence of heat-killed bacteria or bacterial supernatant (SN) would be sufficient, we compared treatment of tumor organoids with live STmΔaroA, heat-killed STmΔaroA, or STmΔaroA SN (prepared using 10 kDa exclusion columns). Live bacteria had the strongest effect on reducing stem cell and EMT marker expression (Figure 6G). Heat-killed bacteria induced a slight reduction in Smoc2 and Vim, while STmΔaroA SN had no effect (Figure 6G), suggesting that secreted products from bacteria are not exerting these antitumor effects (note 10 kDa filters exclude LPS; Supplemental Figure 12D). Furthermore, succinate, one of the metabolites identified as being reduced by STmΔaroA treatment in vivo, was measured, and only live STmΔaroA treatment resulted in reduced levels (Figure 6H), further supporting the idea that live STmΔaroA directly impose metabolic competition.
An effect of STmΔaroA treatment on tumor organoid stem–forming capacity.
As we observed effects of stem cell–related transcripts after STmΔaroA treatment both in vivo and in vitro, we assessed the effect of STmΔaroA treatment on organoid-forming capacity. Colon-derived tumor organoids were either PBS- or STmΔaroA-treated for 24 hours (as described in Figure 6) and were then dissociated into single cells. Cell counts were performed, and equal numbers were reseeded and regrowth was followed over subsequent passages. We found that STmΔaroA-treated organoids had reduced capacity to regrow in the first 2 passage, with reduced cell number (Supplemental Figure 14, A–C) and growth as measured by MTT assay (Figure 7A). STmΔaroA-treated organoids recovered to the same density by late passage 2 into passage 3 (Figure 7A and Supplemental Figure 14, A–C). It is worth noting that STmΔaroA did not persist in the organoids when we dissociated them for reseeding. Infection by Salmonella can lead to inflammasome activation, pyroptosis, and release of LDH into the culture medium (12, 42–44). We first tested cell death first by LDH release, and we found no increase in LDH release over 24 hours in STmΔaroA-treated organoids compared with NT (Figure 7B). We also assessed active caspase 3, as this may be more sensitive to localized cell death. There was a moderate, yet significant, increase in active caspase 3 levels upon STmΔaroA treatment, which was reversible with a pan-caspase inhibitor (Figure 7C). Treatment with staurosporine (STS) showed much more robust induction of caspase 3 (Figure 7C). Thus, while there was a degree of apoptosis occurring with STmΔaroA treatment, it was not completely cytotoxic. This is consistent with the estimate that approximately 1:10–1:100 (experimental variability) cells within the organoid culture are infected. Given that a small number of cells are infected by STm in vivo, and Lgr5+ cells showed greater propensity to be infected and undergo cell death, we assessed Lgr5-GFP tumor organoids by flow cytometry. Similar to what we observed in vivo, mCherry+ cells were more likely to be within the dead gate (Figure 7D), and of mCherry+ cells, there was a higher proportion that were Lgr5+ (Figure 7E). Altogether, these data show that STm treatment resulted in a low level of cell death that appears to predominantly affect the Lgr5+ stem compartment.
Figure 7. STmΔaroA treatment affects tumor organoid stem–forming capacity.
(A) Organoids were infected with STmΔaroA (or control) as in Figure 6 for 24 hours. They were then dissociated into a single cell suspension. An equal number was then reseeded into Matrigel and passaged weekly at an equal density for 3 weeks. MTT assay was performed at the indicated day. Representative images are shown below for the indicated days. Scale bars: 500 μm. Each point indicates an independent well. Two-way Students T-test performed. Representative of 2 experiments, data shown from Apcmin/+,SI tumor line. (B) Measurement of LDH in the cell culture supernatant after 24 hours of infection. Data shown as percentage of cell death compared with wells treated with cell lysis solution. Each data point indicates an independent well. Data are representative of 3 experiments. (C) Active caspase 3 assessed by a plate-based colorimetric assay on organoids infected as in B, with the addition of a pan-caspase inhibitor or staurosporine (STS) alone. Each point is an individual well. One-way ANOVA with Dunnett’s multiple-comparison test. Representative images to the right. Scale bar: 500 μm. Data are representative of 2 independent experiments and shown from Apcmin/+,SI tumor line. (D and E) Tumor organoids derived from Lgr5-GFP reporter mice induced with CAC were infected with mCherry-expressing STmΔaroA for 24 hours, as outlined. Organoids were dissociated into single cells, stained with a live/dead marker, and analyzed by flow cytometry. The percentage of cells that are infected (mCherry+) in the live or dead cell gate (D) and the percentage of cells from the mCherry+ gate that are EpCAM+Lgr5– or EpCAM+Lgr5+ (E) are shown. Data are pooled from 2 independent experiments, and each point is an average from 2 wells. Data are shown as mean ± SD.
A single dose of STmΔaroA in vivo can reduce tumor burden.
Since we see an initial effect of STmΔaroA on tumor organoids that subsides once the bacteria are removed, we surmised that the continual dosing of STm would be required in vivo, since they do decline over time in tumors (Supplemental Figure 1). We treated CAC-induced and Apcmin/+ tumor–bearing mice with either 1 (2 for Apcmin/+) dose, or consecutive weekly dosing as previously. Since these experiments were performed in a different animal facility, we found that overall survival of tumor-bearing mice was reduced compared with previous experiments, with CAC-induced mice developing rectal prolapses due to tumor bulk at the rectum and Apcmin/+ mice developing anemia (pale paws being an ethical end point). We found increased survival in CAC-induced mice treated with either 1 or 6 consecutive doses of STmΔaroA compared with control-treated mice (Figure 8A). Additionally, there was a significant decrease in the tumor burden and tumor size of mice treated with STmΔaroA, in both the 1- and 6-dose groups, compared with control treatment (Figure 8A), indicating that a single dose of STmΔaroA is sufficient to exert antitumor effects. Similar to CAC-induced tumor–bearing mice, Apcmin/+ mice treated 8 doses had a similar reduction in polyp burden as mice given 2 doses (Figure 8B); in this case, mice received 2 doses in the first 2 weeks and then PBS control for the remaining 6 weeks. Control-treated mice also showed a trend toward decreased survival, as seen in the CAC model (Figure 8B); however, this was not statistically significant, likely due to relative underpowering of the groups. We aimed to asses CFU of tumors or polyps from mice given either the short dosing or continuous STmΔaroA dosing. It appears that CFUs have mostly contracted in the 2-week dosing compared with 8 weeks of dosing, which would be consistent with the observation that CFUs diminish at 2 weeks after treatment (Figure 8B; 2 doses yielded just 8 CFU in 1 sample and none in the other). However, we cannot completely exclude colonization below the limit of detection, and despite the resolution of the STmΔaroA by the end of the treatment protocol, there is still effective reduction in tumor burden. The idea that 1 or 2 doses is sufficient to reduce tumor burden might indicate that initial outgrowth of STm within tumors and competition for metabolites are key factors in driving tumor regression, as is induction of cell death in infected stem (and other) cells.
Figure 8. Shorter treatment regimens of STmΔaroA yield similar protection.
(A) AOM/DSS CAC was induced as per Figure 1A in female C57B6/J mice. Mice were then split into no treatment (NT, PBS control) and 1 or 6 doses of STmΔaroA (given once per week via oral gavage). The left is survival from treatment start point (P = 0.0184 Mantel-Cox log-rank test), the middle is the tumor burden, and the right is tumor load. (B) Apcmin/+ mice were treated from 9 weeks of age with either PBS control (NT), 2 doses of STmΔaroA (with PBS control for remaining 4 weeks), or 6 doses of STmΔaroA. The left is survival (Mantel-Cox log-rank test), the middle is number of polyps per mouse (small intestine), and the right shows CFU obtained from polyps at necroscopy. Data are shown as mean ± SD.
Discussion
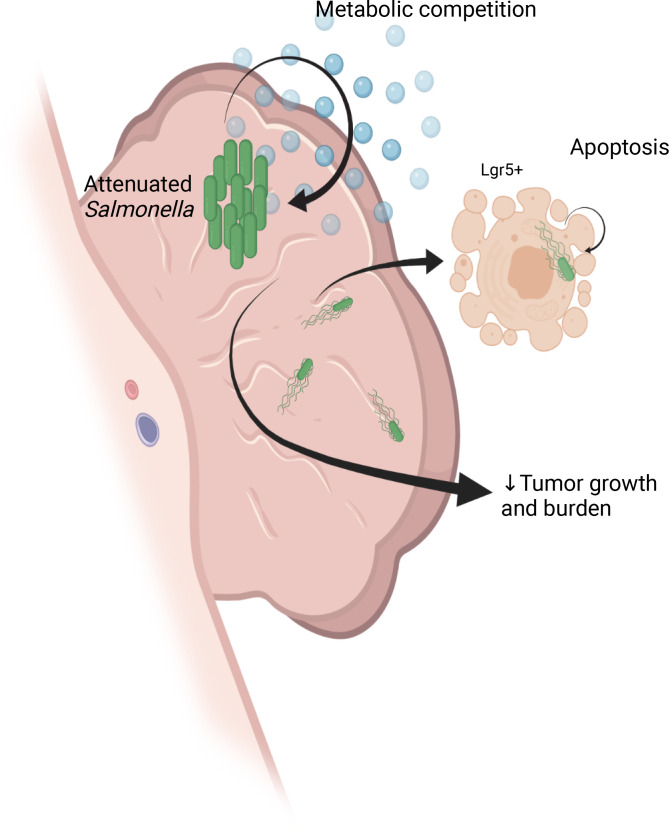
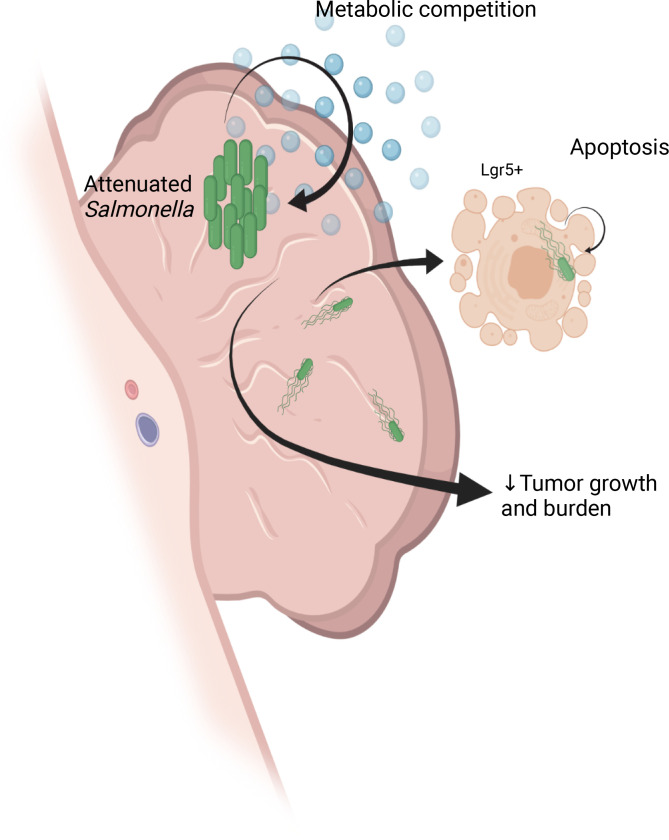
In this study, we present data showing that BCT can be efficacious in in situ models of intestinal cancer, and this is the first study to our knowledge to assess oral delivery of BCT in autochthonous CRC models. Oral delivery of STmΔaroA to colonic or SI tumor–bearing mice induced a strong reduction in tumor number and size. This was preceded by a dramatic shift in the tumor metabolic landscape, which persisted over treatment. Later, reductions in stem cell–associated, cell cycle, and proliferation-related transcripts were observed, along with a reduction in tumor size. In vitro infection of tumor organoids recapitulated effects seen on the tumor metabolome, and reduced stem cell–associated transcripts were associated with delayed regrowth following withdrawal of STmΔaroA. We also observed an overrepresentation of Lgr5+ cells that were infected and dying, both in vivo and in vitro, which may explain the reduction in stem cell–associated transcripts observed. This targeting of tumor stem cells, along with metabolic competition, likely drives nonimmune-mediated effects of STmΔaroA therapy (Figure 9).
Figure 9. Summary.
Attenuated STm therapy shows efficacy in mouse models of autochthonous cancers of the intestine. STm accumulate within 24 hours of oral administration in large intratumoral colonies affecting the tumor metabolic environment. STm also invade a small proportion of tumor and tumor-associated cells, which undergo cell death. Lgr5+ stem cells are preferentially invaded, and accordingly, decreased stem markers can be observed following STm treatment. In summary, this study highlights the feasibility of oral STm therapy of colorectal cancer, and it highlights some previously unappreciated effects of bacterial cancer therapy. Figure created with BioRender.com.
Previous studies have utilized orthotopic or xenograft transplant tumor models, which may not fully recapitulate complex tumor environments in spontaneously formed tumors. Furthermore, studies have delivered bacteria via i.p. or i.v. routes, which, while efficacious in mice, has not been successful in humans. In a phase 1 trial, giving heavily attenuated STm (VNP20009) i.v. resulted in toxicity and poor tumor localization (16), whereas another small trial administering bacteria by intratumoral injection had better tumor localization (54). Lack of chemotactic ability of the VNP20009 strain, due to mutation of the cheY gene, has been suggested to be a limiting factor to its success. Mouse models have shown cheY to be redundant (55), while another study has shown it to be important (47), for tumor localization. Crull et al. (14) hypothesized that tumor invasion in vivo is more passive than in vitro, as the resulting chemokine and cytokine release upon i.v. or i.p. delivery of STm would open tumor vasculature, enabling delivery of bacteria to the tumor. Importantly, the human serum complement system is known to be far more effective than that of mouse (56), and the ΔaroA strain of STm has been shown to have increased sensitivity to complement due to alterations in the LPS structure (22). Thus, i.v. delivery of STm in humans likely leads to rapid clearance of bacteria; therefore, more feasible delivery routes need to be considered to move more BCTs into the clinic.
BCG therapy, the only currently approved BCT, is given directly onto the bladder epithelium via intravesicle delivery, where it is thought to directly affect the bladder epithelium via fibronectin interaction, which precedes immune cell recruitment (10). Additionally, Coley’s original experimental treatment involved direct injection into tumors (1). This suggests that BCT may be more effective where it can be applied more locally. Oral delivery of attenuated STm would feasibly enable targeted colonic tumor delivery while bypassing any i.v. route–associated toxicity. Proof of principle on tolerance and safety of such treatment can be seen with S. Typhi vaccination (15).
We tested whether STmΔaroA treatment affected the composition of the colonic microbiome and found no significant changes. This is in contrast to infection with WT Salmonella (27). One caveat is that we only tested the microbiota at the end point and not early during initial STmΔaroA exposure; thus, it is possible that changes could occur earlier during treatment. However, we did not observe any long-lasting effects on microbiome structure. This is encouraging for therapeutic application, since alteration of the microbiome could have unforeseen consequences. In addition, by testing the treatment in GF mice, we found that there were very strong effects when there was no other competition to colonize the gut, as with SPF mice. However, this very artificial system also demonstrates the importance of the microbiota in protecting the host, as even STmΔaroA could invade and infect systemically. Treating cancer patients with antibiotics prior to surgery is common practice, but in light of the importance of diverse microbiota for controlling cancer (28, 29), it has become apparent that it is not optimal for patient outcome, with antibiotic preconditioning leading to worse outcomes (57–59). Our data do not exclude that there could be certain microbiota compositions that will enable more effective therapy, as with checkpoint blockade therapy (28, 29, 59). Therefore, analyzing microbiomes may be something to consider when starting human clinical trials with orally delivery BCT.
Tumor tissue tropism of attenuated bacteria is thought to be driven by the lack of immune detection within tumors and also the metabolic environment. Previous studies have shown that STm genes involved in ethanolamine catabolism were advantageous for STm within tumors (47) and that STm utilize nutrient-sensing pathways to localize to tumors (48). While the tumor metabolic environment has been suggested to be important for bacterial tumor homing, it was not appreciated what impact BCT might have on the tumor metabolome. We show a dramatic change in the tumor metabolome following STm treatment. As has been previously reported (19), tumors have higher levels of a wide range of metabolites compared with normal tissue, including sugars; central carbon metabolites; and amino acids, including AAs (Phe, Trp, Tyr) (Figure 4 and Supplemental Figures 7 and 8). We found that STmΔaroA form large intratumoral colonies and drastically reshape the tumor metabolome within 24 hours. Multiple metabolic pathways were affected by STmΔaroA treatment, which would impose strong metabolic pressure on tumors cells, and this would possibly make it more difficult for tumors to switch from one pathway to another to meet energy and anabolic requirements. Crucially, these biochemical effects were not seen in surrounding normal tissue. Since STmΔaroA are auxotrophic for AAs, one might expect greater reductions in AAs than we observed. However, it is clear that the levels of AAs, and many other metabolites, are much more abundant in tumor tissue than in the normal colonic tissue, and so there is likely excess levels required for STmΔaroA growth. Targeting tumor metabolism is an important avenue for cancer therapy, with standard chemotherapies taking advantage of metabolic weaknesses (51). However, not all tumors are susceptible, and side effects from inhibiting all fast-dividing cells limit metabolic inhibitor usage. Furthermore, some tumors are able to metabolically adapt if one pathway is blocked (51). BCT may, therefore, be an avenue for introducing metabolic competition; coupling this with other metabolic inhibitors may enable lower doses of these drugs that may otherwise cause severe side effects.
The data presented here show bulk metabolites from tumors that contain STmΔaroA, so it is not possible to decipher which metabolites are bacterial derived and which are host derived. However, the observed decrease in many metabolites from multiple pathways implies that STmΔaroA utilize tumor-derived metabolites. WT STm have been reported to utilize succinate and lactate within the intestinal environment (27, 60), and we found a reduction of succinate in vitro only when live STmΔaroA, and not heat-killed, were present, suggesting active use of tumor metabolites by STmΔaroA. An interesting question raised by our observation of broad reductions in a range of metabolites within STmΔaroA-treated tumors is how this might affect immune responses. Several studies have demonstrated immune cell recruitment following BCT in xenograft/orthotopic models (9, 61, 62). Studies have shown dependence on innate immune subsets, such that depletion of monocytes or deficiency in key innate sensors such as MyD88 ablates therapeutic efficacy (63–65). In contrast, while some studies show the recruitment of adaptive cells, particularly CD8+ T cells (9), many studies have demonstrated treatment efficacy in Rag–/– or nude mice, or upon CD4+ or CD8+ T cell depletion (5, 6, 8, 64, 66), indicating that adaptive immune responses are redundant. Whether the metabolic environment has a role in the apparent T cell redundancy is of interest. On one hand, we observe a reduction in key oncometabolites, such as 2-HG and lactate, which have known roles in immune suppression or promoting Tregs (67–70). On the other hand, the imposed metabolic competition for essential metabolic fuels, such as glucose or amino acids, may further impede T cell responses (71, 72). We would expect that STm acting as an adjuvant would induce immune cell recruitment, which would be important for maintaining tumor control. It will be of interest to determine whether the effect of metabolic competition by STm is a driver of T cell redundancy in these models and what impact that might have on long-term efficacy of BCT.
We show here that STmΔaroA can directly affect tumor environment and tumor cells and that this accounts — at least in part — for the efficacy of BCT. The effect of STm treatment of tumor organoids supports the hypothesis that BCT can directly affect the tumor, independently of effects via the immune system. Previous studies have shown that treatment of xenograft tumors with an AR-1–deficient STm strain could force tumor cells from G0/G1 to S/G2/M phase, sensitizing the tumor to chemotherapy with combined methioninase therapy (73, 74), thus also demonstrating a way by which STm treatment directly affects tumor cells. We provide evidence here of a reduction in tumor stemness characteristics as seen by reduced Lgr5 and Smoc2 in both CAC and Apcmin/+ models, as well as in tumor organoids derived from both models. We also found a slight increase in active caspase 3 following treatment in vitro, and using an Lgr5 reporter mouse, we found an overrepresentation of mCherry-STmΔaroA in Lgr5+ cells both in vivo and in vitro, with a majority of infected cells also appearing in the dead gate. Recent work by Fattinger et al., highlighted that STm infection was capable of inducing mixed cell death pathways in an epithelium-intrinsic manner (42). It is likely that, in our system, STm-mediated inflammasome activation also leads to heterogenous activation of cell death pathways, a process termed PANoptosis (75). Although outside of the scope of this study, it will be of interest to dissect/define the relative requirements for apoptotic, pyroptotic, and necroptotic cell death in the success of this therapy. It appears that STm treatment has a short-term effect on the ability of organoids to regrow. The reduction in stem cell transcripts and increase in cell death, particularly in Lgr5+ cells, would explain this delayed capacity to regrow. It is not surprising that the STmΔaroA-treated organoids recover; as they are passaged, the metabolic pressure that is imposed by STm is removed, so any surviving stem cells could repopulate the niche. However, given that we initially reseed organoids at the same density, it is likely that there are short-term transcriptional effects on the uninfected cells, possibly via the metabolic changes, which are eventually lost.
Our finding that just 1 or 2 doses of STmΔaroA can induce robust reduction in tumor burden (Figure 8), along with the disappearance of STm colonies over time (Supplemental Figure 1 and Figure 8) implies that the early and striking effect on the metabolic landscape, as well as preferential infection of stem cells by STmΔaroA, likely drive an initial antitumor effect of this therapy. Induction of an immune response is then likely to be important for eliciting longer-term and wider antitumor effects. It is important to note that colonization of tumors was characterized by large extracellular colonies, with some individual bacteria dispersed and infecting intracellularly. Only around 2%–7% of cells within a tumor were infected when analyzed by flow cytometry. Thus, it is implausible that STm will reach every tumor cell, or protumorigenic stromal cells of interest, to induce cell death and removal. Therefore, the effect of STm on the metabolic environment and then eliciting an immune response is critical for the success of BCT. This will be important when considering what tumor characteristics are further targeted by BCT.
Several groups are taking the approach of engineering bacteria to deliver drugs or other compounds that can further promote tumor death or immune clearance (63, 76–80). Given that bacteria home specifically to tumors, they are the ideal device to use to ensure tumor-specific drug targeting (3). The data we present here show that BCT does induce tumor regression in autochthonous models of cancer, and we show strong effects on the tumor metabolome and transcriptome. However, it is apparent that STmΔaroA alone does not cure the mice of intestinal tumors, so further engineering of the bacteria and/or cotherapies are required. By understanding the mechanisms of action, we could further improve the engineering of bacteria for BCT — for example, by delivering an engineered bacterium that can better utilize metabolites or by delivery of a cytotoxic compound that can further permeate through the tumor (81). Furthermore, rational selection of tumor types to be targeted, type of bacteria and attenuations, and delivery route are all likely to be important considerations for the success of BCT. This study demonstrates that CRC is an excellent candidate for targeting with BCT via oral delivery of attenuated STm.
Methods
Detailed methodology can be found in the Supplemental Methods.
Data availability.
RNA-Seq data are uploaded and available online (Gene Expression Omnibus, GSE136029).
Statistics.
All data are presented as mean ± SD, unless otherwise indicated. One-way ANOVA, 2-tailed Students t test, or nonparametric statistical tests were used, as indicated for each figure, and were conducted using GraphPad Prism 8. P < 0.05 was considered statistically significant.
Study approval.
All animal experiments were approved by the Institutional Animal Care and Use Committees of RIKEN Yokohama Branch (2018-1[3]) and Yokohama City University (T-A-17-001) or the University of Birmingham local animal welfare and ethical review body and authorized under the authority of Home Office license P06118734 (held by KMM).
Author contributions
KMM conceived the project, designed and performed experiments throughout, analyzed data, and wrote the manuscript. GMM performed organoid experiments and helped write the manuscript. AC performed flow cytometry and helped write the manuscript. MT provided technical assistance throughout and maintained animal breeding. YN performed the GC-MS metabolomics experiments and analyzed data. IE performed organoid experiments. T Kato performed the microbiota 16s RNA-Seq and analysis. H Oda analyzed RNA-Seq data. T Kanaya generated mCherry and GFP-expressing STmΔaroA, helped establish organoid expertise, and sought local (RIKEN) experimental ethics authority. H Ohno advised on the project and provided funding.
Supplementary Material
Acknowledgments
The authors thank David Bending, Adam Cunningham, Jeff Cole, and Nobuo Sasaki for critical reading of the manuscript and suggestions; Sophia Begum and Lisa Scarfe for technical assistance; Calvin Kuo for providing R-spoI producing cells; and Hidenori Matsui for UF20 STm. KMM was supported by a RIKEN Programs for Young Scientists Foreign Postdoctoral Researcher Fellowship and is currently supported by a CRUK CEA (C61638/A27112). The Japan Society for the Promotion of Science KAKENHI (19H01030) and Japan Agency for Medical Research and Development-Core Research for Evolutional Science and Technology (J15652274), awarded to H Ohno, also supported the work.
Version 1. 10/28/2021
In-Press Preview
Version 2. 12/08/2021
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, Mackie et al. This is an open access article published under the terms of the Creative Commons Attribution 4.0 International License.
Reference information: JCI Insight. 2021;6(23):e139900.https://doi.org/10.1172/jci.insight.139900.
Contributor Information
Gillian M. Mackie, Email: G.M.Mackie@bham.ac.uk.
Alastair Copland, Email: A.Copland@bham.ac.uk.
Masumi Takahashi, Email: siplac91@gmail.com.
Yumiko Nakanishi, Email: yumiko.sato.kj@riken.jp.
Isabel Everard, Email: IME722@alumni.bham.ac.uk.
Tamotsu Kato, Email: tamotsu.kato@riken.jp.
Hirotsugu Oda, Email: hirotsugu.oda@nih.gov.
Takashi Kanaya, Email: takashi.kanaya@riken.jp.
Hiroshi Ohno, Email: hiroshi.ohno@riken.jp.
Kendle M. Maslowski, Email: k.m.maslowski@bham.ac.uk.
References
- 1.McCarthy EF. The toxins of William B. Coley and the treatment of bone and soft-tissue sarcomas. Iowa Orthop J. 2006;26:154–158. [PMC free article] [PubMed] [Google Scholar]
- 2.Alexandroff AB, et al. BCG immunotherapy of bladder cancer: 20 years on. Lancet. 1999;353(9165):1689–1694. doi: 10.1016/S0140-6736(98)07422-4. [DOI] [PubMed] [Google Scholar]
- 3.Forbes NS. Engineering the perfect (bacterial) cancer therapy. Nat Rev Cancer. 2010;10(11):785–794. doi: 10.1038/nrc2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hayashi K, et al. Systemic targeting of primary bone tumor and lung metastasis of high-grade osteosarcoma in nude mice with a tumor-selective strain of Salmonella typhimurium. Cell Cycle. 2009;8(6):870–875. doi: 10.4161/cc.8.6.7891. [DOI] [PubMed] [Google Scholar]
- 5.Zhao M, et al. Tumor-targeting bacterial therapy with amino acid auxotrophs of GFP-expressing Salmonella typhimurium. Proc Natl Acad Sci U S A. 2005;102(3):755–760. doi: 10.1073/pnas.0408422102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao M, et al. Monotherapy with a tumor-targeting mutant of Salmonella typhimurium cures orthotopic metastatic mouse models of human prostate cancer. Proc Natl Acad Sci U S A. 2007;104(24):10170–10174. doi: 10.1073/pnas.0703867104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pawelek JM, et al. Tumor-targeted Salmonella as a novel anticancer vector. Cancer Res. 1997;57(20):4537–4544. [PubMed] [Google Scholar]
- 8.Murakami T, et al. Therapeutic efficacy of tumor-targeting Salmonella typhimurium A1-R on human colorectal cancer liver metastasis in orthotopic nude-mouse models. Oncotarget. 2015;6(31):31368–31377. doi: 10.18632/oncotarget.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang WK, et al. The treatment of mouse colorectal cancer by oral delivery tumor-targeting Salmonella. Am J Cancer Res. 2015;5(7):2222–2228. [PMC free article] [PubMed] [Google Scholar]
- 10.Bevers RFM, et al. Role of urothelial cells in BCG immunotherapy for superficial bladder cancer. Br J Cancer. 2004;91(4):607–612. doi: 10.1038/sj.bjc.6602026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kazmierczak RA, et al. Salmonella bacterial monotherapy reduces autochthonous prostate tumor burden in the TRAMP mouse model. PLoS One. 2016;11(8):e0160926. doi: 10.1371/journal.pone.0160926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sellin ME, et al. Epithelium-intrinsic NAIP/NLRC4 inflammasome drives infected enterocyte expulsion to restrict salmonella replication in the intestinal mucosa. Cell Host Microbe. 2014;16(2):237–248. doi: 10.1016/j.chom.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 13.Allam R, et al. Epithelial NAIPs protect against colonic tumorigenesis. J Exp Med. 2015;212(3):369–383. doi: 10.1084/jem.20140474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crull K, et al. Influence of infection route and virulence factors on colonization of solid tumors by Salmonella enterica serovar typhimurium. FEMS Immunol Med Microbiol. 2011;62(1):75–83. doi: 10.1111/j.1574-695X.2011.00790.x. [DOI] [PubMed] [Google Scholar]
- 15.Levine MM, et al. Large-scale field trial of Ty21a live oral typhoid vaccine in enteric-coated capsule formulation. Lancet. 1987;329(8541):1049–1052. doi: 10.1016/S0140-6736(87)90480-6. [DOI] [PubMed] [Google Scholar]
- 16.Toso JF, et al. Phase I study of the intravenous administration of attenuated Salmonella typhimurium to patients with metastatic melanoma. J Clin Oncol. 2002;20(1):142–152. doi: 10.1200/JCO.2002.20.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gulig PA, Doyle TJ. The Salmonella typhimurium virulence plasmid increases the growth rate of salmonellae in mice. Infect Immun. 1993;61(2):504–511. doi: 10.1128/iai.61.2.504-511.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng K, et al. High levels of aromatic amino acids in gastric juice during the early stages of gastric cancer progression. PLoS One. 2012;7(11):e49434. doi: 10.1371/journal.pone.0049434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirayama A, et al. Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time-of-flight mass spectrometry. Cancer Res. 2009;69(11):4918–4925. doi: 10.1158/0008-5472.CAN-08-4806. [DOI] [PubMed] [Google Scholar]
- 20.Hoiseth SK, Stocker BAD. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature. 1981;291(5812):238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- 21.Avogadri F, et al. Intra-tumoral Salmonella typhimurium induces a systemic anti-tumor immune response that is directed by low-dose radiation to treat distal disease. Eur J Immunol. 2008;38(7):1937–1947. doi: 10.1002/eji.200738035. [DOI] [PubMed] [Google Scholar]
- 22.Felgner S, et al. Aroa-deficient Salmonella enterica serovar typhimurium is more than a metabolically attenuated mutant. mBio. 2016;7(5):e01220–16. doi: 10.1128/mBio.01220-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Avogadri F, et al. Cancer immunotherapy based on killing of Salmonella-infected tumor cells. Cancer Res. 2005;65(9):3920–3927. doi: 10.1158/0008-5472.CAN-04-3002. [DOI] [PubMed] [Google Scholar]
- 24.De Robertis M, et al. The AOM/DSS murine model for the study of colon carcinogenesis: from pathways to diagnosis and therapy studies. J Carcinog. 2011;10(1):9. doi: 10.4103/1477-3163.78279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crull K, et al. Biofilm formation by salmonella enterica serovar typhimurium colonizing solid tumours. Cell Microbiol. 2011;13(8):1223–1233. doi: 10.1111/j.1462-5822.2011.01612.x. [DOI] [PubMed] [Google Scholar]
- 26.Taylor WS, et al. MinION Sequencing of colorectal cancer tumour microbiomes-A comparison with amplicon-based and RNA-sequencing. PLoS One. 2020;15(5):e0233170. doi: 10.1371/journal.pone.0233170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gillis CC, et al. Dysbiosis-associated change in host metabolism generates lactate to support Salmonella growth. Cell Host Microbe. 2018;23(1):54–64. doi: 10.1016/j.chom.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sivan A, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350(6264):1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vétizou M, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350(6264):1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maslowski KM, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461(7268):1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang DW, et al. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang DW, et al. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 33.Pfister SP, et al. Uncoupling of invasive bacterial mucosal immunogenicity from pathogenicity. Nat Commun. 2020;11(1):1978. doi: 10.1038/s41467-020-15891-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shvab A, et al. Induction of the intestinal stem cell signature gene SMOC-2 is required for L1-mediated colon cancer progression. Oncogene. 2016;35(5):549–557. doi: 10.1038/onc.2015.127. [DOI] [PubMed] [Google Scholar]
- 35.Muñoz J, et al. The Lgr5 intestinal stem cell signature: Robust expression of proposed quiescent ′ +4′ cell markers. EMBO J. 2012;31(14):3079–3091. doi: 10.1038/emboj.2012.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hirsch D, et al. LGR5 positivity defines stem-like cells in colorectal cancer. Carcinogenesis. 2014;35(4):849–858. doi: 10.1093/carcin/bgt377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barker N, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457(7229):608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 38.Leclerc D, et al. Oncogenic role of PDK4 in human colon cancer cells. Br J Cancer. 2017;116(7):930–936. doi: 10.1038/bjc.2017.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Satelli A, Li S. Vimentin in cancer and its potential as a molecular target for cancer therapy. Cell Mol Life Sci. 2011;68(18):3033–3046. doi: 10.1007/s00018-011-0735-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Makrilia N, et al. Cell adhesion molecules: role and clinical significance in cancer. Cancer Invest. 2009;27(10):1023–1037. doi: 10.3109/07357900902769749. [DOI] [PubMed] [Google Scholar]
- 41.Takeichi M. Dynamic contacts: rearranging adherens junctions to drive epithelial remodelling. Nat Rev Mol Cell Biol. 2014;15(6):397–410. doi: 10.1038/nrm3802. [DOI] [PubMed] [Google Scholar]
- 42.Fattinger SA, et al. Epithelium-autonomous NAIP/NLRC4 prevents TNF-driven inflammatory destruction of the gut epithelial barrier in Salmonella-infected mice. Mucosal Immunol. 2021;14(3):615–629. doi: 10.1038/s41385-021-00381-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Opdenbosch N, et al. Caspase-1 engagement and TLR-induced c-FLIP expression suppress ASC/caspase-8-dependent apoptosis by inflammasome sensors NLRP1b and NLRC4. Cell Rep. 2017;21(12):3427–3444. doi: 10.1016/j.celrep.2017.11.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rauch I, et al. NAIP-NLRC4 inflammasomes coordinate intestinal epithelial cell expulsion with eicosanoid and IL-18 release via activation of caspase-1 and -8. Immunity. 2017;46(4):649–659. doi: 10.1016/j.immuni.2017.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Santos AJM, et al. Preferential invasion of mitotic cells by salmonella reveals that cell surface cholesterol is maximal during metaphase. J Cell Sci. 2013;126(14):2990–2996. doi: 10.1242/jcs.115253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kolb-Mäurer A, et al. Interaction of human hematopoietic stem cells with bacterial pathogens. Blood. 2002;100(10):3703–3709. doi: 10.1182/blood-2002-03-0898. [DOI] [PubMed] [Google Scholar]
- 47.Silva-Valenzuela CA, et al. Solid tumors provide niche-specific conditions that lead to preferential growth of Salmonella. Oncotarget. 2016;7(23):35169–35180. doi: 10.18632/oncotarget.9071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kasinskas RW, Forbes NS. Salmonella typhimurium lacking ribose chemoreceptors localize in tumor quiescence and induce apoptosis. Cancer Res. 2007;67(7):3201–3209. doi: 10.1158/0008-5472.CAN-06-2618. [DOI] [PubMed] [Google Scholar]
- 49.Xia J, et al. MetaboAnalyst 3.0-making metabolomics more meaningful. Nucleic Acids Res. 2015;43(w1):W251–W257. doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xia J, Wishart DS. Using metaboanalyst 3.0 for comprehensive metabolomics data analysis. Curr Protoc Bioinformatics. 2016;55:14.10.1–14.10.91. doi: 10.1002/cpbi.11. [DOI] [PubMed] [Google Scholar]
- 51.Luengo A, et al. Targeting metabolism for cancer therapy. Cell Chem Biol. 2017;24(9):1161–1180. doi: 10.1016/j.chembiol.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sato T, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141(5):1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 53.Singh V, et al. Microbiota-inducible innate immune siderophore binding protein lipocalin 2 is critical for intestinal homeostasis. Cell Mol Gastroenterol Hepatol. 2016;2(4):482–498. doi: 10.1016/j.jcmgh.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cunningham C, Nemunaitis J. A phase I trial of genetically modified Salmonella typhimurium expressing cytosine deaminase (TAPET-CD, VNP20029) administered by intratumoral injection in combination with 5-fluorocytosine for patients with advanced or metastatic cancer. Protocol no: CL-017. Version: April 9, 2001. Hum Gene Ther. 2001;12(12):1594–1596. [PubMed] [Google Scholar]
- 55.Coutermarsh-Ott SL, et al. Effect of Salmonella enterica serovar typhimurium VNP20009 and VNP20009 with restored chemotaxis on 4T1 mouse mammary carcinoma progression. Oncotarget. 2017;8(20):33601–33613. doi: 10.18632/oncotarget.16830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bergman I, et al. Comparison of in vitro antibody-targeted cytotoxicity using mouse, rat and human effectors. Cancer Immunol Immunother. 2000;49(4–5):259–266. doi: 10.1007/s002620000120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lu L, et al. Association of antibiotic exposure with the mortality in metastatic colorectal cancer patients treated with bevacizumab-containing chemotherapy: a hospital-based retrospective cohort study. PLoS One. 2019;14(9):e0221964. doi: 10.1371/journal.pone.0221964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abdel-Rahman O, et al. Outcomes of metastatic colorectal cancer patients in relationship to prior and concurrent antibiotics use; individual patient data analysis of three clinical trials. Clin Transl Oncol. 2020;22(9):1651–1656. doi: 10.1007/s12094-020-02301-1. [DOI] [PubMed] [Google Scholar]
- 59.Elkrief A, et al. The negative impact of antibiotics on outcomes in cancer patients treated with immunotherapy: a new independent prognostic factor? Ann Oncol. 2019;30(10):1572–1579. doi: 10.1093/annonc/mdz206. [DOI] [PubMed] [Google Scholar]
- 60.Spiga L, et al. An oxidative central metabolism enables Salmonella to utilize microbiota-derived succinate. Cell Host Microbe. 2017;22(3):291–301. doi: 10.1016/j.chom.2017.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim JE, et al. Salmonella typhimurium suppresses tumor growth via the pro-inflammatory cytokine interleukin-1β. Theranostics. 2015;5(12):1328–1342. doi: 10.7150/thno.11432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Phan TX, et al. Activation of inflammasome by attenuated Salmonella typhimurium in bacteria-mediated cancer therapy. Microbiol Immunol. 2015;59(11):664–675. doi: 10.1111/1348-0421.12333. [DOI] [PubMed] [Google Scholar]
- 63.Zheng JH, et al. Two-step enhanced cancer immunotherapy with engineered Salmonella typhimurium secreting heterologous flagellin. Sci Transl Med. 2017;9(376):eaak9537. doi: 10.1126/scitranslmed.aak9537. [DOI] [PubMed] [Google Scholar]
- 64.Kaimala S, et al. Salmonella-mediated tumor regression involves targeting of tumor myeloid suppressor cells causing a shift to M1-like phenotype and reduction in suppressive capacity. Cancer Immunol Immunother. 2014;63(6):587–599. doi: 10.1007/s00262-014-1543-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Johnson SA, et al. Monocytes mediate Salmonella typhimurium-induced tumour growth inhibition in a mouse melanoma model. Eur J Immunol. doi: 10.1002/eji.202048913. [published online October 11, 2021]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Luo X, et al. Antitumor effect of VNP20009, an attenuated Salmonella, in murine tumor models. Oncol Res. 2001;12(11–12):501–508. doi: 10.3727/096504001108747512. [DOI] [PubMed] [Google Scholar]
- 67.Brand A, et al. LDHA-associated lactic acid production blunts tumor immunosurveillance by T and NK cells. Cell Metab. 2016;24(5):657–671. doi: 10.1016/j.cmet.2016.08.011. [DOI] [PubMed] [Google Scholar]
- 68.Watson MLJ, et al. Metabolic support of tumour-infiltrating regulatory T cells by lactic acid. Nature. 2021;591(7851):645–651. doi: 10.1038/s41586-020-03045-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kohanbash G, et al. Isocitrate dehydrogenase mutations suppress STAT1 and CD8+ T cell accumulation in gliomas. J Clin Invest. 2017;127(4):1425–1437. doi: 10.1172/JCI90644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Richardson LG, et al. (R)-2-hydroxyglutarate drives immune quiescence in the tumor microenvironment of IDH-mutant gliomas. Transl Cancer Res. 2019;8(suppl 2):S167–S170. doi: 10.21037/tcr.2019.01.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chang CH, et al. Metabolic competition in the tumor microenvironment is a driver of cancer progression. Cell. 2015;162(6):1229–1241. doi: 10.1016/j.cell.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lim AR, et al. The tumor microenvironment as a metabolic barrier to effector T cells and immunotherapy. Elife. 2020;9:e55185. doi: 10.7554/eLife.55185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yano S, et al. Tumor-targeting Salmonella typhimurium A1-R decoys quiescent cancer cells to cycle as visualized by FUCCI imaging and become sensitive to chemotherapy. Cell Cycle. 2014;13(24):3958–3963. doi: 10.4161/15384101.2014.964115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yano S, et al. Tumor-specific cell-cycle decoy by Salmonella typhimurium A1-R combined with tumor-selective cell-cycle trap by methioninase overcome tumor intrinsic chemoresistance as visualized by FUCCI imaging. Cell Cycle. 2016;15(13):1715–1723. doi: 10.1080/15384101.2016.1181240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Malireddi RKS, et al. ZBP1 and TAK1: master regulators of NLRP3 inflammasome/pyroptosis, apoptosis, and necroptosis (PAN-optosis) Front Cell Infect Microbiol. 2019;9:406. doi: 10.3389/fcimb.2019.00406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chowdhury S, et al. Programmable bacteria induce durable tumor regression and systemic antitumor immunity. Nat Med. 2019;25(7):1057–1063. doi: 10.1038/s41591-019-0498-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Loeffler M, et al. Attenuated Salmonella engineered to produce human cytokine LIGHT inhibit tumor growth. Proc Natl Acad Sci U S A. 2007;104(31):12879–12883. doi: 10.1073/pnas.0701959104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jiang SN, et al. Inhibition of tumor growth and metastasis by a combination of Escherichia coli-mediated cytolytic therapy and radiotherapy. Mol Ther. 2010;18(3):635–642. doi: 10.1038/mt.2009.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Din MO, et al. Synchronized cycles of bacterial lysis for in vivo delivery. Nature. 2016;536(7614):81–85. doi: 10.1038/nature18930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chien T, et al. Advances in bacterial cancer therapies using synthetic biology. Curr Opin Syst Biol. 2017;5:1–8. doi: 10.1016/j.coisb.2017.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tan W, et al. Targeting of pancreatic cancer cells and stromal cells using engineered oncolytic Salmonella typhimurium. Mol Ther. doi: 10.1016/j.ymthe.2021.08.023. [published online August 14, 2021]. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-Seq data are uploaded and available online (Gene Expression Omnibus, GSE136029).