Abstract
Ribonucleoprotein granules are ubiquitous features of eukaryotic cells. Several observations argue that the formation of at least some RNP granules can be considered analogous to the formation of unfolded protein aggregates. First, unfolded protein aggregates form from the exposure of promiscuous protein interaction surfaces, while some mRNP granules form, at least in part, by promiscuous intermolecular RNA–RNA interactions due to exposed RNA surfaces when mRNAs are not engaged with ribosomes. Second, analogous to the role of protein chaperones in preventing misfolded protein aggregation, cells contain abundant “RNA chaperones” to limit inappropriate RNA–RNA interactions and prevent mRNP granule formation. Third, analogous to the role of protein aggregates in diseases, situations where RNA aggregation exceeds the capacity of RNA chaperones to disaggregate RNAs may contribute to human disease. Understanding that RNP granules can be considered as promiscuous, reversible RNA aggregation events allow insight into their composition and how cells have evolved functions for RNP granules.
Keywords: RNA aggregates, RNA chaperones, eIF4A, stress granules
RNP GRANULES ARE UBIQUITOUS AND DIVERSE
Ribonucleoprotein (RNP) granules are a ubiquitous feature of eukaryotic cells and exist in both the cytosol, such as stress granules and P-bodies, and the nucleus, such as the nucleolus, paraspeckles, speckles, and Cajal bodies. There are also cell-type specific RNA granules such as maternal mRNP granules in a variety of oocytes and embryos (Buchan 2014), neuronal mRNA transport granules (Dalla Costa et al. 2021), and myogranules in developing muscle (Vogler et al. 2018).
RNP granules contain different subsets of RNAs with paraspeckles primarily consisting of NEAT1 RNA (Clemson et al. 2009); the nucleolus consisting of nascent rRNA transcripts and snoRNAs (Berry et al. 2015); Cajal bodies being enriched in scaRNAs, snRNAs, and snoRNAs (Machyna et al. 2014); and P-bodies and stress granules being enriched in untranslating mRNAs (Hubstenberger et al. 2017; Khong et al. 2017). For each type of RNP granule, there are also characteristic RNA-binding proteins (RBPs) and other constituents that complete their distinct composition (Jain et al. 2016; Markmiller et al. 2018; Youn et al. 2018; Matheny et al. 2019).
Stress granules have been particularly useful to study the composition and assembly parameters of an RNP granule. Stress granules form under conditions where most mRNAs exit translation. For example, during the integrated stress response phosphorylation of the eukaryotic translation, initiation factor-2α (eIF2α) inhibits translation initiation, leading to polysome run-off and condensation of a subset of untranslating mRNPs into stress granules (Kedersha et al. 1999). RNA sequencing of purified stress granules indicates that they contain a highly diverse transcriptome enriched in long, poorly translated mRNAs (Khong et al. 2017; Namkoong et al. 2018; Matheny et al. 2019).
Several observations now argue that stress granules form from the summation of protein–protein, protein–RNA, and RNA–RNA interactions between individual mRNPs (Van Treeck et al. 2018; Van Treeck and Parker 2018; Roden and Gladfelter 2021). Evidence for protein interactions comes from the observation that cell lines lacking the Ras GTPase-activating protein SH3-domain-binding protein 1 and 2 (G3BP1 and G3BP2) paralogs are deficient at stress granule formation under a variety of stresses (Kedersha et al. 2016). Evidence that intermolecular RNA–RNA interactions are important in stress granule formation is that RNA self-assembly in vitro can largely recapitulate the stress granule transcriptome (Van Treeck et al. 2018), and proteins that inhibit RNA self-assembly in vitro limit stress granule formation in cells (Tauber et al. 2020a; Budkina et al. 2021). Thus, stress granule assembly can be understood as ribosome run-off leading to newly exposed RNA sequences, which can allow both additional protein–RNA interactions (Yang et al. 2020) and promiscuous intermolecular RNA–RNA interactions (Van Treeck et al. 2018). The protein–protein or RNA–RNA interactions between individual mRNPs can be thought of as a highly cooperative equilibrium binding reaction leading to a larger assembly.
Multiple observations suggest that the ability of RNA to form assemblies based on promiscuous intermolecular RNA–RNA interactions is robust. First, all RNA homopolymers are able to condense into droplets, tangles, or aggregates in vitro (Van Treeck et al. 2018; Boeynaems et al. 2019). Second, isolated total RNA from yeast forms RNA condensates under physiological conditions (Van Treeck et al. 2018). Third, the estimated concentration of exposed coding regions after ribosome run-off in yeast (170–800 µg/mL) or in mammalian U-2 OS cells (∼180 µg/mL) is in the range that can trigger RNAs to condense (Bounedjah et al. 2012; Van Treeck et al. 2018). Granule-enriched RNAs are on average very long and therefore contain multiple sites for potential interactions (Khong et al. 2017). This allows RNA molecules to have stable interactions within an mRNP granule through the summation of multiple weak interactions (Banani et al. 2016; Matheny et al. 2021).
The specific types of intermolecular RNA–RNA interactions that promote RNA condensation remain to be established in biological conditions. However, one anticipates that any type of interaction between two RNA molecules can contribute to RNA condensation including base-pairing, non-Watson–Crick base interactions, the formation of triple helices, ribose zippers, and co-axial stacking between helices in different RNA molecules (Bevilacqua et al. 2022).
The formation of mRNP granules is a general phenomenon that occurs in multiple biological contexts where there is an increased pool of untranslating mRNPs. For example, P-bodies are constitutive RNP granules that assemble from untranslating mRNPs associated with the mRNA decay machinery (Sheth and Parker 2003) and that increase in the size when the pool of untranslating mRNPs is increased (Teixeira et al. 2005). Similarly, P-granules are present in Caenorhabditis elegans oocytes and embryos that contain pools of untranslating mRNPs and are enhanced upon stress when the pool of untranslating mRNPs is increased (Lee et al. 2020; Parker et al. 2020). Other examples are mRNP granules found in neurons, where untranslating mRNAs condense into granules that are transported to the synapse for their disassembly and local translation (Tsang et al. 2019).
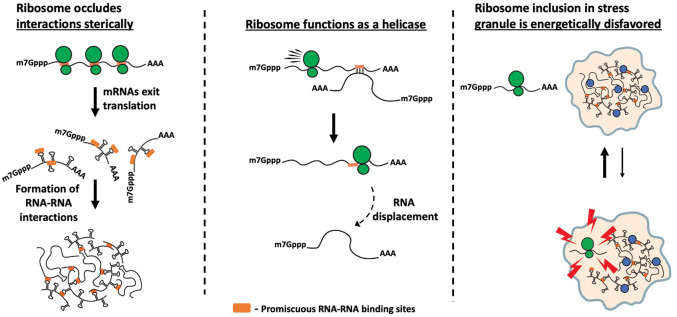
The tendency of untranslating mRNPs to assemble into mRNP granules can be generally understood as the loss of ribosomes causing three alterations to mRNP organization that promotes their condensation (Fig. 1). First, when mRNPs are no longer associated with ribosomes, they expose additional RNA sequences that could be capable of forming intermolecular RNA–RNA interactions, leading to mRNP condensation. Second, ribosomes are highly effective helicases and elongating ribosomes would remove any transient intermolecular RNA–RNA interactions that occur between the coding regions of different mRNPs. Third, we anticipate that the 80S ribosome has evolved to be relatively inert in forming promiscuous intermolecular RNA–RNA interactions and as such would be excluded from the RNP granule, although this final possibility remains to be examined experimentally.
FIGURE 1.
Possible mechanisms by which elongating ribosomes limit stress granule formation. The figure shows three possible mechanisms by which untranslating mRNAs assemble into mRNP granules due to loss of ribosomes: (i) Ribosome runoff leads to exposure of multiple RNA sequences (highlighted in orange) that can form promiscuous interactions. (ii) Ribosome acts as a helicase and removes transient intra- and intermolecular RNA–RNA interactions. (iii) Translating ribosomes do not engage in unspecific interactions and are excluded from RNP granules.
ARE STRESS GRANULES ANALOGOUS TO MISFOLDED PROTEIN AGGREGATES?
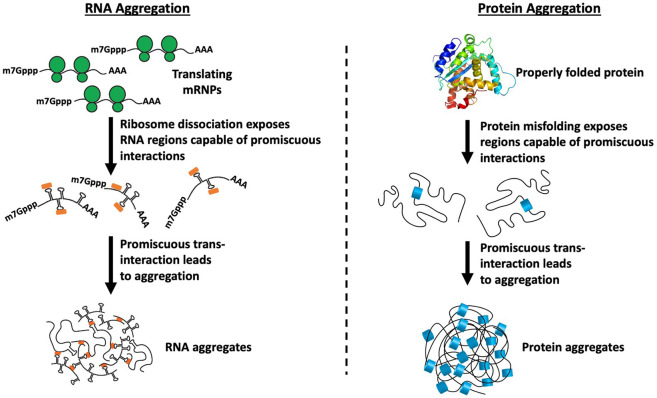
We suggest that promiscuous assembly of untranslating mRNPs into RNP granules can be considered analogous to the formation of misfolded protein aggregates and occurs by the same three critical steps (Fig. 2). First, regions of proteins or RNAs capable of forming promiscuous interactions need to be exposed. For proteins, this happens with misfolding exposing hydrophobic surfaces that can then form aggregates through promiscuous interactions. For mRNAs, we suggest that this occurs when mRNAs are released from translation, exposing the coding region for promiscuous intermolecular RNA–RNA base-pairing and non-Watson–Crick interactions. Second, the exposed promiscuous surfaces need to overwhelm the existing mechanisms that limit nonspecific aggregation. For proteins, this would involve overwhelming protein chaperones, while for RNA aggregation, this would involve overcoming the anti-RNA aggregation effects of abundant RBPs and/or DEAD-box RNA helicases. Third, the initial assembly of proteins or mRNPs into promiscuous assemblies creates a high local concentration that would promote additional intermolecular interactions thereby stabilizing the “aggregate” (Tauber et al. 2020b).
FIGURE 2.
Analogous formation of stress granules and misfolded protein aggregates. Upon ribosome runoff, the newly exposed coding regions are free to form promiscuous intermolecular RNA–RNA interactions. Similarly, upon protein misfolding, hydrophobic surfaces get exposed that can then form aggregates through promiscuous interactions.
Another similarity is that both misfolded proteins in aggregates and RNAs in stress granules show slow exchange rates, while protein chaperones in protein aggregates and many RBPs in stress granules show fast exchange rates (Kim et al. 2002; Winkler et al. 2010; Moon et al. 2019; Tauber et al. 2020b).
THE “RNA CHAPERONE” NETWORK
We suggest that there is an “RNA chaperone” network, which we define as a set of proteins that limits promiscuous intermolecular RNA–RNA interactions. This RNA chaperone network would be analogous to the protein chaperone network that is known to limit misfolded protein aggregation and consists of Heat shock proteins (HSPs) and other components.
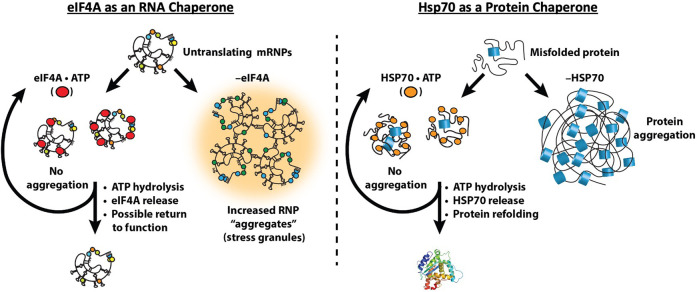
One key component of the RNA chaperone network is the eIF4A protein (Fig. 3). The key observations are that by binding RNA in an ATP-dependent manner, eIF4A can limit the self-assembly of RNA in vitro and limit stress granule formation in vivo (Tauber et al. 2020a). The biochemical properties of eIF4A are ideal for an RNA chaperone. eIF4A is a DEAD-box protein and can bind to, and destabilize, short duplexes (Rogers et al. 1999, 2001), leading to duplex disassembly before ATP hydrolysis and eIF4A release. eIF4A is not efficient at resolving stable duplexes (Rogers et al. 2001) because it does not directly use the energy of ATP hydrolysis to unwind RNA duplexes (Liu et al. 2008). However, the ability to bind and destabilize short RNA duplexes that could form promiscuously is an ideal property for a general RNA chaperone. This suggests that additional DEAD-box and related DEVH-box proteins will play roles in limiting inappropriate RNA–RNA interactions.
FIGURE 3.
Analogous roles of protein and RNA chaperones: The RNA chaperone network consists of DEAD box proteins such as eIF4A to limit self-assembly of RNA in an ATP-dependent manner. Similarly, the protein chaperone HSP70 binds to regions of proteins prone to aggregation to limit protein aggregation and disassemble hydrophobic interactions to allow solubilization and protein refolding.
A role for eIF4A, and other general RNA helicases, in limiting RNA condensation can be considered analogous to protein chaperones, such as HSP70, limiting the aggregation of misfolded proteins (Fig. 3). Multiple protein chaperones, including HSP70 proteins, bind to protein aggregates to disassemble aberrant interactions, thereby allowing for aggregate solubilization and protein refolding, using ATP hydrolysis as a switch for binding (Kampinga and Craig 2010; Mogk et al. 2018). We suggest that RNA condensation and inappropriate aggregation occur when the amount of exposed RNA in the cell exceeds the capacity of the cellular machinery limiting RNA condensation. Thus, the intrinsic aggregation properties of both proteins and RNAs are countered by abundant cellular machinery to keep these macromolecules correctly folded and dispersed for proper function.
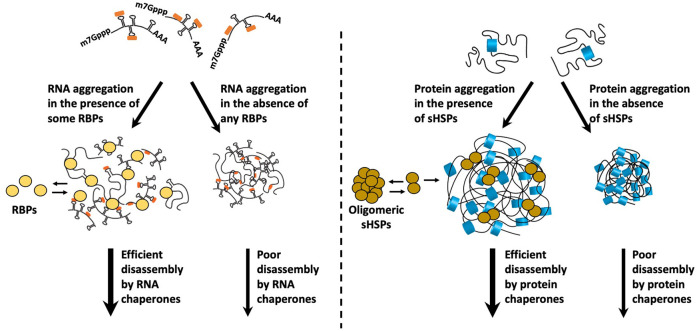
Other members of the RNA chaperone network include abundant RBPs that limit RNA aggregation by binding to single-stranded regions in RNAs and limiting intermolecular RNA–RNA interactions. For example, the abundant RBP YB-1 limits RNA assembly in vitro and, when overexpressed, blocks stress granule formation in cells (Budkina et al. 2021). Such abundant RBPs can be considered analogous to small heat shock proteins (sHsps), a class of protein chaperones promoting the formation of reversible protein aggregates by limiting irreversible protein aggregation, presumably because sHSPs bind, and limit, interactions of aggregation-prone regions in proteins (Fig. 4; Żwirowski et al. 2017; Mogk et al. 2018). By analogy, RBPs could limit irreversible RNA aggregation by limiting the degree of intermolecular RNA–RNA interactions. A prediction of this model is that stress granules formed in the absence of YB-1, or similar RBPs, would show decreased RNA dynamics and prolonged persistence. In this light, G3BP1 has been suggested to prevent stable RNA entanglements by promoting the formation of a less stable, dynamic RNA assembly, although this has only been observed with poly(G) homopolymers (Guillén-Boixet et al. 2020). Since poly(G) homopolymers typically form hyper stable G-quadruplex structures (Williamson et al. 1989), it remains to be seen how G3BP1 affects the condensation of more typical mRNAs.
FIGURE 4.
Do some RNA-binding proteins function analogously to small heat shock proteins (sHSPs)? RNA-binding proteins interact with RNAs, form dynamic assemblies, and facilitate disassembly of promiscuous interactions to limit RNA self-assembly into aggregates. Similarly, sHsps bind to hydrophobic protein regions and form reversible assemblies to prevent their aggregation into insoluble aggregates that are more difficult to disassemble by the protein chaperone network.
Other components of the RNA chaperone network not only remain to be identified but may also include RNA modification enzymes that destabilize RNA–RNA interactions, as well as nucleases that limit the intracellular concentration of RNA.
The importance of the RNA chaperone network will be amplified in some biological contexts. For example, during transcription in eukaryotic cells, a high local concentration of RNA can be produced, which would be prone to intermolecular RNA–RNA interactions potentially creating aberrant RNA-based assemblies. We hypothesize that cells counter this tendency by the higher concentration of RBPs and DEAD-box proteins per RNA in the nucleus as compared to the cytosol (Khong and Parker 2020). This elevated concentration of RBPs may effectively bind nascent RNAs and prevent intermolecular RNA–RNA interactions from forming.
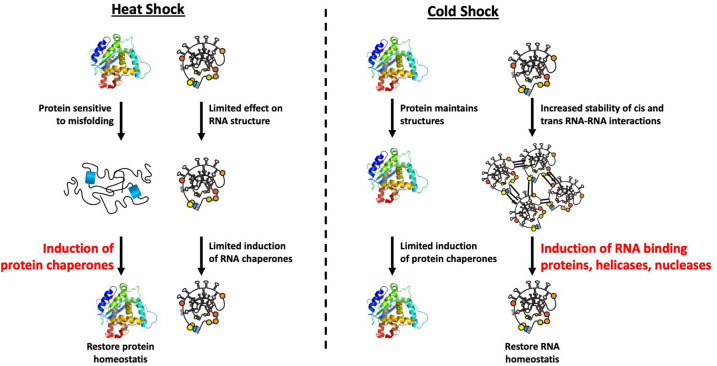
We anticipate that the RNA chaperone network will be important at limiting intermolecular RNA–RNA interactions during cold shock. Moreover, the biophysical properties of proteins and RNAs allow an understanding of the fundamental differences between the heat and cold shock responses (Fig. 5). Specifically, proteins can experience heat-induced misfolding due to temperature increase. This causes cells to induce the protein chaperone network that counters protein misfolding and restores homeostasis. Conversely, compared to proteins, RNA structures are more stable and therefore less likely to be severely perturbed by heat shock. However, RNA structures, as well as inappropriate RNA interactions, will have increased stability in the cold (e.g., Noble and Guthrie 1996; Zhang et al. 2018) increasing the demands for the RNA chaperone network. Notably, the cold shock response, most carefully documented in eubacteria, consists of the induction of RBPs, DEAD-box proteins, including an ortholog of eIF4A, and RNA nucleases (Zhang et al. 2018). This network has been proposed to function in alleviating cis folding of RNAs, but we anticipate an additional contribution in limiting inappropriate trans interactions that would otherwise inhibit RNA function.
FIGURE 5.
Different properties of RNA and protein explain fundamental difference in heat and cold shock response: During heat shock, temperature increase dramatically induces protein unfolding and misfolding, while the reduction in temperature during cold shock on the contrary stabilizes preliminary RNA–RNA contacts in cis or trans. Both lead to different stress signaling cascades and induction of proteins to resolve those promiscuous interactions.
The RNA chaperone network might also be very important in plants under drought conditions, where the intracellular concentration of salt is expected to increase. This is relevant since high intracellular salt would be expected to stabilize RNA duplexes by charge neutralization and higher intracellular salt is known to promote stress granule formation, while low intracellular salt inhibits stress granule formation (Bounedjah et al. 2012). Strikingly, work in a number of plant species has shown that overexpression of eIF4A increases plants’ tolerance to salt stress (e.g., Rao et al. 2017), which might be due to eIF4A limiting RNA aggregation under these conditions. Moreover, plants may limit RNA aggregation by the production of solutes destabilizing RNA–RNA interactions (Bevilacqua et al. 2022).
IMPLICATIONS OF “RNA AGGREGATION” AND THE RNA CHAPERONE NETWORK
The possibility that promiscuous intermolecular RNA–RNA interactions are a prevalent biophysical force within cells, that are generally countered by an “RNA chaperone network,” provides insight into the formation and composition of RNP granules.
There are four basic features of mRNAs that dominate their enrichment in mRNP granules. First, the probability of an mRNA being enriched in an mRNP granule will be enhanced by being translationally repressed, thereby allowing the possibility of more productive intermolecular RNA–RNA interactions (Khong et al. 2017; Matheny et al. 2019; Lee et al. 2020). Second, longer RNAs will in general be preferentially enriched in granules, as increased length allows for multiple weak interactions distributed along an mRNA to act in summation (Matheny et al. 2021). Third, mRNAs with increased binding sites for protein components of mRNP granules will have increased partitioning into the assembly (Matheny et al. 2021). Finally, one anticipates that the mRNAs that fold into structures with more exposed single-stranded RNA (ssRNA) will partition more efficiently than structured mRNAs with less exposed ssRNA regions into mRNP granules due to their presumed ability to form additional intermolecular interactions. This possibility is suggested by the observation that mRNAs predicted to fold into structures with more exposed ssRNA form RNA-based assemblies with more stable structures in vitro than mRNAs with less exposed ssRNA (Ma et al. 2021), although whether these principles will occur in cells has not been demonstrated.
A second important implication is that RNP assembly into RNP granules will generally be dominated by protein–protein interactions under most normal physiological conditions, where RNA aggregation can be effectively countered by the RNA chaperone network. We anticipate that promiscuous intermolecular RNA–RNA interactions will play major roles in two contexts. First, intermolecular RNA–RNA interactions will likely dominate in RNP granule formation when the capacity of the RNA chaperone network capacity is exceeded, such as during the stress response, where the large pools of untranslating mRNAs condense into stress granules (Van Treeck et al. 2018; Tauber et al. 2020a). A similar phenomenon may occur in specialized cell types, such as neurons and oocytes, with larger pools of untranslating mRNPs. We anticipate that promiscuous intermolecular RNA–RNA interactions will also occur once an RNP granule is assembled by protein–protein interactions, due to the formation of a high local concentration of RNA, and thereby reinforce the formation of the RNP granule (Tauber et al. 2020b).
A third point is that promiscuous interactions between untranslating mRNPs can be utilized by cells to build functional assemblies, and this biophysical driving force can explain the otherwise perplexing composition of some RNP granules. One example that can now be understood is the diverse composition of mRNAs in P-granules in C. elegans, which are mRNP granules that segregate specific mRNAs to the developing germline during embryogenesis. Surprisingly, these mRNAs are a very diverse set of mRNAs including mRNA-encoding housekeeping proteins such as ribosomal proteins, and only a few specific mRNAs that are critical to germline development when properly localized (Lee et al. 2020). This composition can be understood as a case where evolution has used the stochastic formation of untranslating mRNAs in bulk to create an RNP granule, which then recruits the specific mRNAs needed for germline development and thereby properly segregates those mRNAs. To ensure that those critical mRNAs are present in P-granules, they are strongly translationally repressed (Lee et al. 2020). Similarly, a wide diversity of mRNAs are localized to dendrites and include mRNAs for housekeeping genes such as ribosomal proteins that have no apparent need to be locally translated in dendrites (Ohashi and Shiina 2020). Such a diversity of mRNAs at dendrites could be explained by these mRNAs being components of an RNP transport granule built by promiscuous interactions between untranslating mRNPs.
One anticipates that evolution will alter the surfaces of RNAs to enhance specific and limit promiscuous intermolecular RNA–RNA interactions. For example, we hypothesize that there will be selective pressure on functional RNAs (such as snRNA, tRNAs, snoRNAs, and rRNAs) to evolve surfaces limited in their ability to form inappropriate intermolecular RNA interactions, through compact RNA secondary and tertiary structure and the binding of proteins. Moreover, evolution can create intermolecular RNA–RNA interactions that target specific mRNAs to mRNP granules. For example, during Drosophila oogenesis, specific trans interactions between the Oskar mRNA (Jambor et al. 2011) or Bicoid mRNA (Ferrandon et al. 1997) 3′ untranslated regions (3′UTRs) allow specific dimerization and target them to distinct RNP granules located at opposite poles of the oocyte. In contrast, mRNAs may also evolve structures that limit promiscuous interactions and therefore influence their assembly into mRNP granules. Such a possibility is suggested by the observations that altering the folding of the Cln3 mRNA in Ashiyba can lead to increased interactions with some distally localized mRNAs in vitro and a corresponding mislocalization of the Cln3 mRNA from RNP granules near the nucleus to distally localized mRNP granules (Langdon et al. 2018).
DOES “RNA AGGREGATION” CONTRIBUTE TO HUMAN DISEASE?
We suggest that in some conditions, RNA aggregation will contribute to human disease progression. For example, stress granule formation, which is driven in part by RNA aggregation, contributes to tumor progression and some degenerative diseases (Anderson et al. 2015; Shukla and Parker 2016; Taylor et al. 2016). In addition, some diseases thought to be based on protein aggregation may in fact be driven by a combination of both protein and RNA aggregation. This is suggested by the observation that tau aggregates in both cell and mouse models of disease are enriched in specific RNAs that may contribute to disease progression (Lester et al. 2021). Moreover, since many of the proteins that aggregate in neurodegenerative diseases are RBPs (e.g., FUS, TDP-43, Annexin-11, Ataxin-2), a role for RNA aggregation contributing to “protein aggregation” may be more prevalent. RNA could contribute to the pathology of protein aggregates both by enhancing the aggregation process and reducing the functional level of key RBPs whose loss of function contributes to disease (Lester et al. 2021).
CONCLUSION
In this study, we hypothesize that stress granules are the RNA equivalent of misfolded protein aggregates and that they form when increased levels of mRNP surfaces are exposed due to widespread mRNA release from ribosomes. The understanding that promiscuous intermolecular RNA–RNA interactions can be a driving force leading to RNA aggregation highlights the need for an “RNA chaperone network” of proteins that act to limit promiscuous RNA aggregation. A fuller understanding of the RNA chaperone network and its role in regulating RNA structure and function remains to be determined.
It is also clear that cells have taken advantage of the propensity of untranslating mRNAs to form RNP granules for function. Untranslating mRNP granules with important biological roles include neuronal granules, which can be important for synaptic plasticity (Bakthavachalu et al. 2018) and maternal mRNP granules, which can be important for early development (Buchan 2014; Lee et al. 2020). Similarly, stress granules have functions in promoting survival during stress (Kedersha et al. 2013) and limiting viral infections (Eiermann et al. 2020), although the specific mechanisms by which stress granules exert these functions remain to be determined. Further understanding of the underlying mechanisms of RNA aggregation, and how cells regulate and utilize that process, should both illuminate new principles of cellular physiology and might provide new insights to develop therapeutic strategies in a number of disease states.
ACKNOWLEDGMENTS
We would like to thank Carolyn Decker (University of Colorado Boulder) for feedback on the manuscript and Anne Webb (University of Colorado Boulder) for assembling the figures. This work was supported by funds from Howard Hughes Medical Institute.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.079000.121.
Freely available online through the RNA Open Access option.
REFERENCES
- Anderson P, Kedersha N, Ivanov P. 2015. Stress granules, P-bodies and cancer. Biochim Biophys Acta 1849: 861–870. 10.1016/j.bbagrm.2014.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakthavachalu B, Huelsmeier J, Sudhakaran IP, Hillebrand J, Singh A, Petrauskas A, Thiagarajan D, Sankaranarayanan M, Mizoue L, Anderson EN, et al. 2018. RNP-granule assembly via ataxin-2 disordered domains is required for long-term memory and neurodegeneration. Neuron 98: 754–766.e4. 10.1016/j.neuron.2018.04.032 [DOI] [PubMed] [Google Scholar]
- Banani SF, Rice AM, Peeples WB, Lin Y, Jain S, Parker R, Rosen MK. 2016. Compositional control of phase-separated cellular bodies. Cell 166: 651–663. 10.1016/j.cell.2016.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry J, Weber SC, Vaidya N, Haataja M, Brangwynne CP. 2015. RNA transcription modulates phase transition-driven nuclear body assembly. Proc Natl Acad Sci 112: E5237–E5245. 10.1073/pnas.1509317112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeynaems S, Holehouse AS, Weinhardt V, Kovacs D, Van Lindt J, Larabell C, Van Den BL, Das R, Tompa PS, Pappu R V, et al. 2019. Spontaneous driving forces give rise to protein−RNA condensates with coexisting phases and complex material properties. Proc Natl Acad Sci 116: 7889–7898. 10.1073/pnas.1821038116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bounedjah O, Hamon L, Savarin P, Desforges B, Curmi PA, Pastré D. 2012. Macromolecular crowding regulates assembly of mRNA stress granules after osmotic stress: new role for compatible osmolytes. J Biol Chem 287: 2446–2458. 10.1074/jbc.M111.292748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevilacqua PC, Williams AM, Chou HL, Assmann SM. 2022. RNA multimerization as an organizing force for liquid–liquid phase separation. RNA 28: 16–26 (this issue). 10.1261/rna.078999.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR. 2014. mRNP granules. Assembly, function, and connections with disease. RNA Biol 11: 1019–1030. 10.4161/15476286.2014.972208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budkina K, El Hage K, Clément M-J, Desforges B, Bouhss A, Joshi V, Maucuer A, Hamon L, Ovchinnikov LP, Lyabin DN, et al. 2021. YB-1 unwinds mRNA secondary structures in vitro and negatively regulates stress granule assembly in HeLa cells. Nucleic Acids Res 49: 10061–10081. 10.1093/nar/gkab748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. 2009. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 33: 717–726. 10.1016/j.molcel.2009.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalla Costa I, Buchanan CN, Zdradzinski MD, Sahoo PK, Smith TP, Thames E, Kar AN, Twiss JL. 2021. The functional organization of axonal mRNA transport and translation. Nat Rev Neurosci 22: 77–91. 10.1038/s41583-020-00407-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiermann N, Haneke K, Sun Z, Stoecklin G, Ruggieri A. 2020. Dance with the devil: stress granules and signaling in antiviral responses. Viruses 12: 984. 10.3390/v12090984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandon D, Koch I, Westhof E, Nüsslein-Volhard C. 1997. RNA–RNA interaction is required for the formation of specific bicoid mRNA 3′ UTR-STAUFEN ribonucleoprotein particles. EMBO J 16: 1751–1758. 10.1093/emboj/16.7.1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillén-Boixet J, Kopach A, Holehouse AS, Wittmann S, Jahnel M, Schlüßler R, Kim K, Trussina IREA, Wang J, Mateju D, et al. 2020. RNA-induced conformational switching and clustering of G3BP drive stress granule assembly by condensation. Cell 181: 346–361.e17. 10.1016/j.cell.2020.03.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubstenberger A, Courel M, Bénard M, Souquere S, Ernoult-Lange M, Chouaib R, Yi Z, Morlot JB, Munier A, Fradet M, et al. 2017. P-body purification reveals the condensation of repressed mRNA regulons. Mol Cell 68: 144–157.e5. 10.1016/j.molcel.2017.09.003 [DOI] [PubMed] [Google Scholar]
- Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A, Parker R. 2016. ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 164: 487–498. 10.1016/j.cell.2015.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambor H, Brunel C, Ephrussi A. 2011. Dimerization of oskar 3′ UTRs promotes hitchhiking for RNA localization in the Drosophila oocyte. RNA 17: 2049–2057. 10.1261/rna.2686411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampinga HH, Craig EA. 2010. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol 11: 579–592. 10.1038/nrm2941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Gupta M, Li W, Miller I, Anderson P. 1999. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2α to the assembly of mammalian stress granules. J Cell Biol 147: 1431–1441. 10.1083/jcb.147.7.1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Ivanov P, Anderson P. 2013. Stress granules and cell signaling: more than just a passing phase? Trends Biochem Sci 38: 494–506. 10.1016/j.tibs.2013.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Panas MD, Achorn CA, Lyons S, Tisdale S, Hickman T, Thomas M, Lieberman J, McInerney GM, Ivanov P, et al. 2016. G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J Cell Biol 212: 845–860. 10.1083/jcb.201508028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khong A, Parker R. 2020. The landscape of eukaryotic mRNPs. RNA 26: 229–239. 10.1261/rna.073601.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khong A, Matheny T, Jain S, Mitchell SF, Wheeler JR, Parker R. 2017. The stress granule transcriptome reveals principles of mRNA accumulation in stress granules. Mol Cell 68: 808–820.e5. 10.1016/j.molcel.2017.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Nollen EAA, Kitagawa K, Bindokas VP, Morimoto RI. 2002. Polyglutamine protein aggregates are dynamic. Nat Cell Biol 4: 826–830. 10.1038/ncb863 [DOI] [PubMed] [Google Scholar]
- Langdon EM, Qiu Y, Ghanbari Niaki A, McLaughlin GA, Weidmann CA, Gerbich TM, Smith JA, Crutchley JM, Termini CM, Weeks KM, et al. 2018. mRNA structure determines specificity of a polyQ-driven phase separation. Science 360: 922–927. 10.1126/science.aar7432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CYS, Putnam A, Lu T, He S, Ouyang JPT, Seydoux G. 2020. Recruitment of mRNAs to P granules by condensation with intrinsically-disordered proteins. Elife 9: e52896. 10.7554/eLife.52896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester E, Ooi FK, Bakkar N, Ayers J, Woerman AL, Wheeler J, Bowser R, Carlson GA, Prusiner SB, Parker R. 2021. Tau aggregates are RNA–protein assemblies that mislocalize multiple nuclear speckle components. Neuron 109: 1675–1691.e9. 10.1016/j.neuron.2021.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Putnam A, Jankowsky E. 2008. ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc Natl Acad Sci 105: 20209–20214. 10.1073/pnas.0811115106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Zheng G, Xie W, Mayr C. 2021. In vivo reconstitution finds multivalent RNA–RNA interactions as drivers of mesh-like condensates. Elife 10: e64252. 10.7554/eLife.64252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machyna M, Kehr S, Straube K, Kappei D, Buchholz F, Butter F, Ule J, Hertel J, Stadler PF, Neugebauer KM. 2014. The coilin interactome identifies hundreds of small noncoding RNAs that traffic through Cajal bodies. Mol Cell 56: 389–399. 10.1016/j.molcel.2014.10.004 [DOI] [PubMed] [Google Scholar]
- Markmiller S, Soltanieh S, Server KL, Mak R, Jin W, Fang MY, Luo EC, Krach F, Yang D, Sen A, et al. 2018. Context-dependent and disease-specific diversity in protein interactions within stress granules. Cell 172: 590–604.e13. 10.1016/j.cell.2017.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheny T, Rao B, Parker R. 2019. Transcriptome-wide comparison of stress granules and P-bodies reveals that translation plays a major role in RNA partitioning. Mol Cell Biol 39: e00313-19. 10.1128/MCB.00313-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheny T, Van Treeck B, Huynh TN, Parker R. 2021. RNA partitioning into stress granules is based on the summation of multiple interactions. RNA 27: 174–189. 10.1261/rna.078204.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogk A, Bukau B, Kampinga HH. 2018. Cellular handling of protein aggregates by disaggregation machines. Mol Cell 69: 214–226. 10.1016/j.molcel.2018.01.004 [DOI] [PubMed] [Google Scholar]
- Moon SL, Morisaki T, Khong A, Lyon K, Parker R, Stasevich TJ. 2019. Multicolour single-molecule tracking of mRNA interactions with RNP granules. Nat Cell Biol 21: 162–168. 10.1038/s41556-018-0263-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namkoong S, Ho A, Woo YM, Kwak H, Lee JH. 2018. Systematic characterization of stress-induced RNA granulation. Mol Cell 70: 175–187. 10.1016/j.molcel.2018.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble SM, Guthrie C. 1996. Identification of novel genes required for yeast pre-mRNA splicing by means of cold-sensitive mutations. Genetics 143: 67–80. 10.1093/genetics/143.1.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi R, Shiina N. 2020. Cataloguing and selection of mRNAs localized to dendrites in neurons and regulated by RNA-binding proteins in RNA granules. Biomolecules 10: 167. 10.3390/biom10020167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker DM, Winkenbach LP, Boyson S, Saxton MN, Daidone C, Al-Mazaydeh ZA, Nishimura MT, Mueller F, Nishimura EO. 2020. mRNA localization is linked to translation regulation in the Caenorhabditis elegans germ lineage. Development 147: dev186817. 10.1242/dev.186817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao TSRB, Naresh JV, Reddy PS, Reddy MK, Mallikarjuna G. 2017. Expression of Pennisetum glaucum eukaryotic translational initiation factor 4A (PgeIF4A) confers improved drought, salinity, and oxidative stress tolerance in groundnut. Front Plant Sci 8: 453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roden C, Gladfelter AS. 2021. RNA contributions to the form and function of biomolecular condensates. Nat Rev Mol Cell Biol 22: 183–195. 10.1038/s41580-020-0264-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers GW, Richter NJ, Merrick WC. 1999. Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J Biol Chem 274: 12236–12244. 10.1074/jbc.274.18.12236 [DOI] [PubMed] [Google Scholar]
- Rogers GW, Lima WF, Merrick WC. 2001. Further characterization of the helicase activity of eIF4A. Substrate specificity. J Biol Chem 276: 12598–12608. 10.1074/jbc.M007560200 [DOI] [PubMed] [Google Scholar]
- Sheth U, Parker R. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300: 805–808. 10.1126/science.1082320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla S, Parker R. 2016. Hypo- and hyper-assembly diseases of RNA—protein complexes. Trends Mol Med 22: 615–628. 10.1016/j.molmed.2016.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauber D, Tauber G, Khong A, Van Treeck B, Pelletier J, Parker R. 2020a. Modulation of RNA condensation by the DEAD-Box protein eIF4A. Cell 180: 411–426.e16. 10.1016/j.cell.2019.12.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauber D, Tauber G, Parker R. 2020b. Mechanisms and regulation of RNA condensation in RNP granule formation. Trends Biochem Sci 45: 764–778. 10.1016/j.tibs.2020.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JP, Brown RH, Cleveland DW. 2016. Decoding ALS: From genes to mechanism. Nature 539: 197–206. 10.1038/nature20413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R. 2005. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 11: 371–382. 10.1261/rna.7258505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang B, Arsenault J, Vernon RM, Lin H, Sonenberg N, Wang L, Bah A, Forman-Kay JD. 2019. Phosphoregulated FMRP phase separation models activity-dependent translation through bidirectional control of mRNA granule formation. Proc Natl Acad Sci 116: 4218–4227. 10.1073/pnas.1814385116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Treeck B, Parker R. 2018. Emerging roles for intermolecular RNA–RNA interactions in RNP assemblies. Cell 174: 791–802. 10.1016/j.cell.2018.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Treeck B, Protter DSW, Matheny T, Khong A, Link CD, Parker R. 2018. RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. Proc Natl Acad Sci 115: 2734–2739. 10.1073/pnas.1800038115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogler TO, Wheeler JR, Nguyen ED, Hughes MP, Britson KA, Lester E, Rao B, Betta ND, Whitney ON, Ewachiw TE, et al. 2018. TDP-43 and RNA form amyloid-like myo-granules in regenerating muscle. Nature 563: 508–513. 10.1038/s41586-018-0665-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson JR, Raghuraman MK, Cech TR. 1989. Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell 59: 871–880. 10.1016/0092-8674(89)90610-7 [DOI] [PubMed] [Google Scholar]
- Winkler J, Seybert A, König L, Pruggnaller S, Haselmann U, Sourjik V, Weiss M, Frangakis AS, Mogk A, Bukau B. 2010. Quantitative and spatio-temporal features of protein aggregation in Escherichia coli and consequences on protein quality control and cellular ageing. EMBO J 29: 910–923. 10.1038/emboj.2009.412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Mathieu C, Kolaitis RM, Zhang P, Messing J, Yurtsever U, Yang Z, Wu J, Li Y, Pan Q, et al. 2020. G3BP1 is a tunable switch that triggers phase separation to assemble stress granules. Cell 181: 325–345.e28. 10.1016/j.cell.2020.03.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn JY, Dunham WH, Hong SJ, Knight JDR, Bashkurov M, Chen GI, Bagci H, Rathod B, MacLeod G, Eng SWM, et al. 2018. High-density proximity mapping reveals the subcellular organization of mRNA-associated granules and bodies. Mol Cell 69: 517–532.e11. 10.1016/j.molcel.2017.12.020 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Burkhardt DH, Rouskin S, Li GW, Weissman JS, Gross CA. 2018. A stress response that monitors and regulates mRNA structure is central to cold shock adaptation. Mol Cell 70: 274–286.e7. 10.1016/j.molcel.2018.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Żwirowski S, Kłosowska A, Obuchowski I, Nillegoda NB, Piróg A, Ziętkiewicz S, Bukau B, Mogk A, Liberek K. 2017. Hsp70 displaces small heat shock proteins from aggregates to initiate protein refolding. EMBO J 36: 783–796. 10.15252/embj.201593378 [DOI] [PMC free article] [PubMed] [Google Scholar]