Abstract
RNA interactions are exceptionally strong and highly redundant. As such, nearly any two RNAs have the potential to interact with one another over relatively short stretches, especially at high RNA concentrations. This is especially true for pairs of RNAs that do not form strong self-structure. Such phenomena can drive liquid–liquid phase separation, either solely from RNA–RNA interactions in the presence of divalent or organic cations, or in concert with proteins. RNA interactions can drive multimerization of RNA strands via both base-pairing and tertiary interactions. In this article, we explore the tendency of RNA to form stable monomers, dimers, and higher order structures as a function of RNA length and sequence through a focus on the intrinsic thermodynamic, kinetic, and structural properties of RNA. The principles we discuss are independent of any specific type of biomolecular condensate, and thus widely applicable. We also speculate how external conditions experienced by living organisms can influence the formation of nonmembranous compartments, again focusing on the physical and structural properties of RNA. Plants, in particular, are subject to diverse abiotic stresses including extreme temperatures, drought, and salinity. These stresses and the cellular responses to them, including changes in the concentrations of small molecules such as polyamines, salts, and compatible solutes, have the potential to regulate condensate formation by melting or strengthening base-pairing. Reversible condensate formation, perhaps including regulation by circadian rhythms, could impact biological processes in plants, and other organisms.
Keywords: biophysics, condensate, plant biology, RNA structure
INTRODUCTION
Life as we know it exists inside of compartments. It is almost impossible to imagine it otherwise. In the simplest terms, compartments keep cellular contents from leaking into the surroundings, but they can do much more, organizing cellular processes and promoting diversity at the single-cell level (Raser and O'Shea 2005; Wu et al. 2017; Stuart and Satija 2019; Seyfferth et al. 2021). Compartments can comprise an entire organism, a cell, a membrane-delimited organelle, or a nonmembranous biomolecular condensate. Nonmembranous compartments, which form through liquid–liquid phase separation (LLPS), are of special interest because they can concentrate biomolecules and metabolites without the complexity of a membrane, and in some cases by purely physical parameters (e.g., temperature and pH) (Nott et al. 2015; Falahati et al. 2016; Shin and Brangwynne 2017). Because of their relative simplicity, nonmembranous compartments may have been especially important in the emergence of life, forming the first cells or “protocells” (Poudyal et al. 2018), which promoted the retention of functional RNAs (Cakmak et al. 2020). In extant organisms, nonmembranous compartments impact gene expression, signaling cascades, and macromolecular stability (Shin and Brangwynne 2017; Poudyal et al. 2018).
Liquid–liquid phase separation happens both inside and outside of cells. Extracellular LLPS has been described as protein-centric (Muiznieks et al. 2018; Chiu et al. 2020), while condensates inside of the cell often involve RNA molecules. Cellular condensates can form in the cytoplasm or the nucleus (Sabari et al. 2020) and more than 20 different types of cellular condensates have been described. Some of the best-known condensates that include RNA are stress granules, which contain RNAs that are translationally inactive, and P-bodies, which are involved in mRNA decay (Buchan 2014; Khong et al. 2017). It is well known that protein–protein and protein–RNA interactions in trans drive assembly of condensates (Gilks et al. 2004; Decker et al. 2007; Nonhoff et al. 2007; Kedersha et al. 2016), and that some proteins, particularly those with intrinsically disordered regions (IDRs), can form liquid droplets without RNA (Lin et al. 2015; Molliex et al. 2015; Patel et al. 2015). Importantly, interactions solely between RNAs can also drive condensate formation, as has been shown in vitro in RNA-only assemblies that form without proteins yet have similar transcript compositions as those in vivo (Van Treeck et al. 2018). Indeed, Ripin and Parker (2022) point out that RNA–RNA interactions will become important in biomolecular condensates when the RNA chaperone network is overwhelmed with RNAs. Condensates can be comprised of riboswitches driven by loop–loop RNA base-pairing (Poudyal et al. 2021), as well as RNAs with CNG repeats (Jain and Vale 2017). For the many condensates that involve RNA–RNA interactions, their formation and dissolution should be responsive to the variables known to affect RNA structure formation, including RNA sequence and concentration, divalent cations, and temperature.
In this article, we focus on how RNA can provide the molecular, thermodynamic, and kinetic forces for liquid–liquid phase separation. We conclude with considerations for how proteins further affect the properties of biomolecular condensates. Our overarching goal is to express general principles of condensate formation in terms of RNA thermodynamics, kinetics, and structure, independently of cellular domains, or species of compartments. There are outstanding articles that describe the contents of specific condensates; our hope is that our articulation in this article of general principles of RNA structure and function vis-à-vis nonmembranous compartments can provoke new insights into biological condensates, their functions, and their regulation.
RNA SECONDARY STRUCTURE IS STRONG AND UBIQUITOUS
Because RNA–RNA interactions can mediate or assist LLPS and biomolecular condensate formation, it is important to appreciate how strong RNA–RNA interactions are. Duplexes can form from stretches of RNA as short as 4 bp. For instance, we measured base-pairing between CUCU and GGAGAA as part of one of our earlier studies (Bevilacqua and Turner 1991). This is a good case to consider, not only because the base-pairing length is short, but because it has a mixture of different base pairs: two GC pairs, an AU pair, and a GU pair, along with a 3′A dangling end stack, which strongly promotes RNA folding. Base-pairing between these two strands gives rise to an experimentally determined ΔH° of −36 kcal/mol, a ΔS° of −103 eu, and a favorable ΔG°37 of −3.9 kcal/mol, corresponding to an association constant, KA, at 37°C of 550 M−1 (Bevilacqua and Turner 1991). Base-pairing is enthalpically driven and entropically opposed, which can be understood because stacking and hydrogen bonds form while the system becomes more ordered upon duplex formation. We note that the free energy values reported here were determined in 50 mM Mg2+/25 mM Na+; however, melting temperatures for model oligonucleotides, albeit DNA, are similar between 50 mM Mg2+/12 mM Na+ and more physiological salt conditions of 1 to 2 mM Mg2+/150 mM Na+ (Williams et al. 1989), suggesting that the parameters used above are reasonable. Nonetheless, there is a clear need to obtain a full set of thermodynamic parameters under physiologically relevant conditions.
The melting temperature (TM) for a duplex formed between two strands of RNA is determined by their ΔH° and ΔS° thermodynamic parameters, as well as by strand concentration (Bloomfield et al. 2000). At a total strand concentration of 8 mM, a TM of 37.5°C is obtained for the above 4 bp duplex (see Supplemental Information, Tab 1), meaning significant base-pairing would be occurring at 37°C and lower, which is relevant to humans and most poikilotherms. A concentration of 8 mM is attainable in a stress granule using published condensate parameters (Protter and Parker 2016) and the following assumptions. One of the defining features of stress granules is their small size. A typical stress granule can be as small as 100 nm (Wolozin and Ivanov 2019). Assuming a spherical shape, the volume of a 100 nm stress granule is less than 1/1000th of a fL (see Supplemental Information, Tab 1). Small containers give rise to high strand concentrations, which drive base-pairing in RNA. If we put one copy each of CUCU and GGAGAA inside, we obtain an RNA strand concentration of ∼6 µM (see Supplemental Information, Tab 1). A typical stress granule has thousands of mRNAs, many of which are thousands of nucleotides in length (Khong et al. 2017). If we take any stretch of four bases, then the probability of finding its Watson–Crick complement is (1/4)4; in the millions of stretches found in the stress granule there would likely be ∼4000 complements, although some will be tied up in base-pairing or protein binding. Long mRNAs, which tend to have long base-pairing tracks, preferentially accumulate in stress granules in vivo and in RNA-only assemblies in vitro (Khong et al. 2017; Van Treeck et al. 2018). Similarly, there would be ∼4000 copies of the original stretch of four nucleotides that would correspond to an effective site concentration of 10 mM and a TM of 40°C. If we take G•U wobbles into account, then the probability of base-pairing between any two positions rises to 6/16, or 37.5%, making short base-paired stretches in RNA probable.
To summarize, interstrand base-pairing is promoted in biomolecular condensates, owing to the strength of stacking and hydrogen bonding, the multitude of canonical and noncanonical interactions across the four bases (see below), and the small volume of the droplets, which concentrates biomolecules. Evidence that trans RNA–RNA base-pairing occurs in coacervates comes from the observation that the subset of mRNAs from total yeast RNA that form RNA-only assemblies in vitro, largely recapitulate the stress granule transcriptome in vivo, as well as from enrichment of antisense ncRNAs that are able to base-pair to longer mRNAs already enriched in stress granules (Van Treeck et al. 2018). Moreover, Tauber and coworkers provide multiple lines of evidence—including stability to dilution, fluorescence recovery after photobleaching (FRAP), and RNA–RNA crosslinking—that recruitment of RNA to condensates leads to enhanced interactions of those RNAs (Tauber et al. 2020a). Additionally, we recently reported that self-complementary as well as non-self-complementary loop–loop interactions drive condensate formation between two riboswitch strands (Poudyal et al. 2021). In these cases, increasing Mg2+ concentrations from 5 mM up to 100 mM led to more and bigger droplets, as well as droplets with more RNA. This fits a theme of LLPS of RNA being favored by cations (see below for organic cations).
Above we took a specific example, one of 4 bp. In an effort to generalize the effects, we next asked what would happen if we altered the number of base pairs in the duplex, here increasing from 4 to 8 bp. To estimate the parameters of the 8 bp duplex, we doubled the ΔH° and ΔS° of the 4 bp stem according to nearest neighbor theory. We note that this approach is conservative since the price for initiating the helix is not paid again according to the zipper model for duplex formation (Bloomfield et al. 2000). In this case, we obtain a ΔH° of −71 kcal/mol, a ΔS° of −205 eu, and a favorable ΔG°37 of −7.8 kcal/mol, corresponding to an association constant, Keq, at 37°C of 3 × 105. Clearly, this is a very strong RNA–RNA interaction. The probability of finding the Watson–Crick complement, however, is only (1/4)8, and in the millions of 8mer stretches in the stress granule, there would likely be only ∼15 complements. This would correspond to an effective concentration of ∼50 µM and a TM of ∼40.5°C, which is essentially the same as found for the 4 bp duplex. In other words, doubling the length of complementarity did not appreciably increase the melting temperature, which is because there is an offset between the gain in stability and the rarity of complementary base-pairing. Nonetheless, this example illustrates that many different stretches of base-pairing, with different lengths and sequences, serve to stabilize interactions between different RNA molecules. Four base pairs was the shortest helix we considered, but it might be possible to have as few as 3 or even 2 bp if the effective concentration was very high and the base-pairing was GC-rich.
It is worth noting that RNA structure formation is so common that it is hard to design an RNA that does not fold. For instance, it was exceptionally difficult to design a structure-free RNA in previous work we conducted on interactions between the RNA-activated protein kinase PKR and single-stranded RNA (Nallagatla et al. 2007). We ended up choosing a sequence rich in A and C because each can have only a single base-pairing partner (A with U, and C with G). The notion that RNA readily adopts structure runs counter to the familiar case of protein folding, wherein isolated secondary structures of α-helices and β-strands are very weak. Indeed, the free energy of protein folding and protein–protein interactions for these secondary structures tend to be relatively modest at about −1 kcal/mol (Yang and Honig 1995a,b). The isolated α-helix is only slightly stable, if at all (Creighton 1993). Moreover, the overall free energy of folding for a globular protein under physiological conditions is modest, being between −5 and −15 kcal/mol. (Pace 1990). The relatively weak folding of proteins suggests a reason for their presence in biomolecular condensates of RNA, which is to temper the strength of interactions in the condensate giving it more liquid-like properties and allowing protein–protein, and perhaps protein–RNA, interactions to form and break more dynamically.
To give a sense of RNA's proclivity for pairing, we generated 100 random 50mers (Stothard 2000) and predicted their secondary structure and free energy for folding using Mfold, a leading secondary structure prediction program (Zuker 2003). We found that on average 47% ± 10% of the bases were paired, with an average free energy of −9 ± 4 kcal/mol, which gives a range of Keq from ∼102 to1010 (Supplemental Information, Tab 2). In other words, RNA molecules are prone to basepair. While this simulation was on intramolecular pairing, it is transferable to intermolecular pairing, especially in the small confines of a condensate where high RNA concentrations would statistically favor encounters between complementary sequences.
Finally, it is noteworthy that two strands of RNA can interact beyond Watson–Crick and wobble pairings. Experimentally, this is supported by the observation that all four homopolymers can form RNA condensates (Aumiller et al. 2016; Van Treeck et al. 2018; Boeynaems et al. 2019). Leontis and Westhof systematically characterized interactions between two strands of RNA, considering all possible interactions of the Watson–Crick, Hoogsteen, and sugar edges of the bases (Leontis and Westhof 2001). These give rise to six pairwise interactions of the bases and two base orientations, or 12 geometric types with at least two hydrogen bonds, with all 12 possibilities experimentally known (Leontis et al. 2002). Because these interactions have stacking and two-hydrogen bond base-pairing, we can expect them to be relatively strong. Overall, we are left with a picture of RNA as generally capable of forming strong and frequent base-pairing interactions with other RNAs, often beyond Watson–Crick base-pairing, even without taking into consideration any evolutionary effects. In the next section, we consider the tendency of RNAs to multimerize via both canonical and noncanonical secondary structure motifs and via tertiary interactions.
RNAs TEND TO MULTIMERIZE VIA SECONDARY AND TERTIARY STRUCTURE INTERACTIONS
The previous section focused on RNA's tendency to form strong and frequent secondary structure interactions. For condensates to form, multiple strands of RNA must interact with one another along their lengths. We begin with a consideration of interactions that lead to RNA dimerization and then move on to consider interactions that lead to higher-order multimerization. In this section, we identify two general ways that RNAs can interact in trans to promote RNA condensation. First, prefolded RNAs can interact without any concomitant unfolding, such as through single-stranded loops, through bases engaging in preformed grooves to form triples and quartets, and by stacking on the end of helices. Since no bonds have to be broken, these should be the most facile class of interactions. Second, RNAs can rearrange to interact. This involves the breaking of preformed stems and so comes with a penalty that is both thermodynamic and kinetic (see below for further considerations). Finally, these interactions can be altered by the presence of proteins such as helicases and the ribosome, which is considered in the next section.
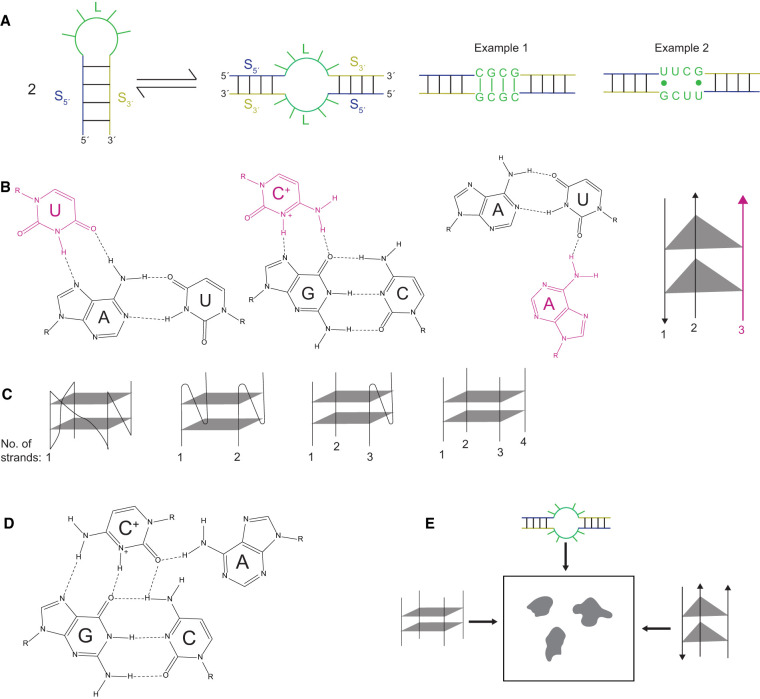
The fundamental building block of RNA structure is the stem–loop, or hairpin. All stems have the intrinsic ability to dimerize with a copy of themselves, which is favored by the high concentrations of RNA found in condensates. Dimerization is illustrated in Figure 1A wherein a 12mer stem–loop with a 4 bp stem and a four-nucleotide loop interacts with a copy of itself. Because there are 8 bp on each side of this equation, this part of the process is thermoneutral; in other words, this equilibrium involves breaking and making the same number and type of base pairs, albeit with somewhat different dangling ends. Thus, what determines whether the hairpin or the dimer state is favored at equilibrium is the nature of the hairpin/internal loops. If the loop sequences are self-complementary, for example in CGCG, then the dimer will be favored, as they can form four strong Watson–Crick base pairs in this state (Fig. 1A, Example 1). However, if the loops form a strong hairpin tetraloop, for example in UUCG (described below), then the hairpin will be favored since it has a stable loop structure while the dimer's internal loop will not as it has only two weak G•U wobbles (Fig. 1A, Example 2). Indeed, we recently demonstrated that strong dimer internal loop interactions drive the formation of RNA condensates (Fig. 2A; Poudyal et al. 2021).
FIGURE 1.
Examples of RNA multimerization. (A) Duplex formation. A 12mer stem–loop interacts with a copy of itself to form a duplex. The 5′- and 3′-strands (S5′ and S3′), as well as the loop (L), are designated and color coded in both the hairpin (left) and duplex (right) states. The image depicts the base-pairing bringing together two different strands of RNA. The first example is of a self-complementary dimer internal loop sequence of CGCG that results in strong base-pairing and favors the duplex. The second example is of a non-self-complementary dimer internal loop sequence that results in weak base-pairing and favors the stable UNCG hairpin. (B) Trimer formation. The first two images show a third strand, denoted in magenta, interacting in the major groove of either an AU, or a GC Watson–Crick base pair. In the case of the GC base pair, the third strand has a protonated C. The third image shows a third strand interacting, denoted in pink, in the minor groove of an AU Watson–Crick base pair. The fourth image depicts two triples (triangles) bringing together three different strands of RNA, with the non-Watson–Crick strand in pink. (C) Quartet formation. The four images depict two G-quartets (squares) bringing together up to four different strands of RNA. (D) Quartets can form in many different ways such as a major groove base triple interacting with a fourth base. Note that here the C+ interacts differently than in the standard triple in panel B. (E) Overview of the myriad RNA–RNA interactions that can lead to LLPS.
FIGURE 2.
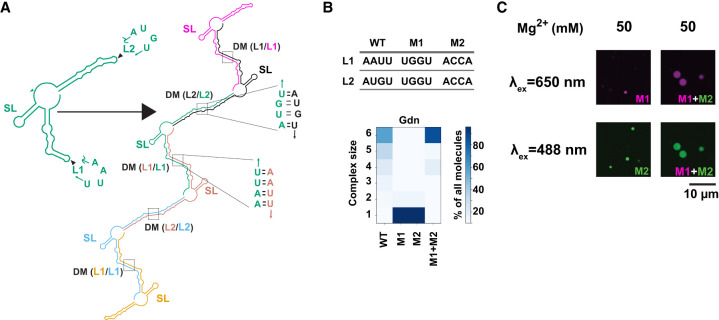
Loop–loop interactions mediate assembly of RNA condensates. (A) NUPACK-predicted architecture for multimerization (n = 6 here) of the guanidine riboswitch from Bacillus subtilis. Self-complementary loops L1 and L2 form trans-strand dimerization motifs (DM). Stem–loop structure (SL) is retained as a cis-strand structure within the multimers. Each copy of the transcript is a separate color and labels are color-matched. (B) Propensity for RNA multimerization of the guanidine riboswitch as predicted by NUPACK for WT (wild-type), M1 (mutant 1), M2 (mutant 2), and M1 + M2 (equimolar mixture of M1 and M2). Sequences of WT, M1, and M2 are provided. We capped NUPACK calculations at complexes of six monomers in this example; calculations for up to 10 monomers are possible (Zadeh et al. 2011) and were conducted, but showed no new modes of interaction (Poudyal et al. 2021). (C) Samples containing both M1 (magenta) and M2 (green) showing condensate formation in 50 mM Mg2+ with complementary loops but not in samples containing M1-only and M2-only. Figure and legend adapted from Poudyal et al. (2021) with permission.
The above observations indicate that stable hairpin loops in RNA would oppose condensate formation or partitioning of RNA into condensates. The most stable hairpin loops are the tetraloops UNCG and GNRA (“N” is any of the four bases and “R” is a purine, A or G) (Antao et al. 1991). However, other classes of stable tetraloops have also been found, including CUUG and the YNMG family, where “Y” is a pyrimidine, C or U, and M is C or A (Proctor et al. 2002). It is notable that positions 1 and 4 of hairpin tetraloops do not undergo Watson–Crick base-pairing while in the hairpin state because it would cause ring strain, and that these loops often adopt well-characterized non-Watson–Crick interactions of purine syn conformations, as well as 2′OH and phosphate hydrogen bonding (Cheong et al. 1990; Heus and Pardi 1991). Triloops are the smallest RNA loops known, and the most stable triloops include UUA, AUU, and GUU with a CG closing base pair (Shu and Bevilacqua 1999). Because these triloops are so strong, an RNA might avoid condensate formation arising from RNA–RNA interaction by having one of these stable hairpin loops. Indeed, we showed that UNCG tetraloops tend to stay as monomeric hairpins even at the high concentrations required for NMR, while CGCG tetraloops form dimers under these conditions (Proctor et al. 2003). We further showed that even the equilibrium of a CGCG tetraloop could be tilted toward the hairpin by inserting bromine at the 8 position of G4 of the loop; this change induced the syn conformation of G in YNMG structure while blocking Watson–Crick base-pairing required for the duplex. Modifications of RNA (or binding of proteins) can modulate these principles adding complexity to condensate formation.
As mentioned above, the TM of a duplex is a function of ΔH°, ΔS°, and strand concentration, while the TM of a hairpin is a function of ΔH° and ΔS° only—not of strand concentration (Bloomfield et al. 2000). Consequently, the position of the equilibrium between duplex and hairpin will depend on strand concentration and will increasingly favor the duplex at increasing RNA concentration. Because condensates have very small volumes, they concentrate RNAs and favor dimerization strongly. Finally, it is worth noting that not all RNA-containing systems reach equilibrium. For instance, a particularly long and GC-rich stem might provide a kinetic barrier to RNA strand multimerization since breaking of strong base-pairing is very slow in RNA. The dissociation of an 8 bp GC duplex was previously calculated to have a half-life of ∼16 h at 37°C (Turner and Bevilacqua 1993), although this barrier can be lessened considerably by toehold/branch migration processes (Auslander and Fussenegger 2014; Zhou et al. 2019). Since RNA tends to fold as it is being transcribed (Chadalavada et al. 2010; Watters et al. 2016; Passalacqua and Lupták 2021), local structures such as these stem–loops are initially favored. Collectively, these kinetic barriers, their lessening by toeholds, and folding during synthesis all add to the complexity of the folding landscape.
Above we considered interactions between two strands, but RNA can form three- and four-stranded interactions as well. These multimeric complexes are especially appealing for condensate formation because they bring together even more strands. Triplexes can form when a pyrimidine strand interacts in the major groove of a duplex homopurine–homopyrimidine stretch to form a base triple (Fig. 1B, images 1 and 2), which can bring up to three different RNA strands together (Fig. 1B, image 4; Devi et al. 2015). In these cases, a U interacts with the Hoogsteen face of A in an AU Watson–Crick pair, while a C+ (protonated) interacts with the Hoogsteen face of G in a GC Watson–Crick base pair. What is the likelihood of such a triplex? For the interaction of four consecutive base triplets, the probability that a 4 bp stretch is homopurine is (1/2)3 or 12.5%, which is reasonably high. The likelihood of a third strand complementing this duplex is smaller at (1/4)4, or 0.4%. However, given evidence that there are a vast number of nucleotides in a condensate, it also is reasonably high. Because the likelihood of their formation is so high, triplexes may be important contributors to condensate formation. One of the appealing features of this type of an interaction is that, unlike in the hairpin-dimer equilibrium discussed above, no Watson–Crick base pairs have to be broken; the triplex forms in the context of preformed base pairs, which may lessen the aforementioned kinetic barriers to structure formation.
Another way triplexes can form is through the A-minor motif, which is quite common, being present in nearly all large RNAs (Devi et al. 2015). In this motif, A residues interact with a preformed duplex via base-base and base-sugar (2′OH) interactions in the wide and shallow (i.e., accessible) minor groove of the duplex (e.g., Fig. 1B, image 3). The A-minor motif is common in tertiary interactions, including structures such as pseudoknots (Shalybkova et al. 2021). This motif is more plastic than the aforementioned major groove triplexes because A's can interact with GC, CG, AU, and UA pairs, as well as with non-Watson–Crick pairs to make base triples in the wide and accessible minor groove of RNA.
Quadruplexes have been well described in RNA as well. These include four-base G-quartets, which can form within the same RNA strand or between two, three, or four different RNA strands to make base quartets (Fig. 1C; Liu et al. 2000; Collie et al. 2010). As such, they could be especially effective in bringing multiple RNA strands together (Khong et al. 2017; Zhang et al. 2019). Stretches of as few as two G's could allow quadruplex formation, and these can be favored by the presence of polyamines, especially the organic cation spermine (Williams et al. 2021). Quartets can form out of bases other than just G as well. For instance, base quartets can form when the pyrimidine in a major groove of a base triple interacts with a fourth base (Fig. 1D; Su et al. 1999). The myriad ways these RNA–RNA interactions conspire to drive LLPS is summarized in the scheme in Figure 1E.
Finally, long RNAs such as mRNAs and lncRNAs can interact along their entire length (Van Treeck et al. 2018; Luo et al. 2021). For instance, one segment of a given RNA might interact with a second RNA, while another segment of the first RNA might interact with a third RNA and so on. Moreover, those interactions could involve base-pairing, which bring two strands together locally, or base triples or quartets, which could bring together three or four strands locally. Given that mRNAs can be thousands of bases long, a given mRNA could have myriad RNA partners inside a condensate, each held in place by numerous two-base, three-base, or four-base interactions. We recently reported such daisy chaining of RNA in the case of a riboswitch (Poudyal et al. 2021) as analyzed by the RNA design software NUPACK (Zadeh et al. 2011) where minimally six-stranded complexes held together by base-pairing were predicted (Fig. 2B). These considerations are consistent with reports that longer mRNAs, which concomitantly have more interactions, tend to accumulate in stress granules in vivo and in RNA-only assembles in vitro (Khong et al. 2017; Van Treeck et al. 2018). These findings and others, which showed a correlation between calculations (complex size by NUPACK) and experiments (phase separation), suggest that RNA design software packages such as NUPACK may be useful for assessing the propensity of RNA to phase separate (Zadeh et al. 2011; Poudyal et al. 2021).
DISRUPTIVE FORCES FOR PHASE SEPARATION
Stress granules are known to undergo reversible formation. One question then is: what could lead to dissolution of the granule? The answer may lie in disrupting RNA–RNA interactions, both by spontaneous breaking of base pairs and by active disruption by helicases. As mentioned above, RNA–RNA interactions can be long-lived, especially for longer base-pairing stretches. Moreover, in vitro FRAP experiments show that RNA-only condensates have much longer half-lives than RNA–protein ones (hours versus seconds) (Van Treeck et al. 2018; Poudyal et al. 2021). Helicases can thus be expected to play a role in the dissolution of condensates (Van Treeck and Parker 2018). Indeed, stress granules contain RNA helicases such as DEAD-box helicases, eIF4A, RHAU, and DDX3 (Chalupnikova et al. 2008; Hilliker et al. 2011; Epling et al. 2015; Valentin-Vega et al. 2016; Kosmacz et al. 2019; Tauber et al. 2020a). In particular, the DEAD-box helicase eIF4A has been shown to suppress stress granule accumulation in human cell lines in a manner dependent on its RNA binding capability (Tauber et al. 2020a). Over-expression of eIF4A is associated with improved drought and salinity tolerance in several plant species (Santosh Rama Bhadra Rao et al. 2017 and references therein), and it would be of interest to assess RNA condensation and stress granule formation in these transgenic plant lines. In addition, translation may disperse stress granules owing to ribosome interactions with exiting mRNAs, preventing their reentrance into the condensate; likewise, mRNA turnover may break up P-bodies due to loss of RNAs themselves (Gilks et al. 2004; Khong et al. 2017). Furthermore, there is in vitro and cellular evidence that RNA degradation within RNP granules can reduce their assembly and size (Teixeira et al. 2005; Burke et al. 2020).
Another factor that could interfere with RNA–RNA interaction and eliminate phase separation is an increase in temperature, particularly in the case of poikilotherms such as micro-organisms, fungi, plants, and invertebrates. This is because RNA–RNA interaction is generally enthalpically driven, as mentioned above, and so disfavored at higher temperatures (Nott et al. 2015; Falahati et al. 2016; Shin and Brangwynne 2017). Indeed, one could imagine droplets forming and dissolving as a function of temperature, perhaps according to the diurnal cycle, or entrained by circadian rhythms. Intriguingly, several proteins of the circadian clock show accumulation in nuclear condensates in plants (Emenecker et al. 2020). Also, in plants, high temperatures as well as the desiccation signaling hormone, abscisic acid, promote redistribution of a transcriptional repressor (Jung et al. 2020) and an RNA binding protein (Li et al. 2002) into nuclear speckles. Other abiotic stress-related factors could also affect condensate formation. Proline, which is a compatible solute synthesized by plants during drought and salt stress (Tester and Davenport 2003; Tack et al. 2020) is a known potent denaturant of RNA structure (Lambert and Draper 2007). Removal of salt stress (Tack et al. 2020) would relieve conventional RNA–RNA base-pairing interactions, which are favored by high salt, which might also dissolve condensates. On other hand, we have found that aggregates of spermine-promoted G-quadruplexes are eliminated by higher salt (Williams et al. 2021), which is the opposite effect, occurring here because the salt competes the spermine away from the RNA. The major takeaway from these studies is that abiotic variables such as temperature and salt can have a multitude of effects on biomolecular condensates such that their dynamics are affected, especially in living systems. This may be important for fine-tuning regulation but can make dynamics difficult to predict.
RNA AS A MOLECULAR CHAPERONE
The biological functions of proteins are governed by their amino acid properties and three-dimensional structure. If proteins misfold, as a result of either mutation or environmental stress, protein aggregation is likely to be one of the major consequences, which can result in cytotoxic events and cell death (Maisonneuve et al. 2008). Environmental challenges (e.g., extreme temperatures, dehydration, and osmotic stress) often expedite protein aggregation and misfolding within the cell (Tyedmers et al. 2010). Under these circumstances, diverse molecular chaperones have been identified with pivotal functions that aid in preventing protein aggregation and in regulating protein folding. These chaperones are involved in maintaining the equilibrium of folded proteins (proteostasis) and proteome integrity (Balch et al. 2008).
Protein-based chaperones have been well studied and are classified into two groups: (i) The ATP-independent type chaperones, often called “holdases,” which bind to unfolded or misfolded proteins directly, minimizing protein aggregation without consuming ATP; (ii) The ATP-dependent type chaperones, often called “foldases,” which utilize ATP to assist proper protein folding (Hartl et al. 2011).
Aside from the above-described protein-based chaperones, chaperoning of proteins by RNAs has been documented in several model systems (Kudlicki et al. 1997; Choi et al. 2008; Son et al. 2015; Docter et al. 2016). Among these studies, sequence specificity and the length of the RNA (as short as 19 bases) have been characterized in vitro for chaperone function (Docter et al. 2016); it is also worth noting that RNA chaperones are more efficient than other types of chaperones (protein-based, polyphospate, and dsDNA) in suppression of protein aggregation (Gray et al. 2014; Docter et al. 2016).
In biomolecular condensates that contain both RNA and proteins, it is thus possible that RNA could chaperone its protein partners, perhaps acting as a holdase. It should be noted that extensive RNA oligomerization or condensation is often associated with cellular stress (Tauber et al. 2020b). These stress conditions could trigger mRNAs to form non-membrane-bound ribonucleoprotein (RNP) complexes, stress granules or P-bodies, wherein some of the protein-binding mRNAs could direct their protein partners to the biomolecular condensate with consequent translation shutoff or protection from protein degradation (Mateju et al. 2017; Chantarachot and Bailey-Serres 2018; Alriquet et al. 2019).
For example, it is known that heat shock proteins are often found in stress granules, and some of them (Hsp70, 110, and 60) can interact with A + U rich elements (AREs) both in vitro and in vivo (Henics et al. 1999; Wilson et al. 2001; Kishor et al. 2013; Docter et al. 2016). Particularly, poly(U) RNA has been reported as a highly effective sequence that, in cooperation with the DnaK chaperone (the homolog of Hsp70), can synergistically increase Hsp protein refolding efficiency (Son et al. 2015; Docter et al. 2016). Notably, the chaperone activity of poly(U) RNA was about 300-fold more effective at preventing luciferase protein aggregation in vitro than the well-studied protein-based chaperone GroEL (Docter et al. 2016). It merits attention that ∼10% of eukaryotic mRNAs have AREs in their 3′ UTR (Halees et al. 2008), thus suggesting possible roles of AREs in rapid stress response via assisting protein folding in cooperation with protein-based chaperones. A related biological question is whether there exist other RNA-based chaperones that aid—via ARE recognition or by other sequences or motifs—in protein or RNA folding, thereby opposing the degradation of their targets. Although most mechanisms remain unknown to date, insights obtained from future studies will undoubtedly further extend and complement the current concept of RNA–protein networks in rapid stress responses.
CONCLUSIONS
Base pairing, both Watson–Crick and non-Watson–Crick, between two RNA strands can lead to the formation of dimers that are both stable and frequent. Three- and four-stranded interactions of RNAs involving major groove and more permissive minor groove interactions are also known and their formation may be more facile in that they do not require disruption of base-pairing. At the same time, interactions that first require the breaking of preformed base pairs could be facilitated by toehold/branch migration processes. RNA multimerization processes are driven by increases in RNA concentration, as would happen in the sub-fL volumes of condensates, and could drive liquid–liquid phase separation. One of the most striking features of the models considered herein is that pairing between RNAs is common and strong, even when it initiates purely by chance. That is not to say that all interactions between RNAs are nonspecific; indeed, there is strong evidence for specific interaction between sense and antisense transcripts, for example (Van Treeck et al. 2018). Nonetheless, given the considerations herein, there is likely to be a stochastic aspect to the formation of RNA–RNA interactions in condensates. Diversity between different droplets of the same condensate could lead to noise in gene expression, as has been observed for single cells, which could in turn lead to a type of robustness (Raser and O'Shea 2005). Probing the composition of biomolecular condensates at the single compartment level is thus an area for further study that could reveal underlying heterogeneity of compartment composition and RNA structure.
In this article, we have taken a decidedly RNA-centric view of condensates. It will be of interest to see if RNA-only condensates are discovered in vivo, since they form readily in vitro. Such condensates would likely be associated with high divalent and organic cation concentrations on the basis of our findings with riboswitches (Poudyal et al. 2021) and G-quadruplexes (Williams et al. 2021). Nonetheless, proteins play critical roles in condensate formation and properties. For instance, arginine-rich peptides, which can bind robustly to RNA, trigger stress granule formation by stabilizing RNA–RNA interactions through relieving anionic repulsion, base-pairing with guanines, and engaging in cation-π interactions (Lee et al. 2013; Boeynaems et al. 2017; Reiss et al. 2017; Van Treeck et al. 2018). Proteins also play important roles in the properties of condensates, perhaps best illustrated by the aforementioned in vitro FRAP experiments which show that RNA-only condensates have very long half-lives (Van Treeck et al. 2018; Poudyal et al. 2021). Like RNA–RNA interactions, it is likely that some RNA–protein and protein–protein interactions are specific, while others are not. Because protein–protein interactions tend to be weaker, they may temper the very strong RNA–RNA interactions and foster rapid dissolution of droplets, which may be needed for an effective biological response. In summary, the physicochemical properties of RNAs provide an important context in which to understand and investigate the behavior of diverse biomolecular condensates.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by the National Institutes of Health grants R35 GM127064 to P.C.B. and National Science Foundation PGRP grant IOS-2122357 to P.C.B. and S.M.A. We thank members of our laboratories for helpful comments on the manuscript and Professor Roy Parker and Dr. Nina Ripin for their helpful review of the manuscript.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.078999.121.
Freely available online through the RNA Open Access option.
REFERENCES
- Alriquet M, Martinez-Limon A, Hanspach G, Hengesbach M, Tartaglia GG, Calloni G, Vabulas RM. 2019. Assembly of proteins by free RNA during the early phase of proteostasis stress. J Proteome Res 18: 2835–2847. 10.1021/acs.jproteome.9b00143 [DOI] [PubMed] [Google Scholar]
- Antao VP, Lai SY, Tinoco I. 1991. A thermodynamic study of unusually stable RNA and DNA hairpins. Nucleic Acids Res 19: 5901–5905. 10.1093/nar/19.21.5901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aumiller WM, Pir-Cakmak F, Davis BW, Keating CD. 2016. RNA-based coacervates as a model for membraneless organelles: formation, properties, and interfacial liposome assembly. Langmuir 32: 10042–10053. 10.1021/acs.langmuir.6b02499 [DOI] [PubMed] [Google Scholar]
- Auslander S, Fussenegger M. 2014. Synthetic biology: toehold gene switches make big footprints. Nature 516: 333–334. 10.1038/516333a [DOI] [PubMed] [Google Scholar]
- Balch WE, Morimoto RI, Dillin A, Kelly JW. 2008. Adapting proteostasis for disease intervention. Science 319: 916–919. 10.1126/science.1141448 [DOI] [PubMed] [Google Scholar]
- Bevilacqua PC, Turner DH. 1991. Comparison of binding of mixed ribose-deoxyribose analogues of CUCU to a ribozyme and to GGAGAA by equilibrium dialysis: evidence for ribozyme specific interactions with 2′ OH groups. Biochemistry 30: 10632–10640. 10.1021/bi00108a005 [DOI] [PubMed] [Google Scholar]
- Bloomfield VA, Crothers DM, Tinoco I. 2000. Nucleic acids: structures, properties, and functions. University Science Books, Sausalito, California. [Google Scholar]
- Boeynaems S, Bogaert E, Kovacs D, Konijnenberg A, Timmerman E, Volkov A, Guharoy M, De Decker M, Jaspers T, Ryan VH, et al. 2017. Phase separation of C9orf72 dipeptide repeats perturbs stress granule dynamics. Mol Cell 65: 1044–1055.e45. 10.1016/j.molcel.2017.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeynaems S, Holehouse AS, Weinhardt V, Kovacs D, Van Lindt J, Larabell C, Van Den Bosch L, Das R, Tompa PS, Pappu RV, et al. 2019. Spontaneous driving forces give rise to protein-RNA condensates with coexisting phases and complex material properties. Proc Natl Acad Sci 116: 7889–7898. 10.1073/pnas.1821038116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR. 2014. mRNP granules. Assembly, function, and connections with disease. RNA Biol 11: 1019–1030. 10.4161/15476286.2014.972208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke JM, Lester ET, Tauber D, Parker R. 2020. RNase l promotes the formation of unique ribonucleoprotein granules distinct from stress granules. J Biol Chem 10.1074/jbc.RA119.011638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cakmak FP, Choi S, Meyer MO, Bevilacqua PC, Keating CD. 2020. Prebiotically-relevant low polyion multivalency can improve functionality of membraneless compartments. Nat Commun 11: 5949. 10.1038/s41467-020-19775-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadalavada DM, Gratton EA, Bevilacqua PC. 2010. The human HDV-like CPEB3 ribozyme is intrinsically fast-reacting. Biochemistry 49: 5321–5330. 10.1021/bi100434c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalupnikova K, Lattmann S, Selak N, Iwamoto F, Fujiki Y, Nagamine Y. 2008. Recruitment of the RNA helicase RHAU to stress granules via a unique RNA-binding domain. J Biol Chem 283: 35186–35198. 10.1074/jbc.M804857200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantarachot T, Bailey-Serres J. 2018. Polysomes, stress granules, and processing bodies: a dynamic triumvirate controlling cytoplasmic mRNA fate and function. Plant Physiol 176: 254–269. 10.1104/pp.17.01468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong C, Varani G, Tinoco I Jr. 1990. Solution structure of an unusually stable RNA hairpin, 5′ggac(uucg)gucc. Nature 346: 680–682. 10.1038/346680a0 [DOI] [PubMed] [Google Scholar]
- Chiu Y-P, Sun Y-C, Qiu D-C, Lin Y-H, Chen Y-Q, Kuo J-C, Huang JR. 2020. Liquid-liquid phase separation and extracellular multivalent interactions in the tale of galectin-3. Nat Commun 11: 1229. 10.1038/s41467-020-15007-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SI, Han KS, Kim CW, Ryu KS, Kim BH, Kim KH, Kim SI, Kang TH, Shin HC, Lim KH, et al. 2008. Protein solubility and folding enhancement by interaction with RNA. PLoS One 3: e2677. 10.1371/journal.pone.0002677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collie GW, Parkinson GN, Neidle S, Rosu F, De Pauw E, Gabelica V. 2010. Electrospray mass spectrometry of telomeric RNA (TERRA) reveals the formation of stable multimeric G-quadruplex structures. J Am Chem Soc 132: 9328–9334. 10.1021/ja100345z [DOI] [PubMed] [Google Scholar]
- Creighton TE. 1993. Proteins: structures and molecular properties. W.H. Freeman and Company, New York. [Google Scholar]
- Decker CJ, Teixeira D, Parker R. 2007. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J Cell Biol 179: 437–449. 10.1083/jcb.200704147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devi G, Zhou Y, Zhong Z, Toh DF, Chen G. 2015. RNA triplexes: from structural principles to biological and biotech applications. Wiley Interdiscip Rev RNA 6: 111–128. 10.1002/wrna.1261 [DOI] [PubMed] [Google Scholar]
- Docter BE, Horowitz S, Gray MJ, Jakob U, Bardwell JC. 2016. Do nucleic acids moonlight as molecular chaperones? Nucleic Acids Res 44: 4835–4845. 10.1093/nar/gkw291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emenecker RJ, Holehouse AS, Strader LC. 2020. Emerging roles for phase separation in plants. Dev Cell 55: 69–83. 10.1016/j.devcel.2020.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epling LB, Grace CR, Lowe BR, Partridge JF, Enemark EJ. 2015. Cancer-associated mutants of RNA helicase DDX3X are defective in RNA-stimulated ATP hydrolysis. J Mol Biol 427: 1779–1796. 10.1016/j.jmb.2015.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falahati H, Pelham-Webb B, Blythe S, Wieschaus E. 2016. Nucleation by rRNA dictates the precision of nucleolus assembly. Curr Biol 26: 277–285. 10.1016/j.cub.2015.11.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. 2004. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell 15: 5383–5398. 10.1091/mbc.e04-08-0715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MJ, Wholey WY, Wagner NO, Cremers CM, Mueller-Schickert A, Hock NT, Krieger AG, Smith EM, Bender RA, Bardwell JC, et al. 2014. Polyphosphate is a primordial chaperone. Mol Cell 53: 689–699. 10.1016/j.molcel.2014.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halees AS, El-Badrawi R, Khabar KS. 2008. ARED organism: expansion of ARED reveals AU-rich element cluster variations between human and mouse. Nucleic Acids Res 36: D137–D140. 10.1093/nar/gkm959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. 2011. Molecular chaperones in protein folding and proteostasis. Nature 475: 324–332. 10.1038/nature10317 [DOI] [PubMed] [Google Scholar]
- Henics T, Nagy E, Oh HJ, Csermely P, von Gabain A, Subjeck JR. 1999. Mammalian hsp70 and hsp110 proteins bind to RNA motifs involved in mRNA stability. J Biol Chem 274: 17318–17324. 10.1074/jbc.274.24.17318 [DOI] [PubMed] [Google Scholar]
- Heus HA, Pardi A. 1991. Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science 253: 191–194. 10.1126/science.1712983 [DOI] [PubMed] [Google Scholar]
- Hilliker A, Gao Z, Jankowsky E, Parker R. 2011. The DEAD-box protein Ded1 modulates translation by the formation and resolution of an eIF4F-mRNA complex. Mol Cell 43: 962–972. 10.1016/j.molcel.2011.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, Vale RD. 2017. RNA phase transitions in repeat expansion disorders. Nature 546: 243–247. 10.1038/nature22386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JH, Barbosa AD, Hutin S, Kumita JR, Gao M, Derwort D, Silva CS, Lai X, Pierre E, Geng F, et al. 2020. A prion-like domain in elf3 functions as a thermosensor in Arabidopsis. Nature 585: 256–260. 10.1038/s41586-020-2644-7 [DOI] [PubMed] [Google Scholar]
- Kedersha N, Panas MD, Achorn CA, Lyons S, Tisdale S, Hickman T, Thomas M, Lieberman J, McInerney GM, Ivanov P, et al. 2016. G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J Cell Biol 212: 845–860. 10.1083/jcb.201508028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khong A, Matheny T, Jain S, Mitchell SF, Wheeler JR, Parker R. 2017. The stress granule transcriptome reveals principles of mRNA accumulation in stress granules. Mol Cell 68: 808–820 e805. 10.1016/j.molcel.2017.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishor A, Tandukar B, Ly YV, Toth EA, Suarez Y, Brewer G, Wilson GM. 2013. Hsp70 is a novel posttranscriptional regulator of gene expression that binds and stabilizes selected mRNAs containing AU-rich elements. Mol Cell Biol 33: 71–84. 10.1128/MCB.01275-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosmacz M, Gorka M, Schmidt S, Luzarowski M, Moreno JC, Szlachetko J, Leniak E, Sokolowska EM, Sofroni K, Schnittger A, et al. 2019. Protein and metabolite composition of Arabidopsis stress granules. New Phytol 222: 1420–1433. 10.1111/nph.15690 [DOI] [PubMed] [Google Scholar]
- Kudlicki W, Coffman A, Kramer G, Hardesty B. 1997. Ribosomes and ribosomal RNA as chaperones for folding of proteins. Fold Des 2: 101–108. 10.1016/S1359-0278(97)00014-X [DOI] [PubMed] [Google Scholar]
- Lambert D, Draper DE. 2007. Effects of osmolytes on RNA secondary and tertiary structure stabilities and RNA-Mg2+ interactions. J Mol Biol 370: 993–1005. 10.1016/j.jmb.2007.03.080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YB, Chen HJ, Peres JN, Gomez-Deza J, Attig J, Stalekar M, Troakes C, Nishimura AL, Scotter EL, Vance C, et al. 2013. Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep 5: 1178–1186. 10.1016/j.celrep.2013.10.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis NB, Westhof E. 2001. Geometric nomenclature and classification of RNA base pairs. RNA 7: 499–512. 10.1017/S1355838201002515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis NB, Stombaugh J, Westhof E. 2002. The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Res 30: 3497–3531. 10.1093/nar/gkf481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Kinoshita T, Pandey S, Ng CK, Gygi SP, Shimazaki K, Assmann SM. 2002. Modulation of an RNA-binding protein by abscisic-acid-activated protein kinase. Nature 418: 793–797. 10.1038/nature00936 [DOI] [PubMed] [Google Scholar]
- Lin Y, Protter DS, Rosen MK, Parker R. 2015. Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol Cell 60: 208–219. 10.1016/j.molcel.2015.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Kanagawa M, Matsugami A, Tanaka Y, Katahira M, Uesugi S. 2000. NMR study of a novel RNA quadruplex structure. Nucleic Acids Symp Ser 65–66. 10.1093/nass/44.1.65 [DOI] [PubMed] [Google Scholar]
- Luo J, Qu L, Gao F, Lin J, Liu J, Lin A. 2021. LncRNAs: architectural scaffolds or more potential roles in phase separation. Front Genet 12: 369. 10.3389/fgene.2021.626234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisonneuve E, Ezraty B, Dukan S. 2008. Protein aggregates: an aging factor involved in cell death. J Bacteriol 190: 6070–6075. 10.1128/JB.00736-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateju D, Franzmann TM, Patel A, Kopach A, Boczek EE, Maharana S, Lee HO, Carra S, Hyman AA, Alberti S. 2017. An aberrant phase transition of stress granules triggered by misfolded protein and prevented by chaperone function. EMBO J 36: 1669–1687. 10.15252/embj.201695957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, Taylor JP. 2015. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163: 123–133. 10.1016/j.cell.2015.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muiznieks LD, Sharpe S, Pomes R, Keeley FW. 2018. Role of liquid-liquid phase separation in assembly of elastin and other extracellular matrix proteins. J Mol Biol 430: 4741–4753. 10.1016/j.jmb.2018.06.010 [DOI] [PubMed] [Google Scholar]
- Nallagatla SR, Hwang J, Toroney R, Zheng X, Cameron CE, Bevilacqua PC. 2007. 5'-triphosphate-dependent activation of PKR by RNAs with short stem-loops. Science 318: 1455–1458. 10.1126/science.1147347 [DOI] [PubMed] [Google Scholar]
- Nonhoff U, Ralser M, Welzel F, Piccini I, Balzereit D, Yaspo ML, Lehrach H, Krobitsch S. 2007. Ataxin-2 interacts with the DEAD/H-box RNA helicase DDX6 and interferes with P-bodies and stress granules. Mol Biol Cell 18: 1385–1396. 10.1091/mbc.e06-12-1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott TJ, Petsalaki E, Farber P, Jervis D, Fussner E, Plochowietz A, Craggs TD, Bazett-Jones DP, Pawson T, Forman-Kay JD, et al. 2015. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol Cell 57: 936–947. 10.1016/j.molcel.2015.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace CN. 1990. Conformational stability of globular proteins. Trends Biochem Sci 15: 14–17. 10.1016/0968-0004(90)90124-t [DOI] [PubMed] [Google Scholar]
- Passalacqua LFM, Lupták A. 2021. Co-transcriptional analysis of self-cleaving ribozymes and their ligand dependence. In Ribozymes: methods and protocols (ed. Scarborough RJ, Gatignol A), pp. 13–24. Springer US, New York. [DOI] [PubMed] [Google Scholar]
- Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, et al. 2015. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162: 1066–1077. 10.1016/j.cell.2015.07.047 [DOI] [PubMed] [Google Scholar]
- Poudyal RR, Pir Cakmak F, Keating CD, Bevilacqua PC. 2018. Physical principles and extant biology reveal roles for RNA-containing membraneless compartments in origins of life chemistry. Biochemistry 57: 2509–2519. 10.1021/acs.biochem.8b00081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poudyal RR, Sieg JP, Portz B, Keating CD, Bevilacqua PC. 2021. RNA sequence and structure control assembly and function of RNA condensates. RNA 27: 1589–1601. 10.1261/rna.078875.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor DJ, Schaak JE, Bevilacqua JM, Falzone CJ, Bevilacqua PC. 2002. Isolation and characterization of a family of stable RNA tetraloops with the motif YNMG that participate in tertiary interactions. Biochemistry 41: 12062–12075. 10.1021/bi026201s [DOI] [PubMed] [Google Scholar]
- Proctor DJ, Kierzek E, Kierzek R, Bevilacqua PC. 2003. Restricting the conformational heterogeneity of RNA by specific incorporation of 8-bromoguanosine. J Am Chem Soc 125: 2390–2391. 10.1021/ja029176m [DOI] [PubMed] [Google Scholar]
- Protter DSW, Parker R. 2016. Principles and properties of stress granules. Trends Cell Biol 26: 668–679. 10.1016/j.tcb.2016.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raser JM, O'Shea EK. 2005. Noise in gene expression: origins, consequences, and control. Science 309: 2010–2013. 10.1126/science.1105891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss CW, Xiong Y, Strobel SA. 2017. Structural basis for ligand binding to the Guanidine-I riboswitch. Structure 25: 195–202. 10.1016/j.str.2016.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripin N, Parker R. 2022. Are stress granules the RNA analogs of misfolded protein aggregates? RNA 28: 67–75 (this issue). 10.1261/rna.079000.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabari BR, Dall'Agnese A, Young RA. 2020. Biomolecular condensates in the nucleus. Trends Biochem Sci 45: 961–977. 10.1016/j.tibs.2020.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santosh Rama Bhadra Rao T, Vijaya Naresh J, Sudhakar Reddy P, Reddy MK, Mallikarjuna G. 2017. Expression of Pennisetum glaucum eukaryotic translational initiation factor 4A (PgeIF4A) confers improved drought, salinity, and oxidative stress tolerance in groundnut. Front Plant Sci 8: 453. 10.3389/fpls.2017.00453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyfferth C, Renema J, Wendrich JR, Eekhout T, Seurinck R, Vandamme N, Blob B, Saeys Y, Helariutta Y, Birnbaum KD, et al. 2021. Advances and opportunities in single-cell transcriptomics for plant research. Annu Rev Plant Biol 72: 847–866. 10.1146/annurev-arplant-081720-010120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalybkova AA, Mikhailova DS, Kulakovskiy IV, Fakhranurova LI, Baulin EF. 2021. Annotation of the local context of the RNA secondary structure improves the classification and prediction of A-minors. RNA 27: 907–919. 10.1261/rna.078535.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y, Brangwynne CP. 2017. Liquid phase condensation in cell physiology and disease. Science 357: eaaf4382. 10.1126/science.aaf4382 [DOI] [PubMed] [Google Scholar]
- Shu Z, Bevilacqua PC. 1999. Isolation and characterization of thermodynamically stable and unstable RNA hairpins from a triloop combinatorial library. Biochemistry 38: 15369–15379. 10.1021/bi991774z [DOI] [PubMed] [Google Scholar]
- Son A, Choi SI, Han G, Seong BL. 2015. M1 RNA is important for the in-cell solubility of its cognate C5 protein: implications for RNA-mediated protein folding. RNA Biol 12: 1198–1208. 10.1080/15476286.2015.1096487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stothard P. 2000. The sequence manipulation suite: Javascript programs for analyzing and formatting protein and DNA sequences. BioTechniques 28: 1102–1104. 10.2144/00286ir01 [DOI] [PubMed] [Google Scholar]
- Stuart T, Satija R. 2019. Integrative single-cell analysis. Nat Rev Genet 20: 257–272. 10.1038/s41576-019-0093-7 [DOI] [PubMed] [Google Scholar]
- Su L, Chen L, Egli M, Berger JM, Rich A. 1999. Minor groove RNA triplex in the crystal structure of a ribosomal frameshifting viral pseudoknot. Nat Struct Biol 6: 285–292. 10.1038/6722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tack DC, Su Z, Yu Y, Bevilacqua PC, Assmann SM. 2020. Tissue-specific changes in the RNA structurome mediate salinity response in Arabidopsis. RNA 26: 492–511. 10.1261/rna.072850.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauber D, Tauber G, Khong A, Van Treeck B, Pelletier J, Parker R. 2020a. Modulation of RNA condensation by the DEAD-box protein eIF4A. Cell 180: 411–426 e416. 10.1016/j.cell.2019.12.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauber D, Tauber G, Parker R. 2020b. Mechanisms and regulation of RNA condensation in RNP granule formation. Trends Biochem Sci 45: 764–778. 10.1016/j.tibs.2020.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R. 2005. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 11: 371–382. 10.1261/rna.7258505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tester M, Davenport R. 2003. Na+ tolerance and Na+ transport in higher plants. Ann Bot 91: 503–527. 10.1093/aob/mcg058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner DH, Bevilacqua PC. 1993. Thermodynamic considerations for evolution by RNA. In The RNA world (ed. Gesteland RF, Atkins JF), pp. 447–464. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Tyedmers J, Mogk A, Bukau B. 2010. Cellular strategies for controlling protein aggregation. Nat Rev Mol Cell Biol 11: 777–788. 10.1038/nrm2993 [DOI] [PubMed] [Google Scholar]
- Valentin-Vega YA, Wang YD, Parker M, Patmore DM, Kanagaraj A, Moore J, Rusch M, Finkelstein D, Ellison DW, Gilbertson RJ, et al. 2016. Cancer-associated DDX3X mutations drive stress granule assembly and impair global translation. Sci Rep 6: 25996. 10.1038/srep25996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Treeck B, Parker R. 2018. Emerging roles for intermolecular RNA–RNA interactions in RNP assemblies. Cell 174: 791–802. 10.1016/j.cell.2018.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Treeck B, Protter DSW, Matheny T, Khong A, Link CD, Parker R. 2018. RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. Proc Natl Acad Sci 115: 2734–2739. 10.1073/pnas.1800038115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watters KE, Strobel EJ, Yu AM, Lis JT, Lucks JB. 2016. Cotranscriptional folding of a riboswitch at nucleotide resolution. Nat Struct Mol Biol 23: 1124–1131. 10.1038/nsmb.3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams AP, Longfellow CE, Freier SM, Kierzek R, Turner DH. 1989. Laser temperature-jump, spectroscopic, and thermodynamic study of salt effects on duplex formation by dGCATGC. Biochemistry 28: 4283–4291. 10.1021/bi00436a025 [DOI] [PubMed] [Google Scholar]
- Williams AM, Poudyal RR, Bevilacqua PC. 2021. Long tracts of guanines drive aggregation of RNA g-quadruplexes in the presence of spermine. Biochemistry 60: 2715–2726. 10.1021/acs.biochem.1c00467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson GM, Sutphen K, Bolikal S, Chuang KY, Brewer G. 2001. Thermodynamics and kinetics of hsp70 association with A+U-rich mRNA-destabilizing sequences. J Biol Chem 276: 44450–44456. 10.1074/jbc.M108521200 [DOI] [PubMed] [Google Scholar]
- Wolozin B, Ivanov P. 2019. Stress granules and neurodegeneration. Nat Rev Neurosci 20: 649–666. 10.1038/s41583-019-0222-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu AR, Wang J, Streets AM, Huang Y. 2017. Single-cell transcriptional analysis. Annu Rev Anal Chem (Palo Alto Calif) 10: 439–462. 10.1146/annurev-anchem-061516-045228 [DOI] [PubMed] [Google Scholar]
- Yang A-S, Honig B. 1995a. Free energy determinants of secondary structure formation: I. α-helices. J Mol Biol 252: 351–365. 10.1006/jmbi.1995.0502 [DOI] [PubMed] [Google Scholar]
- Yang A-S, Honig B. 1995b. Free energy determinants of secondary structure formation: II. Antiparallel β-sheets. J Mol Biol 252: 366–376. 10.1006/jmbi.1995.0503 [DOI] [PubMed] [Google Scholar]
- Zadeh JN, Steenberg CD, Bois JS, Wolfe BR, Pierce MB, Khan AR, Dirks RM, Pierce NA. 2011. Nupack: analysis and design of nucleic acid systems. J Comput Chem 32: 170–173. 10.1002/jcc.21596 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yang M, Duncan S, Yang X, Abdelhamid MAS, Huang L, Zhang H, Benfey PN, Waller ZAE, Ding Y. 2019. G-quadruplex structures trigger RNA phase separation. Nucleic Acids Res 47: 11746–11754. 10.1093/nar/gkz978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Kim S, Ho KH, O'Flaherty DK, Giurgiu C, Wright TH, Szostak JW. 2019. Non-enzymatic primer extension with strand displacement. Elife 8: e51888. 10.7554/eLife.51888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415. 10.1093/nar/gkg595 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.