Abstract
Substrates of cyclin-cdk2 kinases contain two distinct primary sequence motifs: a cyclin-binding RXL motif and one or more phosphoacceptor sites (consensus S/TPXK/R or S/TP). To identify novel cyclin-cdk2 substrates, we searched the database for proteins containing both of these motifs. One such protein is human HIRA, the homologue of two cell cycle-regulated repressors of histone gene expression in Saccharomyces cerevisiae, Hir1p and Hir2p. Here we demonstrate that human HIRA is an in vivo substrate of a cyclin-cdk2 kinase. First, HIRA bound to and was phosphorylated by cyclin A- and E-cdk2 in vitro in an RXL-dependent manner. Second, HIRA was phosphorylated in vivo on two consensus cyclin-cdk2 phosphoacceptor sites and at least one of these, threonine 555, was phosphorylated by cyclin A-cdk2 in vitro. Third, phosphorylation of HIRA in vivo was blocked by cyclin-cdk2 inhibitor p21cip1. Fourth, HIRA became phosphorylated on threonine 555 in S phase when cyclin-cdk2 kinases are active. Fifth, HIRA was localized preferentially to the nucleus, where active cyclin A- and E-cdk2 are located. Finally, ectopic expression of HIRA in cells caused arrest in S phase and this is consistent with the notion that it is a cyclin-cdk2 substrate that has a role in control of the cell cycle.
The cyclin-dependent kinases (cdks) are key regulators of progression through the eukaryotic cell cycle (21, 37). For example, cyclin E- and A-cdk2 promote progression of the cell from G1 phase into and through S phase. Periodic activation of the different cyclin-cdk complexes is largely responsible for the characteristic sequence of cell cycle events, such as mitosis, DNA synthesis, chromatin assembly, and other biosynthetic processes. However, at present we do not fully understand how periodic cyclin-cdk activation is translated into other events of the cell cycle, e.g., S-phase-specific expression of histone genes. This is largely due to a failure to identify many of the key cyclin-cdk substrates.
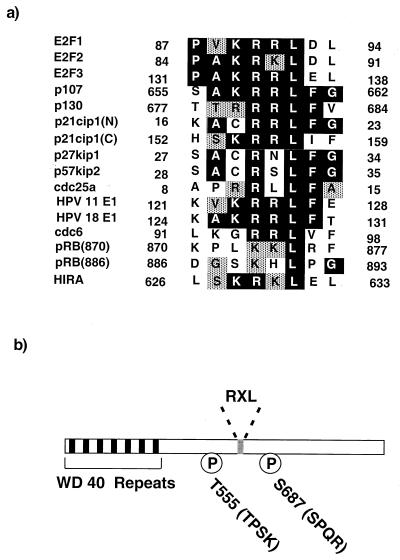
Previous studies have shown that cyclin-cdk2 kinases phosphorylate serine and threonine residues found within the consensus S/TPXK/R, although the K or R at position +3 relative to the phosphoacceptor site is not absolutely required (38, 55, 57, 62). However, it has been appreciated for some time that the cyclin subunit plays a role in further controlling and restricting substrate specificity beyond this simple requirement (43). For example, cyclin A but not cyclin E binds to the E2F1 transcription factor (13, 26, 61), allowing cyclin A-cdk2 to phosphorylate E2F1's heterodimeric partner, DP1. Recently, we and others gained novel insight into the role played by the cyclin in substrate recognition. Specifically, we identified a short sequence motif that is present in a number of cyclin-cdk2 substrates and is both necessary for and promotes the phosphorylation of these substrates (1, 6, 65). This motif, which has a conserved RXL at its core, is entirely distinct from the S/TPXK/R phosphoacceptor site. Figure 1a shows a list of proteins that have been shown to have functional RXL cyclin-cdk2-binding sequences (1, 2, 6, 14, 30, 33, 46, 47, 65). This list includes many substrates, such as E2F1, p107, pRB, cdc6, and the human papillomavirus E1 protein.
FIG. 1.
HIRA contains a putative cyclin-cdk2-binding sequence. (a) Alignment of the RXL motifs of cell cycle control proteins. Black boxes, identical residues; grey boxes, conserved residues. The numbers at the N- and C-terminal ends of each sequence indicate the numbers of the residues in the protein's full-length primary sequence. There are two RXL motifs in p21cip1 (N and C) and at least two in pRB (870 and 886). (b) Schematic of human HIRA primary sequence. Black boxes, 7 WD 40 repeats; grey box, RXL motif (residues 629 to 631), Ps, two potential cyclin-cdk2 phosphorylation sites that conform to the best-fit consensus of S/TPXK/R: threonine 555 (TPSK) and serine 687 (SPQR).
Figure 1a indicates that a number of cyclin-cdk2-inhibitory proteins, namely, p21cip1, p27kip1, and p57kip2, also contain RXL motifs that are required for efficient binding to the cyclin-cdk2 complex and inhibition of kinase activity (1, 6, 65). The RXL motifs derived from substrates and cyclin-cdk2 inhibitors appear to be functionally equivalent. For example, when a 10-amino-acid sequence containing the RXL motif of either p21cip1 or E2F1 was fused to the C terminus of a poorly phosphorylated retinoblastoma tumor suppressor protein mutant (pRB) which lacks its own RXL motif, phosphorylation was restored (2). Based on these and other biochemical studies it was proposed that the RXL motifs of both substrates and inhibitors serve as docking sites for the binding of the cyclin-cdk2 kinase. This was confirmed by X-ray crystallographic structural data for cyclin A-cdk2 complexed to p27kip1 or a p107-derived peptide (5, 45). Both crystal structures and subsequent studies in which mutations were introduced into cyclin A showed that the RXL motif and surrounding residues interact with a hydrophobic patch on cyclin A (5, 45, 50). The fact that both substrates and inhibitors have a common sequence that is required for interaction with the cyclin-cdk2 complex implies that the inhibitors function, at least in part, by competing with substrates for access to the kinase.
We performed a database search to identify other proteins that contain putative RXL cyclin-cdk2-targeting sequences and S/TPXK/R phosphoacceptor sites. One gene product identified through this screen was a protein previously named HIRA (28). The primary sequence of HIRA contains an RXL cyclin-cdk2-binding motif, two potential cyclin-cdk2 phosphorylation sites that conform to the S/TPXK/R best-fit consensus, and another 13 S/TP motifs that might also be targets of cyclin-cdk2 kinases. The HIRA gene was originally cloned due to its presence within a region of human chromosome 22q11.2 that is deleted in most patients with DiGeorge syndrome (DGS), a syndrome that results from haploinsufficiency of one or more gene products (28). The name HIRA is derived from its amino-acid sequence homology to the products of two Saccharomyces cerevisiae genes, HIR1 and HIR2, that encode cell cycle-regulated transcriptional repressors of histone gene expression (11, 52, 56).
Here we present data to show that human HIRA is a cyclin-cdk2 substrate that is phosphorylated in S phase. In addition, consistent with the notion that human HIRA is a cell cycle regulator, we have shown that ectopic expression of HIRA causes cell cycle arrest in S phase. Taken together, these results indicate that HIRA is a novel cyclin-cdk2 substrate and cell cycle regulator.
MATERIALS AND METHODS
Cell culture and transfections.
U2OS cells were cultured and transfected as described previously (1).
Plasmid construction.
Wild-type (WT) pGEX2T-E2F1, pGEX2T-E2F1[Δ24], pRC-CMVp21, pGEX2T-RB[792–928], pGEX2T-RB[792–829]E2F1Cy, and pGEX2T-RB[792–829]E2F1CyMut have been described previously (1, 2). pBS-Cyclin A was a gift of Ed Harlow.
pGEX2T-HIRA[421–729], pGEX2T-HIRA[421–729]ΔRXL, pGEX2T-RB[792–829]HIRACy, pGEX2T-RB[792–829]HIRACyMut, pcDNA3-HAHIRA[1–1017], pcDNA3-HAHIRA[421–729], pcDNA3-HAHIRA[421–729]ΔRXL, pcDNA3-HAHIRA[421–729]T555A, pcDNA3-HAHIRA[520–1017], and pcDNA3-HA-HIRA[1–1017] were made by standard molecular-biology techniques. All plasmids were confirmed by direct sequencing and/or restriction digestion. Construction methods are available on request.
Peptides.
Synthetic peptides were purchased from Biosynthesis, Inc. (Lewisville, Tex.).
Antibodies.
The anti-simian virus 40 (SV40) T-antigen antibody (419) and the anti-cyclin A antibody (C160) were gifts from Ed Harlow. The antihemagglutinin (HA) antibody (12CA5) was purchased from Roche. The anti-cdk2 antibody (M2) and the anti-p21cip1 (sc-187) antibody were purchased from Santa Cruz Biotechnology Inc. (Santa Cruz, Calif.). The anti-phosphothreonine-proline antibody (P-thr-pro-101) was purchased from Cell Signaling Technology (Beverly, Mass.).
The anti-HIRA monoclonal antibodies (WC15, WC19, WC117, and WC119) were raised against glutathione S-transferase (GST)-HIRA[421–729] in mice according to standard protocols (19). The anti-HIRA polyclonal antisera (D32 and D34) were raised in rabbits against GST-HIRA[421–729] according to standard protocols (19).
The anti-phosphothreonine 555 antiserum (D44) was raised in a rabbit. The phosphopeptide containing threonine 555 (CLSPSVLTpTPSK; synthesized by the Howard Hughes Medical Institute peptide synthesis facility at Harvard Medical School) was coupled to keyhole limpet hemocyanin (KLH) using a Pierce kit. Antiserum to the KLH-conjugated phosphopeptide was generated according to standard protocols. The rabbit serum was used without further purification.
GST fusion protein purification and binding assays.
Recovery and purification of GST fusion proteins for use in kinase assays was as described previously (1).
Recovery and purification of GST fusion proteins for use in assays of binding to radiolabeled in vitro translation products were as described previously (1), except that HD buffer (20 mM HEPES [pH 7.3], 110 mM potassium acetate, 5 mM sodium acetate, 2 mM magnesium acetate, 0.5 mM EGTA, 0.05% digitonin, 1 mM dithiothreitol, 5 μg of leupeptin/ml, 10 μg of aprotinin/ml) was substituted for NETN (120 mM NaCl, 50 mM Tris [pH 8], 1 mM EDTA, 0.5% NP-40).
In vitro kinase assays.
Cyclin A and cdk2 immunoprecipitates and kinase assays were performed essentially as described previously (1).
Recombinant cyclin A-cdk2 complexes were expressed in and purified from insect Sf9 cells essentially as described previously (7).
Immunoprecipitation, Western blotting, and treatment with λ-phosphatase.
Immunoprecipitations were performed essentially as described previously (1, 2), except that in immunoprecipitations of endogenous cellular HIRA the cells were lysed in 1 ml of EBC (50 mM Tris [pH 8], 0.5% Nonidet P-40, 100 mM NaF, 0.2 mM sodium orthovanadate, 50 μg of phenylmethylsulfonyl fluoride/ml, 10 μg of aprotinin/ml, 5 μg of leupeptin/ml)–500 mM NaCl per 10 cm-diameter plate of cells.
Western blotting was performed essentially as described previously (1).
Immunoprecipitates were treated with λ-phosphatase as described previously (2).
MS analysis.
Immunoprecipitated HIRA protein was purified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and stained with Coomassie brilliant blue R250. HIRA was excised, destained, equilibrated in 0.1 M NH4HCO3, reduced with dithiothreitol, alkylated with iodoacetamide, and hydrolyzed in 0.02 μg of trypsin (porcine; Promega)/ml in 0.05 M NH4HCO3. Tryptic peptides were extracted with 50% CH3CN–5% formic acid. The supernatants were reduced in vacuo and applied to a 0.32 (inner diameter)- by 150-mm C18 column (Symmetry 300; Waters Corporation) equilibrated with 5% CH3CN–0.1% HO-acetic acid. A linear gradient (5 to 80% CH3CN) was developed with a Waters Alliance 3690 separation module. Output from the column was directed to the LCQ quadrupole ion trap mass spectrometer (Finnigan) equipped with the stock microelectrospray ion source. The mass spectrometry (MS)/MS data were analyzed using the SEQUEST program.
FACS.
Two-color fluorescence-activated cell sorting (FACS) to determine the DNA content of cells transiently transfected with a plasmid encoding CD19 was performed as described previously (1). Two-color FACS to assay 5′-bromodeoxyuridine (BrdU) incorporation of cells transiently transfected with CD19 was performed essentially as described previously (59). Briefly, for 1 h prior to harvesting cells were pulsed with 10 μM 5′-BrdU. Cells were harvested, stained with phycoerythrin-conjugated CD19, and fixed in 70% ethanol essentially as described previously. Fixed cells were washed in phosphate-buffered saline (PBS) fixed again in PBS–1% paraformaldehyde–0.01% Tween 20, and then treated with DNase I and stained with anti-5′-BrdU fluorescein isothiocyanate. Samples were analyzed on a Becton Dickinson FACScan.
Immunofluorescence.
For staining of endogenous HIRA, cells grown on coverslips were washed once in EBC–120 mM NaCl and twice in PBS and then fixed in PBS–4% paraformaldehyde. The staining of ectopically expressed HIRA after cells were transiently transfected was performed similarly except that cells grown on coverslips were washed twice in PBS and then fixed directly in PBS–4% paraformaldehyde. Staining with primary and secondary antibodies and DAPI (4′,6′-diamidino-2-phenylindole) was essentially as described previously (19).
RESULTS
To identify novel substrates of cyclin A- or E-cdk2 kinases, we performed a search of the SWISSPROT database for proteins that contain a cyclin-cdk2-docking motif (RXL) and consensus cyclin-cdk2 phosphorylation sites (S/TPXK/R). This search was carried out using the PatScan program (www-unix.mcs.anl.gov/compbio/PatScan/HTML/patscan.html). One protein identified through this search, HIRA, was of particular interest due to its previously described homology to two S. cerevisiae proteins, Hir1p and Hir2p, that are known to play a role in control of cell cycle-regulated transcription of histone genes. Sequence comparisons indicate that HIRA is the best candidate identified to date to be a human ortholog (functional equivalent) of Hir1p and Hir2p. Figure 1 a shows an alignment of the putative cyclin-cdk2-binding motif of HIRA (amino acids 626 to 633) with the previously characterized cyclin-cdk2-binding motifs of other human cell cycle control proteins. In addition to the RXL motif, the HIRA primary sequence contains 2 putative cyclin-cdk2 phosphorylation sites that conform to the consensus S/TPXK/R (threonine 555 and serine 687), 13 other S/TP motifs that might also serve as cyclin-cdk2 phosphorylation sites, and 7 WD repeats (Fig. 1b) (28).
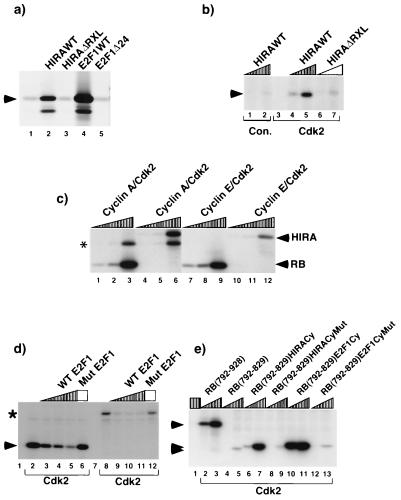
Several RXL-containing cyclin-cdk2 substrates stably bind to cyclin-cdk2 complexes in a manner that requires the RXL motif (1, 6, 14, 33, 46, 47, 65). To determine whether HIRA similarly binds to cyclin A, GST fused to residues 421 to 729 of HIRA (GST-HIRA[421–729]) was tested for binding to in vitro-translated 35S-labeled cyclin A. Residues 421 to 729 of HIRA contain the RXL motif and both S/TPXK/R cyclin-cdk2 phosphorylation sites (Fig. 1b). As shown in Fig. 2a, GST-HIRA[421–729] efficiently bound to cyclin A whereas GST alone or a HIRA mutant containing a four-alanine substitution in place of the KRKL of the RXL (GST-HIRA[421–729]ΔRXL) did not. Similarly, and as described previously, WT GST-E2F1, but not a mutant lacking the RXL motif (GST-E2F1Δ24), bound to cyclin A in this assay (26). All of the WT and mutant proteins were present in the assay mixture at comparable levels (data not shown). Thus, like that of E2F1, HIRA binding to cyclin A was dependent on an intact RXL cyclin-cdk2-binding motif.
FIG. 2.
The RXL motif of HIRA directs binding to and phosphorylation by cyclin-cdk2 kinases. (a) HIRA binds to cyclin A, and this requires the RXL motif. In vitro-translated 35S-labeled cyclin A was incubated with GST (lane 1), GST-HIRA[421–729] (lane 2), GST-HIRA[421–729]ΔRXL (lane 3), GST-E2F1 (lane 4), and -GST-E2F1Δ24 (lane 5). The bound proteins were fractionated by SDS-PAGE and visualized by autoradiography. Arrowhead, cyclin A. (b) HIRA is phosphorylated by cyclin-cdk2 kinases, and this requires the RXL motif. Extracts of U2OS cells were immunoprecipitated with antibodies to cdk2 (lane 3 to 7) or SV40 large T antigen (control [con.]; lanes 1 and 2) and used in kinase assays with 0.1 or 1 μg of GST-HIRA[421–729] or GST-HIRA[421–729]ΔRXL as substrates, as indicated. The phosphoproteins were fractionated by SDS-PAGE and visualized by autoradiography. Arrowhead, phosphorylated GST-HIRA[421–729]. (c) Phosphorylation of HIRA by purified recombinant cyclin A- and E-cdk2 in vitro. Cyclin A- and E-cdk2 were expressed in and purified from Sf9 cells, and increasing amounts were used to phosphorylate 1 μg of GST-RB[792–928] (lanes 1 to 3 and 7 to 9) and GST-HIRA[421–729] (lanes 4 to 6 and 10 to 12), as indicated. The reactions were stopped by addition of 3× Laemmli sample buffer, and the phosphoproteins were separated by SDS-PAGE. Arrowheads, phosphorylated HIRA and RB; asterisk, autophosphorylated cyclin A. (d) Phosphorylation of HIRA by cyclin-cdk2 is blocked by a peptide containing the RXL motif of E2F1. Extracts of U2OS cells were immunoprecipitated with antibodies to cdk2 (lanes 2 to 6 and 8 to 12) or SV40 large T antigen (control; lanes 1 and 7) and used in kinase assays with 1 μg of GST-HIRA[421–729] (lanes 7 to 12) or GST-RB[792–928] (lanes 1 to 6) as the substrate. Kinase assays were performed in the presence of 0.1, 1, or 10 μg of a 10-residue synthetic peptide that spans the cyclin-cdk2-binding sequence of E2F1 (WT E2F1; PPVKRRLDLE) or in the absence of the peptide, as indicated. As controls, assays were performed in the presence of 10 μg of a peptide of identical amino acid composition but scrambled sequence (Mut E2F1; lanes 6 and 12). The phosphoproteins were fractionated by SDS-PAGE and visualized by autoradiography. Arrowhead, GST-pRB[792–928]; asterisk, GST-HIRA[421–729]. (e) The RXL motif of HIRA potentiates the phosphorylation of another poorly phosphorylated substrate when fused to the C terminus of that substrate. Extracts of U2OS cells were immunoprecipitated with antibodies to cdk2 (lanes 2 to 13) or SV40 large T antigen (control; lane 1) and used in kinase assays with 0.1 or 1 μg of the indicated protein fused to GST as the substrate. The phosphoproteins were fractionated by SDS-PAGE and visualized by autoradiography. Arrowheads, relevant GST-pRB fusion proteins.
Efficient phosphorylation of p107 and E2F1 in vitro by cyclin-cdk2 kinase requires that each substrate have an intact RXL motif (1). We next asked whether HIRA was phosphorylated in vitro by cyclin-cdk2 kinases and whether this too required an intact RXL cyclin-cdk2-binding motif. Cyclin-cdk2 kinase was immunopurified from asynchronously growing U2OS cells and tested for its ability to phosphorylate equal amounts of purified GST-HIRA[421–729] and GST-HIRA[421–729]ΔRXL. Cyclin-cdk2 kinase efficiently phosphorylated the former substrate but not the latter (Fig. 2b). Thus, cyclin-cdk2 kinase phosphorylates HIRA in vitro and, as for p107 and E2F1, this requires an intact RXL motif. However, cdk2 complexes immunopurified from asynchronously growing cells consist of cyclin A-cdk2 and cyclin E-cdk2. Therefore we directly compared the abilities of purified recombinant cyclin A-cdk2 and cyclin E-cdk2 expressed in Sf9 cells to phosphorylate GST-HIRA[421–729] and, as a control, GST-RB[792–928]. Although under these particular conditions the ability of each kinase to phosphorylate GST-HIRA[421–729] was less then its ability to phosphorylate GST-RB[792–928], both cyclin A- and E-cdk2 efficiently phosphorylated GST-HIRA[421–729] in vitro (Fig. 2c).
We have previously shown that short synthetic peptides containing functional RXL motifs will compete with the full-length proteins for binding to the cyclin-cdk2 complex (1, 6). If the RXL motif of HIRA serves to target the protein to the cyclin-cdk2 kinase in a manner similar to that for the RXL motifs of E2F1 and p107, then a peptide containing the RXL motif of E2F1 should block phosphorylation of HIRA in vitro. To test this, kinase assays were performed with cyclin-cdk2 complexes and GST-HIRA[421–729] or GST-RB[792-928] as a substrate in the absence or presence of a short synthetic peptide encompassing the RXL cyclin-cdk2-binding motif of E2F1. The peptide efficiently blocked phosphorylation of both substrates (Fig. 2d). In contrast, a peptide of identical amino acid composition but of scrambled sequence blocked phosphorylation of neither substrate. Thus the RXL motifs of HIRA and E2F1 compete with one another, suggesting that they are functionally equivalent.
Previously, Harlow and coworkers showed that the cyclin-cdk2-binding domain of E2F1 potentiates phosphorylation of E2F4 (14). Likewise, we showed that the RXL motifs of E2F1 and p21cip1 potentiated the phosphorylation of a poorly phosphorylated variant of the retinoblastoma tumor suppressor protein (pRB) that is mutated to remove all of its RXL motifs (pRB[792–829]) (2). Therefore, as a final test of the functionality of the HIRA RXL motif we tested whether it would also promote phosphorylation of pRB[792–829]. As shown previously, when pRB was expressed and purified as a GST fusion protein, its C terminus (GST-RB[792–928]) was efficiently phosphorylated by cyclin-cdk2 in vitro whereas GST-RB[792–829] was not (2). Phosphorylation was restored when 10 amino acids encompassing the RXL motif of either E2F1 or HIRA were fused to the C terminus of GST-RB[792–829] (GST-RB[798–829]E2F1Cy and GST-RB[792–829]HIRACy, respectively) (Fig. 2e). In contrast, fusion of either peptide with a mutation in the RXL motif did not restore phosphorylation (GST-RB[792–829]E2F1CyMut and GST-RB[792–829]HIRACyMut). Thus, the RXL of HIRA can promote phosphorylation of a heterologous protein. Taken together these studies clearly show that, at least in vitro, the HIRA RXL is functionally similar to the RXL of a known cyclin-cdk2 substrate, E2F1.
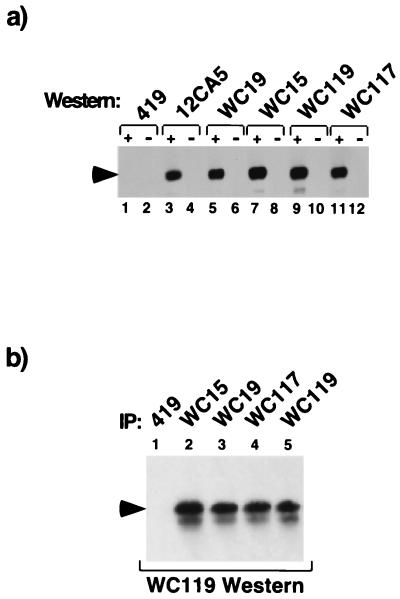
To facilitate subsequent studies of the endogenous protein in vivo, antibodies were raised to HIRA as described in Materials and Methods. Four monoclonal (WC15, 19, 117, and 119) and two polyclonal antibodies (D32 and D34) were obtained. As determined by Western blotting, each of these efficiently detected HA-tagged HIRA[421–729] that was ectopically expressed in U2OS cells (Fig. 3a and data not shown). Furthermore, in extracts of untransfected cells, each antibody detected a polypeptide with a molecular weight corresponding to that predicted for HIRA. Immunoprecipitation followed by Western blot analysis showed that each antibody reacted with the same polypeptide (Fig. 3b and data not shown), confirming that each antibody recognizes HIRA.
FIG. 3.
Characterization of antibodies to HIRA. (a) Monoclonal antibodies to HIRA detect HIRA by Western blotting. U2OS cells were transfected with a plasmid encoding HA-HIRA[421–729] (odd-numbered lanes) or with empty vector (even-numbered lanes). Cell extracts were prepared, and 15 μg of soluble protein per lane was fractionated by SDS-PAGE. The proteins were Western blotted with anti-SV40 T antigen (419), anti-HA (12CA5), or anti-HIRA (WC15, WC19, WC117, or WC119), as indicated. Arrowhead, HA-HIRA[421-729]. (b) Monoclonal antibodies to HIRA detect endogenous HIRA by immunoprecipitation and Western blotting. Extracts were prepared from U2OS cells and immunoprecipitated with anti-SV40 large T antigen (419) or anti-HIRA (WC15, WC19, WC117, or WC119), as indicated. The proteins were fractionated by SDS-PAGE and Western blotted with anti-HIRA monoclonal antibody WC119. Arrowhead, position of HIRA.
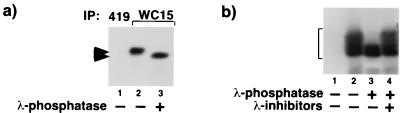
Using these antibodies we tested whether endogenous HIRA was phosphorylated in vivo. Extracts derived from U2OS cells were immunoprecipitated with anti-HIRA monoclonal antibody WC15 and the washed immunoprecipitates were treated with or without λ-phosphatase and fractionated by SDS-PAGE. Western blotting with WC119 showed that treatment with λ-phosphatase resulted in increased mobility of HIRA, indicating that the protein was phosphorylated (Fig. 4a). When HA-HIRA[421–729] was ectopically expressed in U2OS cells, it too was phosphorylated, as determined by a λ-phosphatase-dependent increase in mobility in SDS-PAGE (Fig. 4b).
FIG. 4.
HIRA is phosphorylated in vivo. (a) Endogenous cellular HIRA is a phosphoprotein. Extracts were prepared from asynchronously growing U2OS cells and immunoprecipitated with anti-SV40 large T antigen (419; lane 1) or anti-HIRA (WC15; lanes 2 and 3). The immunoprecipitates were treated with (lane 3) or without (lanes 1 and 2) λ-phosphatase, fractionated by SDS-PAGE, and Western blotted with anti-HIRA (WC119). Upper arrowhead, position of phosphorylated HIRA; lower arrowhead, position of unphosphorylated HIRA. (b) Ectopically expressed HA-HIRA[421–729] is a phosphoprotein. U2OS cells were transiently transfected with a plasmid encoding HA-HIRA[421–729] (lanes 2 to 4) or empty vector (lane 1), and extracts were prepared and immunoprecipitated with anti-HA (12CA5). The washed immunoprecipitates were treated with (lanes 3 and 4) or without (lanes 1 and 2) λ-phosphatase in the absence (lanes 1 to 3) or presence (lane 4) of phosphatase inhibitors, fractionated by SDS-PAGE, and Western blotted with anti-HA (12CA5). Bracket, position of HA-HIRA[421–729].
If HIRA is an in vivo substrate of cyclin-cdk2 kinases, then it should be phosphorylated in vivo on residues that are consensus cyclin-cdk2 phosphorylation sites and that are phosphorylated by cyclin-cdk2 kinase in vitro. To test this, we raised a rabbit polyclonal serum (D44) against a phosphopeptide containing a phosphothreonine that corresponds to threonine 555 (one of the two residues that conforms to the cyclin-cdk2 consensus, S/TPXK/R). An enzyme-linked immunosorbent assay showed that the rabbit immune, but not preimmune, serum specifically reacted with the phosphorylated but not the unphosphorylated peptide (data not shown). Subsequently, immunoprecipitation analysis showed that the anti-phosphothreonine 555 serum reacted with GST-HIRA[421–729] only after the protein had been incubated with cyclin A-cdk2 and magnesium-ATP (Fig. 5a). Thus, the anti-phosphothreonine 555 serum reacted only with GST-HIRA[421–729] phosphorylated by cyclin A-cdk2, presumably on threonine 555. To determine whether threonine 555 was phosphorylated on HIRA in vivo and whether this phosphorylation was dependent on the RXL motif, U2OS cells were transfected with a plasmid encoding HA-HIRA[421–729] or a mutant either lacking the RXL motif (HA-HIRA[421–729]ΔRXL) or containing a substitution of alanine for threonine 555 (HA-HIRA[421–729]T555A). As shown in Fig. 5b, phosphorylation of HA-HIRA[421–729] on threonine 555 was readily detectable. In contrast, phosphorylation was undetectable on HA-HIRA[421–729]ΔRXL or HIRA[421–729]T555A. Thus ectopically expressed HIRA was phosphorylated on threonine 555 and, like phosphorylation by cyclin A-cdk2 in vitro, this required an intact RXL cyclin-cdk2-binding motif.
FIG. 5.
HIRA is phosphorylated on two consensus cyclin-cdk2 phosphorylation sites in vivo. (a) Specificity of phosphothreonine 555-specific antibody D44. GST-HIRA[421–729] (100 ng) was incubated without (lanes 1 and 4) or with (lanes 2, 3, and 5 to 8) recombinant cyclin A-cdk2 in the presence of MgCl2 and ATP. The relative amount of kinase is indicated (titration over a 10-fold range). The reaction mixtures were diluted and immunoprecipitated (IP) with anti-HIRA (WC15; lanes 1 to 3), anti-phosphothreonine 555 (D44; lanes 4 to 6), or control antibodies (419; lane 7; D44 preimmune, lane 8). The immunoprecipitates were fractionated by SDS-PAGE and then Western blotted with anti-HIRA (WC119). Arrowhead, GST-HIRA[421–729]. (b) HA-HIRA[421–729] is phosphorylated in vivo on threonine 555. U2OS cells were transfected with plasmids encoding HA-HIRA[421–729] (lanes 2, 5, and 6), HA-HIRA[421–729]ΔRXL (lanes 3 and 7), or HA-HIRA[421–729]T555A (lanes 4 and 8) or with the empty vector (lane 1). Extracts were prepared and immunoprecipitated with anti-HA (12CA5; lanes 1 to 4), anti-phosphothreonine 555 (D44; lanes 6 to 8), or the control antibody (D44 preimmune; lane 5). Immunoprecipitates were fractionated by SDS-PAGE and then Western blotted with anti-HA (12CA5). Arrowhead, HA-HIRA[421–729] and mutants thereof. (c) Endogenous HIRA is phosphorylated on threonine 555 in vivo. Extracts were prepared from asynchronously growing U2OS cells and immunoprecipitated with anti-HIRA (WC15; lane 2), anti-phosphothreonine 555 (D44; lane 4), or control antibodies (419, lane 1; D44 preimmune, lane 3). Arrowhead, HIRA. (d) HA-HIRA[421–729] is phosphorylated on serine 687 in vivo. U2OS cells were transfected with a plasmid encoding HA-HIRA[421–729], extracts were immunoprecipitated with anti-HA (12CA5), and the protein was further purified by SDS-PAGE. The protein band was excised, digested with trypsin, and processed for LC-MS/MS mass spectrometry. The major ions derived from collision-induced dissociation of tryptic phosphopeptide LPIPpS687PQR (where pS687 represents phosphoserine) are indicated as y- and b-series ions (for definitions of these ions, see reference 22). The difference in the mass/charge (m/z) ratios of ions b1-5 and b1-4 and y1-3 and y1-4 corresponds to phosphoserine rather than serine, showing that the peptide is phosphorylated at this site.
Next we asked whether the endogenous HIRA protein was phosphorylated on threonine 555. Extracts from U2OS cells were immunoprecipitated with an antibody to total HIRA, the anti-phosphothreonine 555-specific serum, or control antibodies. As shown in Fig. 5c, the phosphoamino acid-specific antibody immunoprecipitated the endogenous protein. However, the phosphoamino acid-specific antibody precipitated a relatively small proportion of total HIRA protein, and this is consistent with the notion that HIRA is phosphorylated on threonine 555 only under certain conditions, e.g., in specific phases of the cell cycle. Taken together, these results show that HIRA is phosphorylated by cyclin A-cdk2 in vitro on a site that is phosphorylated in vivo, threonine 555.
As an alternative approach to identify the residues of HIRA that are phosphorylated in vivo, mass spectrometry was utilized. In particular, we were interested in determining whether other consensus cyclin-cdk2 phosphorylation sites, e.g., serine 687, were phosphorylated. Asynchronously growing U2OS cells were transfected with a plasmid encoding HA-HIRA[421–729], and the ectopically expressed protein was immunopurified with an anti-HA antibody. The protein was purified by SDS-PAGE, extracted from the gel, proteolytically digested with trypsin, and subjected to matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) and liquid chromatography (LC)-MS/MS mass spectrometry. Both approaches showed that the protein was phosphorylated on the residue that corresponds to serine 687 of the full-length protein (Fig. 5d and data not shown). In this experiment we failed to obtain unambiguous data on the phosphorylation status of threonine 555. This could be due to a number of technical aspects of the mass spectrometry approach and cannot be taken as evidence that this site is not phosphorylated. Taken together, the mass spectrometry- and immunology-based approaches indicate that HIRA is phosphorylated in vivo on both of the consensus cyclin-cdk2 phosphorylation sites, threonine 555 and serine 687.
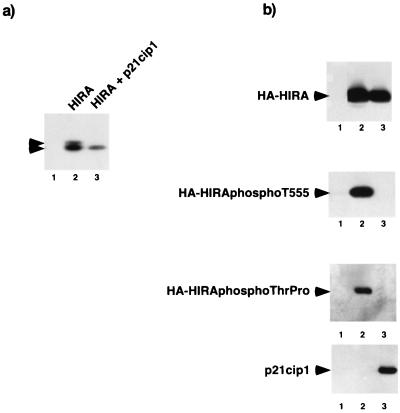
If HIRA is a substrate of a cyclin-cdk2 kinase in vivo, then its phosphorylation should be dependent on cyclin-cdk2 kinase activity. To test whether this is the case, U2OS cells were transiently transfected with plasmids encoding HA-HIRA[421–729] alone or together with a plasmid encoding cyclin-cdk2 inhibitor p21cip1. When expressed in the absence of p21cip1, HA-HIRA[421–729] migrated as a doublet (Fig. 6a). The more slowly migrating band results from phosphorylation (Fig. 4b and data not shown). However, when expressed in the presence of p21cip1, HA-HIRA[421–729] migrated as a single band of higher mobility, indicating that the cyclin-cdk2 inhibitor blocks the phosphorylation of HIRA (Fig. 6a).
FIG. 6.
Phosphorylation of HIRA requires cyclin-cdk2 kinase activity. (a) Phosphorylation of HIRA is inhibited by cyclin-cdk2 inhibitor p21cip1. U2OS cells were transiently transfected with a plasmid encoding HA-HIRA[421–729] (lanes 2 and 3) or empty vector (lane 1) in the absence (lanes 1 and 2) or presence (lane 3) of a plasmid encoding p21cip1. Extracts were immunoprecipitated with anti-HA (12CA5), fractionated by SDS-PAGE, and Western blotted with anti-HA (12CA5). Upper arrowhead, phosphorylated HA-HIRA[421–729]; lower arrowhead, unphosphorylated HA-HIRA[421–729]. (b) Phosphorylation of HIRA on threonine 555 and threonine-proline motifs is inhibited by cyclin-cdk2 inhibitor p21cip1. U2OS cells were mock-transfected (lane 1) or transiently transfected with a plasmid encoding HA-HIRA[520–1017] (lanes 2 and 3) in the absence (lanes 1 and 2) or presence (lane 3) of a plasmid encoding p21cip1. Extracts were prepared and immunoprecipitated with anti-HA (12CA5; top), anti-phosphothreonine 555 (D44; second from top) or anti-phosphothreonine-proline (P-thr-pro-101; third from top). The immunoprecipitates were fractionated by SDS-PAGE and then Western blotted with anti-HA (12CA5). Bottom, Western blot of the crude cell extracts with the anti-p21cip (SC-187). Arrowheads, HA-HIRA[520–1017] and p21cip1.
Next we tested whether inhibition of cyclin-cdk2 kinase activity inhibited phosphorylation of the mapped cyclin-cdk2 phosphorylation site, threonine 555, and some of the other potential cdk2 phosphoacceptor sites (specifically threonine residues followed by proline). U2OS cells were transfected with a plasmid encoding residues 520 to 1017 of HIRA (HA-HIRA[520–1017]) in the absence or presence of a plasmid encoding p21cip1. Residues 520 to 1017 were utilized in this experiment because, unlike full-length HIRA and residues 421 to 729, this region of the protein does not perturb the cell cycle (see Fig. 9; data not shown). Phosphorylation of threonine 555 was dramatically inhibited by coexpression of p21cip1, as revealed by immunoprecipitation with the phosphothreonine 555-specific antiserum followed by Western blotting with anti-HA (Fig. 6b). Likewise, p21cip1 inhibited phosphorylation of HA-HIRA[520–1017], as detected by an anti-phosphothreonine-proline-specific antibody (Fig. 6b). This antibody reacts with most phosphothreonine residues that are followed by proline. Including T555, there are four such threonine residues in HA-HIRA[520–1017]. Taken together, these experiments show that phosphorylation of HIRA in vivo requires cyclin-cdk2 kinase activity, as would be expected if HIRA is a substrate of a cyclin-cdk2 kinase.
FIG. 9.
Ectopic expression of HIRA causes arrest in S phase of the cell cycle. (a) U2OS cells were transfected without (−HIRA) or with (+HIRA) a plasmid encoding WT HA-HIRA[1–1017] together with a plasmid encoding cell surface marker pCD19. Two days after transfection the cells were processed for two-color FACS analysis to determine the DNA content (propidium iodide staining) of the transfected cells (CD19 positive). The dot plots show DNA content of transfected and untransfected cells. Transfected cells (CD19 positive) are boxed (R1). The histograms show DNA content of only the transfected cells. FITC, fluorescein isothiocyanate. (b) U2OS cells were transfected without (−HIRA) or with (+HIRA) a plasmid encoding WT HA-HIRA[1–1017] together with a plasmid encoding cell surface marker pCD19. Two days after transfection the cells were pulse-labeled for 1 h with 5′-BrdU and then processed for two-color FACS analysis to determine whether the transfected cells were actively synthesizing DNA (5′-BrdU-positive cells). The dot plots show 5′-BrdU labeling of transfected and untransfected cells. Transfected cells (CD19 positive) are boxed (R1). The histograms show 5′-BrdU labeling of only the transfected cells.
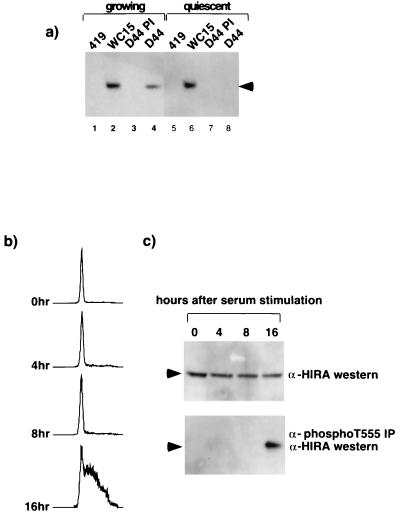
Another prediction, if HIRA is an in vivo substrate of a cyclin-cdk2 kinase, is that the protein should be phosphorylated when these kinases are active. Both cyclin A- and E-cdk2 are active in growing cells but not in quiescent cells (12, 18, 25, 44). Therefore, we asked whether HIRA was phosphorylated on threonine 555, a known in vitro phosphorylation site of cyclin-cdk2 (Fig. 5a), in growing but not quiescent cells. Extracts derived from asynchronously growing NIH 3T3 cells and cells that had been deprived of serum for 48 h to induce quiescence were immunoprecipitated with the anti-phosphothreonine 555-specific antiserum (D44) and then Western blotted with the anti-HIRA antibody (WC119). Although the total amounts of HIRA protein in growing and quiescent cells were the same, the protein was phosphorylated on threonine 555 in growing cells only (Fig. 7a). Next, we asked whether HIRA is phosphorylated on threonine 555 at a time in the cell cycle consistent with it being a cyclin A- or E-cdk2 substrate. Cyclin A- and E-cdk2 are activated in late G1 and S phases after quiescent cells are stimulated to reenter the cell cycle (18, 25, 39, 44). Extracts derived from quiescent NIH 3T3 cells or cells refed with serum for 4, 8, or 16 h were analyzed for the abundance of HIRA phosphorylated on threonine 555. Although, the total amounts of HIRA in quiescent and stimulated cells did not vary (Fig. 7c, top), phosphorylated HIRA was only detected 16 h after serum stimulation (Fig. 7c, bottom). FACS analysis indicated that phosphorylation temporally coincided with entry of the cells into S phase (Fig. 7b) and, presumably, activation of cyclin A- and E-cdk2.
FIG. 7.
HIRA is phosphorylated on threonine 555 in S phase of the cell cycle. (a) Extracts were prepared from asynchronous NIH 3T3 cells growing in 10% fetal bovine serum (FBS) (lanes 1 to 4) or cells grown in 0.1% FBS for 48 h (lanes 5 to 8) and immunoprecipitated with the antibody to HIRA (WC15; lanes 2 and 6), phosphothreonine 555 (D44 immune serum; lanes 4 and 8), or the controls (419 and D44 preimmune serum; lanes 1, 3, 5, and 7) as indicated. Immunoprecipitates were Western blotted with the anti-HIRA antibody (WC119). Arrowhead, HIRA. (b) FACS analysis to determine the DNA content of propidium iodide-stained NIH 3T3 cells grown in 0.1% FBS for 48 h and then refed with 10% FBS for 0, 4, 8, or 16 h, as indicated. (c) Extracts prepared from the NIH 3T3 cells used for panel b were immunoprecipitated with D44 immune serum and then Western blotted with WC119 (bottom). An aliquot of each cell lysate (150 μg of protein) was Western blotted directly with WC119 (top). Arrowheads, position of HIRA.
The active forms of cyclin A- and E-cdk2 are localized predominantly in the cell nucleus (39, 41). Therefore, if HIRA is an in vivo substrate of these kinases, then it too should be localized, at least in part, to the cell nucleus. To test whether this is the case, U2OS cells grown on coverslips were fixed and stained with monoclonal antibodies to HIRA or a control antibody. Three separate monoclonal antibodies to endogenous HIRA (WC15, 19, and 119) revealed a specific punctate nuclear staining pattern (Fig. 8a, b, e, and f and data not shown). To confirm this staining pattern, U2OS cells were transiently transfected with a plasmid encoding HA-HIRA[1–1017] and stained with anti-HA antibody 12CA5. Ectopically expressed HA-HIRA[1–1017] was present predominantly in the nucleus, although in some cells cytoplasmic staining was also observed (Fig. 8c, d, g, and h). This observation is consistent with previous studies by Lorain and coworkers, who detected endogenous HIRA as a primarily nuclear protein in various other cell lines (32). Taken together, these observations confirm that HIRA is a predominantly nuclear protein, and this is consistent with it being an in vivo substrate of cyclin A- or E-cdk2.
FIG. 8.
HIRA is a nuclear protein. Asynchronously growing U2OS cells grown on coverslips were stained with a control immunglobulin G1 (IgG1) monoclonal antibody (a) or two anti-HIRA monoclonal antibodies together, WC19 and WC119 (both IgG1) (e). DNA was visualized with DAPI (b and f). U2OS cells grown on coverslips were transfected with a plasmid encoding HA-HIRA[1–1017] and then stained with anti-HA antibody 12CA5 (d and h). DNA was visualized with DAPI (c and g).
These data clearly show that HIRA is an in vivo substrate of cyclin A- or E-cdk2. As such, it is a likely regulator of progression through the cell cycle. Accordingly, we next tested whether HIRA was able to modulate progression through the cell cycle. Asynchronously growing U2OS cells were transiently transfected with a plasmid encoding HA-tagged HIRA[1–1017], together with a plasmid encoding cell surface marker CD19, to allow identification of the transfected cells. FACS analysis showed that ectopic expression of HA-HIRA[1–1017] in cells caused the percentage of cells in S phase to increase from 45 to 75% (Fig. 9a). To confirm that this S-phase accumulation was due to an arrest in S phase, cells were again transiently transfected with plasmids encoding HA-HIRA[1–1017] and CD19 and 1 h prior to harvesting they were pulse-labeled with the thymidine analogue 5′-BrdU. FACS analysis was performed to determine whether the transfected cells had incorporated 5′-BrdU. As shown in Fig. 9b, approximately 41% of the cells transfected with the empty vector and the plasmid encoding CD19 were 5′-BrdU positive. This percentage of cells in S phase is comparable to the value of 45% that was obtained by propidium iodide staining (Fig. 9a). In contrast, only 3% of the cells ectopically expressing HA-HIRA[1–1017] were 5′-BrdU positive. Thus, although ectopic expression of HA-HIRA[1–1017] caused a dramatic accumulation of cells with DNA content corresponding to that found in S phase (Fig. 9a), these cells did not incorporate 5′-BrdU (Fig. 9b). Thus, they were not actively synthesizing DNA and were arrested in S phase.
DISCUSSION
HIRA is named after its homology to Hir1p and Hir2p of S. cerevisiae (28). Homologues have also been reported in mice, chickens, Drosophila melanogaster, Caenorhabditis elegans, and Schizosaccharomyces pombe (24, 31, 42, 48). Based on comparison of the entire S. cerevisiae genomic sequence with the Homo sapiens genome that has been sequenced to date, HIRA is the best candidate so far to be an ortholog (functional equivalent) of Hir1p and Hir2p. In yeast, Hir1p and Hir2p control histone gene expression through the cell cycle. In WT yeast, histones are transcribed periodically throughout the cell cycle, with expression peaking in S phase. However, hir1 and hir2 null mutants show constitutive expression of the HTA1-HTB1 locus that encodes histones H2A and H2B (40, 52). Accounting for this phenotype, Spector and coworkers reported that Hir1p and Hir2p are cell cycle-regulated transcriptional repressors of histone genes (56). In addition, during S phase Hir1p and Hir2p are required to recruit components of the SWI-SNF chromatin-remodeling complex to yeast histone promoters, and this is required to overcome the G1-phase repression of the promoters (11). Interestingly, relief of Hir1p- and Hir2p-mediated transcriptional repression requires the yeast cdk, CDC28 (56). Thus, genetic evidence from yeast suggests that Hir1p and Hir2p might couple cyclical cdk activity to periodic histone gene expression.
A number of lines of evidence indicate that HIRA is an in vivo substrate of a cyclin-cdk2 kinase. First, HIRA bound to and was phosphorylated by cyclin-cdk2 kinases in vitro. Importantly, both binding and phosphorylation required an intact RXL cyclin-cdk2-binding motif. Furthermore, the RXL of HIRA is clearly functional in directing substrate phosphorylation since an E2F1-derived RXL-containing peptide blocked phosphorylation of HIRA in vitro and the RXL motif from HIRA strongly potentiated the phosphorylation of a poorly phosphorylated pRB mutant when it was fused to the C terminus of that substrate. It is important to note that in these in vitro assays HIRA behaves in a manner identical to that of other previously characterized cyclin-cdk2 substrates, such as E2F1, p107, and pRB (1, 2, 6, 14, 30, 33, 46, 47, 65). Second, we showed that both endogenous cellular HIRA and ectopically expressed HIRA are phosphorylated proteins in vivo. Moreover, we showed that HIRA is phosphorylated in vivo on two consensus cyclin-cdk2 phosphorylation sites, threonine 555 and serine 687. At least one of these sites, threonine 555, was phosphorylated by cyclin A-cdk2 in vitro, consistent with the notion that a cyclin-cdk2 kinase is responsible for phosphorylation in vivo. Like phosphorylation by cyclin-cdk2 in vitro, phosphorylation in vivo on threonine 555 required the RXL motif. Third, when cellular cyclin-cdk kinase activity was inhibited by cyclin-cdk2 inhibitor p21cip1, the phosphorylation of HIRA was blocked. This is consistent with the notion that HIRA is phosphorylated directly by a cyclin-cdk2 kinase. Fourth, we showed that HIRA's phosphorylation was altered in a cell cycle-dependent manner and that the protein is phosphorylated on threonine 555 in S phase, a time in the cell cycle when cyclin A- and E-cdk2 kinases are active. Fifth, as shown previously by others (32), we showed that HIRA is localized to the nucleus where active cyclin-cdk2 complexes are found. In summary, we have satisfied all of the necessary criteria to propose that HIRA is an in vivo substrate of a cyclin-cdk2 kinase. At this point, our data are consistent with HIRA being a substrate of either cyclin A-cdk2, cyclin E-cdk2, or both. A detailed analysis of the cell cycle kinetics of HIRA phosphorylation and a more detailed comparison of the sites phosphorylated in vivo with those phosphorylated by these kinases in vitro will contribute to a distinction being drawn between these two kinases.
Many cyclin-cdk2 substrates are regulators of cell cycle progression. Consistent with HIRA also being a cell cycle regulator, we show here that, at least when ectopically expressed in cells, HIRA is able to modulate cell cycle progression. Specifically, ectopic expression of HIRA in cells caused an arrest of those cells in S phase. The molecular mechanism underlying this arrest and the extent to which it reflects a physiological function of HIRA remain to be established. However, the joint observations that ectopic expression of HIRA arrests cells in S phase and that HIRA becomes phosphorylated at this time are consistent with the notion that HIRA normally regulates progression through S phase and that this activity is modulated by phosphorylation. If so, then mutation of HIRA phosphorylation sites to nonphosphorylatable residues might be expected to modulate the ability of HIRA to cause S-phase arrest. For example, HIRA might normally restrain progression through S phase and phosphorylation might overcome this and facilitate S-phase progression. If so, nonphosphorylatable HIRA mutants might more potently arrest cells in S phase. To date we have been unable to show an effect of phosphorylation on the ability of HIRA to regulate the cell cycle. Mutation of both T555 and S687 to nonphosphorylatable alanine residues did not affect HIRA activity. However, in addition to T555 and S687, HIRA contains another 13 S/TP motif that might also be phosphorylated by cyclin-cdk2 kinase. Indeed, mutation of T555 and S687 to nonphosphorylatable alanine did not affect the apparent phosphorylation of the protein as detected in one-dimensional SDS-PAGE (Fig. 4b and data not shown), suggesting that HIRA is phosphorylated on additional and/or alternative sites. Current efforts are directed toward identifying other phosphorylation sites on HIRA and determining which of these, either alone or in combination, regulate HIRA activity.
One possible mechanism underlying the S-phase arrest is suggested by the homology of HIRA to Hir1p and Hir2p of yeast. Like Hir1p and Hir2p, HIRA might function as a regulator of histone gene expression. In proliferating mammalian cells, expression of histones peaks in S phase of the cell cycle as a result of both increased gene transcription and posttranscriptional regulation (4, 9, 20, 53, 58). Cis-acting promoter-specific sequences (or subtype-specific consensus sequences [SSCEs]) in the gene promoters mediate cell cycle periodicity of transcription (3, 9, 15, 27, 36). For example, in the promoter of at least one copy of the human histone H2B gene an Oct-1 site is required for S-phase-specific activation, although Oct-1 DNA binding activity remains unchanged from G1 to S phase (27, 51). Recently, the cyclin E-cdk2 substrate NPAT was shown to play a role in mediating cyclin-cdk2-dependent activation of the histone H2B and H4 promoters (34, 64). NPAT activation of the histone H4 and H2B promoters is dependent on the SSCE in each case and requires phosphorylation of NPAT by cyclin E-cdk2. Therefore, NPAT is at least part of the link between cyclin E-cdk2 activity and histone gene transcription. However, although chromatin immunoprecipitation assays and immunofluorescence showed that NPAT is located at histone promoters in vivo, it has not been shown to bind directly to promoter DNA, and so whether it functions directly in transcription activation or more indirectly is not clear. Regardless, undoubtedly additional regulators of histone expression in mammalian cells await identification. HIRA might be one such regulator. If, like Hir1p and Hir2p, HIRA is a repressor of histone gene expression, then the HIRA-mediated S-phase arrest might be a consequence of depletion of free histones. This would presumably prevent the incorporation of newly replicated DNA into chromatin, which might be expected to block S-phase progression. However, when drawing parallels between the function of yeast Hir1p and Hir2p and that of human HIRA, it should be noted that there are important differences between the regulation of histones in yeast and that in humans. For example, although Hir1p and Hir2p serve as regulated repressors of histones through the cell cycle (40, 52, 56), most studies of human histone promoters have found little evidence for regulation of transcription repression. Instead, most human histone promoters appear to be regulated through activation of transcription (3, 9, 15, 27, 36). Therefore, HIRA might have other functions in S phase that are responsible for the arrest. Interestingly, HIRA also has homology to the 60-kDa subunit of CAF1 (chromatin assembly factor 1), a protein that assembles newly replicated DNA and histones into chromatin (23, 54). Thus, HIRA might couple cyclin-cdk2 kinase activity and formation of chromatin in S phase.
The gene encoding HIRA (HIRA) was originally cloned as a candidate for a gene whose haploinsufficiency could contribute to the human developmental disorder DGS (28). DGS is characterized by conotruncal and cardiac abnormalities and hypoplasia of the thymus and parathyroid glands (49). These defects are thought to result from abnormality of the third and fourth pharyngeal pouches, and this in turn is thought to be a consequence of defective early migration of neural crest cells (49). Proper spatial development of a multicellular organism requires fine integration of cellular proliferation and cell migration. As a cell cycle control protein, HIRA could play a role in such processes. A number of lines of evidence are consistent with haploinsufficiency of HIRA being responsible for at least some of the symptoms of DGS. First, the HIRA gene falls within the region of chromosome 22q11.2 that is commonly deleted in DGS (the DGS critical region). Second, the protein product of the HIRA gene is expressed at high levels in the tissues of developing mouse and chicken embryos, which are thought to be the sites of the primary developmental defect; that is, the neural crest and neural-crest-derived tissues (42, 60). Third, when explanted premigratory chicken neural crest tissue was treated with antisense oligonucleotides designed to ablate HIRA mRNA and then reimplanted into the chicken embryo, persistent truncus arteriosus, one of the conotruncal abnormalities characteristic of DGS patients, was observed (17). Finally, HIRA has been reported to interact with transcription factor Pax3, and mice lacking Pax3 die in utero with a phenotype reminiscent of that observed in DGS patients (35). However, to date, neither HIRA nor any other gene has been definitively characterized as a DGS gene, and it appears that haploinsufficiency of more than one gene at 22q11.2 could be implicated (29).
In summary, we have shown that HIRA is a novel in vivo substrate of a cyclin-cdk2 kinase and a likely regulator of the cell cycle. As a cyclin-cdk2 substrate, HIRA keeps company with a relatively small number of other known proteins. Future studies of HIRA will address the function of HIRA in cell cycle control and might shed light on the molecular basis of the developmental abnormalities characteristic of DGS.
ACKNOWLEDGMENTS
The early part of this work was supported by an NIH grant (1R01 CA76120) to the laboratory of William G. Kaelin, Jr., at the Dana-Farber Cancer Institute and Harvard Medical School, Boston. This work was supported by grants to P.D.A. from the W. W. Smith charitable trust and the V-foundation. Work in the laboratory of M.L. was supported by a grant from the Human Frontiers Science and the Association pour la Recherche sur le Cancer.
We thank Jianmin Gan for excellent technical assistance in generation of antibodies to HIRA. We thank Jon Chernoff and Randy Strich for critical reading of the manuscript and Wade Harper and Ken Zaret for discussion during the course of this work. P.D.A. thanks W.G.K. for his teaching and generous support.
REFERENCES.
- 1.Adams P D, Sellers W R, Sharma S K, Wu A D, Nalin C M, Kaelin W G., Jr Identification of a cyclin-cdk2 recognition motif present in substrates and p21-like cyclin-dependent kinase inhibitors. Mol Cell Biol. 1996;16:6623–6633. doi: 10.1128/mcb.16.12.6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams P D, Li X, Sellers W R, Baker K, Leng X, Harper J W, Taya Y, Kaelin W G., Jr The retinoblastoma protein contains a C-terminal motif that targets it for phosphorylation by cyclin/cdk complexes. Mol Cell Biol. 1999;19:1068–1080. doi: 10.1128/mcb.19.2.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aziz F, van Wijnen A J, Stein J L, Stein G S. HiNF-D (CDP-cut/CDC2/cyclin A/pRB-complex) influences the timing of IRF-2-dependent cell cycle activation of human histone H4 gene transcription at the G1/S phase transition. Cell Physiol. 1998;177:453–464. doi: 10.1002/(SICI)1097-4652(199812)177:3<453::AID-JCP8>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 4.Baumbach L L, Stein G S, Stein J L. Regulation of human histone gene expression: transcriptional and posttranscriptional control in the coupling of histone messenger RNA stability with DNA replication. Biochemistry. 1987;26:6178–6187. doi: 10.1021/bi00393a034. [DOI] [PubMed] [Google Scholar]
- 5.Brown N R, Noble M E, Endicott J A, Johnson L N. The structural basis for specificity of substrate and recruitment peptides for cyclin-dependent kinases. Nat Cell Biol. 1999;1:438–443. doi: 10.1038/15674. [DOI] [PubMed] [Google Scholar]
- 6.Chen J, Saha P, Kornbluth S, Dynlacht B D, Dutta A. Cyclin-binding motifs are essential for the function of p21cip1. Mol Cell Biol. 1996;16:4673–4682. doi: 10.1128/mcb.16.9.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coleman T R, Tang Z, Dunphy W G. Negative regulation of the wee1 protein kinase by direct action of the nim1/cdr1 mitotic inducer. Cell. 1993;72:919–929. doi: 10.1016/0092-8674(93)90580-j. [DOI] [PubMed] [Google Scholar]
- 8.Dallapiccola B, Pizzuti A, Novelli G. How many breaks do we need to CATCH on 22q11? Am J Hum Genet. 1996;59:7–11. [PMC free article] [PubMed] [Google Scholar]
- 9.Dalton S, Wells J R. A gene-specific promoter element is required for optimal expression of the histone H1 gene in S-phase. EMBO J. 1988;7:49–56. doi: 10.1002/j.1460-2075.1988.tb02782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeLisle A J, Graves R A, Marzluff W F, Johnson L F. Regulation of histone mRNA production and stability in serum-stimulated mouse 3T6 fibroblasts. Mol Cell Biol. 1983;3:1920–1929. doi: 10.1128/mcb.3.11.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dimova D, Nackerdien Z, Furgeson S, Eguchi S, Osley M A. A role for transcriptional repressors in targeting the yeast Swi/Snf complex. Mol Cell. 1999;4:75–83. doi: 10.1016/s1097-2765(00)80189-6. [DOI] [PubMed] [Google Scholar]
- 12.Dulic V, Kaufmann W K, Wilson S J, Tlsty T D, Lees E, Harper J W, Elledge S J, Reed S I. p53-dependent inhibition of cyclin-dependent kinase activities in human fibroblasts during radiation-induced G1 arrest. Cell. 1994;76:1013–1023. doi: 10.1016/0092-8674(94)90379-4. [DOI] [PubMed] [Google Scholar]
- 13.Dynlacht B D, Flores O, Lees J A, Harlow E. Differential regulation of E2F transactivation by cyclin/cdk2 complexes. Genes Dev. 1994;8:1772–1786. doi: 10.1101/gad.8.15.1772. [DOI] [PubMed] [Google Scholar]
- 14.Dynlacht B D, Moberg K, Lees J A, Harlow E, Zhu L. Specific regulation of E2F family members by cyclin-dependent kinases. Mol Cell Biol. 1997;17:3867–3875. doi: 10.1128/mcb.17.7.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eilers A, Bouterfa H, Triebe S, Doenecke D. Role of a distal promoter element in the S-phase control of the human H1.2 histone gene transcription. Eur J Biochem. 1994;223:567–574. doi: 10.1111/j.1432-1033.1994.tb19026.x. [DOI] [PubMed] [Google Scholar]
- 16.Ewen M E, Sluss H K, Sherr C J, Matsushime H, Kato J, Livingston D M. Functional interactions of the retinoblastoma protein with mammalian D-type cyclins. Cell. 1993;73:487–497. doi: 10.1016/0092-8674(93)90136-e. [DOI] [PubMed] [Google Scholar]
- 17.Farrell M J, Stadt H, Wallis K T, Scambler P, Hixon R L, Wolfe R, Leatherbury L, Kirby M L. HIRA, a DiGeorge syndrome candidate gene, is required for cardiac outflow tract septation. Circ Res. 1999;84:127–135. doi: 10.1161/01.res.84.2.127. [DOI] [PubMed] [Google Scholar]
- 18.Girard F, Strausfeld U, Fernandez A, Lamb N J. Cyclin A is required for the onset of DNA replication in mammalian fibroblasts. Cell. 1991;67:1169–1179. doi: 10.1016/0092-8674(91)90293-8. [DOI] [PubMed] [Google Scholar]
- 19.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1988. [Google Scholar]
- 20.Heintz N. The regulation of histone gene expression during the cell cycle. Biochim Biophys Acta. 1991;1088:327–339. doi: 10.1016/0167-4781(91)90122-3. [DOI] [PubMed] [Google Scholar]
- 21.Hunter T, Pines J. Cyclins and cancer. II. Cyclin D and CDK inhibitors come of age. Cell. 1994;79:573–582. doi: 10.1016/0092-8674(94)90543-6. [DOI] [PubMed] [Google Scholar]
- 22.Jimenez C R, Huang L, Qiu Y, Burlingame L A. Mass spectrometry. In: Coligan J E, Dunn B M, Ploegh H L, Speicher D W, Wingfield P T, editors. Current protocols in protein science. New York, N.Y: John Wiley & Sons, Inc.; 1999. pp. 16.3.1–16.7.3. [Google Scholar]
- 23.Kaufman P D, Cohen J L, Osley M A. Hir proteins are required for position-dependent gene silencing in Saccharomyces cerevisiae in the absence of chromatin assembly factor I. Mol Cell Biol. 1998;18:4793–4806. doi: 10.1128/mcb.18.8.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirov N, Shtilbans A, Rushlow C. Isolation and characterization of a new gene encoding a member of the HIRA family of proteins from Drosophila melanogaster. Gene. 1998;212:323–332. doi: 10.1016/s0378-1119(98)00143-7. [DOI] [PubMed] [Google Scholar]
- 25.Koff A, Ohtsuki M, Polyak K, Roberts J M, Massague J. Negative regulation of G1 in mammalian cells: inhibition of cyclin E-dependent kinase by TGF-beta. Science. 1993;260:536–539. doi: 10.1126/science.8475385. [DOI] [PubMed] [Google Scholar]
- 26.Krek W, Ewen M E, Shirodkar S, Arany Z, Kaelin W G, Jr, Livingston D M. Negative regulation of the growth-promoting transcription factor E2F-1 by a stably bound cyclin A-dependent protein kinase. Cell. 1994;78:161–172. doi: 10.1016/0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- 27.LaBella F, Sive H L, Roeder R G, Heintz N. Cell-cycle regulation of a human histone H2b gene is mediated by the H2b subtype-specific consensus element. Genes Dev. 1988;2:32–39. doi: 10.1101/gad.2.1.32. [DOI] [PubMed] [Google Scholar]
- 28.Lamour V, Lecluse Y, Desmaze C, Spector M, Bodescot M, Aurias A, Osley M A, Lipinski M. A human homolog of the S. cerevisiae HIR1 and HIR2 transcriptional repressors cloned from the DiGeorge syndrome critical region. Hum Mol Genet. 1995;4:791–799. doi: 10.1093/hmg/4.5.791. [DOI] [PubMed] [Google Scholar]
- 29.Lindsay E A, Botta A, Jurecic V, Carattini-Rivera S, Cheah Y C, Rosenblatt H M, Bradley A, Baldini A. Congenital heart disease in mice deficient for the DiGeorge syndrome region. Nature. 1999;401:379–383. doi: 10.1038/43900. [DOI] [PubMed] [Google Scholar]
- 30.Lisztwan J, Marti A, Sutterluty H, Gstaiger M, Wirbelauer C, Krek W. Association of human CUL-1 and ubiquitin-conjugating enzyme CDC34 with the F-box protein p45(SKP2): evidence for evolutionary conservation in the subunit composition of the CDC34-SCF pathway. EMBO J. 1998;17:368–383. doi: 10.1093/emboj/17.2.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Llevadot R, Marques G, Pritchard M, Estivill X, Ferrus A, Scambler P. Cloning, chromosome mapping and expression analysis of the HIRA gene from Drosophila melanogaster. Biochem Biophys Res Commun. 1998;249:486–491. doi: 10.1006/bbrc.1998.9165. [DOI] [PubMed] [Google Scholar]
- 32.Lorain S, Quivy J P, Monier-Gavelle F, Scamps C, Lecluse Y, Almouzni G, Lipinski M. Core histones and HIRIP3, a novel histone-binding protein, directly interact with WD repeat protein HIRA. Mol Cell Biol. 1998;18:5546–5556. doi: 10.1128/mcb.18.9.5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma T, Zou N, Lin B Y, Chow L T, Harper J W. Interaction between cyclin-dependent kinases and human papillomavirus replication-initiation protein E1 is required for efficient viral replication. Proc Natl Acad Sci USA. 1999;96:382–387. doi: 10.1073/pnas.96.2.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma T, van Tine B A, Wei Y, Garrett M D, Nelson D, Adams P D, Wang J, Qin J, Chow L T, Harper J W. Cell cycle-regulated phosphorylation of p220(NPAT) by cyclin E/Cdk2 in Cajal bodies promotes histone gene transcription. Genes Dev. 2000;14:2298–2313. doi: 10.1101/gad.829500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Magnaghi P, Roberts C, Lorain S, Lipinski M, Scambler P J. HIRA, a mammalian homologue of Saccharomyces cerevisiae transcriptional co-repressors, interacts with Pax3. Nat Genet. 1998;20:74–77. doi: 10.1038/1739. [DOI] [PubMed] [Google Scholar]
- 36.Meergans T, Albig W, Doenecke D. Conserved sequence elements in human main type-H1 histone gene promoters: their role in H1 gene expression. Eur J Biochem. 1998;256:436–446. doi: 10.1046/j.1432-1327.1998.2560436.x. [DOI] [PubMed] [Google Scholar]
- 37.Morgan D O. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu Rev Cell Dev Biol. 1997;13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- 38.Nigg E. The substrates of the cdc2 kinase. Semin Cell Biol. 1991;2:261–270. [PubMed] [Google Scholar]
- 39.Ohtsubo M, Theodoras A M, Schumacher J, Roberts J M, Pagano M. Human cyclin E, a nuclear protein essential for the G1-to-S phase transition. Mol Cell Biol. 1995;15:2612–2624. doi: 10.1128/mcb.15.5.2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Osley M A, Lycan D. Trans-acting regulatory mutations that alter transcription of Saccharomyces cerevisiae histone genes. Mol Cell Biol. 1987;7:4204–4210. doi: 10.1128/mcb.7.12.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pines J, Hunter T. Human cyclins A and B1 are differentially located in the cell and undergo cell cycle-dependent nuclear transport. J Cell Biol. 1991;115:1–17. doi: 10.1083/jcb.115.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roberts C, Daw S C, Halford S, Scambler P J. Cloning and developmental expression analysis of chick Hira (Chira), a candidate gene for DiGeorge syndrome. Hum Mol Genet. 1997;6:237–245. doi: 10.1093/hmg/6.2.237. [DOI] [PubMed] [Google Scholar]
- 43.Roberts J M. Evolving ideas about cyclins. Cell. 1999;98:129–132. doi: 10.1016/s0092-8674(00)81007-7. [DOI] [PubMed] [Google Scholar]
- 44.Rosenblatt J, Gu Y, Morgan D O. Human cyclin-dependent kinase 2 is activated during the S and G2 phases of the cell cycle and associates with cyclin A. Proc Natl Acad Sci USA. 1992;89:2824–2828. doi: 10.1073/pnas.89.7.2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Russo A A, Jeffrey P D, Pattern A K, Massague J, Pavletich N P. Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A-Cdk2 complex. Nature. 1996;382:325–331. doi: 10.1038/382325a0. [DOI] [PubMed] [Google Scholar]
- 46.Saha P, Eichbaum Q, Silberman E D, Mayer B J, Dutta A. p21cip1 and Cdc25A: competition between an inhibitor and an activator of cyclin-dependent kinases. Mol Cell Biol. 1997;17:4338–4345. doi: 10.1128/mcb.17.8.4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saha P, Chen J, Thome K C, Lawlis S J, Hou Z H, Hendricks M, Parvin J D, Dutta A. Human CDC6/Cdc18 associates with Orc1 and cyclin-cdk and is selectively eliminated from the nucleus at the onset of S phase. Mol Cell Biol. 1998;18:2758–2767. doi: 10.1128/mcb.18.5.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scamps C, Lorain S, Lamour V, Lipinski M. The HIR protein family: isolation and characterization of a complete murine cDNA. Biochim Biophys Acta. 1996;1306:5–8. doi: 10.1016/0167-4781(96)00010-3. [DOI] [PubMed] [Google Scholar]
- 49.Schinke M, Izumo S. Getting to the heart of DiGeorge syndrome. Nat Med. 1999;10:1120–1121. doi: 10.1038/13438. [DOI] [PubMed] [Google Scholar]
- 50.Schulman B A, Lindstrom D L, Harlow E. Substrate recruitment to cyclin-dependent kinase 2 by a multipurpose docking site on cyclin A. Proc Natl Acad Sci USA. 1998;95:10453–10458. doi: 10.1073/pnas.95.18.10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Segil N, Roberts S B, Heintz N. Mitotic phosphorylation of the Oct-1 homeodomain and regulation of Oct-1 DNA binding activity. Science. 1991;254:1814–1816. doi: 10.1126/science.1684878. [DOI] [PubMed] [Google Scholar]
- 52.Sherwood P W, Tsang S V M, Osley M A. Characterization of HIR1 and HIR2, two genes required for regulation of histone gene transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:28–38. doi: 10.1128/mcb.13.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sive H L, Heintz N, Roeder R G. Regulation of human histone gene expression during the HeLa cell cycle requires protein synthesis. Mol Cell Biol. 1984;12:2723–2734. doi: 10.1128/mcb.4.12.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith S, Stillman Purification and characterization of CAF-1, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell. 1989;58:15–25. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- 55.Songyang Z, Blechner S, Hoagland N, Hoekstra M F, Piwnica-Worms H, Cantley L C. Use of an orientated peptide library to determine the optimum substrates of protein kinases. Curr Biol. 1994;4:973–982. doi: 10.1016/s0960-9822(00)00221-9. [DOI] [PubMed] [Google Scholar]
- 56.Spector M S, Raff A, DeSilva H, Lee K, Osley M A. Hir1p and Hir2p function as transcriptional corepressors to regulate histone gene transcription in the Saccharomyces cerevisiae cell cycle. Mol Cell Biol. 1997;17:545–552. doi: 10.1128/mcb.17.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Srinivasan J, Koszelak M, Mendelow M, Kwon Y-G, Lawrence D S. The design of peptide based substrates for the cdc2 protein kinase. Biochem J. 1995;309:927–931. doi: 10.1042/bj3090927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stein G S, Stein J L, Van Wijnen A J, Lian J B. Transcriptional control of cell cycle progression: the histone gene is a paradigm for the G1/S phase and proliferation/differentiation transitions. Cell Biol Int. 1996;20:41–49. doi: 10.1006/cbir.1996.0007. [DOI] [PubMed] [Google Scholar]
- 59.Tough D F, Sprent J. Measurement of T and B cell turnover with 5-BrdU, supplement 18. In: Coligan J E, Kruisbeek A M, Margulies D H, Shevach E M, Strober W, editors. Current protocols in immunology. New York, N.Y: John Wiley & Sons, Inc.; 1996. pp. 4.7.1–4.7.6. [Google Scholar]
- 60.Wilming L G, Snoeren C A, van Rijswijk A, Grosveld F, Meijers C. The murine homologue of HIRA, a DiGeorge syndrome candidate gene, is expressed in embryonic structures affected in human CATCH22 patients. Hum Mol Genet. 1997;6:247–258. doi: 10.1093/hmg/6.2.247. [DOI] [PubMed] [Google Scholar]
- 61.Xu M, Sheppard K A, Peng C Y, Yee A S, Piwnica-Worms H. Cyclin A/CDK2 binds directly to E2F-1 and inhibits the DNA-binding activity of E2F-1/DP-1 by phosphorylation. Mol Cell Biol. 1994;14:8420–8431. doi: 10.1128/mcb.14.12.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang J, Sanchez R J, Wang S, Guarnaccia C, Tossi A, Zahariev S, Pongor S. Substrate specificity of cdc2 kinase from human HeLa cells as determined with synthetic peptides and molecular modelling. Arch Biochem Biophys. 1994;315:415–424. doi: 10.1006/abbi.1994.1519. [DOI] [PubMed] [Google Scholar]
- 63.Zhao J, Kennedy B K, Lawrence B D, Barbie D A, Matera A G, Fletcher J A, Harlow E. NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev. 2000;14:2283–2297. [PMC free article] [PubMed] [Google Scholar]
- 64.Zhao J, Dynlacht, Imai T, Hori T, Harlow E. Expression of NPAT, a novel substrate of cyclin E-CDK2, promotes S-phase entry. Genes Dev. 1998;12:456–461. doi: 10.1101/gad.12.4.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu L, Harlow E, Dynlacht B D. p107 uses a p21cip1-related domain to bind cyclin/cdk2 and regulate interactions with E2F. Genes Dev. 1995;9:1740–1752. doi: 10.1101/gad.9.14.1740. [DOI] [PubMed] [Google Scholar]