Abstract
This review covers research findings reported over the past decade concerning the ability of low complexity (LC) domains to self-associate in a manner leading to their phase separation from aqueous solution. We focus our message upon the reductionist use of two forms of phase separation as biochemical assays to study how LC domains might function in living cells. Cells and their varied compartments represent extreme examples of material condensates. Over the past half century, biochemists, structural biologists, and molecular biologists have resolved the mechanisms driving innumerable forms of macromolecular condensation. In contrast, we remain largely ignorant as to how 10%–20% of our proteins actually work to assist in cell organization. This enigmatic 10%–20% of the proteome corresponds to gibberish-like LC sequences. We contend that many of these LC sequences move in and out of a structurally ordered, self-associated state as a means of offering a combination of organizational specificity and dynamic pliability to living cells. Finally, we speculate that ancient proteins may have behaved similarly, helping to condense, organize, and protect RNA early during evolution.
Keywords: cross-β polymers, hydrogel, low-complexity sequences, neurodegenerative diseases, phase separation
INTRODUCTION
We have focused this review upon the thesis that certain protein domains of low sequence complexity function by adopting labile and reversible structural order. The pathway by which we stumbled over this controversial discovery has been guided by serendipity. Our journey began with the discovery of a chemical that selectively binds LC domains only upon crystallization, and has profited substantially from the unanticipated finding that purified LC domains become phase separated into liquid-like droplet and hydrogel states upon incubation at high concentrations. Prior to the observation of LC domain phase separation, we lacked functional assays for biochemical studies of these enigmatic proteins. Over the past decade, we have used two related assays of phase separation to perform reductionist, test tube experiments on LC domains.
We do not delve into the broad topic of phase separation in the context of cell biology—recent issues of Nature Reviews Molecular Cell Biology contained no less than 44 such articles. Test tube examples of phase separation have now been observed for RNA in isolation (Jain and Vale 2017), folded proteins (Li et al. 2012), and innumerable other examples of biological macromolecules. We instead restrict the focus of this review to studies of LC domain self-association performed both in vitro and in vivo. Our sole concern is to ask whether what we are learning from test tube biochemical experiments is relevant to the manner in which LC domains function in living cells.
LOW COMPLEXITY SEQUENCES WERE FIRST RECOGNIZED AS THE ACTIVATION DOMAINS OF TRANSCRIPTION FACTORS
The earliest descriptions of eukaryotic transcription factors were reported three to four decades ago. These gene-specific transcription factors were prototypically composed of two domains. One domain allowed for avid recognition of specific DNA sequences representing cis-regulatory sites required for selective gene activation or repression. Biochemical and structural studies revealed how these DNA binding domains, including homeoboxes, zinc fingers and leucine zippers, achieve binding selectivity (Pabo and Sauer 1992). A second, unanticipated domain associated with gene-specific transcription factors was discovered as being required for transcriptional activation. When studied in living cells, removal of activation domains abrogated the regulatory function of gene-specific transcription factors.
Unlike DNA binding domains that fold into specific, three-dimensional shapes, activation domains appeared to function in the absence of molecular structure. This surprising and enigmatic fact was concordant with the chemical nature of activation domains. Instead of deploying the 20 amino acids normally required for proteins to adopt a specific, three-dimensional shape, activation domains were of low sequence complexity. The activation domain characterized by Robert Tjian and colleagues in their studies of the SP1 transcription factor was composed almost exclusively of glutamine residues (Courey et al. 1989). Other activation domains, including that of the yeast Gal4 protein studied by Mark Ptashne, were also of low sequence complexity—the Gal4 activation domain was enriched in acidic residues (Ma and Ptashne 1987). A similar enrichment in acidic amino acids also characterized the VP16 protein studied in the McKnight laboratory (Triezenberg et al. 1988), and the Hap4 activation domain studied by Leonard Guarente (Forsburg and Guarente 1989). Despite intense interest and three decades of research, little has been learned as to how activation domains function in a mechanistic sense. If a protein assumes no form of molecular structure, it is inordinately difficult to understand how it performs its biological task.
LOW COMPLEXITY DOMAINS ARE UBIQUITOUS CONSTITUENTS OF EUKARYOTIC PROTEOMES
Whereas first uncovered in studies of gene-specific transcription factors, it is now recognized that eukaryotic cells are composed of thousands of different low complexity domains. LC domains are present on the vast majority of RNA binding proteins. They also sit at either end of the α-helical, coiled-coil rod domains of our 75 intermediate filament proteins. They fill the permeability gate of nucleopores. They decorate the exterior surface of neuronal vesicles. They elaborate membrane-bound proteins of the Golgi apparatus and mitochondria. Low complexity domains are deployed liberally in all aspects of cell biology.
An entire field has emerged dedicated to the study of low complexity domains, which are also termed intrinsically disordered proteins. Biomedical interest in these proteins has been prompted by the fact that mutations in coding sequences for low complexity domains often cause neurological disease. Such mutations sometimes expand the length of low complexity domains, whereas others are simple missense mutations. If we are to properly understand neurological and neurodegenerative disease pathophysiology, we must first develop a mechanistic understanding of how low complexity domains function.
LOW COMPLEXITY DOMAINS CAN PHASE SEPARATE IN THE FORM OF HYDROGELS
Fifteen years ago, the laboratory of Dirk Görlich reported the inspired discovery that the low complexity domains of certain nucleoporin proteins can form translucent hydrogels (Frey and Görlich 2007; Frey et al. 2006; ). The relevant domain required for gel formation coincided with the phenylalanine:glycine (FG) repeats that fill the permeability channel of the nucleopore. Evidence favoring the biological validity of FG hydrogels came from binding assays using the β-importin nuclear transport protein. When applied to FG hydrogels, β-importin selectively penetrated FG hydrogels relative to control protein samples by a factor of 1000. The Görlich discoveries represented a huge step forward for studies of low complexity domains. First, they showed that FG repeats must be capable of self-associating to elicit gel formation. Second, they allowed biochemical reconstitution of the activity of an LC domain. Not only did FG hydrogels selectively bind β-importin, but they facilitated the efficiency of importin-mediated cargo influx by more than 20,000-fold.
The chemical basis of FG domain self-association in hydrogels has yet to be established. Solid state NMR studies of FG hydrogels gave clear evidence of amyloid-like cross-β interactions (Ader et al. 2010), yet the favored interpretation of FG domain self-association was that of extended polypeptides forming a mesh-like matrix. Phenylalanine residues were hypothesized to reside at the nexus of polypeptide contacts. Thus, despite evidence of cross-β interactions within FG domain hydrogels, this mode of self-association was dismissed.
Some five to six years after the groundbreaking studies of Görlich and colleagues, the McKnight laboratory reported a second example of a hydrogel-forming low complexity domain. This domain was specified by the amino-terminal 214 residues of the fused in sarcoma (FUS) RNA binding protein (Han et al. 2012; Kato et al. 2012). Unlike the phenylalanine:glycine-rich LC domain of nucleoporins, the LC domain of FUS contains triplet repeats (27) of the sequence G/S-Y-G/S. For all intents and purposes, the sequence of the FUS LC domain can be considered a series of YG repeats in place of the nucleoporin FG repeats—FUS simply replaces phenylalanine side chains with another aromatic side chain (that of tyrosine).
Not unlike Görlich's FG domain hydrogels, those made from the FUS LC domain become phase separated upon some form of self-association. Studies of FUS hydrogels revealed amyloid-like polymers as deduced by electron microscopy and X-ray diffraction. Surprisingly, unlike pathogenic amyloid polymers, those formed from the FUS LC domain were found to be labile to disassembly. A molecular structure of the cross-β core of FUS polymers has been resolved by solid state NMR spectroscopy (Murray et al. 2017). Of the 214 residue LC domain of FUS, self-association was mapped to a region between residues 37 and 95 that we designate as the amino-terminal cross-β core.
The molecular structure FUS LC domain polymers revealed two differences from pathogenic amyloids that are inordinately stable, such as α-synuclein and Aβ polymers found in the brain tissue of Alzheimer's patients. First, FUS polymers are monomorphic—the same structure is repeatedly formed from the FUS LC domain. In contrast, α-synuclein and Aβ polymers form a variety of structures. Second, the latter polymers contain between 20 and 30 hydrophobic residues per protomer at the polymer interface. FUS polymers contain but a single hydrophobic side chain, that of proline residue 72, within the entire polymer interface. The lability of FUS cross-β interactions is likely attributable to this paucity of hydrophobic residues at the subunit interface. Structural studies of FUS hydrogels show unequivocally that in this form of phase separation, protein self-association is dictated by structurally specific cross-β interactions (Murray et al. 2017).
As with the Görlich demonstration of reconstituted activity of FG domains in his hydrogel preparations, including selective β-importin binding and the dramatic enhancement of importin-mediated cargo penetration, studies of FUS hydrogels also revealed evidence of biological relevance. When linked to the DNA binding domain of any of a number of gene-specific transcription factors, the FUS LC domain functions as a potent transcriptional activation domain. Capture assays using FUS hydrogels revealed binding of the largest subunit of RNA polymerase II. This binding was traced to the carboxy-terminal domain (CTD) of RNA polymerase II, and could be reconstituted using purified components. CTD binding was also observed for hydrogels prepared from the LC domains of the Ewing sarcoma (EWS) and TAF15 proteins that represent paralogs of FUS. The CTD was released from FUS, EWS or TAF15 hydrogels upon exposure of the preparations to ATP and either of the two related cyclin-dependent kinase enzymes known to phosphorylate the CTD. The Supplemental Movie S1 shows phosphorylation-mediated release of the CTD from TAF15 hydrogels.
Perhaps importantly, or perhaps coincidentally, unbiased mutagenesis of the FUS LC domain led to the discovery of a dual serine substitution that enhanced CTD binding to FUS hydrogels and simultaneously enhanced the capacity of the FUS LC domain to activate transcription in living cells (Kwon et al. 2013). These findings argue that the self-associated state of the FUS LC domain existing within phase separated hydrogels may be biologically valid, and have been independently repeated (Janke et al. 2018; Guo et al. 2019). Importantly, evidence of direct interaction between FUS and the CTD of RNA polymerase II was contemporaneously and independently discovered by Schwartz et al. (2012, 2013).
LIQUID-LIKE DROPLETS COALESCE PRIOR TO FORMATION OF HYDROGELS
Eight papers were published in 2015 showing that the low complexity domains of a variety of different RNA binding proteins, upon incubation at high concentration, become phase separated in the form of liquid-like droplets. These included the LC domain of FUS (Altmeyer et al. 2015; Burke et al. 2015; Patel et al. 2015), the LC domain of hnRNPA2 (Lin et al. 2015; Molliex et al. 2015; Xiang et al. 2015), the LC domain of an RNA helicase enzyme (Nott et al. 2015), and the LC domain of the LAF-1 P-granule protein (Elbaum-Garfinkle et al. 2015). Several of these papers further reported that upon prolonged incubation, liquid-like droplets formed from the LC domains of various RNA binding proteins transitioned into hydrogels. Figure 1 offers schematic representations of liquid-like droplets and hydrogels.
FIGURE 1.
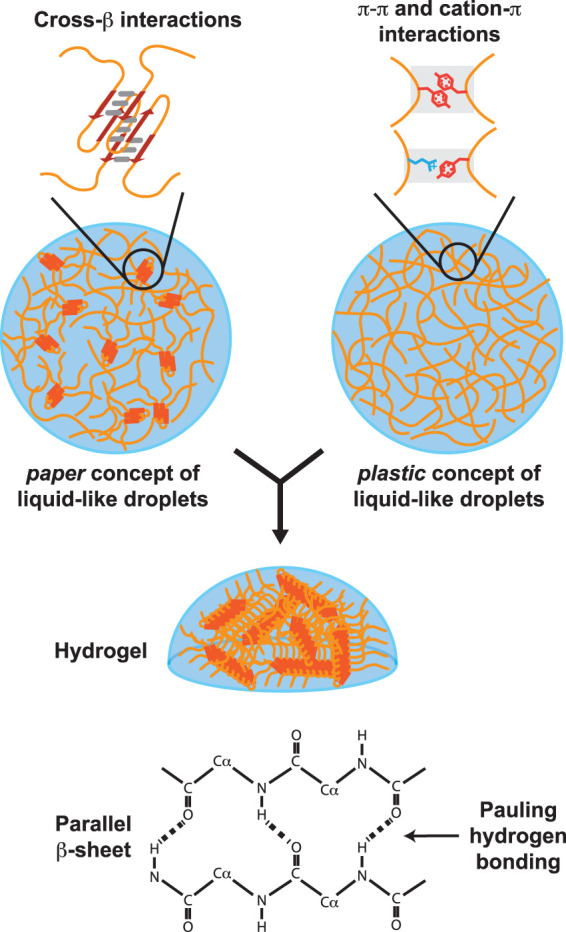
Schematic representation of low complexity domain self-association in liquid-like droplets and hydrogels. Top left image shows schematic diagram of a liquid-like droplet wherein LC domains are self-associated via labile cross-β interactions. Top right image shows diagram of liquid-like droplets assembled via structurally disordered polypeptides adhered by either π:π or cation:π interactions. Both types of droplets mature with time to a hydrogel state composed of elongated cross-β polymers. Bottom diagram shows chemical basis of cross-β self-association as described 70 years ago by Pauling and Corey (1951).
Seven of these eight papers came to the conclusion that self-association of the FUS, hnRNPA2, LAF-1, and DDX4 LC domains does not involve the formation of labile, cross-β structure (Altmeyer et al. 2015; Burke et al. 2015; Elbaum-Garfinkle et al. 2015; Lin et al. 2015; Molliex et al. 2015; Nott et al. 2015; Patel et al. 2015). It was instead concluded that in the absence of any form of protein structure, extended polypeptide chains coalesce via side chain-mediated π:π, cation:π, electrostatic and other forms of transiently attractive interaction. The proposed interactions viewed the main polypeptide chain as an inert noodle, and described self-association in a “stickers-and-spacers” model amenable to the concepts of polymer theory. This structure-independent view of LC domain phase separation has been reviewed recently by field-leading scientists including Rohit Pappu, Paul Taylor, Julie Forman-Kay, Simon Alberti, Amy Gladfelter, Tanja Mittag, Anthony Hyman, Mike Rosen, and Clifford Brangwynne (Alberti et al. 2019; Mathieu et al. 2020; Tsang et al. 2020; Alberti and Hyman 2021; Lafontaine et al. 2021; Lyon et al. 2021).
One of the eight 2015 papers describing liquid-like droplets formed from an LC domain came to a different conclusion, instead contending that the forces responsible for liquid-like droplet formation by the hnRNPA2 LC domain were structure-dependent (Xiang et al. 2015). The structure was argued to be specified by cross-β interactions indistinguishable from those responsible for formation of hnRNPA2 hydrogels.
This hypothesis was supported by two lines of investigation. First, each of the 25 tyrosine and phenylalanine residues of the hnRNPA2 LC domain was individually replaced with serine. All 25 variants were tested for incorporation into either hydrogels formed from the hnRNPA2 LC domain, which are composed of monomorphic cross-β polymers, or liquid-like droplets formed from the domain. Whereas the mechanistic effects of the 25 substitutions were variable, closely matching patterns were observed in the two-phase separation assays. We thus concluded that the same chemical forces facilitate self-association of the protein in the two settings. The location of the cross-β core of the hnRNPA2 LC domain is now known from both solid-state NMR and cryo-EM studies of labile polymers (Murray et al. 2018; Lu et al. 2020). Not surprisingly, the tyrosine-to-serine and phenylalanine-to-serine variants that most severely impeded both hydrogel binding and partitioning into liquid-like droplets map directly upon the cross-β core of the hnRNPA2 LC domain.
The second means of comparing the basis for hnRNPA2 LC domain self-association in the settings of hydrogels and liquid-like droplets involved a method of chemical footprinting. N-acetylimidazole (NAI) is a simple, water-soluble chemical capable of acetylating serine, tyrosine, lysine, threonine, arginine and asparagine side chains (Riordan et al. 1965; Timasheff and Gorbunoff 1967). When exposed to denatured protein, NAI acetylates these six side chains indiscriminately. In contrast, its application to folded proteins leads to preferential acetylation of surface-exposed residues. Relative to surface-exposed amino acids, NAI acetylation of residues buried within folded proteins is reduced. By use of mass spectrometry, the difference pattern of NAI acetylation in a denatured protein sample relative to a folded sample allowed visualization of a chemical “footprint” of the folded state. The utility of the NAI method of structure footprinting was validated using two proteins of known structure, glutathione S-transferase and poly-ADP-ribose polymerase (Xiang et al. 2015).
In hydrogel preparations of the protein, the LC domain of the hnRNPA2 protein exists in the folded state as monomorphic cross-β polymers. Knowing this, the authors established a footprint of this folded state for the hnRNPA2 LC domain relative to the denatured, unfolded protein. It was then possible to use the same methods to probe the structural state of the hnRNPA2 LC domain in liquid-like droplets. When assayed immediately upon formation of liquid-like droplets, the same NAI footprint was observed as seen in hydrogel polymers. The intensity of the hnRNPA2 NAI footprint became enhanced as a function of droplet maturation to the hydrogel state, presumably due to elongation of polymer length. These findings, coupled with the aforementioned serine substitution experiments, were interpreted to reflect the fact that hnRNPA2 self-association causative of phase separation of liquid-like droplets is driven by cross-β interactions that are labile, structurally specific and monomorphic.
Several years subsequent to characterization of this NAI footprint (Xiang et al. 2015), the organization of hnRNPA2 LC domain polymers was revealed both by solid state NMR methods (Murray et al. 2018) and cryo-electron microscopy (Lu et al. 2020). It was satisfying to observe that the location of the NAI footprint coincides precisely with the structural core responsible for forming hnRNPA2 cross-β polymers.
The NAI footprinting method was extended to probe the structural state of the hnRNPA2 LC domain within nuclei of living, mammalian cells (Xiang et al. 2015). Importantly, the protein being probed in these studies was produced at the normal level from the endogenous hnRNPA2 gene. That the in vivo footprinting assay yields the same NAI footprint for a biologically relevant sample of the hnRNPA2 protein as had been observed in both hydrogels and liquid-like droplets offers validation to test tube, phase separation assays. Later in this review we will return to a second example of in vivo footprinting that has been used to probe LC domain structure in living cells. To our knowledge, other than studies of pathogenic prions, these two studies represent the sole examples of scientific inquiry into the structural state of LC domains in living cells.
THOUGHTS ON THE PAPER VERSUS PLASTIC CONTROVERSY
Not withstanding valuable help from David Eisenberg, Robert Tycko, Dylan Murray, Glen Liszczak, Ben Tu, and Sina Ghaemmaghami, the McKnight laboratory has largely been isolated in its belief that LC domain self-association is driven by bona fide structural interactions. Our thesis is simple. We believe that LC domains self-associate via the formation of labile cross-β interactions that are structurally specific. Our thoughts rest equally on the shoulders of Linus Pauling, who taught us the chemistry of hydrogen bonding and its driving role in the formation of β-strand secondary structure in proteins (Pauling and Corey 1951), and Max Perutz, one of the fathers of modern structural biology. Just as we are now experiencing disbelief from the phase separation community that LC domains might function by folding into biologically relevant structural states, protein chemists initially rejected Perutz's contention that hemoglobin proteins work in the discretely folded state (Perutz et al. 1998). It is also notable that late in his career, Perutz devoted considerable focus to self-association of glutamine homopolymers (Perutz 1994). As such, we will designate our concept of LC domain self-association as the paper model in honor of Linus Pauling (pa) and Max Perutz (per). The sole difference in our thinking about LC domain self-association and conventionally structured proteins is that the folding of LC domains is dynamic and reversible.
A different model for LC domain self-association has been embraced by most other scientists actively engaged in the phase separation field. This alternative model eschews protein structure and ignores any participation of the polypeptide main chain, including the network of Pauling hydrogen bonding absolutely central to our paper model. The model broadly favored by the phase separation field views LC domains in the way chemists and physicists have long viewed hydrocarbon polymers for successful exploitation by the plastics industry. Over the same six to seven decade span during which scientists sorted out how polymers work in biology, industrial chemists and physicists were equally successful in figuring out the emergent properties of their own polymers. Artificial polymers could be predictably engineered into teflon, silly putty, and an endless variety of plastics.
The essential tenets of how the plastics industry profited so remarkably are to be found in the concepts embraced by the leaders of the phase separation field. LC domains are composed of “stickers and spacers.” The stickers of LC domains represent amino acid side chains allowing for favorable protein:protein interaction, including the phenylalanine residues shown by Görlich to be critical for phase separation of FG repeats (Frey et al. 2006), and the tyrosine residues shown by the McKnight laboratory to be critical for phase separation of FUS (Han et al. 2012; Kato et al. 2012). The spacers in LC domains are inert regions between stickers, including the main chain of the polypeptide itself. The contention of those favoring the plastic model is simple. Evolution has deposited the proper distribution of stickers across LC domains to enable structure-free interactions. Two aspects of this model are particularly appealing. First, it is entirely novel to biological systems. Second, it gives promise that biology may be probed and tamed in the same manner that soft matter physicists and polymer chemists have successfully ruled the plastics industry since the 1940's and 1950's.
We emphasize that our paper model of LC domain self-association embraces the importance of side chain interactions. The transient structures formed by self-associated cross-β interactions would have no opportunity for specificity without side chain interactions. We further appreciate that the aromatic phenylalanine residues critical for self-association of nucleoporin FG repeats, and tyrosine residues critical for self-association of the FUS and hnRNP LC domains, likely facilitate events of intermolecular collision helpful along the pathway toward formation of transient cross-β structures. We equate π:π, cation:π and favorable electrostatic interactions to intermediate events in protein folding. Furthermore, we consider these weak interactions as being useful for intramolecular interactions that allow lone LC domains to collapse upon themselves in the unliganded state. Indeed, we offer that published evidence of these interactions from numerous laboratories are likely to reflect intramolecular side chain contacts.
Whereas we do not seek to resolve the paper versus plastics controversy in this review, we end this discussion with several thoughts. First, is there any need for specificity in the self-association of LC domains? If so, the paper concept will likely prevail. If not, the plastics model may offer numerous advantages. Second, can either model prove to be superior in helping us understand human genetic studies that have pinpointed scores of mutations in LC domains?
What, at this point, can be said of disease-causing mutations in LC domains? Such mutations are biased toward the cause of neurological or neurodegenerative disease, and they tend to be autosomal dominant, as if the mutations are yielding a gain-of-function activity. Paradoxically, the locations and nature of these mutations tend to be surprisingly idiosyncratic. Over and over again they change the exact same amino acid within an LC domain, such as the P8 and P22 mutations within the neurofilament light chain polypeptide (Jordanova et al. 2003; Shin et al. 2008), the P301 mutation in Tau (Rizu et al. 2000), the P298 mutation in hnRNPA2 (Qi et al. 2017), or replacements of evolutionarily conserved aspartic acid residues within the LC domains of hnRNPA1, hnRNPA2 or hnRNPDL (Kim et al. 2013; Vieira et al. 2014). We anticipate that further research on these mutated LC domains will eventually yield knowledge that comports far better with either the paper or plastic model.
We have little doubt that one of these two models will prevail in relatively short order. The phase separation field is highly active and much is at stake—the concepts being probed are relevant to many aspects of modern biomedical science. Cryo-EM methods are accelerating the pace of structural resolution of labile cross-β polymers (Gui et al. 2019; Lee et al. 2020b; Lu et al. 2020; Li et al. 2021). It took the Tycko/McKnight laboratories five years to solve the atomic structure of FUS polymers by solid state NMR methods. The same end can now be accomplished in a matter of months by cryo-electron microscopy.
We are also encouraged that machine learning methods developed for protein structure prediction (AlphaFold) can already be mined for confident prediction of β-strand structure within the LC domains located on the carboxy-terminal sides of the coiled-coil rod domains of the desmin, vimentin, peripherin, GFAP and keratin intermediate filament proteins (Jumper et al. 2021). With somewhat less confidence, AlphaFold has also predicted the presence of β-strands within all six of the intermediate filament head domains that we have experimentally shown to form labile cross-β polymers (Lin et al. 2016; Zhou et al. 2021). It is encouraging to anticipate that as databases become populated by an increasing number of experimentally validated structures of labile LC domain polymers, the impact of the AlphaFold machine learning methods will flourish.
Finally, we have no problem in facing skepticism from the plastics faction that currently dominates the phase separation field. Controversy is inherently useful as any new field probes the scientific unknown, its absence is reflective of either well-trodden ground or a lack of vitality and excitement.
METHIONINES INSTEAD OF AROMATIC SIDE CHAINS
Can our concept of labile structural order for LC domain self-association open new perspectives to otherwise mysterious facets of biology? We offer that an affirmative answer to this question derives from recent studies of a methionine-rich LC domain found within a baker's yeast protein called ataxin-2.
First, a bit of background. Yeast cells execute a fundamental change in metabolism upon shifting from growth on rich to nutrient-poor culture medium. Instead of free provision of glucose and amino acids, nutrient-deprived yeast cells must undertake the de novo synthesis of complex metabolites on their own. Ben Tu and colleagues have identified a signal produced in mitochondria—reactive oxygen species—that prevents yeast cells from activating autophagy under conditions of nutritional stress. They have used traditional forward genetic methods to identify genes essential for activation of autophagy under conditions of starvation. One such gene is the yeast ortholog of the mammalian ataxin-2 protein (Yang et al. 2019). In the absence of ataxin-2, yeast cells are unable to sense the mitochondrial signal and properly regulate autophagy upon changes in the supply of growth nutrients.
Ben Tu and his students proceeded to discover a low complexity domain located within the carboxy-terminal 150 residues of the yeast ataxin-2 protein. Variants of the protein lacking this LC domain are incapable of activating autophagy. Owing to proximity between the Tu and McKnight laboratories, we teamed up to demonstrate phase separation by the yeast ataxin-2 LC domain. Like the LC domains of FUS, hnRNP, and numerous intermediate filament head domains studied in the McKnight laboratory over the past decade, the yeast ataxin-2 LC domain rapidly formed liquid-like droplets upon incubation at high concentration in the purified state. The ataxin-2 droplets matured into hydrogels composed of uniform, amyloid-like polymers that were labile to disassembly.
Inspection of the yeast ataxin-2 LC domain revealed a striking enrichment in methionine residues. Twenty-four evolutionarily conserved methionines are housed within the terminal 150 residues of the protein, qualifying ataxin-2 as one of the three most methionine-rich members of the yeast proteome. Recognizing the chemical liability of methionine to oxidation, we exposed yeast ataxin-2 liquid-like droplets to hydrogen peroxide (H2O2). The oxidant melted the droplets in a manner fully reversed by the two methionine sulfoxide reductase (MSR) enzymes beautifully characterized decades ago by Earl Stadtman (Moskovitz et al. 1995, 1997). The Supplemental Movie S2 shows the melting of ataxin-2 liquid-like droplets upon exposure to H2O2, followed by droplet reformation subsequent to exposure of the melted material to a combination of the Stadtman MSR enzymes, thioredoxin, thioredoxin reductase, and NADPH (Kato et al. 2019). Liquid-like droplets made from mutated variants of yeast ataxin-2 bearing methionine-to-tyrosine or methionine-to-phenylalanine substitutions were resistant to H2O2-mediated melting. Finally, and perhaps most importantly, when the latter variants were used to replace the endogenous ataxin-2 gene, yeast cells failed to respond to the mitochondrion-generated signal normally used to regulate autophagy (Yang et al. 2019).
The Tu/Kato experiments give evidence of an expanded utility of LC domains. That LC domain self-association can be regulated by phosphorylation has been documented for FUS (Murray et al. 2017), and the head domains of the neurofilament light (NFL) and desmin intermediate filament proteins (Zhou et al. 2021). We have also found that interaction between the CTD of RNA polymerase II and self-associated FUS can be reversed by cyclin-dependent kinase-mediated phosphorylation (Kwon et al. 2013). The Tu/Kato studies of yeast ataxin-2 demonstrate methionine oxidation as an additional means of regulating LC domain self-association. Future structural studies promise to resolve the chemical basis as to how methionine oxidation can weaken self-association of the ataxin-2 LC domain, allowing us to understand how a mitochondrion-specified chemical signal can be transduced to a fundamental change in cellular metabolism.
Inspired by Ben Tu's work on yeast ataxin-2, the McKnight laboratory recognized the LC domain of TDP43 to be methionine rich. TDP43 is an RNA binding protein frequently aggregated in the brain tissue of patients suffering from neurodegenerative disease (Neumann et al. 2006). Human genetic studies have revealed scores of missense mutations causative of neurodegenerative disease in the TDP43 LC domain (Buratti 2015). Following the Tu/Kato playbook, we quickly observed liquid-like droplets made from the TDP43 LC domain to be melted by H2O2 and reformed following exposure to the Stadtman methionine sulfoxide reductase enzymes, thioredoxin, thioredoxin reductase, and NADPH (Lin et al. 2020). It was likewise observed by transmission electron microscopy that labile, cross-β polymers composed of the TDP43 LC domain were dissolved in response to H2O2, as contrasted with no such effects on polymers composed of the LC domains of either FUS or hnRNPA2.
Not having the benefit of understanding the regulatory pathway controlled by TDP43, we have yet to resolve the biological utility of methionine oxidation of its LC domain in a manner commensurate to what Ben Tu and colleagues have accomplished with the yeast ataxin-2 protein. In contrast, we did have access to David Eisenberg's molecular structures of the polymerized TDP43 LC domain (Cao et al. 2019). To this end, we proceeded to generate a structure-based footprint useful for asking whether the TDP43 LC domain self-associates in living cells.
In the case of TDP43, we lightly treated either denatured or polymeric samples of the TDP43 LC domain with H2O2. The samples were then trypsinized and evaluated by mass spectrometry by Sina Ghaemmaghami in search of methionine residues that might be protected from oxidation in the structurally ordered state. These efforts yielded a footprint characterized by structure-dependent protection from oxidation for methionine residues 322, 323, 336, 337, and 339. All five of these methionine residues reside within a structurally ordered region according to two independent cryo-EM studies of TDP43 LC domain polymers (Cao et al. 2019; Li et al. 2021).
The same footprint was observed in hydrogel polymers made from the TDP43 LC domain, liquid-like droplets and living cells (Lin et al. 2020). Our in vivo studies of TDP43 were carried out on cells bearing normal levels of the protein expressed from the endogenous gene. These experiments confirm that the LC domain of TDP43 is capable of self-associating in a structurally specific manner in living cells. We hypothesize that this form of oxidation-sensitive self-association is of biological utility and reflective of the fact that in the 500M years of evolutionary divergence between fish and humans, there has not been a single sequence alteration within the 25 residue, structure-forming region of the TDP43 LC domain. What remains of distinct intrigue is why evolution has crafted redox-sensitivity into the labile, cross-β structure formed by the TDP43 LC domain.
PRIMITIVE PROTEINS
Profound changes in concepts of the origin of life came upon recognition that RNA polymers can fold in a manner yielding enzymes endowed with discrete, catalytic activities (Doudna and Cech 2002). These discoveries, coupled with Harry Noller's astounding prediction that the peptidyl transferase activity of ribosomes is primarily driven by ribosomal RNA rather than ribosomal proteins (Noller et al. 1992), prompted conceptualization of an “RNA world” in which RNA polymers preceded protein polymers to form the earliest living organisms (Gilbert 1986).
If evolving subsequent to their “RNA masters,” in what way might the earliest of protein polymers have helped advance evolution? We parsimoniously speculate that the most primitive proteins were either homopolymeric or modestly heteropolymeric. If so, what might have been the earliest amino acid side chains composing these polymers? Given the acidic nature of nucleic acid polymers, at least those of present-day life held together by phosphodiester bonds, we imagine that the earliest proteins may have used basic amino acid side chains.
It is impossible to know whether the earliest proteins used side chains represented in extant proteins. Whereas evolutionarily primitive proteins may have utilized amino acids chemically distinct from the modern set of 20 amino acids, for the purpose of simplicity we start by considering homopolymers composed exclusively of arginine, histidine or lysine side chains. We imagine that strings of basic amino acids may have interacted with RNA to bring about condensation of granules reminiscent of present-day intracellular structures found in the form of P-bodies, Cajal bodies, nuclear speckles, neuronal granules, histone locus bodies (Fig. 2) and any of a number of other cytoplasmic and nuclear puncta enriched in both RNA and protein.
FIGURE 2.
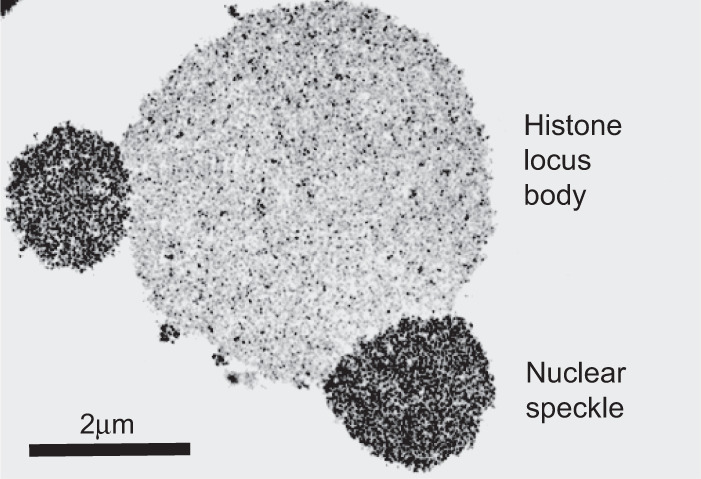
Transmission electron micrograph of two types of RNA granules within germinal vesicles of amphibian oocytes. Larger, central structure represents a histone locus body. Smaller granules fused at 5 and 9 o'clock positions upon the central histone locus body represent nuclear speckles. Scale bar = 2 µm. Photograph reproduced from Gall (2000).
The present-day RNA bodies were first described over a century ago by Ramon Y. Cajal. Dozens of these RNA-enriched subcellular puncta have been intensively studied over the past two to three decades by Ruth Lehmann, Anne Ephrussi, Joe Gall, Roy Parker, Geraldine Seydoux, Paul Taylor, and many other scientists. RNA granules are not surrounded by lipid membranes, but instead organized via the interaction of RNA binding proteins and resident RNA. As first shown by the experiments of Joe Gall in his studies of RNA granules within the germinal vesicles of amphibian oocytes, these structures are not static deposits within cells, but instead allow for rapid exchange of their constituent materials (Gall 2000). The beautiful live cell imaging experiments of Brangwynne and Hyman revealing the ease of deformability of RNA granules (Brangwynne et al. 2009) are satisfyingly consistent with Joe Gall's mechanistic evidence favoring the rapid flux of materials in and out of RNA granules. Despite lacking the organizational utility of investing membranes, and despite being both dynamic and deformable, RNA granules are not structurally amorphic. They instead display clear evidence of morphological order. This aspect, bordering upon crystallinity, is revealed in Joe Gall's transmission electron micrograph of Figure 2 showing the image of a histone locus body fused with two nuclear speckles.
The organization of extant RNA granules is co-reliant upon both RNA and protein constituents. In certain cases, removal of a single RNA binding protein can eliminate the granule in which it resides (Ephrussi et al. 1991; Marlow and Mullins 2008; Lee et al. 2020a; Yang et al. 2020). The domain architecture of prototypic RNA binding proteins is similar to that of gene-specific transcription factors. In place of homeobox, leucine zipper, or zinc finger DNA binding domains, RNA binding proteins use folded RNA recognition motifs (RRMs), KH domains or pumilio domains to facilitate direct interaction with RNA. Like gene-specific transcription factors, RNA binding proteins are also endowed with low complexity domains. Granule formation is understood to result from a combination of multivalent interaction reflective of the coating of RNA by RNA binding proteins, and the self-associative properties of LC domains. As will be articulated in the final section of this review, chemical agents that selectively impede LC domain self-association also melt RNA granules. As such, condensation of RNA granules is understood to be driven both by molecular forces that are largely understood (the interaction of RNA binding domains with RNA), as well as forces that remain controversial (self-association of LC domains).
If the earliest proteins in evolution were indeed RNA binding proteins, we can be confident that they were not endowed with structured RNA binding domains. Assuming that ancient proteins used basic amino acid side chains to interact with electrostatically acidic RNA, how might they have assisted in the formation of condensed, protective granules? We speculate that evolutionarily primitive proteins utilized Pauling-like hydrogen bonding mediated through the chemistry of the peptide backbone to facilitate condensation (Fig. 1).
This concept is reminiscent of how we envision the paper model of phase separation. One side of the earliest proteins was endowed with chemical properties having an electrostatic complementarity to RNA. The other side, we speculate was equally complex in a chemical sense, and optimally crafted for self-association. If these molecules and interactions preceded the advent of lipid membranes, they may have played a pivotal role in evolution, helping condense and protect the RNA genetic material from an otherwise harsh environment. Even if their temporal advent was subsequent to the evolution of protective lipid membranes, it is not hard to imagine the organizational benefit of RNA binding proteins that allowed condensation of life's earliest genetic material.
The persistence of LC domains through evolution is enigmatic. Our best guess as to the modern utility of LC domains is reflective of the inherent weakness of their propensity to self-associate. If they constantly move in and out of the folded state, virtually every amino acid within an LC domain is at some point labile to chemical modification via phosphorylation, oxidation, methylation, acetylation or any form of post-translational modification. This property differs radically from the majority of modern proteins that fold stably, thus masking buried amino acid side chains from chemical modification. We offer that the weakness and reversibility of LC domain folding/self-association may be at the heart of many dynamic aspects of cell organization. Simply put, weakness may sometimes be virtuous in the complexity of biology.
A TALE OF TWO CHEMICALS
The concluding section of this review is focused on two enigmatic chemicals, 1,6-hexanediol (1,6-HD) and a biotinylated isoxazole (b-isox). Both of these chemicals have played central roles in conceptualization of our paper model of LC domain function.
The experimental utility of aliphatic alcohols was first discovered according to their ability to weaken the permeability barrier of nucleopores (Ribbeck and Görlich 2002; Patel et al. 2007; Updike et al. 2011). Knowing that the constituent proteins of both nuclear pores and RNA granules contain domains of low sequence complexity, the McKnight laboratory performed correlative studies measuring the capacities of a series of aliphatic alcohols to melt hydrogel polymers, liquid-like droplets and cellular structures enriched in LC domains (Lin et al. 2016). Our study investigated 1,6-HD, 1,5-pentanediol (1,5-PD), 2,5-hexanediol (2,5-HD) and 1,4-butanediol (1,4-BD). Two of these aliphatic alcohols, 1,6-HD and 1,5-PD, melted hydrogels composed of FUS polymers, and two did not (2,5-HD and 1,4-BD). This same pattern of melting was observed for suspension preparations of FUS LC domain polymers, as well as liquid-like droplets formed from the LC domains of FUS and hnRNPA2. Turning to living cells, the same hierarchical pattern of effects was observed for the melting of cytoplasmic RNA granules containing the TIA1 RNA binding protein, as well as nuclear speckles and Cajal bodies.
These experiments were interpreted to reflect the involvement of labile cross-β interactions present within hydrogels, liquid-like droplets, and three different forms of intracellular puncta believed to commonly rely on LC domain self-association. When self-associated, the many LC domains within various intracellular puncta are interpreted to exist in a cross-β conformation. Upon exposure to 1,6-HD, either in vitro or in vivo, LC domain self-association is theorized to dissolve. This simple idea assigns a detergent-like effect to 1,6-HD, postulating that the chemical might loosely adhere to the free polypeptide chain in a manner impeding formation of cross-β interactions.
Recognizing the blunt nature of these aliphatic alcohols, we asked whether they might simply represent generic hydrophobic denaturants. To this end, we treated cells with the same four chemicals at the same concentrations required for the melting of RNA granules (6%–8%). None of the four aliphatic alcohols affected the integrity of either actin or tubulin polymers within cells. Much to our surprise, however, both 1,6-HD and 1,5-PD led to the collapse of intermediate filaments, including vimentin, desmin, keratin, and a number of other members of this class of cytoskeletal proteins. The two aliphatic alcohols that failed to melt RNA granules, 2,5-HD and 1,4-BD, also failed to affect the assembly state of intermediate filaments. Supplemental Movies S3 and S4 show cultured mammalian cells expressing a GFP-tagged form of vimentin (Gan et al. 2016) following exposure to 8% levels of either 1,6-HD (Supplemental Movie S3) or 2,5-HD (Supplemental Movie S4). The former aliphatic alcohol rapidly melted vimentin intermediate filaments, whereas the latter did not.
McKnight should have anticipated these observations due to advice he received years ago from Vann Bennett, a longtime friend from Duke Medical School. After hearing a McKnight seminar at Duke, Bennett told McKnight that the phenomena of phase separation we were observing for the LC domains of RNA binding proteins should also hold for the LC domains located on either end of the coiled-coil rod domains of intermediate filament (IF) proteins. Whereas IF proteins can form both dimers and tetramers via their isolated coiled coil domains, assembly of mature, cylindrical filaments is reliant on flanking LC domains (Godsel et al. 2008; Kornreich et al. 2015). We tested the Bennett prediction and quickly came to realize that the head domains of at least six different IF proteins were functionally indistinguishable from the LC domains of FUS, EWS, TAF15, hnRNPA2, and TDP43. Upon incubation at high concentration, these IF head domains phase separated into hydrogels that were melted by 1,6-HD and 1,5-PD, but not 2,5-HD and 1,4-BD.
These serendipitous observations on IF proteins launched what we consider to be our most incisive experiments conducted over the past three to four years. In collaboration with Dylan Murray and Rob Tycko, we obtained solid state NMR spectra on labile, cross-β polymers formed from the functionally essential head domains of the desmin and neurofilament light chain (NFL) proteins, as well as the functionally essential tail domain of the TM1-I/C IF protein of fruit flies. By use of intein chemistry, we then ligated the 13C/15N-labeled head domains of desmin and NFL onto the isotopically normal bodies of the proteins. Similarly, we ligated the isotopically labeled tail domain of TM1-I/C onto its unlabeled body.
These methods allowed execution of what we hoped might be a definitive experiment. After assembling the segmentally labeled proteins into properly organized intermediate filaments, we packed them into NMR rotors and collected spectra. Three outcomes were envisioned, two of which would have been inconsistent with our paper concept of LC domain function. To wit, the properly assembled IF proteins might reveal spectra diagnostic of no molecular order whatsoever, or spectra diagnostic of a distinctly different structural state. Alternatively, the spectra of the segmentally labeled proteins might correspond to the spectra observed for cross-β polymers of the isolated head and tail domains, thus bolstering the biological validity and functional utility of these interactions. Much to our delight, the spectra of the desmin and NFL head domains overlapped beautifully when evaluated in either the hydrogel polymer state or that of properly assembled intermediate filaments (Zhou et al. 2021). Likewise, the spectrum of the TM1-I/C tail domain was indistinguishable upon comparison of the two states (Sysoev et al. 2020). Together with the footprints’ diagnostic of cross-β structure of the hnRNPA2 and TDP43 LC domains in vivo, these studies of IF head and tail domains represent our strongest evidence that self-association of LC domains indeed involves the formation of bona fide structural order.
Returning to the aliphatic alcohols that melt β-strands, how might they work in a mechanistic sense? All four alcohols evaluated in our studies contain two chemical features—an aliphatic body flanked by hydroxyl groups. As shown in Figure 3, we envision that the aliphatic body of 1,6-HD and 1,5-PD may interact with hydrophobic aspects of the polypeptide main chain, with flanking hydroxyl groups donating hydrogen bonds to carbonyl oxygen atoms of the polypeptide backbone normally used for Pauling hydrogen bonding. Whereas the two hydroxyl groups are predicted to be acceptably disposed in 1,6-HD and 1,5-PD, we simplistically theorize that they are too closely disposed in both 2,5-HD and 1,4-PD for concomitant hydrogen bonding to adjacent carbonyl oxygen atoms. Importantly, whereas hydrophobicity of the aliphatic chain is likely critical for the detergent-like properties of these aliphatic alcohols, hydrophobicity is alone insufficient to describe their activity. 1,5-PD is less hydrophobic than 2,5-HD, yet melts cross-β interactions at concentrations where 2,5-HD is inactive.
FIGURE 3.
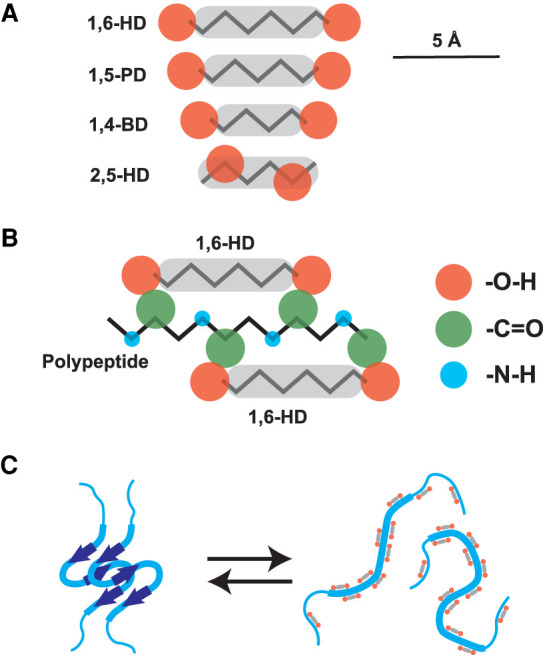
Schematized conceptualization of the mode by which aliphatic alcohols might bind disordered polypeptides and impede formation of cross-β structural interactions. Panel A shows chemical structures of four aliphatic alcohols, 1,6-hexanediol (1,6-HD), 1,5-pentanediol (1,5-PD), 1,4-butanediol (1,4-BD), and 2,5-hexanediol (2,5-HD). Panel B shows hypothetical interaction between 1,6-HD and an extended polypeptide chain. Panel C shows hypothetical basis for the melting of cross-β structures by active aliphatic alcohols (1,6-HD and 1,5-PD) but not by inactive alcohols (1,4-BD and 2,5-HD). The displayed distances between hydroxyl groups (orange) of each aliphatic alcohol represent the extreme width of separation. Flexibility of the aliphatic chains allows the prediction that the hydroxyl groups of each molecule are, on average, slightly closer together than distances displayed.
Our interpretation of how aliphatic alcohols melt labile cross-β structures predicts that any cellular structure melted by 1,6-HD, but not by the regioisomeric 2,5-HD chemical, will use LC domain self-association as described according to our paper concept. This thesis is speculative. At best we are biochemists practicing without a chemistry license. If transferred to the hands of properly trained protein chemists, our ideas should be easily validated or refuted.
The second chemical useful to elaboration of our concept of structure-based LC domain self-association is the biotinylated isoxazole shown in Figure 4A. Upon suspension into cold aqueous buffer the biotinylated isoxazole (b-isox) crystallizes. When incubated with cellular lysates, b-isox crystals coprecipitate a distribution of proteins virtually identical to the complete parts list of RNA granules (Kato et al. 2012). Simple molecular biology experiments quickly showed that the determinants of RNA binding proteins critical for b-isox coprecipitation were their LC domains. When the LC domain of an RNA binding protein was removed, b-isox precipitation was neutralized. When fused to GFP, any of a number of isolated LC domains were sufficient for b-isox precipitation.
FIGURE 4.
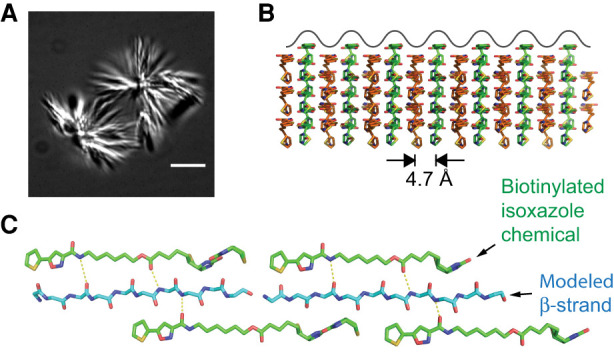
Morphological and structural properties of microcrystals formed from biotinylated isoxazole chemical. Panel A shows light microscopic image of biotinylated isoxazole microcrystals formed upon dilution of the chemical into cold aqueous buffer. Panel B shows side view of molecular lattice of biotinylated isoxazole crystals as deduced from an X-ray diffraction structure solved at 0.9 Å resolution (Kato et al. 2012). Crystalline surface is depicted by waves of alternative peaks and troughs 4.7 Å in width. Panel C shows magnified, top-down view of biotinylated isoxazole crystalline surface. An extended β-strand is modeled into the central trough, including proposed hydrogen bonds (dashed lines) between the polypeptide main chain and nitrogen and oxygen atoms of the biotinylated isoxazole chemical. Figure adapted from Kato and McKnight (2018).
How do b-isox crystals work their magic? An X-ray structure of the crystals resolved at 0.9 Å resolution offers a possible answer to this question (Kato et al. 2012). The surface of the crystals contains longitudinal arrays of mounds and troughs (Fig. 4B). The troughs are 4.7 Å in width, a size matching the width of the polypeptide chain in an extended, β conformation. Even more fortuitously, the two sides of the troughs display an iterative array of inward-pointing hydrogen bond donors and acceptors disposed with nearly perfect fit to the carbonyl oxygen and peptide nitrogen atoms displayed by extended β strands as taught to us 70 years ago by Linus Pauling. Figure 4C models an extended β strand into the 4.7 Å trough of b-isox crystals, showing the putative hydrogen bonding network between the sides of the crystal troughs and the peptide backbone.
Our interpretations as to how aliphatic alcohols melt self-associated LC domains, and how b-isox crystals selectively precipitate LC domains from cell lysates, are speculative. If correct, these ideas align favorably with our belief that LC domain self-association involves the formation of weak structural order. Minimally, however, both 1,6-HD and the b-isox chemical have played important, serendipitous roles in almost all topics considered in this review. If better interpretations of the modes of action of these chemicals can be advanced, we are all ears.
CONCLUDING COMMENTS
The science described in this review has been conducted over the past decade by Masato Kato and Steve McKnight with help from many trainees, two wonderful technicians—Leeju Wu and Lillian Sutherland—and a handful of significant collaborators including Betsy Goldsmith, David Eisenberg, Dylan Murray, Robert Tycko, Joe Gall, Glen Lisczcak, Ben Tu, and Sina Ghaemmaghami. Given that our findings have largely been dismissed by the phase separation field, readers should understand that few of our experiments have been repeated by other scientists. Other than the spectacular work reported by Dirk Görlich on FG domain hydrogels, the science covered in this review derives almost exclusively from our own experiments. Throughout the past decade we have enjoyed valuable intellectual and emotional support from Deepak Nijhawan, Glen Lisczcak, Joe Goldstein, Mike Brown, Rich Losick, Bruce Alberts, Art Horwich, David Eisenberg, Peter St. George-Hyslop, Peter Walter, Roger Kornberg, and Stan Prusiner. Without our friends, trainees, and collaborators, this scientific story would have withered long ago.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Deepak Nijhawan, Glen Liszczak, Joe Goldstein, Mike Brown, Rich Losick, Dirk Görlich, Robert Tycko, and Roger Kornberg for reading the first draft of this review and offering valuable comments. We also thank Andrea Roth for extensive administrative help. Research on low complexity domain self-association in the McKnight laboratory over the past decade has been generously supported by an anonymous donor. Our research has also been funded by grants from the National Institute of General Medical Science (R35GM130358) and the National Cancer Institute (U54CA231649).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.078990.121.
Freely available online through the RNA Open Access option.
REFERENCES
- Ader C, Frey S, Maas W, Schmidt HB, Görlich D, Baldus M. 2010. Amyloid-like interactions within nucleoporin FG hydrogels. Proc Natl Acad Sci 107: 6281–6285. 10.1073/pnas.0910163107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti S, Hyman AA. 2021. Biomolecular condensates at the nexus of cellular stress, protein aggregation disease and ageing. Nat Rev Mol Cell Biol 22: 196–213. 10.1038/s41580-020-00326-6 [DOI] [PubMed] [Google Scholar]
- Alberti S, Gladfelter A, Mittag T. 2019. Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell 176: 419–434. 10.1016/j.cell.2018.12.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmeyer M, Neelsen KJ, Teloni F, Pozdnyakova I, Pellegrino S, Grofte M, Rask MD, Streicher W, Jungmichel S, Nielsen ML, et al. 2015. Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose). Nat Commun 6: 8088. 10.1038/ncomms9088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Julicher F, Hyman AA. 2009. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324: 1729–1732. 10.1126/science.1172046 [DOI] [PubMed] [Google Scholar]
- Buratti E. 2015. Functional significance of TDP-43 mutations in disease. Adv Genet 91: 1–53. 10.1016/bs.adgen.2015.07.001 [DOI] [PubMed] [Google Scholar]
- Burke KA, Janke AM, Rhine CL, Fawzi NL. 2015. Residue-by-residue view of in vitro FUS granules that bind the C-terminal domain of RNA polymerase II. Mol Cell 60: 231–241. 10.1016/j.molcel.2015.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Q, Boyer DR, Sawaya MR, Ge P, Eisenberg DS. 2019. Cryo-EM structures of four polymorphic TDP-43 amyloid cores. Nat Struct Mol Biol 26: 619–627. 10.1101/601120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courey AJ, Holtzman DA, Jackson SP, Tjian R. 1989. Synergistic activation by the glutamine-rich domains of human transcription factor Sp1. Cell 59: 827–836. 10.1016/0092-8674(89)90606-5 [DOI] [PubMed] [Google Scholar]
- Doudna JA, Cech TR. 2002. The chemical repertoire of natural ribozymes. Nature 418: 222–228. 10.1038/418222a [DOI] [PubMed] [Google Scholar]
- Elbaum-Garfinkle S, Kim Y, Szczepaniak K, Chen CC, Eckmann CR, Myong S, Brangwynne CP. 2015. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc Natl Acad Sci 112: 7189–7194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ephrussi A, Dickinson LK, Lehmann R. 1991. oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell 66: 37–50. [DOI] [PubMed] [Google Scholar]
- Forsburg SL, Guarente L. 1989. Identification and characterization of HAP4: a third component of the CCAAT-bound HAP2/HAP3 heteromer. Genes Dev 3: 1166–1178. 10.1101/gad.3.8.1166 [DOI] [PubMed] [Google Scholar]
- Frey S, Görlich D. 2007. A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell 130: 512–523. 10.1016/j.cell.2007.06.024 [DOI] [PubMed] [Google Scholar]
- Frey S, Richter RP, Görlich D. 2006. FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science 314: 815–817. 10.1126/science.1132516 [DOI] [PubMed] [Google Scholar]
- Gall JG. 2000. Cajal bodies: the first 100 years. Annu Rev Cell Dev Biol 16: 273–300. [DOI] [PubMed] [Google Scholar]
- Gan Z, Ding L, Burckhardt CJ, Lowery J, Zaritsky A, Sitterley K, Mota A, Costigliola N, Starker CG, Voytas DF, et al. 2016. Vimentin intermediate filaments template microtubule networks to enhance persistence in cell polarity and directed migration. Cell Syst 3: 252–263.e8. 10.1016/j.cels.2016.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W. 1986. Origin of life: the RNA world. Nature 319: 618. 10.1038/319618a0 [DOI] [Google Scholar]
- Godsel LM, Hobbs RP, Green KJ. 2008. Intermediate filament assembly: dynamics to disease. Trends Cell Biol 18: 28–37. 10.1016/j.tcb.2007.11.004 [DOI] [PubMed] [Google Scholar]
- Gui X, Luo F, Li Y, Zhou H, Qin Z, Liu Z, Gu J, Xie M, Zhao K, Dai B, et al. 2019. Structural basis for reversible amyloids of hnRNPA1 elucidates their role in stress granule assembly. Nat Commun 10: 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo YE, Manteiga JC, Henninger JE, Sabari BR, Dall'Agnese A, Hannett NM, Spille JH, Afeyan LK, Zamudio AV, Shrinivas K, et al. 2019. Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature 572: 543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han TW, Kato M, Xie S, Wu LC, Mirzaei H, Pei J, Chen M, Xie Y, Allen J, Xiao G, et al. 2012. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell 149: 768–779. 10.1016/j.cell.2012.04.016 [DOI] [PubMed] [Google Scholar]
- Jain A, Vale RD. 2017. RNA phase transitions in repeat expansion disorders. Nature 546: 243–247. 10.1038/nature22386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke AM, Seo DH, Rahmanian V, Conicella AE, Mathews KL, Burke KA, Mittal J, Fawzi NL. 2018. Lysines in the RNA polymerase II C-terminal domain contribute to TAF15 fibril recruitment. Biochemistry 57: 2549–2563. 10.1021/acs.biochem.7b00310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordanova A, De Jonghe P, Boerkoel CF, Takashima H, De Vriendt E, Ceuterick C, Martin JJ, Butler IJ, Mancias P, Papasozomenos S, et al. 2003. Mutations in the neurofilament light chain gene (NEFL) cause early onset severe Charcot-Marie-Tooth disease. Brain 126: 590–597. 10.1093/brain/awg059 [DOI] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Zidek A, Potapenko A, et al. 2021. Highly accurate protein structure prediction with AlphaFold. Nature 596: 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, McKnight SL. 2018. A solid-state conceptualization of information transfer from gene to message to protein. Annu Rev Biochem 87: 351–390. 10.1146/annurev-biochem-061516-044700 [DOI] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. 2012. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149: 753–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Yang YS, Sutter BM, Wang Y, McKnight SL, Tu BP. 2019. Redox state controls phase separation of the yeast ataxin-2 protein via reversible oxidation of its methionine-rich low-complexity domain. Cell 177: 711–721.e718. 10.1016/j.cell.2019.02.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Kim NC, Wang YD, Scarborough EA, Moore J, Diaz Z, MacLea KS, Freibaum B, Li S, Molliex A, et al. 2013. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 495: 467–473. 10.1038/nature11922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornreich M, Avinery R, Malka-Gibor E, Laser-Azogui A, Beck R. 2015. Order and disorder in intermediate filament proteins. FEBS Lett 589: 2464–2476. 10.1016/j.febslet.2015.07.024 [DOI] [PubMed] [Google Scholar]
- Kwon I, Kato M, Xiang S, Wu L, Theodoropoulos P, Mirzaei H, Han T, Xie S, Corden JL, McKnight SL. 2013. Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell 155: 1049–1060. 10.1016/j.cell.2013.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine DLJ, Riback JA, Bascetin R, Brangwynne CP. 2021. The nucleolus as a multiphase liquid condensate. Nat Rev Mol Cell Biol 22: 165–182. 10.1038/s41580-020-0272-6 [DOI] [PubMed] [Google Scholar]
- Lee CS, Putnam A, Lu T, He S, Ouyang JPT, Seydoux G. 2020a. Recruitment of mRNAs to P granules by condensation with intrinsically-disordered proteins. Elife 9: e52896. 10.7554/elife.52896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Ghosh U, Thurber KR, Kato M, Tycko R. 2020b. Molecular structure and interactions within amyloid-like fibrils formed by a low-complexity protein sequence from FUS. Nat Commun 11: 5735. 10.1038/s41467-020-19512-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Banjade S, Cheng HC, Kim S, Chen B, Guo L, Llaguno M, Hollingsworth JV, King DS, Banani SF, et al. 2012. Phase transitions in the assembly of multivalent signalling proteins. Nature 483: 336–340. 10.1038/nature10879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Babinchak WM, Surewicz WK. 2021. Cryo-EM structure of amyloid fibrils formed by the entire low complexity domain of TDP-43. Nat Commun 12: 1620. 10.1101/2020.12.14.422689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Protter DS, Rosen MK, Parker R. 2015. Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol Cell 60: 208–219. 10.1016/j.molcel.2015.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Mori E, Kato M, Xiang S, Wu L, Kwon I, McKnight SL. 2016. Toxic PR poly-dipeptides encoded by the C9orf72 repeat expansion target LC domain polymers. Cell 167: 789–802.e712. 10.1016/j.cell.2016.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Zhou X, Kato M, Liu D, Ghaemmaghami S, Tu BP, McKnight SL. 2020. Redox-mediated regulation of an evolutionarily conserved cross-β structure formed by the TDP43 low complexity domain. Proc Natl Acad Sci 117: 28727–28734. 10.1073/pnas.2012216117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Cao Q, Hughes MP, Sawaya MR, Boyer DR, Cascio D, Eisenberg DS. 2020. CryoEM structure of the low-complexity domain of hnRNPA2 and its conversion to pathogenic amyloid. Nat Commun 11: 4090. 10.2210/pdb6wqk/pdb [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon AS, Peeples WB, Rosen MK. 2021. A framework for understanding the functions of biomolecular condensates across scales. Nat Rev Mol Cell Biol 22: 215–235. 10.1038/s41580-020-00303-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Ptashne M. 1987. A new class of yeast transcriptional activators. Cell 51: 113–119. 10.1016/0092-8674(87)90015-8 [DOI] [PubMed] [Google Scholar]
- Marlow FL, Mullins MC. 2008. Bucky ball functions in Balbiani body assembly and animal-vegetal polarity in the oocyte and follicle cell layer in zebrafish. Dev Biol 321: 40–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu C, Pappu RV, Taylor JP. 2020. Beyond aggregation: pathological phase transitions in neurodegenerative disease. Science 370: 56–60. 10.1126/science.abb8032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, Taylor JP. 2015. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163: 123–133. 10.1016/j.cell.2015.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskovitz J, Rahman MA, Strassman J, Yancey SO, Kushner SR, Brot N, Weissbach H. 1995. Escherichia coli peptide methionine sulfoxide reductase gene: regulation of expression and role in protecting against oxidative damage. J Bacteriol 177: 502–507. 10.1128/jb.177.3.502-507.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskovitz J, Berlett BS, Poston JM, Stadtman ER. 1997. The yeast peptide-methionine sulfoxide reductase functions as an antioxidant in vivo. Proc Natl Acad Sci 94: 9585–9589. 10.1073/pnas.94.18.9585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray DT, Kato M, Lin Y, Thurber KR, Hung I, McKnight SL, Tycko R. 2017. Structure of FUS protein fibrils and its relevance to self-assembly and phase separation of low-complexity domains. Cell 171: 615–627 e616. 10.1016/j.cell.2017.08.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray DT, Zhou X, Kato M, Xiang S, Tycko R, McKnight SL. 2018. Structural characterization of the D290V mutation site in hnRNPA2 low-complexity-domain polymers. Proc Natl Acad Sci 115: E9782–E9791. 10.1073/pnas.1806174115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, et al. 2006. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314: 130–133. [DOI] [PubMed] [Google Scholar]
- Noller HF, Hoffarth V, Zimniak L. 1992. Unusual resistance of peptidyl transferase to protein extraction procedures. Science 256: 1416–1419. [DOI] [PubMed] [Google Scholar]
- Nott TJ, Petsalaki E, Farber P, Jervis D, Fussner E, Plochowietz A, Craggs TD, Bazett-Jones DP, Pawson T, Forman-Kay JD, et al. 2015. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol Cell 57: 936–947. 10.1016/j.molcel.2015.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabo CO, Sauer RT. 1992. Transcription factors: structural families and principles of DNA recognition. Annu Rev Biochem 61: 1053–1095. 10.1146/annurev.bi.61.070192.005201 [DOI] [PubMed] [Google Scholar]
- Patel SS, Belmont BJ, Sante JM, Rexach MF. 2007. Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell 129: 83–96. 10.1016/j.cell.2007.01.044 [DOI] [PubMed] [Google Scholar]
- Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, et al. 2015. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162: 1066–1077. 10.1016/j.cell.2015.07.047 [DOI] [PubMed] [Google Scholar]
- Pauling L, Corey RB. 1951. The pleated sheet, a new layer configuration of polypeptide chains. Proc Natl Acad Sci 37: 251–256. 10.1073/pnas.37.5.251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perutz M. 1994. Polar zippers: their role in human disease. Protein Sci 3: 1629–1637. 10.1002/pro.5560031002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perutz MF, Wilkinson AJ, Paoli M, Dodson GG. 1998. The stereochemical mechanism of the cooperative effects in hemoglobin revisited. Annu Rev Biophys Biomol Struct 27: 1–34. 10.1146/annurev.biophys.27.1.1 [DOI] [PubMed] [Google Scholar]
- Qi X, Pang Q, Wang J, Zhao Z, Wang O, Xu L, Mao J, Jiang Y, Li M, Xing X, et al. 2017. Familial early-onset Paget's disease of bone associated with a novel hnRNPA2B1 mutation. Calcif Tissue Int 101: 159–169. 10.1007/s00223-017-0269-0 [DOI] [PubMed] [Google Scholar]
- Ribbeck K, Görlich D. 2002. The permeability barrier of nuclear pore complexes appears to operate via hydrophobic exclusion. EMBO J 21: 2664–2671. 10.1093/emboj/21.11.2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riordan JF, Wacker WEC, Vallee BL. 1965. N-acetylimidazole: a reagent for determination of “free” tyrosyl residues of proteins. Biochemistry 4: 1758–1765. 10.1021/bi00885a012 [DOI] [Google Scholar]
- Rizzu P, Joosse M, Ravid R, Hoogeveen A, Kamphorst W, van Swieten JC, Willemsen R, Heutink P. 2000. Mutation-dependent aggregation of tau protein and its selective depletion from the soluble fraction in brain of P301L FTDP-17 patients. Hum Mol Genet 9: 3075–3082. 10.1093/hmg/9.20.3075 [DOI] [PubMed] [Google Scholar]
- Schwartz JC, Ebmeier CC, Podell ER, Heimiller J, Taatjes DJ, Cech TR. 2012. FUS binds the CTD of RNA polymerase II and regulates its phosphorylation at Ser2. Genes Dev 26: 2690–2695. 10.1101/gad.204602.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz JC, Wang X, Podell ER, Cech TR. 2013. RNA seeds higher-order assembly of FUS protein. Cell Rep 5: 918–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sysoev VO, Kato M, Sutherland L, Hu R, McKnight SL, Murray DT. 2020. Dynamic structural order of a low-complexity domain facilitates assembly of intermediate filaments. Proc Natl Acad Sci 117: 23510–23518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JS, Chung KW, Cho SY, Yun J, Hwang SJ, Kang SH, Cho EM, Kim SM, Choi BO. 2008. NEFL Pro22Arg mutation in Charcot-Marie-Tooth disease type 1. J Hum Genet 53: 936–940. 10.1007/s10038-008-0333-8 [DOI] [PubMed] [Google Scholar]
- Timasheff SN, Gorbunoff MJ. 1967. Conformation of proteins. Annu Rev Biochem 36: 13–54. 10.1146/annurev.bi.36.070167.000305 [DOI] [PubMed] [Google Scholar]
- Triezenberg SJ, Kingsbury RC, McKnight SL. 1988. Functional dissection of VP16, the trans-activator of herpes simplex virus immediate early gene expression. Genes Dev 2: 718–729. 10.1101/gad.2.6.718 [DOI] [PubMed] [Google Scholar]
- Tsang B, Pritisanac I, Scherer SW, Moses AM, Forman-Kay JD. 2020. Phase separation as a missing mechanism for interpretation of disease mutations. Cell 183: 1742–1756. 10.1016/j.cell.2020.11.050 [DOI] [PubMed] [Google Scholar]
- Updike DL, Hachey SJ, Kreher J, Strome S. 2011. P granules extend the nuclear pore complex environment in the C. elegans germ line. J Cell Biol 192: 939–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira NM, Naslavsky MS, Licinio L, Kok F, Schlesinger D, Vainzof M, Sanchez N, Kitajima JP, Gal L, Cavaçana N, et al. 2014. A defect in the RNA-processing protein HNRPDL causes limb-girdle muscular dystrophy 1G (LGMD1G). Hum Mol Genet 23: 4103–4110. 10.1093/hmg/ddu127 [DOI] [PubMed] [Google Scholar]
- Xiang S, Kato M, Wu LC, Lin Y, Ding M, Zhang Y, Yu Y, McKnight SL. 2015. The LC domain of hnRNPA2 adopts similar conformations in hydrogel polymers, liquid-like droplets, and nuclei. Cell 163: 829–839. 10.1016/j.cell.2015.10.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YS, Kato M, Wu X, Litsios A, Sutter BM, Wang Y, Hsu CH, Wood NE, Lemoff A, Mirzaei H, et al. 2019. Yeast ataxin-2 forms an intracellular condensate required for the inhibition of TORC1 signaling during respiratory growth. Cell 177: 697–710.e17. 10.1016/j.cell.2019.02.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Mathieu C, Kolaitis RM, Zhang P, Messing J, Yurtsever U, Yang Z, Wu J, Li Y, Pan Q, et al. 2020. G3BP1 is a tunable switch that triggers phase separation to assemble stress granules. Cell 181: 325–345.e28. 10.1016/j.cell.2020.03.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Lin Y, Kato M, Mori E, Liszczak G, Sutherland L, Sysoev VO, Murray DT, Tycko R, McKnight SL. 2021. Transiently structured head domains control intermediate filament assembly. Proc Natl Acad Sci 118: e2022121118. 10.1073/pnas.2022121118 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


