Abstract
In this short Perspective, we discuss how recent dynamic live-cell imaging experiments have challenged our understanding of mechanisms driving functional molecular interactions in vivo. While we have generally considered the formation of functional biomolecular complexes as resulting from the stable assembly of two or more partner molecules, here we entertain the possibility that function may actually be maintained while molecules are rapidly exchanged within a complex. We postulate that at high effective concentrations, even very weak interactions can lead to strong binding site occupancy and thereby mediate function in a highly dynamic fashion. This new perspective in our definition of what represents a functional complex in living cells and in vivo could significantly alter how we define the nature of molecular transactions critical for mediating regulation in the cellular context. These less conventional principles also allow a broadening of the mechanistic options we should explore when interpreting essential biological processes such as gene regulation.
Our understanding of the molecular interactions, chemical reactions, and remarkable regulatory mechanisms that sustain a living cell remains stubbornly incomplete. Different fields have investigated aspects of this fundamental problem and established some overarching principles to frame our understanding of cell biology. Geneticists have elegantly demonstrated dense and highly interconnected gene networks. Biochemists and molecular biologists have toiled diligently to uncover similar and partly overlapping networks of molecular interactions with painstaking in vitro reconstitution studies. Cell biologists have beautifully mapped how different components of cellular systems are spatially organized by organelles and other subcellular compartments, including membraneless phase-separated condensates. At much higher resolution, biochemists and structural biologists have provided key mechanistic frameworks and structural details about how biological macromolecules interact and enzymes function. This illuminating yet restricted window into molecular mechanisms of macromolecular transactions has largely been limited to strong stoichiometric interactions that allow for purification and subsequent reconstitution. A major hurdle that is just now beginning to yield to analysis is the fact that within a living cell, biomolecules exist in a complex milieu, diffuse very rapidly, and are at much higher effective concentrations than can be achieved in a test tube. Consequently, it has been very difficult to learn the rules of biomolecular interactions in vivo—for example, the requirements for strength of interaction, how specificity is achieved—and their functional relevance to a particular biological process or regulatory system.
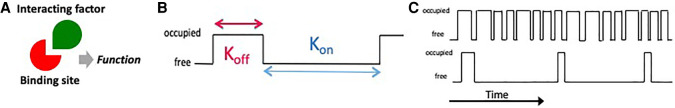
For example, when considering a stochiometric assembly between two factors (Fig. 1A), we typically assess the biophysical properties of the interaction in terms of an association rate (ON-rate) and a dissociation rate (OFF-rate) (Fig. 1B). Dissociation rates have generally been well accepted as an important potentially rate-limiting step that can be measured in reconstituted systems outside their native cellular environment. On the other hand, association rates (ON-rate) have proven to be far more difficult to measure and consequently, less appreciated as a potentially powerful regulatory mechanism. ON-rates, in addition to being dependent on concentrations of the interacting partners of interest, are also highly influenced by the concentrations of many other cellular factors able to compete with the cognate interacting partners. ON-rates also include a potential entropic component that includes both the entropic penalty for complex formation as well as the potential entropic gain from displacing bound water molecules or other solution components. It is therefore essential to consider and measure the ON-rate for a particular assembly in the context of the multiple competing interactions the target molecules invariably are engaged with inside a cell at relevant concentrations at any given time.
FIGURE 1.
(A) When studying an interaction between two molecules, one generally considers the fast-moving particle as the explorer (green interacting factor) and the slower or less mobile one, the binding site or target (in red). Neither the explorer nor the target has any function alone, but they can potentiate a function when assembled into a complex. (B) Complex formation is defined by an OFF-rate that drives how long every formed complex will last in time and reflects an intrinsic property of the interaction in question. The ON-rate is not just an intrinsic parameter dictating frequency of the queried interaction but is also influenced by the diffusion coefficient and effective concentration of the explorer. (C) For relatively weak interactions with a high OFF-rate, the frequency at which the interactions take place will determine how often the target will be occupied, integrated over time. In this ON-rate-dominated setting, how long a particular molecule binds becomes less relevant and the rate-limiting parameter becomes how often the target is visited.
In recent years, weak, multivalent and transient interactions between biomolecules (proteins, RNA, etc.) have been recognized as playing an important role in cellular mechanisms, especially in complex regulatory processes. As an example, this became apparent from our own research when we found that classically defined sequence-specific DNA-binding transcription factors like Oct4 and Sox2 bind to cognate regulatory DNA sites for less than 10 sec on average in live cells, and RNA Pol II forms local concentration clusters with dwell times of only ∼6 sec (Cisse et al. 2013; Chen et al. 2014). Among many such examples emerging from live-cell single-particle imaging, one of the most striking cases is seen during early fly development where the dwell time of the morphogen Bicoid on is less than 1 sec (Mir et al. 2017). This striking finding made us realize that the length of time (residence time) a particular individual trans-acting factor binds stably to its regulatory site may not be the critical regulatory parameter. Instead, it seems likely that what is driving molecular regulatory events may actually be how often the cis-acting DNA site is visited and transiently occupied. Substantial time-averaged occupancy at the site might be the result not of stable binding (low OFF-rate), but of many rapid binding events executed by different molecules of the cognate species (high ON-rate) (Fig. 1C).
It is therefore essential to recognize how the strength and specificity of interactions between molecules in cells coevolved. Biomolecules are decorated with complex surfaces bearing different intrinsic or induced properties (charge, hydrophobicity, etc.) and they are never inert to the surrounding water or the multitude of other macromolecules present in a living cell. While each molecule may engage in a few specific and strong interactions, we must also consider the innumerable weaker interactions, especially as the number and diversity of interacting molecules increases with the complexity and size of genomes. Moreover, proteins typically do not consist only of the well-structured domains amenable to structural biology that give rise to strong and stoichiometric binding interactions, but often contain long intrinsically disordered regions (IDRs). We have also increasingly come to appreciate and recognize the as-yet poorly understood functional contributions of repetitive polymers such as RNA as well as a class of IDRs called low-complexity domains (LCD) whose inherent multivalency offers multiple opportunities to engage in weak interactions. The picture emerging from these findings is one in which macromolecules in vivo are likely perpetually sampling numerous distinct surfaces in their surrounding environment, giving rise to a diversity of weak multivalent interactions that effectively compete with interactions of greater affinity in what we refer to as a strength versus numbers model.
We are especially interested in these weak but numerous interactions because they offer greater potential for regulatory efficiency than the more conventional high-affinity “lock and key” interactions, since their inherently labile nature (high OFF-rates) enhances the sampling of their environment by allowing rapid release from nonproductive, competing interactions. We also envision that protein low complexity domains (LCDs), and nucleic acids (especially RNA) may play a central role in nucleating such weak interactions because of their intrinsic multivalent nature. This concept has been classically defined in one prominent case that can be studied at single-molecule resolution in vitro: sequence-specific DNA-binding proteins interacting nonspecifically and sliding along the surface of bacterial DNA (that is, adsorbed onto the polymer) targeted to their sequence-specific recognition sites in a reduced-dimensionality search. A similar mechanism is also at play in the CRISPR/Cas9 target search (Koh et al. 2012; Lee and Myong 2021). Although a similar phenomenon has yet to be experimentally demonstrated with protein LCDs, we speculate that such a mechanism is in essence similar to “heterogeneous catalysis” in chemistry in which adsorption to a surface facilitates molecular encounters (Kang and Weinberg 1995) and could well be driving protein:protein transactions in vivo. This same concept has also been independently formalized in theoretical physics (ben-Avraham and Havlin 2000; Bénichou et al. 2010). Adsorption could be a powerful lens for understanding molecular motion in the cell. Molecules in cells rarely exhibit properties of free diffusion (classical Brownian motion) and often display strong anomalous diffusion coefficients representative of constraints in their sampling space (Woringer and Darzacq 2018). A revealing case of such “anisotropic behavior” can be seen with the CTCF protein binding to chromatin as a result of its interaction with RNA in cells (Hansen et al. 2020) or the compact nuclear exploration by enzymes like the protein kinase complex P-TEFb involved in transcription elongation (Izeddin et al. 2014). Such phenomena could be, in part, the result of molecular adsorption on nucleic acid or repetitive low complexity peptide surfaces restricting the search space in the cellular environment. Figure 2 depicts different strategies to control effective concentrations in cells.
FIGURE 2.
How numbers of molecules can compete with the strength of an interaction. (A) Different membranous or membraneless organelles can locally confine particular factors and therefore modulate their effective concentration. In addition to controlling endogenous protein levels via expression, such local concentration effects provide alternative mechanisms to control concentrations at different spatial and temporal scales. (B) In addition to modulating the number of potential partner molecules, the space and dimensionality a particular molecule is able to explore can be regulated. This can be achieved either by controlling the size/volume of organelles/compartments or by restricting the space accessible to the molecule during exploration—for example with features within the cell that will reduce the exploration volume or by presenting adsorptive surfaces such as DNA or RNA to restrict the dimensionality of an exploration. (C) Upper diagram—cellular compartmentalization serves as one means to control concentration. Lower diagram—Sliding on a surface like a nucleic acid polymer (DNA or RNA) can also serve to reduce sampling dimensionality and thereby facilitate the guiding of molecules to their targets.
The multivalent nature of LCDs and nucleic acids in recent years has been a subject of great interest because of their ability to undergo or nucleate a physical phenomenon called phase separation, in which interacting molecules separate into distinct phases like oil in water (Banani et al. 2017). While this phenomenon is well established to play an important role in cellular compartmentalization of nonmembranous organelles such as the nucleolus, in many cases the functional consequences of phase separation in vivo are less clear. For example, phase partitioning is very seductive and has been widely speculated to play an active role in transcription regulation but with scant evidence to establish functional causality (Bhat et al. 2021).
The recent appeal of phase-separation models derives from the notion that it can create domains of stable high local concentration where macromolecules that have copartitioned will sample each other more often while being sequestered from other molecules that remain in solution. The question we would like to contemplate here is how often does phase separation constitute a mechanism that is physiologically relevant for driving essential biological function per se? Or could it be that in many cases, phase separation is largely a natural and inevitable, if extreme, consequence of the multiple weak interactions intrinsic to the behavior of most biological macromolecules and not necessarily a mechanism that drives the function of the phase-separated components? We believe that this question needs to be addressed for every system engaging in multiple weak and transient interactions. However, at higher concentrations, multivalent interactions will likely lead to molecular aggregation and/or phase separation regardless of functional consequences. It also seemed to us that formation of phase-separated compartments would not be particularly advantageous for regulatory processes where dynamic flexibility must be at a premium. In short, the link between cause and effect for liquid–liquid phase separation remains largely unresolved in most cases that have been studied in live cells or in vivo.
We suspect that in some cases, these weak and transient interactions will instead drive function via molecular adsorption as seen with proteins sliding on DNA—a mechanism quite distinct from phase separation since it is a property that a single molecule can adopt interacting with its environment, whereas a phase is a bulk property requiring large numbers of interactors. One example of the cumulative strength of such behavior in vivo can be seen with the sequestration of RNA Pol II in the replication compartments of HSV infected cells (McSwiggen et al. 2019). Indeed, on first inspection, these replication compartments show an uncanny resemblance to phase-partitioned droplets, but the underlying driver actually turned out to be naked DNA that served as a multivalent adsorptive surface ideal for depleting the nucleoplasm of DNA-binding factors. In other cases, especially at higher concentrations, phase separation may result in dysfunction by inhibiting critical processes via factor sequestration. Alternatively, condensates could serve as storage organelles that reduce macromolecule concentrations that might otherwise be detrimental to cell health and viability. While we recognize that causal relations are rather difficult to isolate from the natural consequences of multivalent weak interactions and should be pursued, we advocate for caution in claiming causal functionality based on a physical phenomenon, especially in situations where phase separation is observed in vitro or after artificial manipulations in vivo. In several instances, when these obligate multivalent weak interactions are enhanced or exacerbated by overproduction of a partner component, the outcome is more commonly counterproductive in terms of the mechanism driving a particular endogenous function under physiologically relevant contexts.
Consequently, the formation of phase-separated droplets is often the result of artificially overexpressing factors and, interestingly, can lead to cell death—a phenomenon frequently exploited by laboratories as a method for selecting and establishing low-expressing stable cell lines. We therefore suspect that in many cases, such observed or implied correlations of phase separation with function could just be a consequence of abundant multivalent interactors rather than the causal mechanism required for driving biological processes such as transcription activation. We suggest that other properties of multivalent biomolecules beyond their propensity to phase partition at high concentrations should be studied along with their impact on function.
In summary, we close with these questions: How often is biological phase partitioning actually a cause rather than an inevitable consequence of the ubiquitous weak multivalent molecular transactions that govern biomolecular processes and biological regulation? In those cases where phase partitioning is not the functional driver, what mechanisms are functionally relevant to mediating critical biological reactions and processes?
ACKNOWLEDGMENTS
We are grateful to Sally Assmann, Phil Bevilacqua, Michael Botchan, Claire Dugast-Darzacq, Sarah Hymowitz, Garry Karpen, Jonathan Karr, John Lis, and Donald Rio as well as the entire Tjian-Darzacq laboratory for discussions and suggestions on this text.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.079004.121.
Freely available online through the RNA Open Access option.
REFERENCES
- Banani SF, Lee HO, Hyman AA, Rosen MK. 2017. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol 18: 285–298. 10.1038/nrm.2017.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ben-Avraham D, Havlin S. 2000. Classical models of reactions. In Diffusion and reactions in fractals and disordered systems, pp. 157–166. Cambridge University Press, New York. [Google Scholar]
- Bénichou O, Chevalier C, Klafter J, Meyer B, Voituriez R. 2010. Geometry-controlled kinetics. Nat Chem 2: 472–477. 10.1038/nchem.622 [DOI] [PubMed] [Google Scholar]
- Bhat P, Honson D, Guttman M. 2021. Nuclear compartmentalization as a mechanism of quantitative control of gene expression. Nat Rev Mol Cell Biol 22: 653–670. 10.1038/s41580-021-00387-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zhang Z, Li L, Chen B-C, Revyakin A, Hajj B, Legant W, Dahan M, Lionnet T, Betzig E, et al. 2014. Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell 156: 1274–1285. 10.1016/j.cell.2014.01.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisse II, Izeddin I, Causse SZ, Boudarene L, Senecal A, Muresan L, Dugast-Darzacq C, Hajj B, Dahan M, Darzacq X. 2013. Real-time dynamics of RNA polymerase II clustering in live human cells. Science 341: 664–667. 10.1126/science.1239053 [DOI] [PubMed] [Google Scholar]
- Hansen AS, Amitai A, Cattoglio C, Tjian R, Darzacq X. 2020. Guided nuclear exploration increases CTCF target search efficiency. Nat Chem Biol 16: 257–266. 10.1038/s41589-019-0422-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izeddin I, Récamier V, Bosanac L, Cissé II, Boudarene L, Dugast-Darzacq C, Proux F, Bénichou O, Voituriez R, Bensaude O, et al. 2014. Single-molecule tracking in live cells reveals distinct target-search strategies of transcription factors in the nucleus. Elife 3: e02230. 10.7554/eLife.02230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HC, Weinberg WH. 1995. Modeling the kinetics of heterogeneous catalysis. Chem Rev 95: 667–676. 10.1021/cr00035a010 [DOI] [Google Scholar]
- Koh HR, Kidwell MA, Ragunathan K, Doudna JA, Myong S. 2012. ATP-independent diffusion of double-stranded RNA binding proteins. Proc Natl Acad Sci 110: 151–156. 10.1073/pnas.1212917110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C-Y, Myong S. 2021. Probing steps in DNA transcription using single-molecule methods. J Biol Chem 297: 101086. 10.1016/j.jbc.2021.101086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSwiggen DT, Hansen ASAS, Teves SSSS, Marie-Nelly H, Hao Y, Heckert ABAB, Umemoto KKK, Dugast-Darzacq C, Tjian R, Darzacq X. 2019. Evidence for DNA-mediated nuclear compartmentalization distinct from phase separation. Elife 8: e47098. 10.7554/eLife.47098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir M, Reimer A, Haines JE, Li XY, Stadler M, Garcia H, Eisen MB, Darzacq X. 2017. Dense bicoid hubs accentuate binding along the morphogen gradient. Genes Dev 31: 1784–1794. 10.1101/gad.305078.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woringer M, Darzacq X. 2018. Protein motion in the nucleus: from anomalous diffusion to weak interactions. Biochem Soc Trans 46: 945–956. 10.1042/BST20170310 [DOI] [PMC free article] [PubMed] [Google Scholar]