ABSTRACT
Influenza A virus (IAV) is passively surveilled in swine in the United States through a U.S. Department of Agriculture administered surveillance system. We present an interactive Web tool to visualize and explore trends in the genetic and geographic diversity of IAV derived from the surveillance system.
ANNOUNCEMENT
Influenza A virus (IAV) is a negative-sense, single-stranded, enveloped RNA virus of the Orthomyxoviridae family. Subtypes H1N1, H1N2, and H3N2 are endemic in swine in the United States, and the major surface proteins, hemagglutinin (HA) and neuraminidase (NA), exhibit significant genetic and antigenic diversity (1). The other 6 internal gene segments also display genetic diversity (2, 3). There are 6 HA evolutionary lineages, 5 NA evolutionary lineages, and 3 internal gene evolutionary lineages in U.S. swine IAV. Within each of these lineages, multiple genetic clades cocirculate and are the result of bidirectional transmission between humans, avian species, and swine, followed by periods of antigenic drift and shift in the swine host.
In the United States, IAV in swine is passively monitored through a U.S. Department of Agriculture (USDA) administered surveillance system. The system was established in 2009 (4) and has since tested over 178,000 samples from more than 55,000 swine diagnostic submissions, resulting in more than 9,000 publicly available virus isolates and genetic sequences (https://www.aphis.usda.gov/aphis/ourfocus/animalhealth/animal-disease-information/swine-disease-information/influenza-a-virus). When initiated, the hemagglutinin (HA), neuraminidase (NA), and matrix (M) genes were sequenced from IAV-positive diagnostic case submissions to participating members of the National Animal Health Laboratory Network. This process was changed to focus on HA and NA, with a randomly selected subset of strains identified for whole-genome sequencing (WGS) to capture the M and remaining internal genes by stratifying the data by subtype and genetic clade representation. All genetic sequence data are published in NCBI GenBank (5), and virus isolates are stored and available to the influenza community at the National Veterinary Services Laboratories, USDA-APHIS (https://www.aphis.usda.gov/aphis/ourfocus/animalhealth/lab-info-services/sa_reagents/ct_reagents). However, analyses of sequence data that quantify changes in genetic diversity to inform control efforts by veterinarians and public health agencies existed only in static publications or in online quarterly reports.
We generated a systematic approach for analyzing and publicly reporting the USDA IAV surveillance sequencing efforts at the single-gene and whole-virus genome levels. A consistent and continued assessment of the genetic diversity of IAV collected as part of the surveillance system identifies spatial and temporal trends in diversity and novel viruses that require additional phenotypic characterization. octoFLUshow is an interactive visualization platform built within the R Shiny framework (6). It offers a searchable overview of all IAV in swine strains collected in the surveillance system from 2009 to present. These data may be refined by collection date, collection location, subtype, genetic clade (U.S. and global nomenclature) (7), and whole-genome constellation. Following refinement, these data are visualized, and graphs can be downloaded to explore spatial and temporal patterns in genetic diversity; heat maps are generated to quantify the relative proportions of HA-NA and whole-genome genetic patterns (Fig. 1). octoFLUshow uses octoFLU (8) for gene classification and octoFLUdb (https://github.com/flu-crew/octofludb) for synthesizing and organizing data on IAV in swine. octoFLUdb is a Python package that parses public data from NCBI GenBank into a graph database with an ontology (9, 10). The tool links metadata such as the collection location, collection date, phylogenetic clade, and whole-genome constellation to the genetic sequence and generates the input files required for octoFLUshow. These modules ingest USDA surveillance data on IAV in swine, provide objective descriptors of the genetic diversity, and allow IAV stakeholders to make informed decisions on vaccine design or use or about the selection of relevant viruses circulating in U.S. swine herds for further characterization.
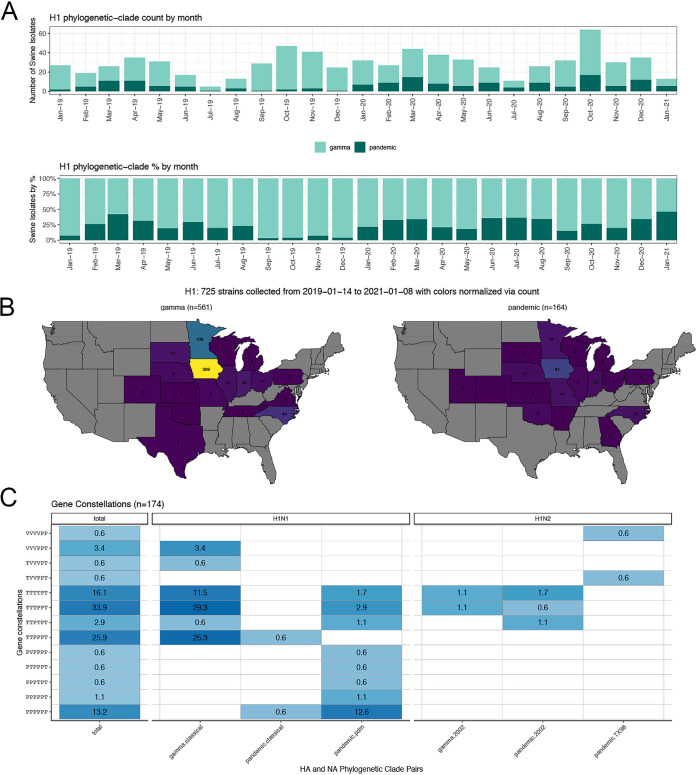
FIG 1.
Example use case of octoFLUshow at https://flu-crew.org/, where a user can select a date range and IAV clades of interest to track their associated detection distribution across time, location, and whole-genome patterns. (A) Detection frequency of two H1 HA clades (1A.3.3.2/pandemic in dark green and 1A.3.3.3/gamma in light green) from January 2019 to January 2021; (B) spatial distribution of each H1 HA clade with the raw count plotted within each U.S. state and colored according to the detection frequency of the HA clade plotted (in this figure, the scale range is 1 to 309); and (C) relative proportions of whole-genome patterns for the selected H1 HA clades, with darker shades of blue indicating higher proportions, the x axis reflecting the HA-NA pairing, and the y axis representing the genome constellation in the order of polymerase basic gene 2, polymerase basic gene 1, polymerase acidic gene, nucleoprotein gene, matrix gene, and nonstructural gene (PB2-PB1-PA-NP-M-NS) and reflecting combinations of either live attenuated influenza virus vaccine lineage (V), the triple reassortant (T) lineage, or lineage H1N1pdm09 (P). Maps were created using R and ggplot2.
Data availability.
The gene segment sequences are available in the Influenza Research Database (11). The tool can be accessed at https://flu-crew.org/ and is hosted on Amazon Web Services as part of the USDA-ARS data science initiative SCINet. The octoFLUshow and octoFLUdb source code is hosted on GitHub (https://github.com/flu-crew/octoflushow and https://github.com/flu-crew/octofludb, respectively). octoFLUdb is also hosted as a Python package on PyPi.
ACKNOWLEDGMENTS
We thank the pork producers, swine veterinarians, and laboratories for participating in the USDA influenza A virus in swine surveillance system and publicly sharing sequences.
This work was supported in part by the USDA-ARS (ARS project number 5030-32000-231-000D), USDA-APHIS (ARS project number 5030-32000-231-080-I), the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services (contract number 75N93021C00015), the USDA-ARS Research Participation Program of the Oak Ridge Institute for Science and Education (ORISE) through an interagency agreement between the U.S. Department of Energy (DOE) and USDA-ARS (contract number DE-AC05-06OR23100), and the SCINet project of the USDA-ARS (ARS project number 0500-00093-001-00-D).
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA, DOE, or ORISE. The USDA is an equal opportunity provider and employer.
Contributor Information
Tavis K. Anderson, Email: tavis.anderson@usda.gov.
John J. Dennehy, Queens College CUNY
REFERENCES
- 1.Anderson TK, Chang J, Arendsee ZW, Venkatesh D, Souza CK, Kimble JB, Lewis NS, Davis CT, Vincent AL. 2021. Swine influenza A viruses and the tangled relationship with humans. Cold Spring Harb Perspect Med 11:a038737. doi: 10.1101/cshperspect.a038737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rajao DS, Walia RR, Campbell B, Gauger PC, Janas-Martindale A, Killian ML, Vincent AL. 2017. Reassortment between swine H3N2 and 2009 pandemic H1N1 in the United States resulted in influenza A viruses with diverse genetic constellations with variable virulence in pigs. J Virol 91:e01763-16. doi: 10.1128/JVI.01763-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao S, Anderson TK, Walia RR, Dorman KS, Janas-Martindale A, Vincent AL. 2017. The genomic evolution of H1 influenza A viruses from swine detected in the United States between 2009 and 2016. J Gen Virol 98:2001–2010. doi: 10.1099/jgv.0.000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson TK, Nelson MI, Kitikoon P, Swenson SL, Korslund JA, Vincent AL. 2013. Population dynamics of cocirculating swine influenza A viruses in the United States from 2009 to 2012. Influenza Other Respir Viruses 7 Suppl 4:42–51. doi: 10.1111/irv.12193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. 2005. GenBank. Nucleic Acids Res 33:D34–D38. doi: 10.1093/nar/gki063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sievert C. 2020. Interactive Web-based data visualization with R, Plotly, and Shiny. CRC Press, Boca Raton, FL. [Google Scholar]
- 7.Anderson TK, Macken CA, Lewis NS, Scheuermann RH, Van Reeth K, Brown IH, Swenson SL, Simon G, Saito T, Berhane Y, Ciacci-Zanella J, Pereda A, Davis CT, Donis RO, Webby RJ, Vincent AL. 2016. A phylogeny-based global nomenclature system and automated annotation tool for H1 hemagglutinin genes from swine influenza A viruses. mSphere 1:e00275-16. doi: 10.1128/mSphere.00275-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang J, Anderson TK, Zeller MA, Gauger PC, Vincent AL. 2019. octoFLU: automated classification for the evolutionary origin of influenza A virus gene sequences detected in US swine. Microbiol Resour Announc 8:e00673-19. doi: 10.1128/MRA.00673-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kohler S, Bauer S, Mungall CJ, Carletti G, Smith CL, Schofield P, Gkoutos GV, Robinson PN. 2011. Improving ontologies by automatic reasoning and evaluation of logical definitions. BMC Bioinformatics 12:418. doi: 10.1186/1471-2105-12-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, Goldberg LJ, Eilbeck K, Ireland A, Mungall CJ, Leontis N, Rocca-Serra P, Ruttenberg A, Sansone SA, Scheuermann RH, Shah N, Whetzel PL, Lewis S, OBI Consortium . 2007. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat Biotechnol 25:1251–1255. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Aevermann BD, Anderson TK, Burke DF, Dauphin G, Gu Z, He S, Kumar S, Larsen CN, Lee AJ, Li X, Macken C, Mahaffey C, Pickett BE, Reardon B, Smith T, Stewart L, Suloway C, Sun G, Tong L, Vincent AL, Walters B, Zaremba S, Zhao H, Zhou L, Zmasek C, Klem EB, Scheuermann RH. 2017. Influenza Research Database: an integrated bioinformatics resource for influenza virus research. Nucleic Acids Res 45:D466–D474. doi: 10.1093/nar/gkw857. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The gene segment sequences are available in the Influenza Research Database (11). The tool can be accessed at https://flu-crew.org/ and is hosted on Amazon Web Services as part of the USDA-ARS data science initiative SCINet. The octoFLUshow and octoFLUdb source code is hosted on GitHub (https://github.com/flu-crew/octoflushow and https://github.com/flu-crew/octofludb, respectively). octoFLUdb is also hosted as a Python package on PyPi.