Abstract
The goal of the Enhancing Neuroimaging Genetics through Meta‐Analysis (ENIGMA) Stroke Recovery working group is to understand brain and behavior relationships using well‐powered meta‐ and mega‐analytic approaches. ENIGMA Stroke Recovery has data from over 2,100 stroke patients collected across 39 research studies and 10 countries around the world, comprising the largest multisite retrospective stroke data collaboration to date. This article outlines the efforts taken by the ENIGMA Stroke Recovery working group to develop neuroinformatics protocols and methods to manage multisite stroke brain magnetic resonance imaging, behavioral and demographics data. Specifically, the processes for scalable data intake and preprocessing, multisite data harmonization, and large‐scale stroke lesion analysis are described, and challenges unique to this type of big data collaboration in stroke research are discussed. Finally, future directions and limitations, as well as recommendations for improved data harmonization through prospective data collection and data management, are provided.
Keywords: big data, lesions, MRI, neuroinformatics, stroke
1. INTRODUCTION
Stroke results in neural damage to the brain and subsequent physical, cognitive, and emotional deficits (Dimyan & Cohen, 2011; Meyer et al., 2015; Ward, 2017). Globally, there are over 13.7 million new strokes each year, and stroke is a leading cause of adult long‐term disability (Benjamin et al., 2018; Johnson et al., 2019; Lindsay et al., 2019). Since stroke is a vascular disease, there is vast heterogeneity in both brain and behavioral changes across patients, posing a significant challenge to the development of effective stroke neurorehabilitation strategies (Bernhardt et al., 2016). The recovery process also contributes to additional interindividual variability in the time course and extent of the resolution of neural damage and behavioral deficits.
Given the heterogeneity in both injury and recovery, large samples of diverse patients are needed to accurately study processes related to and supporting stroke recovery. However, a recent systematic review suggested that most studies seeking to determine the best predictors of stroke recovery are underpowered (Kim & Winstein, 2017). This is in part because acquiring large datasets of this nature is logistically and financially prohibitive for a single stroke research site. Recognizing these challenges, the Stroke Recovery and Rehabilitation Roundtable (SRRR) task force developed consensus statements on how to move stroke recovery research forward (Bernhardt et al., 2016; Bernhardt et al., 2019). Two key priority areas include: (a) a better understanding of the neurobiology of spontaneous and treatment‐dependent recovery in humans; and (b) characterizing different stroke recovery phenotypes (Bernhardt et al., 2017), with a long‐term goal of identifying potential biomarkers that predict stroke recovery (Boyd et al., 2017). Here, we describe the steps that the Enhancing Neuroimaging Genetics through Meta‐Analysis (ENIGMA) Stroke Recovery working group has taken to address these priorities using a unique, big data approach with an emphasis on reliable and reproducible methods.
The ENIGMA worldwide consortium consists of over 1,400 researchers from 45 countries who have come together to form 26 different disease working groups as well as 24 additional working groups to study healthy human variation and develop novel, robust methods for neuroimaging and genetic meta‐analyses (Thompson, 2019; Thompson et al., 2019; Thompson et al., 2020). Formed in 2009, ENIGMA has resulted in significant, large‐scale contributions on the neural correlates of major depression (Schmaal et al., 2016), schizophrenia (Van Erp et al., 2018), bipolar disorder (Hibar et al., 2016), obsessive–compulsive disorder (Boedhoe et al., 2016), and epilepsy (Whelan et al., 2018). ENIGMA methods, discussed in this article, have also resulted in some of the largest studies of genetic correlates of specific brain characteristics, such as genetic variations determining subcortical, hippocampal, and intracranial brain volumes (Grasby et al., 2020; Hibar et al., 2015; Stein et al., 2012). In order to analyze multisite data in these large studies, ENIGMA typically uses a meta‐analytic approach in which analysis methods, which are tested for reliability across diverse research sites, are openly shared. Participating research sites can analyze their brain magnetic resonance imaging (MRI) and genetic data locally and share either their site results (meta‐analysis) or individual participant results (mega‐analysis) with the rest of the working group (Zugman et al., 2020). In this way, many research sites can be brought together to meta‐ and mega‐analyze retrospective MRI and genetic data collected across different research sites around the world. The developed methods perform consistently despite high variability in MRI scanner manufacturers, scanner strengths, and pulse sequences, and the same methods can be applied across different disease working groups, allowing for cross‐disorder comparisons despite using different acquisition protocols (Boedhoe et al., 2019).
The ENIGMA Stroke Recovery working group is one of the 26 disease working groups within the ENIGMA consortium. Initially developed by a small group of researchers in 2015, ENIGMA Stroke Recovery has grown to over 100 members and has amassed a dataset of 2,137 MRI scans and counting from 39 research studies across 10 countries (see Table 1 for a summary of the number of scans by institution and geographical region at the time of publication). A primary goal of ENIGMA Stroke Recovery is to provide a reliable infrastructure for the collection and analysis of large, diverse datasets of poststroke brain MRI and behavioral data across research laboratories worldwide. The focus of the ENIGMA Stroke Recovery working group was initially on understanding the neural correlations of poststroke sensorimotor performance, which is affected in up to 80% of individuals after stroke (Rathore, Hinn, Cooper, Tyroler, & Rosamond, 2002). However, this focus has diversified as new members have joined. ENIGMA Stroke Recovery currently includes measures of sensorimotor, cognitive, emotional, and psychosocial behavior. Both ischemic and hemorrhagic stroke are included in the database, along with demographic, clinical and bedside diagnostic information. ENIGMA Stroke Recovery uses pipelines developed by several of the ENIGMA methods working groups (for a recent review, see Thompson et al., 2020, as well as other widely used software packages (Fischl et al., 2002; Fischl & Dale, 2000; Smith et al., 2004; Smith et al., 2007). Given the diversity and the unique challenges associated with stroke‐specific brain imaging data, discussed further in the article, the ENIGMA Stroke Recovery working group also requests raw MRI data when available. The raw data are processed by ENIGMA Stroke Recovery research staffs who perform additional quality control and lesion segmentation steps. Reliable and robust systems for data collection and maintenance are thus critical for the success of this collaborative approach.
TABLE 1.
Number of T1‐weighted MRI scans by geographical region/institute
| Country | City | Institute | Number of scans |
|---|---|---|---|
| Australia | Melbourne | The Florey Institute of Neuroscience and Mental Health | 317 |
| Brazil | Sao Paolo | University of Sao Paolo | 28 |
| Brazil | Sao Paolo | Albert Einstein Israeli Hospital | 15 |
| Canada | Toronto | University of Toronto/Sunnybrook Research Institute | 29 |
| China | Tianjin | Tianjin Medical University General Hospital | 65 |
| Germany | Griefswald | University of Griefswald | 68 |
| Germany | Tübingen | University of Tübingen | 175 |
| Italy | Rome | IRCCS Santa Lucia Foundation | 192 |
| New Zealand | Auckland | University of Auckland | 104 |
| Norway | Oslo | University of Oslo | 265 |
| UK | London | University College London | 50 |
| USA | Atlanta | Emory University | 110 |
| USA | Charleston | Medical University of South Carolina | 174 |
| USA | College Park | University of Maryland | 128 |
| USA | Irvine | University of California, Irvine | 191 |
| USA | Los Angeles | University of Southern California | 189 |
| USA | Philadelphia | University of the Sciences | 37 |
| Total scans | 2,137 | ||
Note: The total number of T1‐weighted MRI scans (N = 2,137) includes data from both individuals with stroke (n = 1,918, or 89.8% of the total dataset) and healthy individuals (n = 219, or 10.2% of the total dataset). A subset of the scans also includes repeated MRIs from the same individual (e.g., longitudinal data; n = 672 scans, or 31.4% of the total dataset).
The ENIGMA Stroke Recovery database can be used for many different purposes. First, the data can be used to test whether brain–behavior relationships identified in smaller, more homogeneous samples also exist in larger, more diverse samples. A specific example might be examining whether individuals with left hemisphere stroke show better or worse outcomes than those with right hemisphere stroke, as reports from the literature on this topic are inconsistent (Beuscher et al., 2017; Macciocchi, Diamond, Alves, & Mertz, 1998; Ween, Alexander, D'Esposito, & Roberts, 1996; Wu et al., 2015). Second, these data can be used to identify characteristics, such as specific lesion locations that affect stroke outcomes of interest. For example, the data could be used to identify whether depression is more common following a stroke that impacts the left frontal cortex, or whether injury to specific cortical regions, such as the right dorsal premotor region, is associated with differences in functional outcomes. The data could also be used in a voxel‐wise analysis to identify voxels (i.e., three‐dimensional pixels) in the poststroke brain that are related to a specific behavior, such as spasticity, mild cognitive impairment or poststroke fatigue. This voxel‐wise whole‐brain approach provides a granular way to examine entire vascular territories, not limited by specific anatomical boundaries. A third use of the data is in generating new data‐driven hypotheses using machine‐learning methods. The large size of the dataset makes it uniquely powered for machine‐learning methods, which require big datasets to train and test computer algorithms to identify patterns. In particular, this dataset is well suited for unsupervised machine‐learning techniques, which can be used to identify clusters, or subgroups, of people who show similar patterns of recovery, and then examine what brain traits are common within those specific subgroups. Notably, as all of the data provided in ENIGMA Stroke Recovery is collected for specific research studies, the secondary use of these data in ENIGMA Stroke Recovery reduces research waste and improves the efficiency and speed with which we can test the reproducibility of existing research findings, identify novel brain–behavior relationships, and generate data‐driven hypotheses.
In this article, we provide a comprehensive description of the ENIGMA Stroke Recovery working group's approach to large‐scale analyses of multisite retrospective poststroke brain MRI and behavioral data for the broad neuroscience community. Specifically, this article will discuss methods and protocols developed for: (a) data intake, (b) data harmonization, and (c) lesion analysis. In each section, we also highlight future directions and recommendations to facilitate collaborative data sharing (see also Box 1).
BOX 1. ENIGMA Stroke Recovery Working Group recommendations for prospective data collection, data management, and data sharing .
Add language in ethics protocols and informed consent forms for data sharing (see Appendix 2).
Use standardized protocols for MRI data collection, following either the Human Connectome Project's Lifespan Aging protocol (https://www.humanconnectome.org/study/hcp‐lifespan‐aging/project‐protocol/imaging‐protocols‐hcp‐aging) or the Alzheimer's Disease Neuroimaging Initiative protocol (http://adni.loni.usc.edu/methods/mri‐tool/mri‐analysis/) where possible.
Collect Stroke Recovery and Rehabilitation Roundtable (SRRR) task force consensus‐based demographic variables and behavioral measures as prescribed (e.g., do not modify the measure to exclude certain items or include extra items). See Kwakkel et al. (2017) for a complete list of sensorimotor measures and McDonald et al. (2019) for recommendations for cognitive measures (McDonald et al., 2019).
Use the Brain Imaging Data Structure (BIDS) format to name, describe, organize and store all data (https://bids.neuroimaging.io/ ; Gorgolewski et al., 2016).
Share full research protocols on an open platform like Open Science Framework to improve the replicability of experiments (https://osf.io/).
Consider openly sharing data to increase the reach and impact of any collected data (Nichols et al., 2017).
2. DATA INTAKE
2.1. Data components
Key components for the initial ENIGMA Stroke Recovery analyses rely on a T1‐weighted (T1w) anatomical brain MR image and at least one poststroke behavioral outcome measure. These two elements comprise the minimum data required to join ENIGMA Stroke Recovery. Demographic data are also strongly requested, although not required. A summary of all data components is included in Table 2. Each component is described in more depth below.
TABLE 2.
Data elements collected by the ENIGMA Stroke Recovery working group
| MRI | Behavior | Demographics | |
|---|---|---|---|
| Required |
T1‐weighted structural MRI Scanner strength, brand, and model Alternative if unable to share raw T1 MRIs: A spreadsheet with FreeSurfer cortical and subcortical measurements, quality control 2D image slices, and lesion masks (registered to a standardized template) |
At least 1 behavioral outcome measure
Most common measures at time of publication:
|
None required
Most common demographics at time of publication:
|
| Recommended |
|
We suggest collecting measures recommended by the Stroke Recovery and Rehabilitation Roundtable (Bernhardt et al., 2017) Current recommendations for sensorimotor outcomes can be found in Kwakkel et al. (2017) Current recommendations for cognitive outcomes can be found in McDonald et al. (2019) |
|
Note: The data elements are divided into three main components: MRI, behavioral measures, and demographic data, and further separated into required versus recommended elements.
2.1.1. Magnetic resonance imaging
The high‐resolution T1w MRI is used to quantify anatomical variation in regions across the brain, which is then related to the behavioral measures. We use a T1w MRI instead of other MR sequences, which might be more sensitive to the stroke lesion acutely, because high‐resolution T1w MRIs are routinely collected during research studies and can be used with FreeSurfer, a software used by all ENIGMA working groups for structural MRI analysis. FreeSurfer performs automated cortical and subcortical brain segmentation (Fischl et al., 2002; Fischl & Dale, 2000) and has reliable performance across different scanner characteristics and T1w acquisition parameters (Han et al., 2006). However, we acknowledge that the T1w MRI is not the best choice for identifying the lesion volume in acute and subacute patients. Therefore, we also collect additional MRI data that can be received and combined for analysis, such as diffusion MRI, T2‐weighted fluid‐attenuated inversion recovery (FLAIR), as well as other modalities of interest, such as resting‐state functional MRI (fMRI).
If raw MRI data cannot be shared, research sites can choose to analyze the data locally using processing scripts found on the ENIGMA website (http://enigma.ini.usc.edu/protocols/); results can then be sent for subsequent analyses (outputs include a spreadsheet with measures of cortical thickness, cortical surface area, and subcortical volumes following FreeSurfer segmentation). However, the ENIGMA Stroke Recovery group has the complicating issue of lesions within the data (see Section 4). Therefore, more stringent quality control, as well as delineation of stroke lesion boundaries, is required for more fine‐grained analyses of the lesion's impact on subsequent brain structure and behavior. For this reason, sites not sending raw data are requested to send the FreeSurfer segmentation measures, along with quality control images generated by our in‐house code (see Section 2.2 for more detail), and lesion masks normalized to a standard template brain when possible.
2.1.2. Behavioral data
ENIGMA Stroke Recovery collects many different types of behavioral data. As mentioned previously, the focus of the initial research question is on the neural correlates of poststroke sensorimotor behavior. To this end, over 80% of the dataset includes measures of sensorimotor performance. However, the growth and diversification of the ENIGMA Stroke Recovery group has led to the collection of additional types of behavioral data, including measures of cognition, mood, dysphagia, and psychosocial well‐being (see Section 3 for more information).
2.1.3. Demographic data
Demographic information on the participants is also collected, such as age, sex, time from last known well to imaging, type of stroke, number of previous strokes, stroke location, risk factors, and comorbidities (see Section 3 for more information). There are no exclusionary demographic criteria for entry into the database. There is growing interest in characterizing rehabilitation history, although this is an extremely complex variable to harmonize across research sites and encode. In general, the reported demographic variables can be included as covariates or inclusion/exclusion criteria for specific analyses.
2.2. Data intake workflow
When a research site joins ENIGMA Stroke Recovery, the site securely transfers de‐identified MRI data as well as a comma‐separated values (CSV) spreadsheet with all demographic and behavioral outcomes. If the raw MRI data are not available or cannot be shared, the site can run analyses scripts locally (https://github.com/npnl/ENIGMA-Wrapper-Scripts) and send FreeSurfer results in a CSV file along with the behavioral/demographic CSV file. Transfer can be accomplished via a secure file transfer protocol to an ENIGMA Stroke Recovery dedicated Linux server, or by the research site's preferred secure transfer method (e.g., using Box). The complete process, from data intake to data analysis, is displayed in Figure 1 and described in brief below. The scripts and code developed for these processes are freely and publicly available on Github (https://www.github.com/npnl).
FIGURE 1.
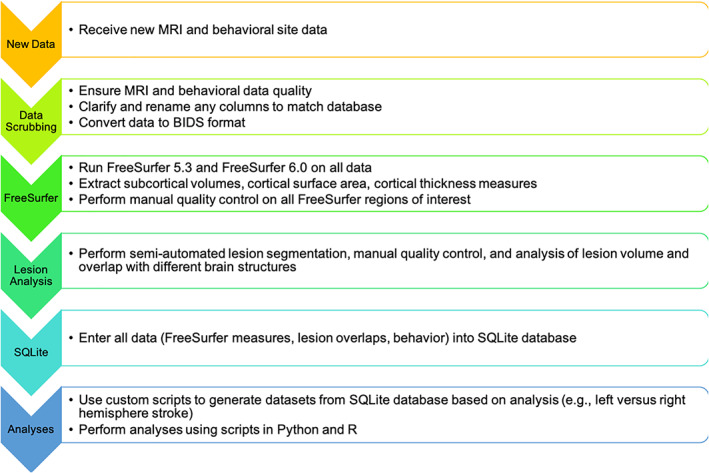
ENIGMA Stroke Recovery workflow. Workflow for ENIGMA Stroke Recovery from data intake to data analysis
First, the received data are manually inspected to ensure: (a) good MRI data quality, and (b) appropriate values for the behavioral data. For MRI data quality, we visually inspect the images to ensure there are not large motion artifacts or other visible sources of noise in the data. Data that are of poor quality are excluded from the database. More recently, to standardize this process, we have begun to use the University of Southern California's Laboratory of Neuroimaging (LONI) Quality Control system, which is a freely available, semiautomated, web‐based system for quantitatively evaluating MRI image quality (Kim et al., 2019). We ensure that the behavioral data are appropriately coded according to our ENIGMA Stroke Recovery database conventions. For instance, we convert and record time since stroke in units of days and denote the lesioned hemisphere by an integer value where left = 1, right = 2, both = 3, and other = 4. The behavioral data are also inspected for values within the normal range for each measure. For the initial analysis of sensorimotor outcomes, a “primary” sensorimotor outcome that most closely aligns with the existing ENIGMA dataset is selected (see Section 3 for details). Altogether, these steps ensure that the incoming data are consistent with the existing database for subsequent analysis.
Next, we write a site‐specific script to reformat the data to conform to the Brain Imaging Data Structure (BIDS; https://bids.neuroimaging.io/) (Gorgolewski et al., 2016). BIDS provides a standardized way to organize and describe neuroimaging and behavioral data. If the data are not consistently named and formatted within each site, we manually reformat the data to conform to the BIDS standard. Having all of the data in BIDS format allows us to quickly and easily analyze ENIGMA Stroke Recovery data using a variety of software tools, many of which are written with the assumption that the data are in BIDS format (Gorgolewski et al., 2017). It also allows all of the code and scripts created for ENIGMA Stroke Recovery analyses to be easily used by others who use BIDS format, thereby increasing the transparency, reproducibility, and impact of this work.
Third, we run FreeSurfer, a brain imaging software package developed to analyze MRI scans, which segments the brain into anatomical regions for morphometric analysis (Fischl et al., 2002). We run two versions of FreeSurfer (version 5.3 and version 6.0) on the MRI data using scripts that automate the processing of all participants in parallel on the LONI computing cluster. FreeSurfer version 5.3 is currently the primary analysis version, which is used to maintain consistency with recent ENIGMA working group analyses. We also analyze all raw data using FreeSurfer version 6.0 because this more recent version has been shown to have improved performance and more accurate brain segmentations compared to FreeSurfer version 5.3. In addition, FreeSurfer version 6.0 also provides improved hippocampal segmentations and analysis of hippocampal subfields in stroke (Khlif et al., 2018), which is an area of research interest given recent findings from our ENIGMA Stroke Recovery working group showing a relationship between poststroke sensorimotor behavior and hippocampal volumes (Zavaliangos‐Petropulu et al., 2019).
We then use the ENIGMA scripts mentioned previously to extract standard measures of subcortical volume (from eight subcortical regions of interest, bilaterally), and cortical thickness and cortical surface area (from 34 cortical regions of interest, bilaterally; https://github.com/npnl/ENIGMA-Wrapper-Scripts). All regions are from the standard Desikan‐Killany atlas implemented in FreeSurfer (Desikan et al., 2006). These measures provide insight into brain morphometry for both subcortical and cortical regions. ENIGMA scripts are used to generate images of the overlap between each segmented region of interest and the underlying brain and put these into a webpage that can be visually inspected for segmentation accuracy. This is done for each brain. Although FreeSurfer has shown acceptable poststroke brain segmentation performance (Li et al., 2015), it may occasionally fail to create accurate segmentations on lower quality data (e.g., MRIs with lower spatial resolution or greater noise from movement) or on data with lesions that disrupt its surface‐based algorithms. In our ENIGMA Stroke Recovery dataset, FreeSurfer fails to produce accurate segmentations in 10–20% of the data. To address this, trained researchers manually inspect each region of interest for each brain MRI. Any regions that are not properly segmented by FreeSurfer are marked as “failed” and excluded from the analyses (see Appendix 1 for additional information about the ENIGMA Stroke Recovery FreeSurfer quality control protocol).
Finally, we use an in‐house script that pulls all of each site's FreeSurfer outputs and behavioral data and enters them into a relational database using Structured Query Language (SQL). Data are entered into a SQLite database (https://sqlite.org), a free, relational database engine that allows for simple and intuitive data storage using the SQL language (Owens, 2006). Key benefits of a SQL‐based environment—compared to other database options, such as REDCap (Harris et al., 2009) (https://www.project-redcap.org/)—are that it is freely available (not institutionally constrained) and widely used across both research and industry, leading to many options for integrating SQL analyses with popular programming environments such as Python, and statistical packages such as R. SQLite runs quickly and does not require specialized computing resources, and SQLite databases can be easily shared. SQL‐based databases are also being used in other large‐scale data sharing projects, such as the NIH‐funded All of Us research initiative, which aims to gather health‐related data about one million or more people living in the United States (Klann, Joss, Embree, & Murphy, 2019). Given these factors, SQLite provides an accessible option for scalable data analysis and for future data sharing and is aligned with existing big data initiatives. Within the SQLite database, there are seven tables that hold information about demographics, behavioral measures, brain measures (two tables, one for each of the FreeSurfer analysis versions), MR scanner information, research site information, and metadata about the database itself. This database is queried to extract the specific data used in each analysis.
2.3. Future directions for improving data intake
A key future priority for updating the data intake process is using artificial intelligence to improve the manual quality control process, capitalizing on the efforts from other ENIGMA working groups (Petrov et al., 2017; Petrov et al., 2018). Currently, performing visual quality control of each segmented FreeSurfer region is a significant bottleneck in our data intake pipeline. Each individual brain has a total of 84 subcortical and cortical regions that should be inspected by a trained investigator, which takes on average about 20 min per brain; performing quality control on so many regions manually is not easy to scale up. In addition, visual quality control is an inherently subjective process. Investigators are intensively trained to ensure good inter‐rater reliability, but there is still the potential for human error. Machine‐learning algorithms to reliably perform quality control of the segmentations would remove an enormous barrier to performing this work. To facilitate the development of an automated quality control process, we have manually generated ratings on the subcortical volumes of over 1,000 poststroke brain MRIs to date. We anticipate that these data can be used to train and test deep learning algorithms (such as convolutional neural networks) to identify failures in segmentation.
3. DATA HARMONIZATION
A second key challenge that ENIGMA Stroke Recovery faces is harmonizing data collected using different MRI scanners and scanning protocols, as well as using different behavioral outcome measures. Data harmonization refers to all efforts to combine data collected across different sources and with different formats, naming conventions, and measures into one cohesive dataset. Optimizing data harmonization is important because MRI data collected using different scanners and/or scanning protocols can significantly affect analyses and results [e.g., (Zavaliangos‐Petropulu et al., 2019)]. For behavioral data, different measures may focus on measuring specific aspects of sensorimotor impairment, function, or quality, and maybe more or less sensitive to a certain range of performance values. Importantly, the harmonization of methods also provides an opportunity to explore the effects of inter‐site differences on both MRI and behavioral data analysis, which is difficult to examine otherwise. Here, we describe efforts to address each of these challenges so that we can combine imaging scans and behavioral data obtained at different centers.
3.1. MRI harmonization
ENIGMA working groups have developed robust methods for analyzing T1w anatomical MRIs, diffusion MRI, and resting‐state fMRI in a way that is reproducible and reliable across multiple research sites (Acheson et al., 2017; Jahanshad et al., 2013; Pizzagalli et al., 2019). While the scans themselves are not directly combined, a number of specific metrics are extracted from each type of imaging sequence so that data across centers can be used in mega‐ and meta‐analyses. The key measures extracted for each sequence are described below. In addition, we provide guidelines for quality control and harmonization in order to ensure that all segmentations are representative of the desired anatomy. In this way, even if the scans themselves are not combined, statistical approaches for combining these extracted features in a meta‐analytical or mega‐analytical manner can be taken. Detailed instructions and code for analysis and quality control for each method may be found on the ENIGMA website ( http://enigma.ini.usc.edu/protocols/imaging-protocols/).
3.1.1. T1‐weighted MRI
As noted previously, a T1w MRI is a key component of the initial ENIGMA Stroke Recovery analyses. T1w MRIs are processed using ENIGMA's structural image processing protocols (publicly available in the link above), which include several methods for analyzing T1w MRIs including cortical and subcortical volume and surface area, sulcal geometry analysis, and vertex‐wise subcortical shape analysis. The ENIGMA structural protocols all utilize outputs from FreeSurfer, a brain imaging software package developed to analyze MRI scans of brain tissue, which segments and labels neuroanatomical structures in the data (Fischl et al., 2002). Importantly, FreeSurfer is reliable across research sites and demonstrates good test–retest reliability across scanner manufacturers and field strengths (Han et al., 2006). In addition, as mentioned above in Data Intake (Section 2.2), we perform manual quality control on all FreeSurfer segmentations, so that subsequent structural analyses use only quality‐controlled inputs. Trained researchers manually inspect each region of interest for each structural brain MRI. Regions that are not properly segmented by FreeSurfer are marked as “failed” and excluded from further analysis (see Appendix 1 for the full ENIGMA Stroke Recovery FreeSurfer quality control protocol).
3.1.2. Diffusion MRI
The ENIGMA‐DTI protocol is used to study fractional anisotropy and diffusion tensor imaging‐derived diffusivity measures of whole brain and atlas‐defined regions of interest in the white matter, based on the tract‐based spatial statistics method implemented in FMRIB Software Library (Smith et al., 2007). While preprocessing diffusion MRI can vary by data quality, the ENIGMA‐DTI protocol provides suggestions including steps for motion correction, echo‐planar imaging distortion correction, and tensor fitting. The ENIGMA‐DTI protocol can be found on the ENIGMA website ( http://enigma.ini.usc.edu/protocols/dti-protocols/) and is detailed elsewhere (Jahanshad et al., 2013). This protocol has been shown to have excellent reproducibility between scanners for the analysis of white matter microstructure (Acheson et al., 2017). The ENIGMA‐DTI working group also works on methods for harmonizing multisite diffusion MRI (Zhu, Moyer, Nir, Thompson, & Jahanshad, 2018), and the ENIGMA Stroke Recovery group will use recently recommended mega‐analytic methods for diffusion MRI analyses (Boedhoe et al., 2019).
3.1.3. Resting‐state fMRI
Resting‐state fMRI (rs‐fMRI) offers an approach to understand patterns of synchronized brain activity in the resting state, which can further be decomposed into networks with known functions (e.g., default mode, salience, attention networks) (Biswal et al., 2010; Biswal, Yetkin, Haughton, & Hyde, 1995). Harmonized processing of rs‐fMRI in ENIGMA has used one of two pipelines: (a) an echo‐planar imaging‐based pipeline, based on the Analysis of Functional NeuroImages software, which does not require the use of a co‐registered anatomical MRI dataset (Adhikari et al., 2018; Adhikari et al., 2019; Adhikari et al., 2019); and (b) a pipeline known as fMRIprep+, based on the fMRIprep approach (Esteban et al., 2019), which can also be used for the analysis of multisite task‐based fMRI (Veer, Waller, Lett, Erk, & Walter, 2019).
3.1.4. Future Directions in Brain Imaging Harmonization
Future directions include the harmonization of stroke electroencephalography (EEG) data, as the ENIGMA EEG working group is currently developing methods for analyzing resting‐state EEG (Smit, 2020; Smit et al., 2016). EEG has garnered growing attention in stroke rehabilitation over the years due to its portability, safety, and lower cost, compared to MRI or fMRI. Advancing our capability to use EEG was also identified as a developmental priority in the SRRR taskforce on biomarkers (Boyd et al., 2017). EEG measures, such as hemispheric asymmetry or frontoparietal coherence, have also been related to and/or predictive of stroke outcomes (Ramos‐Murguialday et al., 2013; Zhou et al., 2018).
Finally, although all our existing MRI pipelines offer methods for handling previously collected data, the ideal scenario is a harmonized prospective data collection. We recommend that individual research groups consider using MRI sequences that match the publicly available sequences from the NIH‐funded Lifespan Human Connectome Project Aging group https://www.humanconnectome.org/study/hcp-lifespan-aging/project-protocol/imaging-protocols-hcp-aging) or the NIH‐funded Alzheimer's Disease Neuroimaging Initiative (ADNI; (http://adni.loni.usc.edu/methods/mri-tool/mri-analysis/). Doing so would expand the reach of one's individual research data by allowing for comparison with large, existing datasets, and help align prospective scanning efforts in ENIGMA Stroke Recovery with that of other groups. Future work may also try to align the ENIGMA diffusion MRI and rs‐fMRI data processing protocols with the UK Biobank imaging efforts, which aim to provide multimodal brain imaging data on 100,000 individuals living in the United Kingdom (Alfaro‐Almagro et al., 2018).
3.2. Behavioral and demographic data harmonization
3.2.1. Behavioral data
Stroke researchers test a broad range of hypotheses, and different behavioral outcomes are selected to address specific hypotheses. In addition, the same behavior may be assessed using multiple measures. Thus, although many researchers in the ENIGMA Stroke Recovery working group study poststroke arm performance, the current ENIGMA Stroke Recovery database has more than 75 unique behavioral measures. This large number of behavioral measures is in line with a study reporting that 144 different outcome measures were used to study poststroke arm rehabilitation across 243 clinical trials (Duncan Millar, van Wijck, Pollock, & Ali, 2019).
In order to organize the many different types of behavioral data, ENIGMA Stroke Recovery relies on the International Classification of Functioning, Disability and Health (ICF) (World Health Organization, 2001), which provides a framework that can be used to categorize different assessments of poststroke outcomes. The ICF model has three levels that can be used to conceptualize behavioral measurements about a person: (a) body functions and structures (measuring the person's impairment), (b) activities (measuring function at the level of the person), and (c) participation (measuring function of the person as a member of society). The ICF framework is used by the SRRR task force to categorize core measures for all stroke recovery trials to collect (Kwakkel et al., 2017). Specifically, the SRRR recommends collecting the Fugl‐Meyer Assessment (FMA) (Fugl‐Meyer, Jaasko, Leyman, Olsson, & Steglind, 1975) to measure body function and structure for the upper and lower limbs, and the Action Research Arm Test (ARAT) (Lyle, 1981) and a 10‐min walk test to measure activity limitations in the upper and lower limbs. Although there was no consensus recommendation on participation, there were recommendations to collect the EuroQoL 5 dimension scale (EQ‐5D) as a measure of quality of life (Brooks & Group, 1996) and the modified Rankin Scale as a measure of global disability (Van Swieten, Koudstaal, Visser, Schouten, & Van Gijn, 1988). Finally, the National Institutes of Health Stroke Scale (NIHSS) (Brott et al., 1989) was recommended to measure stroke severity (i.e., global impairment across multiple domains), but not as an outcome measure.
These ICF‐based and SRRR‐recommended measures are well represented in the ENIGMA Stroke Recovery database. The most common measures include (listed in order of frequency in the database with percent frequency in parentheses): (a) FMA‐UE (72%), (b) NIHSS (19%), (c) Motor Activity Log (MAL; 16%,) (Uswatte, Taub, Morris, Vignolo, & McCulloch, 2005), (d) modified Ashworth measure of spasticity (12%) (Bohannon & Smith, 1987), (e) ARAT (11%), and (f) Wolf Motor Function Test (WMFT; 9%) (Wolf et al., 2001). Notably, the most common measures focus on sensorimotor performance of the upper extremity after stroke, which reflects the research focus of the many ENIGMA Stroke Recovery working group members who study this topic. As the working group grows more diverse, so too do the behavioral measures. At present, the database also includes measures of gait and balance, such as the 6‐min walk test (Butland, Pang, Gross, Woodcock, & Geddes, 1982), cardiovascular fitness, such as VO2 max (Shephard et al., 1968), cognition (e.g., Mini‐Mental State Examination (Folstein, Robins, & Helzer, 1983), Montreal Cognitive Assessment (Nasreddine et al., 2005), and mood [e.g., the Hospital Anxiety and Depression Scale (Zigmond & Snaith, 1983)].
Prospectively, we recommend that stroke researchers collect all of the measures recommended by the SRRR task force (Kwakkel et al., 2017), with a particular emphasis on the FMA, which is contained in 72% of entries in our database and thus the most well‐represented measure. However, given that ENIGMA Stroke Recovery currently contains only retrospective data, there are several ways that we maximize this data, despite the different behavioral measures.
For our initial study of sensorimotor outcomes, for each site, we define a primary sensorimotor score, which is the measure from the site that is the most widely reported measure in the database. For instance, if a site collected the ARAT, nine‐hole pegboard test, and grip strength, we would use the ARAT as the primary sensorimotor score, since, of the three collected by that site, it is the best represented within the ENIGMA Stroke Recovery database across all sites. Using this measure, we then take a percentage of the maximum possible score for that measure, where 0% is the worst sensorimotor outcome and 100% is the best. For example, if someone received a 33 on the ARAT (where the maximum score is 57), their primary sensorimotor score would be 58%. Doing this also addresses the variability in how people collect even standardized measures. For instance, some research sites do not collect the full FMA‐UE because they do not agree with the reflex measures. The maximum possible score for the site might then be 60 instead of the full FMA‐UE scale of 66. Calculating a percentage of the maximum possible sensorimotor score allows for the normalization of different behavioral scores, enabling a comparison across participants from different research studies. A limitation of our approach is that it mixes measures of sensorimotor impairment, function, and participation, examiner‐ versus patient‐reported outcomes, and different times after stroke that may be more or less ideal for a specific behavioral measure. We attempt to overcome this limitation by also grouping measures by the level of measurement for analyses when possible: impairment (e.g., FMA‐UE, grip strength), function (e.g., ARAT, WMFT), and participation (e.g., MAL, Stroke Impact Scale). Lastly, we analyze the most commonly reported measures separately. Although this last approach limits the sample size for these analyses, it allows for a more rigorous examination of the neural correlates of specific measures (e.g., to study neural correlates of impairment versus function or to examine differences between examiner‐ versus patient‐reported outcomes).
3.2.2. Demographic variables
Many different demographic variables, such as age, sex, hemiparetic side, and time since stroke, have been shown to strongly influence poststroke behavioral outcomes (Appelros, Stegmayr, & Terént, 2009; Chang, Chang, Cragg, & Cramer, 2013; Jongbloed, 1986; Stewart, Gordon, & Winstein, 2014; Stinear et al., 2006; Stinear et al., 2017). Similar to the behavioral outcome measures, the demographic variables collected across research sites and studies are also wide‐ranging. Within ENIGMA Stroke Recovery, the most consistently reported variables are age, race, sex, time since stroke, and lesioned hemisphere. Not surprisingly, these variables are reported with different levels of granularity across research sites. For instance, time since stroke is often reported in units of days, weeks, months, or even years. To maximize precision in at least a subset of the data, we convert time since stroke to days for all (e.g., 2 months after stroke would be converted to 61 days, calculated as 2 × 30.5 days on average per month). Notably, this reduces the precision of the data, especially if the time since stroke is reported in years for some research studies, in which case the value “2 years” could actually represent anywhere from 730 to 1,094 days. To this end, we strongly recommend collecting the smallest unit of measurement possible. Additional variables are highly useful as well, although less frequently reported, such as hand dominance prior to the stroke, therapy received, risk factors for cardiovascular disease such as hypertension, obesity, diabetes and smoking, and comorbidities (including several of the aforementioned risk factors). The latter variables have been shown to affect the brain beyond the lesion and may result in decreased white matter integrity and increased structural atrophy.
3.2.3. Future directions in behavioral measure and demographic variable harmonization
We recommend that researchers prospectively conducting research follow the SRRR task force recommendations regarding both behavioral measurements and demographic variables, as well as the precision with which to collect them (Kwakkel et al., 2017). As data collection of these measures can be subjective, we also highly recommend standardized training for the collection of behavioral measures. For example, previous research has shown that training on how to administer and measure the FMA‐UE (See et al., 2013) can greatly improve measurement reliability across different research sites. ENIGMA Stroke Recovery would also benefit from increased reporting of behavioral measures beyond sensorimotor outcomes, such as measures of gait, balance, cognition, and psychosocial health, as well as neuropsychiatric outcomes. Future directions for this aspect of the dataset include harmonizing and modeling longitudinal data aggregated across research sites. This important analysis would allow us to not only discuss sensorimotor behavior at a single, cross‐sectional timepoint but also sensorimotor recovery, which by definition requires an examination of longitudinal changes over multiple timepoints.
4. LESIONS
Finally, in addition to harmonizing the measurement of brain volumes and behavior across research sites, ENIGMA Stroke Recovery also has to account for different types of brain lesions. There are two primary types of brain lesions encountered in stroke MRI data: those that are a direct result of the stroke, and white matter lesions, also known as white matter hyperintensities, which represent small vessel disease. Both types of lesions are important to capture in stroke analyses, as both have previously been associated with stroke outcomes across cognitive and motor domains (Arsava et al., 2009; Auriat et al., 2019; Boyd et al., 2017; Feng et al., 2015; Riley et al., 2011). While white matter hyperintensities can be measured using automated software (Ramirez et al., 2011), software to accurately and automatically identify stroke lesions still poses a significant challenge. This is because the size, shape, and location of acute stroke lesions are more variable, and thus less predictable, than white matter lesions, which typically occur in the periventricular or deep white matter and are of a characteristic shape and size. There is a strong scientific interest in understanding how stroke lesions both directly and indirectly (e.g., through secondary degeneration) affect brain structure and, subsequently, behavior. Therefore, the ENIGMA Stroke Recovery working group has developed methods for accurately capturing the stroke lesion to characterize both direct versus indirect effects of the lesion on the brain. This section will detail the steps we have taken to develop a complete neuroinformatics pipeline for large‐scale, semiautomated stroke lesion segmentation.
4.1. An open‐source stroke lesion dataset
The current gold standard for lesion segmentation using T1w MRIs is manual segmentation (Ito, Kim, & Liew, 2019). When ENIGMA Stroke Recovery started in 2015, there were few publicly available automated lesion segmentation methods for T1w MRIs. A key reason for the small number of T1w lesion segmentation methods was the limited availability of open‐source stroke MRI data with lesion masks that could be used to develop, train, and test algorithms. Of the available methods, most were developed on small datasets of individuals with stroke (e.g., 8–60 patients) collected at one or two local research sites (Griffis, Allendorfer, & Szaflarski, 2016; Pustina et al., 2016; Seghier, Ramlackhansingh, Crinion, Leff, & Price, 2008). This lack of a large public stroke lesion dataset has not only resulted in limited methods for lesion segmentation but also resulted in methods that may have limited generalizability to more diverse stroke datasets.
To address this problem, we developed a large, manually segmented dataset of N = 304 stroke lesion masks and T1w brain MRIs collected at eight different research sites around the world, which we publicly released (Liew et al., 2018). The Anatomical Tracings of Lesions After Stroke (ATLAS) dataset is the largest open‐source dataset of stroke anatomical MRIs and manually segmented lesion masks (see Box 2). It is shared on two public repositories (ICPSR: https://www.icpsr.umich.edu/icpsrweb/ADDEP/studies/36684; and FCP/INDI: http://fcon_1000.projects.nitrc.org/indi/retro/atlas.html). The procedure and training protocols for lesion segmentation are publicly available (Liew et al., 2018). The purpose of releasing this dataset was to provide a large, standardized test dataset, or benchmark, for researchers to compare the performance of their lesion segmentation algorithms with those of other research groups. Releasing this dataset has now achieved the goal of improving lesion segmentation algorithms and much more. It has been downloaded over 650 times by individuals across more than 30 countries around the world and has been used for educational, research, and industry projects. Toward the primary goal, it has been published in papers reporting improved automated lesion segmentation performance of more than 90% accuracy (Sharique, Pundarikaksha, Sridar, Krishnan, & Krishnakumar, 2019). However, it has also been used in creative and unexpected ways. It has been used as an example dataset in numerous university classes on machine learning and computer vision and as a dataset for masters and PhD theses. It has also led to the creation of new stroke tools, such as probabilistic stroke atlas (Wang, Juliano, Liew, McKinney, & Payabvash, 2019) and an in silico head model for stroke simulation (Bing, Garcia‐Gonzalez, Voets, & Jérusalem, 2020). These unexpected uses highlight a key benefit of data sharing, which is that sharing data can not only move research forward in ways that are intended but can also lead to novel methods, data discovery and powerful educational opportunities for trainees.
BOX 2. ENIGMA Stroke Recovery Working Group open‐source tools for lesion quality control and segmentation .
- ATLAS (Anatomical Tracings of Lesions After Stroke) dataset—the largest open‐source dataset of stroke anatomical MRIs and manually segmented lesion masks (Liew et al., 2018). It can be downloaded from two public repositories:
- ICPSR (raw data): https://www.icpsr.umich.edu/icpsrweb/ADDEP/studies/36684
- INDI (preprocessed data): http://fcon_1000.projects.nitrc.org/indi/retro/atlas.html
- PALS (Pipeline for Analyzing Lesions after Stroke) Toolbox—an open‐source toolbox for lesion analysis (Pipeline for Analyzing Lesions after Stroke; PALS) (Ito et al., 2018).
- The PALS toolbox can be downloaded from Github: https://www.github.com/npnl/PALS
- Braindrles—a web‐based platform for crowd‐sourcing manual quality control of lesion segmentations (Liew et al., 2019).
- Braindrles can be played at https://braindrles.us/
- Source code for Braindrles can be found at https://github.com/npnl/braindrles
4.2. Comparing automated lesion segmentation methods
Using ATLAS, we systematically compared the existing, publicly available methods for lesion segmentation using T1w MRIs (Ito et al., 2019). Although there are new methods constantly emerging for lesion segmentation, most of these are not publicly available. Of the existing methods available, we determined that Lesion Identification with Neighborhood Data Analysis (LINDA) performed the best (Pustina et al., 2016), with a median value on the Dice coefficient, or measure of similarity between the LINDA segmentations and the gold standard manual segmentations, of 0.5. LINDA performs extremely well on larger lesions, which are the most time‐consuming lesions to manually segment. In ENIGMA Stroke Recovery, we therefore use LINDA to generate initial lesion masks on all the data, and then perform manual quality control and manual correction on the resulting lesion masks (see Box 2 for more details).
A key question regarding the adoption of automated lesion segmentation techniques is: When is the automated lesion segmentation method good enough to use? The answer to this question is determined by the research topic of interest and the research group. For ENIGMA Stroke Recovery, a primary research focus is on understanding how lesion overlap with different brain structures relates to specific behaviors. This can be performed by calculating a lesion load (e.g., lesion overlap) with specific regions of interest, or at the voxel‐level, using methods like voxel‐lesion symptom mapping to examine whole‐brain correlates of lesioned tissue within a stroke cohort. Both of these approaches require precise lesion boundaries, and we are therefore aiming for an automated lesion segmentation method that performs with over 90% accuracy in the ENIGMA Stroke Recovery dataset. As of December 2019, no publicly available automated lesion segmentation method has been able to meet this threshold. When available, we request working group members to submit their expert‐drawn lesion masks with their MRI data. When lesion masks do not exist, ENIGMA Stroke Recovery research staff run LINDA on the MRIs and manually correct the resulting segmentations. In all cases, we perform a careful visual inspection to validate the quality of the segmentations.
Finally, as many recent methods have reported improved performance (e.g., >90% accuracy on the ATLAS dataset) but have not openly shared their methods, we are creating a web‐based challenge for research groups to evaluate their novel lesion segmentation methods on a test dataset, for which the labeled data are hidden from the contestants, similar to previous lesion challenges (Maier et al., 2017). This challenge website is being built in collaboration with researchers at the Paris‐Saclay Center for Data Science using the Rapid Analytics and Model Prototyping framework (https://ramp.studio/), which will require the open sharing of the lesion segmentation method for entry, in order to encourage collaboration, transparency, and reproducibility (Kégl et al., 2018).
4.3. Quality control and analysis of automated lesion segmentation outputs
In order to manage the large volume of stroke lesion masks, we also created an open‐source toolbox for lesion analysis (Pipeline for Analyzing Lesions after Stroke; PALS) (Ito, Kumar, Zavaliangos‐Petropulu, Cramer, & Liew, 2018). The PALS toolbox can be downloaded from Github (https://www.github.com/npnl/PALS) and installed locally (see also Box 2). It has a function that creates a local quality control webpage that shows each brain's lesion mask overlaid on the T1w MRI. The webpage is interactive, and each lesion mask can be rated as “good,” “maybe,” or “fail.” This sorts the lesions into the respective folders, and any lesions that fall into the “maybe” or “fail” category are manually corrected. In this way, a person can review hundreds of lesion masks quickly and efficiently, without needing to open and overlay each mask on each brain individually.
We also created a crowd‐sourcing web‐based platform called Braindrles to engage citizen scientists in performing manual quality control of automated lesion segmentation outputs (https://braindrles.us/) (Liew et al., 2019; Box 2). Braindrles is built on the same web‐based platform as its predecessor, Braindr (Keshavan, Yeatman, & Rokem, 2018). Braindrles presents dynamic gifs of a lesion mask overlaid on a stroke brain, and users swipe right if they believe the lesion mask is correct, and left if they believe it is incorrect or inaccurate. There is also an option to “chat” about the lesion mask and ask questions. To encourage player engagement, Braindrles is gamified with a leaderboard and levels based on the number of swipes. Anyone in the public is welcome to play, and all users are given a tutorial as well as feedback about their accuracy on a subset of 100 stroke lesion masks. Because not all users are expected to perform similarly (e.g., a lay person with no brain anatomy knowledge versus an experienced neuroradiologist), a machine learning boosted tree‐based algorithm called XGboost (Chen & Guestrin, 2016) is used to identify and heavily weight raters who are the most accurate. XGboost is also used to generate an aggregate probability score from 0 (fail) to 1 (pass) for each lesion mask, based on the weighted rater scores. To date, Braindrles contains 1,464 lesion masks generated by a mix of both automated and manual segmentations and has garnered over 18,000 votes from over 100 users. Once fine‐tuned, this platform will allow for a higher volume of quality control decisions on lesion segmentation outputs and should provide a scalable solution as the project grows.
4.4. Future directions for stroke lesion segmentation
In the future, we aim to create an automated lesion segmentation pipeline with embedded, automated quality control (see Figure 2). This is critical because in addition to FreeSurfer segmentation quality control (mentioned previously), manual lesion segmentation and lesion quality control are the other major bottlenecks in the analysis of large‐scale stroke data. Manual lesion segmentation and quality control is tedious, slow, and requires extensive expertise, and it cannot be easily scaled up for thousands of stroke brains. Thus, an automated pipeline in which manual input is only needed to correct failed lesion masks would greatly accelerate the pace of stroke research and could be used for small and large studies alike.
FIGURE 2.
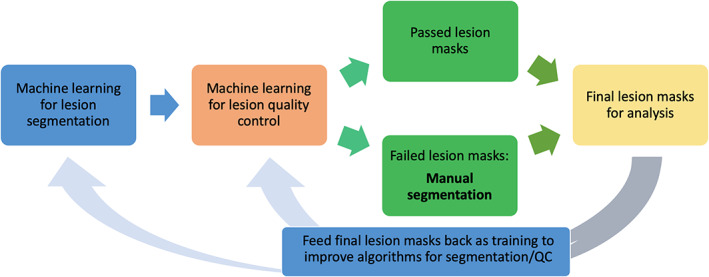
Optimized lesion segmentation pipeline. Example of a future neuroinformatics system for lesion segmentation, with only one point of manual input (manual segmentation of failed lesion masks, indicated in bold)
5. DISCUSSION AND FUTURE DIRECTIONS
In this article, we have described the approaches and methods that the ENIGMA Stroke Recovery working group has taken to facilitate the establishment of the first large‐scale collaborative multisite stroke brain MRI and behavioral data analyses. We hope that these efforts are useful in addressing key priorities in stroke research, including better characterizing the neurobiology of stroke recovery in humans, and identifying different stroke recovery phenotypes (Bernhardt et al., 2017), with an ultimate goal of discovering biomarkers that predict stroke recovery (Boyd et al., 2017).
The work presented here is designed to ultimately improve our ability to: (a) test whether brain–behavior relationships identified in smaller, more homogeneous samples also exist in larger, more diverse samples and (b) identify characteristics, such as specific lesion locations or specific demographics, which affect stroke outcomes of interest. Toward these two aims, the data generated in ENIGMA Stroke Recovery provide a valuable, standardized sample for researchers to test findings reported in smaller studies on a larger, heterogeneous dataset. Doing so may inform researchers of specific demographics or eligibility criteria that may be critical for specific brain–behavior relationships—a hypothetical example is finding that lesion overlap with the parietal cortex may be significantly related to motor behavior but primarily in people with chronic, right‐hemisphere stroke. In this way, ENIGMA Stroke Recovery's large data approach allows researchers to ask not only “Is this true?” but also “For whom is this true?” The importance of the latter question is that it accepts that what is true for one person may not be true for another and strives to identify individual differences in poststroke neural relationships underlying sensorimotor performance. This approach aligns well with the NIH's Precision Medicine Initiative, which recognizes that there are individual differences in health and recovery and places importance on the personalization of healthcare wherever possible (Collins & Varmus, 2015).
In addition, the exceptionally large size of this database affords the statistical power to begin using supervised machine‐learning techniques to test and train models to predict sensorimotor outcomes and to develop data‐driven hypotheses. Unsupervised machine learning can also be developed to identify clusters, or subgroups, of individuals who show similar behavioral outcomes. These subgroups can then be examined for similar traits that may relate to these outcomes. Although the current size of the ENIGMA Stroke Recovery database is relatively small compared to existing machine‐learning datasets, such as those used in Kaggle competitions (https://www.kaggle.com/), it provides a beginning point for developing these methods for stroke data, and will become more powerful as it grows.
Finally, while we have initially applied this framework toward understanding the structural brain correlates of sensorimotor performance, the infrastructure and tools created can now support many additional questions and analyses. As noted throughout this article, we have begun to expand the types of imaging sequences and modalities collected across sites, as well as the behavioral measures, including cognitive, affective, psychosocial, and interpersonal questionnaires. Scientists who are participating members of ENIGMA Stroke Recovery can propose secondary analyses of the data and partner with other members to ask questions that may require a certain type of data or a specific behavioral outcome measure. ENIGMA Stroke Recovery provides a dynamic and growing platform for international collaborations across diverse topics. Furthermore, these collaborations are bound to extend beyond the datasets with prespecified MRI characteristics as additional methodologies emerge in the future to reconcile the variability across sequences and to extract the data relevant to stroke recovery. For instance, clinical MRI scans obtained during emergency hospitalization for acute ischemic stroke provide an abundance of data related to poststroke outcomes when analyzed using novel machine‐learning methods for stroke lesion segmentation on diffusion‐weighted imaging (Wu et al., 2019). Future artificial intelligence‐powered methodologies will allow researchers to optimize the analysis of these clinical scans and reconcile different types of data and approaches for lesion segmentation. This will further enable the growth and expansion of the stroke recovery database capabilities and data utilization, ranging from “real‐life” clinical scans to research‐protocol driven scans.
There are several current limitations of ENIGMA Stroke Recovery's methods, which we are working to address. First, there are still several manual steps that create a bottleneck in the processing pipelines and limit the speed at which we can perform analyses, specifically regarding the quality control of brain segmentations (from both FreeSurfer and the lesion masks). For instance, performing quality control of just the FreeSurfer subcortical regions requires the manual review of 16 regions per individual—or 16,000 regions for a dataset of N = 1,000. Performing quality control of the FreeSurfer cortical regions will require the review of an additional 68 regions per individual. We anticipate that with enough initial effort, we will be able to generate sufficient data to effectively train machine‐learning algorithms to perform these tasks; however, at present, we have limited the analyses to subcortical regions of interest and lesion analyses. Second, the current methods (e.g., FreeSurfer) work best on high‐resolution scans (e.g., dimensions of isotropic 1 mm3 voxels) with minimal distortions or noise. However, we have access to over 14,000 clinically acquired stroke MRI scans, which typically have much lower resolution (e.g., dimensions of 1 × 1 × 5 mm voxels); we thus need to develop robust methods to organize and analyze these scans. Although FreeSurfer does not work well on this type of data, other methods, such as those examining white matter hyperintensities, ventricular asymmetries, and total brain volume (Etherton et al., 2017; Rost et al., 2018), as well as lesion overlap with standardized template structures, may provide insights into the relationship between specific disease states indicated by these measures and mortality and recovery. Finally, the ENIGMA Stroke Recovery measures are limited to primarily cross‐sectional MRI and behavioral data, with a limited subset of longitudinal data. Questions regarding neural recovery, as well as the neural effects of specific interventions, are more difficult to address because the duration between two time points and the nature of the interventions administered varies widely across research sites. Even within one intervention category, such as brain–computer interfaces or robotic interventions, there are different eligibility criteria for entry into each study. Studies also vary in the total length of time and number of treatment sessions during the study, as well as the specific content of treatment (e.g., specific type of neurofeedback or robotic intervention provided).
While there are significant challenges to harmonizing multisite brain MRI and behavioral data after stroke, there are also many new opportunities for the development of novel methods related to automated quality control and lesion segmentation, robust low‐resolution data analysis, and harmonized longitudinal analysis that may ultimately improve our understanding of neural recovery after stroke. By openly sharing our protocols and methods, we hope to provide useful tools not only for ENIGMA Stroke Recovery but also for any stroke researchers wishing to collaborate with others in the field. Overall, we hope to improve the reproducibility of stroke research by reducing barriers to collaboration and to accelerate the innovation and discovery of more effective, personalized rehabilitation strategies for individuals after stroke.
CONFLICT OF INTEREST
Neda Jahanshad and Paul Thompson are MPIs of a research‐related grant from Biogen, Inc. for work unrelated to the contents of this manuscript. Steven C. Cramer is a consultant for Abbvie, Constant Therapeutics, MicroTransponder, Neurolutions, Regenera, SanBio, Stemedica, Fujifilm Toyama Chemical Co., Biogen, and TRCare.
APPENDIX 1. ENIGMA stroke recovery quality control (QC) protocol
Quality control (QC) is done according to the ENIGMA subcortical protocol for visual inspection. While other ENIGMA cohorts use outlier detection to identify failed subcortical segmentations, FreeSurfer segmentations of stroke participants merit additional caution. Large stroke lesions may interfere with the image registration process (Yang et al., 2016) that takes place in FreeSurfer and may cause poor subcortical segmentations that result in a skewed distribution of volume estimates. For ENIGMA Stroke Recovery, expert raters manually inspect subcortical segmentations for every participant. Screenshots of nine slices of the brain (three coronal, three axial, and three sagittal) are generated with bilateral segmentations overlaid onto the T1w MRI (left segmentation appears as transparent blue, right segmentation as transparent red) for every subcortical region for each subject. These screenshots are compiled into eight separate web‐based html files (one for each subcortical region). These html files are then used to inspect segmentations for quality.
Segmentations are scored as either PASS or FAIL depending on segmentation quality (for examples, see Figure A1). The rater documents the scores for every subcortical region in an excel spreadsheet. Image quality is taken into consideration in the QC process; excess motion or low resolution typically results in unreliable segmentations and are more likely to be scored as FAIL. Segmentations that underestimate the volume by not capturing the boundaries are scored as FAIL. FreeSurfer tends to overestimate segmentations (Schoemaker et al., 2016; Perlaki et al., 2017; Khlif et al., 2018), so underestimated segmentations are typically severe and easy to detect. Segmentations that overestimate by including neighboring regions are also scored as FAIL. An example of a common overestimation that includes a neighboring structure is a segmentation of the thalamus that reaches into the adjacent third ventricle to capture choroid plexus, the septum pellucidum, or the thalamus of the opposite hemisphere. Transparency of the segmentation during the QC process is crucial for detecting these kinds of overestimations. Segmentations the rater is unsure of are opened in FreeSurfer viewer tkm‐edit to inspect the entire volume more closely.
All three views (coronal, axial, and sagittal) are considered in determining the over‐ or under‐estimation in order to consider the segmentation as a three‐dimensional structure. The coronal and axial slices are primarily used for determining segmentation quality for all subcortical segmentations. The sagittal view is mostly used as a supplementary assessment of the quality of the caudate and the hippocampus. The angle of the head is also taken into consideration during QC‐ when the head appears to be tilted, bilateral segmentations in the coronal view may appear heavily asymmetrical. By drawing an imaginary horizontal line across the bilateral segmentations in one slice (a coronal slice, for example) and predicting how the segmentations should appear in another slice (an axial slice), the rater can determine if the asymmetry is due to poor segmentation or head placement.
Segmentations that overlap with lesions are scored as FAIL—the intensity of voxels containing lesions and gray matter make it difficult to differentiate gray matter boundaries. Lesions within close proximity to subcortical regions are at times mistaken for a boundary by the FreeSurfer algorithm so the segmentation will overestimate to capture part of the lesion—this is scored as FAIL. Segmentations that capture punctate lesions or perivascular spaces are also scored as FAIL.
No more than 50 participants are inspected for each subcortical region at a time to keep raters consistent. Training a new rater takes approximately 1 month. During the first week, the rater is trained on 25 pre‐identified participants with minimal pathology to gain familiarity with good quality segmentations. During the second and third weeks, they QC 50 additional participants with a range of stroke pathology, flagging segmentations they are unsure of for discussion. All 75 participants are then QCed a second time during the fourth week to ensure intra‐rater reliability. An expert rater will check the new rater's QC ratings for the next month until consistent inter‐rater reliability greater than 0.9 is achieved.
FIGURE A1.

Examples of quality control decisions. (A) An axial view of a participant's left (blue) and right (red) caudate that are both scored as PASS. (B) An axial view of an under segmented right caudate (FAIL). Lesion overlap in the caudate caused ambiguous gray matter boundaries, resulting in a failed segmentation. (C) FreeSurfer can fail to segment correctly even without the presence of a lesion. This is an axial view of under segmented left and right caudate segmentation that were scored as FAIL. (D) A coronal view of a participant's left and right caudate that are both scored as PASS. (E) A coronal view of a participant with a lesion that overlaps with the left caudate creates ambiguous boundaries. In this case, FreeSurfer overestimated the left caudate, mistaking the lesion as a gray matter boundary (FAIL)
APPENDIX 2. Language for data sharing in the ethics protocol and informed consent form
Example Protocol Language
Data Collection and Monitoring
The researchers intend to keep the de‐identified research data indefinitely. This de‐identified data may be shared with other researchers for future analysis or shared in archives or databases. Any data shared with other researchers will not include personal identifying information.
Example Informed Consent Form Language
Data Storage and Retention
Research data will be maintained in paper format in a secure location at the institution or electronically on secure, password‐protected computers and servers. Only authorized individuals will have access to it, and all electronic data will be de‐identified. The researchers intend to keep the de‐identified research data indefinitely. Other researchers may have access to the de‐identified data for future research, and the de‐identified data may be included in future repositories or archives for use by other researchers. Your information that is collected as part of this research will be used or distributed for future research studies without your additional informed consent. Any information that identifies you (such as your name) will be removed from your private information before being shared with others.
Liew S‐L, Zavaliangos‐Petropulu A, Jahanshad N, et al. The ENIGMA Stroke Recovery Working Group: Big data neuroimaging to study brain–behavior relationships after stroke. Hum Brain Mapp. 2022;43:129–148. 10.1002/hbm.25015
Funding information American Heart Association; AMORSA, Grant/Award Number: FKZ 16SV7754; Brain and Behavior Research Foundation, Grant/Award Numbers: NARSAD Young Investigator Grant, P&S Fund Investigator; BrightFocus Foundation, Grant/Award Number: A2019052S; Canadian Institutes of Health Research, Grant/Award Number: PJT‐153330; Canadian Partnership for Stroke Recovery; Center for Integrated Healthcare, U.S. Department of Veterans Affairs, Grant/Award Numbers: IO1RX001667, N‐1667; Deutsche Forschungsgemeinschaft, Grant/Award Numbers: LO795/22‐1, LO795/5‐1; Einstein Stiftung Berlin; Fortüne‐Program of the University of Tübingen, Grant/Award Number: 2422‐0‐1; H2020 European Research Council, Grant/Award Numbers: ERC‐2017‐STG‐759370, ERC‐STG‐802998; Health Research Council of New Zealand, Grant/Award Numbers: 09/164R, 11/270, 14/136; Italian Ministry of Health, Grant/Award Number: RC 15‐16‐17‐18‐19/A; Leon Levy Foundation Fellowship; Lone Star Stroke Research Consortium; Max‐Planck‐Gesellschaft; National Health and Medical Research Council, Grant/Award Numbers: 1020526, 1088449, 1094974; National Institutes of Health, Grant/Award Numbers: 5P2CHD086851, HD065438, HD086844, K01HD091283, K23NS088107, P20 GM109040, P2CHD06570, R00HD091375, R01AG059874, R01HD065438, R01HD095137, R01MH117601, R01NR015591, R01NS076348, R01NS082285, R01NS086905, R01NS090677, R01NS115845, R21HD067906, R56‐NS100528, U19NS115388, U54EB020403; National Key Research and Development Program of China, Grant/Award Number: 2018YFC1314300; Norges Forskningsråd, Grant/Award Number: 249795; Norwegian ExtraFoundation for Health and Rehabilitation, Grant/Award Number: 2015/FO5146; South‐Eastern Norway Regional Health Authority, Grant/Award Number: 2018076; Stroke Association, Grant/Award Number: TSA 2017/04; the Bundesministerium für Bildung und Forschung BMBF MOTORBIC, Grant/Award Number: FKZ 13GW0053
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
REFERENCES
- Acheson, A. , Wijtenburg, S. A. , Rowland, L. M. , Winkler, A. , Mathias, C. W. , Hong, L. E. , … McGuire, S. A. (2017). Reproducibility of tract‐based white matter microstructural measures using the ENIGMA‐DTI protocol. Brain and Behavior, 7(2), e00615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikari, B. M. , Dukart, J. , Hipp, J. F. , Forsyth, A. , McMillan, R. , Muthukumaraswamy, S. D. , … Jahandshad, N. (2019). Effects of ketamine and midazolam on resting state connectivity and comparison with ENIGMA connectivity deficit patterns in schizophrenia. Human Brain Mapping, 41(3), 767–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikari, B. M. , Jahanshad, N. , Shukla, D. , Glahn, D. C. , Blangero, J. , Reynolds, R. C. , … Novikov, D. S. (2018). Heritability estimates on resting state fMRI data using ENIGMA analysis pipeline. Pacific Symposium on Biocomputing, 23, 307–318. [PMC free article] [PubMed] [Google Scholar]
- Adhikari, B. M. , Jahanshad, N. , Shukla, D. , Turner, J. , Grotegerd, D. , Dannlowski, U. , … Krug, A. (2019). A resting state fMRI analysis pipeline for pooling inference across diverse cohorts: An ENIGMA rs‐fMRI protocol. Brain Imaging and Behavior, 13(5), 1453–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfaro‐Almagro, F. , Jenkinson, M. , Bangerter, N. K. , Andersson, J. L. , Griffanti, L. , Douaud, G. , … Vallee, E. (2018). Image processing and quality control for the first 10,000 brain imaging datasets from UKBiobank. NeuroImage, 166, 400–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appelros, P. , Stegmayr, B. , & Terént, A. (2009). Sex differences in stroke epidemiology: A systematic review. Stroke, 40(4), 1082–1090. [DOI] [PubMed] [Google Scholar]
- Arsava, E. , Rahman, R. , Rosand, J. , Lu, J. , Smith, E. , Rost, N. , … Koroshetz, W. (2009). Severity of leukoaraiosis correlates with clinical outcome after ischemic stroke. Neurology, 72(16), 1403–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auriat, A. M. , Ferris, J. K. , Peters, S. , Ramirez, J. , Black, S. E. , Jacova, C. , & Boyd, L. A. (2019). The impact of covert lacunar infarcts and white matter hyperintensities on cognitive and motor outcomes after stroke. Journal of Stroke and Cerebrovascular Diseases, 28(2), 381–388. [DOI] [PubMed] [Google Scholar]
- Benjamin, E. J. , Virani, S. S. , Callaway, C. W. , Chamberlain, A. M. , Chang, A. R. , Cheng, S. , … Deo, R. (2018). Heart disease and stroke statistics‐2018 update: A report from the American Heart Association. Circulation, 137(12), e67. [DOI] [PubMed] [Google Scholar]
- Bernhardt, J. , Borschmann, K. , Boyd, L. , Thomas Carmichael, S. , Corbett, D. , Cramer, S. C. , … Saposnik, G. (2016). Moving rehabilitation research forward: Developing consensus statements for rehabilitation and recovery research. International Journal of Stroke, 11(4), 454–458. [DOI] [PubMed] [Google Scholar]
- Bernhardt, J. , Hayward, K. S. , Dancause, N. , Lannin, N. A. , Ward, N. S. , Nudo, R. J. , … Jones, T. A. (2019). A stroke recovery trial development framework: Consensus‐based core recommendations from the second stroke recovery and rehabilitation roundtable. International Journal of Stroke, 14(8), 792–802. [DOI] [PubMed] [Google Scholar]
- Bernhardt, J. , Hayward, K. S. , Kwakkel, G. , Ward, N. S. , Wolf, S. L. , Borschmann, K. , … Corbett, D. (2017). Agreed definitions and a shared vision for new standards in stroke recovery research: The Stroke Recovery And Rehabilitation Roundtable taskforce. International Journal of Stroke, 12(5), 444–450. [DOI] [PubMed] [Google Scholar]
- Beuscher, V. D. , Kuramatsu, J. B. , Gerner, S. T. , Köhn, J. , Lücking, H. , Kloska, S. P. , & Huttner, H. B. (2017). Functional long‐term outcome after left‐versus right‐sided intracerebral hemorrhage. Cerebrovascular Diseases, 43(3–4), 117–123. [DOI] [PubMed] [Google Scholar]
- Bing, Y. , Garcia‐Gonzalez, D. , Voets, N. , & Jérusalem, A. (2020). Medical imaging based in silico head model for ischaemic stroke simulation. Journal of the Mechanical Behavior of Biomedical Materials, 101, 103442. [DOI] [PubMed] [Google Scholar]
- Biswal, B. , Yetkin, F. Z. , Haughton, V. M. , & Hyde, J. S. (1995). Functional connectivity in the motor cortex of resting human brain using echo‐planar MRI. Magnetic Resonance in Medicine, 34(4), 537–541. [DOI] [PubMed] [Google Scholar]
- Biswal, B. B. , Mennes, M. , Zuo, X.‐N. , Gohel, S. , Kelly, C. , Smith, S. M. , … Colcombe, S. (2010). Toward discovery science of human brain function. Proceedings of the National Academy of Sciences, 107(10), 4734–4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boedhoe, P. S. , Heymans, M. W. , Schmaal, L. , Abe, Y. , Alonso, P. , Ameis, S. H. , … Benedetti, F. (2019). An empirical comparison of meta‐and mega‐analysis with data from the ENIGMA obsessive–compulsive Disorder Working Group. Frontiers in Neuroinformatics, 12, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boedhoe, P. S. , Schmaal, L. , Abe, Y. , Ameis, S. H. , Arnold, P. D. , Batistuzzo, M. C. , … Bose, A. (2016). Distinct subcortical volume alterations in pediatric and adult OCD: A worldwide meta‐and mega‐analysis. American Journal of Psychiatry, 174(1), 60–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boedhoe, P. S. , van Rooij, D. , Hoogman, M. , Thompson, P. M. , Stein, D. J. , Buitelaar, J. K. , … van den Heuvel Odile A. (2019). Subcortical brain volume, regional cortical thickness and cortical surface area across attention‐deficit/hyperactivity disorder (ADHD), autism spectrum disorder (ASD), and obsessive–compulsive disorder (OCD): findings from the ENIGMA‐ADHD,‐ASD, and‐OCD working groups.
- Bohannon, R , Smith, M. (1987). Inter rater reliability of a modified Ashworth Scale of muscle spasticity. Physical Therapy, 67, 206–307. [DOI] [PubMed] [Google Scholar]
- Boyd, L. A. , Hayward, K. S. , Ward, N. S. , Stinear, C. M. , Rosso, C. , Fisher, R. J. , … Carey, L. M. (2017). Biomarkers of stroke recovery: Consensus‐based core recommendations from the stroke recovery and rehabilitation roundtable. Neurorehabilitation and Neural Repair, 31(10–11), 864–876. [DOI] [PubMed] [Google Scholar]
- Brooks, R. , & Group, E. (1996). EuroQol: The current state of play. Health Policy, 37(1), 53–72. [DOI] [PubMed] [Google Scholar]
- Brott, T. , Adams, H. P. , Olinger, C. P. , Marler, J. R. , Barsan, W. G. , Biller, J. , … Hertzberg, V. (1989). Measurements of acute cerebral infarction: A clinical examination scale. Stroke, 20(7), 864–870. [DOI] [PubMed] [Google Scholar]
- Butland, R. , Pang, J. , Gross, E. , Woodcock, A. , & Geddes, D. (1982). Two‐, six‐, and 12‐minute walking tests in respiratory disease. British Medical Journal (Clinical Research Ed.), 284(6329), 1607–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, E. Y. , Chang, E. , Cragg, S. , & Cramer, S. C. (2013). Predictors of gains during inpatient rehabilitation in patients with stroke: A review. Critical Review in Physical and Rehabilitation Medicine, 25(3–4), 203–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, T. , & Guestrin, C. (2016). Xgboost: A scalable tree boosting system. Paper presented at the Proceedings of the 22nd acm sigkdd international conference on knowledge discovery and data mining.
- Collins, F. S. , & Varmus, H. (2015). A new initiative on precision medicine. New England Journal of Medicine, 372(9), 793–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan, R. S. , Ségonne, F. , Fischl, B. , Quinn, B. T. , Dickerson, B. C. , Blacker, D. , … Hyman, B. T. (2006). An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. NeuroImage, 31(3), 968–980. [DOI] [PubMed] [Google Scholar]
- Dimyan, M. A. , & Cohen, L. G. (2011). Neuroplasticity in the context of motor rehabilitation after stroke. Nature Reviews Neurology, 7(2), 76–85. 10.1038/nrneurol.2010.200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan Millar, J. , van Wijck, F. , Pollock, A. , & Ali, M. (2019). Outcome measures in post‐stroke arm rehabilitation trials: Do existing measures capture outcomes that are important to stroke survivors, carers, and clinicians? Clinical Rehabilitation, 33(4), 737–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban, O. , Markiewicz, C. J. , Blair, R. W. , Moodie, C. A. , Isik, A. I. , Erramuzpe, A. , … Snyder, M. (2019). fMRIPrep: A robust preprocessing pipeline for functional MRI. Nature Methods, 16(1), 111–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etherton, M. R. , Wu, O. , Cougo, P. , Giese, A.‐K. , Cloonan, L. , Fitzpatrick, K. M. , … Lauer, A. (2017). Integrity of normal‐appearing white matter and functional outcomes after acute ischemic stroke. Neurology, 88(18), 1701–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, W. , Wang, J. , Chhatbar, P. Y. , Doughty, C. , Landsittel, D. , Lioutas, V. A. , … Schlaug, G. (2015). Corticospinal tract lesion load: An imaging biomarker for stroke motor outcomes. Annals of Neurology, 78(6), 860–870. 10.1002/ana.24510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl, B. , & Dale, A. M. (2000). Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proceedings of the National Academy of Sciences of the United States of America, 97, 11050–11055. 10.1073/pnas.200033797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl, B. , Salat, D. H. , Busa, E. , Albert, M. , Dieterich, M. , Haselgrove, C. , … Klaveness, S. (2002). Whole brain segmentation: Automated labeling of neuroanatomical structures in the human brain. Neuron, 33(3), 341–355. [DOI] [PubMed] [Google Scholar]
- Folstein, M. F. , Robins, L. N. , & Helzer, J. E. (1983). The mini‐mental state examination. Archives of General Psychiatry, 40(7), 812–812. [DOI] [PubMed] [Google Scholar]
- Fugl‐Meyer, A. R. , Jaasko, L. , Leyman, I. , Olsson, S. , & Steglind, S. (1975). The post‐stroke hemiplegic patient. 1. A method for evaluation of physical performance. Scandinavian Journal of Rehabilitation Medicine, 7(1), 13–31. [PubMed] [Google Scholar]
- Gorgolewski, K. J. , Alfaro‐Almagro, F. , Auer, T. , Bellec, P. , Capotă, M. , Chakravarty, M. M. , … Devenyi, G. A. (2017). BIDS apps: Improving ease of use, accessibility, and reproducibility of neuroimaging data analysis methods. PLoS Computational Biology, 13(3), e1005209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgolewski, K. J. , Auer, T. , Calhoun, V. D. , Craddock, R. C. , Das, S. , Duff, E. P. , … Halchenko, Y. O. (2016). The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Scientific Data, 3, 160044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasby, K. L. , Jahanshad, N. , Painter, J. N. , Colodro‐Conde, L. , Bralten, J. , Hibar, D. P. , … McMahon, M. A. B. (2020). The genetic architecture of the human cerebral cortex. Science, 367(6484), eaay6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis, J. C. , Allendorfer, J. B. , & Szaflarski, J. P. (2016). Voxel‐based Gaussian naïve Bayes classification of ischemic stroke lesions in individual T1‐weighted MRI scans. Journal of Neuroscience Methods, 257, 97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, X. , Jovicich, J. , Salat, D. , van der Kouwe, A. , Quinn, B. , Czanner, S. , … Killiany, R. (2006). Reliability of MRI‐derived measurements of human cerebral cortical thickness: The effects of field strength, scanner upgrade and manufacturer. NeuroImage, 32(1), 180–194. [DOI] [PubMed] [Google Scholar]
- Harris, P. A. , Taylor, R. , Thielke, R. , Payne, J. , Gonzalez, N. , & Conde, J. G. (2009). Research electronic data capture (REDCap)—A metadata‐driven methodology and workflow process for providing translational research informatics support. Journal of Biomedical Informatics, 42(2), 377–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibar, D. P. , Stein, J. L. , Renteria, M. E. , Arias‐Vasquez, A. , Desrivières, S. , Jahanshad, N. , … Andersson, M. (2015). Common genetic variants influence human subcortical brain structures. Nature, 520(7546), 224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibar, D. P. , Westlye, L. T. , van Erp, T. G. , Rasmussen, J. , Leonardo, C. D. , Faskowitz, J. , … Agartz, I. (2016). Subcortical volumetric abnormalities in bipolar disorder. Molecular Psychiatry, 21(12), 1710–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, K. L. , Kim, H. , & Liew, S. L. (2019). A comparison of automated lesion segmentation approaches for chronic stroke T1‐weighted MRI data. Human Brain Mapping, 40(16), 4669–4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, K. L. , Kumar, A. , Zavaliangos‐Petropulu, A. , Cramer, S. C. , & Liew, S.‐L. (2018). Pipeline for analyzing lesions after stroke (PALS). Frontiers in Neuroinformatics, 12, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahanshad, N. , Kochunov, P. V. , Sprooten, E. , Mandl, R. C. , Nichols, T. E. , Almasy, L. , … de Zubicaray, G. I. (2013). Multi‐site genetic analysis of diffusion images and voxelwise heritability analysis: A pilot project of the ENIGMA–DTI working group. NeuroImage, 81, 455–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, C. O. , Nguyen, M. , Roth, G. A. , Nichols, E. , Alam, T. , Abate, D. , … Abu‐Rmeileh, N. M. (2019). Global, regional, and national burden of stroke, 1990–2016: A systematic analysis for the global burden of disease study 2016. The Lancet Neurology, 18(5), 439–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongbloed, L. Y. N. (1986). Prediction of function after stroke: A critical review. Stroke, 17, 765–776. [DOI] [PubMed] [Google Scholar]
- Kégl, B. , Boucaud, A. , Cherti, M. , Kazakci, A. , Gramfort, A. , Lemaitre, G. , … Marini, C. (2018). The RAMP framework: from reproducibility to transparency in the design and optimization of scientific workflows.
- Keshavan, A. , Yeatman, J. , & Rokem, A. (2018). Combining citizen science and deep learning to amplify expertise in neuroimaging. [DOI] [PMC free article] [PubMed]
- Khlif, M. S. , Egorova, N. , Werden, E. , Redolfi, A. , Bocardi, M. , DeCarli, C. S. , … Bird, L. (2018). A comparison of automated segmentation and manual tracing in estimating hippocampal volume in ischemic stroke and healthy control participants. NeuroImage: Clinical, 21, 101581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, B. , & Winstein, C. (2017). Can neurological biomarkers of brain impairment be used to predict poststroke motor recovery? A systematic review. Neurorehabilitation and Neural Repair, 31(1), 3–24. [DOI] [PubMed] [Google Scholar]
- Kim, H. , Irimia, A. , Hobel, S. M. , Esquivel Castelo‐Blanco, R. I. , Duffy, B. , Zhao, L. , … Law, M. (2019). LONI QC system: A semi‐automated, web‐based and freely‐available environment for the comprehensive quality control of neuroimaging data. Frontiers in Neuroinformatics, 13, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klann, J. G. , Joss, M. A. , Embree, K. , & Murphy, S. N. (2019). Data model harmonization for the All Of Us Research Program: Transforming i2b2 data into the OMOP common data model. PLoS One, 14(2), e0212463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwakkel, G. , Lannin, N. A. , Borschmann, K. , English, C. , Ali, M. , Churilov, L. , … Wolf, S. L. (2017). Standardized measurement of sensorimotor recovery in stroke trials: Consensus‐based core recommendations from the Stroke Recovery And Rehabilitation Roundtable. Neurorehabilitation and Neural Repair, 31(9), 784–792. [DOI] [PubMed] [Google Scholar]
- Li, Q. , Pardoe, H. , Lichter, R. , Werden, E. , Raffelt, A. , Cumming, T. , & Brodtmann, A. (2015). Cortical thickness estimation in longitudinal stroke studies: A comparison of 3 measurement methods. NeuroImage: Clinical, 8, 526–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew, S.‐L. , Anglin, J. M. , Banks, N. W. , Sondag, M. , Ito, K. L. , Kim, H. , … Khoshab, N. (2018). A large, open source dataset of stroke anatomical brain images and manual lesion segmentations. Scientific Data, 5, 180011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew, S.‐L. , Kumar, A. , Suri, A. , Notter, M. P. , Ito, K. , Raamana, P. , & Keshavan, A. (2019). Braindrles: A crowd‐sourcing tool for stroke lesion segmentation quality control. Paper presented at the Society for Neuroscience, Chicago, IL.
- Lindsay, M. P. , Norrving, B. , Sacco, R. L. , Brainin, M. , Hacke, W. , Martins, S. , … Feigin, V. (2019). World stroke organization (WSO): Global stroke fact sheet 2019. London, England: SAGE Publications. [DOI] [PubMed] [Google Scholar]
- Lyle, R. C. (1981). A performance test for assessment of upper limb function in physical rehabilitation treatment and research. International Journal of Rehabilitation Research, 4(4), 483–492. [DOI] [PubMed] [Google Scholar]
- Macciocchi, S. N. , Diamond, P. T. , Alves, W. M. , & Mertz, T. (1998). Ischemic stroke: Relation of age, lesion location, and initial neurologic deficit to functional outcome. Archives of Physical Medicine and Rehabilitation, 79(10), 1255–1257. [DOI] [PubMed] [Google Scholar]
- Maier, O. , Menze, B. H. , von der Gablentz, J. , Häni, L. , Heinrich, M. P. , Liebrand, M. , … Reyes, M. (2017). ISLES 2015 ‐ A public evaluation benchmark for ischemic stroke lesion segmentation from multispectral MRI. Medical Image Analysis, 35, 250–269. 10.1016/j.media.2016.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald, M. W. , Black, S. E. , Copland, D. A. , Corbett, D. , Dijkhuizen, R. M. , Farr, T. D. , … Leff, A. P. (2019). Cognition in stroke rehabilitation and recovery research: Consensus‐based core recommendations from the second stroke recovery and rehabilitation roundtable. International Journal of Stroke, 14(8), 774–782. [DOI] [PubMed] [Google Scholar]
- Meyer, M. J. , Pereira, S. , McClure, A. , Teasell, R. , Thind, A. , Koval, J. , … Speechley, M. (2015). A systematic review of studies reporting multivariable models to predict functional outcomes after post‐stroke inpatient rehabilitation. Disability and Rehabilitation, 37(15), 1316–1323. [DOI] [PubMed] [Google Scholar]
- Nasreddine, Z. S. , Phillips, N. A. , Bédirian, V. , Charbonneau, S. , Whitehead, V. , Collin, I. , … Chertkow, H. (2005). The Montreal Cognitive Assessment, MoCA: A brief screening tool for mild cognitive impairment. Journal of the American Geriatrics Society, 53(4), 695–699. [DOI] [PubMed] [Google Scholar]
- Nichols, T. E. , Das, S. , Eickhoff, S. B. , Evans, A. C. , Glatard, T. , Hanke, M. , … Poline, J.‐B. (2017). Best practices in data analysis and sharing in neuroimaging using MRI. Nature Neuroscience, 20(3), 299–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens, M. (2006). The definitive guide to SQLite. Berkeley, CA: Apress. [Google Scholar]
- Petrov, D. , Gutman, B. A. , Kuznetsov, E. , Ching, C. R. , Alpert, K. , Zavaliangos‐Petropulu, A. , … Wang, L. (2018). Deep Learning for Quality Control of Subcortical Brain 3D Shape Models. Paper presented at the International Workshop on Shape in Medical Imaging.
- Petrov, D. , Gutman, B. A. , Yu, S.‐H. J. , Alpert, K. , Zavaliangos‐Petropulu, A. , Isaev, D. , … Schmaal, L. (2017). Machine learning for large‐scale quality control of 3d shape models in neuroimaging. Paper presented at the International Workshop on Machine Learning in Medical Imaging. [DOI] [PMC free article] [PubMed]
- Perlaki, G. , Horvath, R. , Nagy, S. A. , Bogner, P. , Doczi, T. , Janszky, J. , Pizzagalli, F. , Auzias, G. , Yang, Q. , Mathias, S. R. , Faskowitz, J. , Boyd, J. , … de Zubicaray, G. I. (2019). The reliability and heritability of cortical folds and their genetic correlations across hemispheres. [DOI] [PMC free article] [PubMed]
- Pustina, D. , Coslett, H. B. , Turkeltaub, P. E. , Tustison, N. , Schwartz, M. F. , & Avants, B. (2016). Automated segmentation of chronic stroke lesions using LINDA: Lesion identification with neighborhood data analysis. Human Brain Mapping, 37, 1405–1421. 10.1002/hbm.23110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez, J. , Gibson, E. , Quddus, A. , Lobaugh, N. J. , Feinstein, A. , Levine, B. , … Black, S. E. (2011). Lesion explorer: A comprehensive segmentation and parcellation package to obtain regional volumetrics for subcortical hyperintensities and intracranial tissue. NeuroImage, 54(2), 963–973. [DOI] [PubMed] [Google Scholar]
- Ramos‐Murguialday, A. , Broetz, D. , Rea, M. , Läer, L. , Yilmaz, Ö. , Brasil, F. L. , … Vyziotis, A. (2013). Brain–machine interface in chronic stroke rehabilitation: A controlled study. Annals of Neurology, 74(1), 100–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathore, S. S. , Hinn, A. R. , Cooper, L. S. , Tyroler, H. A. , & Rosamond, W. D. (2002). Characterization of incident stroke signs and symptoms: Findings from the atherosclerosis risk in communities study. Stroke, 33(11), 2718–2721. [DOI] [PubMed] [Google Scholar]
- Riley, J. D. , Le, V. , Der‐Yeghiaian, L. , See, J. , Newton, J. M. , Ward, N. S. , & Cramer, S. C. (2011). Anatomy of stroke injury predicts gains from therapy. Stroke, 42, 421–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost, N. S. , Cougo, P. , Lorenzano, S. , Li, H. , Cloonan, L. , Bouts, M. J. , … Musolino, P. L. (2018). Diffuse microvascular dysfunction and loss of white matter integrity predict poor outcomes in patients with acute ischemic stroke. Journal of Cerebral Blood Flow & Metabolism, 38(1), 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmaal, L. , Veltman, D. J. , van Erp, T. G. , Sämann, P. , Frodl, T. , Jahanshad, N. , … Niessen, W. (2016). Subcortical brain alterations in major depressive disorder: Findings from the ENIGMA major depressive disorder working group. Molecular Psychiatry, 21(6), 806–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoemaker, D. , Buss, C. , Head, K. , Sandman, C. A. , Davis, E. P. , Chakravarty, M. M. , … See, J. , Dodakian, L. , Chou, C. , Chan, V. , McKenzie, A. , Reinkensmeyer, D. J. , & Cramer, S. C. (2013). A standardized approach to the Fugl‐Meyer assessment and its implications for clinical trials. Neurorehabilitation and Neural Repair, 27(8), 732–741. [DOI] [PubMed] [Google Scholar]
- Seghier, M. L. , Ramlackhansingh, A. , Crinion, J. , Leff, A. P. , & Price, C. J. (2008). Lesion identification using unified segmentation‐normalisation models and fuzzy clustering. NeuroImage, 41(4), 1253–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharique, M. , Pundarikaksha, B. U. , Sridar, P. , Krishnan, R. R. , & Krishnakumar, R. (2019). Parallel capsule net for ischemic stroke segmentation.
- Shephard, R. J. , Allen, C. , Benade, A. , Davies, C. , Di Prampero, P. , Hedman, R. , … Simmons, R. (1968). The maximum oxygen intake: An international reference standard of cardio‐respiratory fitness. Bulletin of the World Health Organization, 38(5), 757–764. [PMC free article] [PubMed] [Google Scholar]
- Smit, D. (2020). Linking electrophysiological brain activity to neurological and psychiatric liability genes: Large‐scale collaborative studies by the ENIGMA‐EEG group. [DOI] [PMC free article] [PubMed]
- Smit, D. , Malone, S. , Burwell, S. , de Geus, E. , Hibar, D. , Thompson, P. , … Porjesz, B. (2016). ENIGMA‐EEG: GWAS of brain function. Paper presented at the Behavior genetics.
- Smith, S. M. , Jenkinson, M. , Woolrich, M. W. , Beckmann, C. F. , Behrens, T. E. , Johansen‐Berg, H. , … Flitney, D. E. (2004). Advances in functional and structural MR image analysis and implementation as FSL. NeuroImage, 23, S208–S219. [DOI] [PubMed] [Google Scholar]
- Smith, S. M. , Johansen‐Berg, H. , Jenkinson, M. , Rueckert, D. , Nichols, T. E. , Miller, K. L. , … Bartsch, A. J. (2007). Acquisition and voxelwise analysis of multi‐subject diffusion data with tract‐based spatial statistics. Nature Protocols, 2(3), 499–503. [DOI] [PubMed] [Google Scholar]
- Stein, J. L. , Medland, S. E. , Vasquez, A. A. , Hibar, D. P. , Senstad, R. E. , Winkler, A. M. , … Bergmann, Ø. (2012). Identification of common variants associated with human hippocampal and intracranial volumes. Nature Genetics, 44(5), 552–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, J. C. , Gordon, J. , & Winstein, C. J. (2014). Control of reach extent with the paretic and nonparetic arms after unilateral sensorimotor stroke II: Planning and adjustments to control movement distance. Experimental Brain Research, 232(11), 3431–3443. [DOI] [PubMed] [Google Scholar]
- Stinear, C. M. , Barber, P. A. , Smale, P. R. , Coxon, J. P. , Fleming, M. K. , & Byblow, W. D. (2006). Functional potential in chronic stroke patients depends on corticospinal tract integrity. Brain, 130(1), 170–180. [DOI] [PubMed] [Google Scholar]
- Stinear, C. M. , Byblow, W. D. , Ackerley, S. J. , Smith, M. C. , Borges, V. M. , & Barber, P. A. (2017). PREP2: A biomarker‐based algorithm for predicting upper limb function after stroke. Annals of Clinical Translational Neurology, 4(11), 811–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, P. (2019). Enigma, big data, and neuroimaging genetics in 50,000 people from 35 countries: Challenges and lessons learned. European Neuropsychopharmacology, 29, S769–S770. [Google Scholar]
- Thompson, P. , Jahanshad, N. , Ching, C. R. , Salminen, L. E. , Thomopoulos, S. I. , Bright, J. , … Bruin, W. B. (2019). ENIGMA and global neuroscience: A decade of large‐scale studies of the brain in health and disease across more than 40 countries. Translational Psychiatry, 10(1), 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, P. M. , Jahanshad, N. , Ching, C. R. K. , Salminen, L. E. , Thomopoulos, S. I. , Bright, J. , … ENIGMA Consortium . (2020). ENIGMA and global neuroscience: A decade of large‐scale studies of the brain in health and disease across more than 40 countries. Translational Psychiatry, 10(1), 100. 10.1038/s41398-020-0705-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uswatte, G. , Taub, E. , Morris, D. , Vignolo, M. , & McCulloch, K. (2005). Reliability and validity of the upper‐extremity motor activity Log‐14 for measuring real‐world arm use. Stroke, 36(11), 2493–2496. [DOI] [PubMed] [Google Scholar]
- Van Erp, T. G. , Walton, E. , Hibar, D. P. , Schmaal, L. , Jiang, W. , Glahn, D. C. , … Hashimoto, R. (2018). Cortical brain abnormalities in 4474 individuals with schizophrenia and 5,098 control subjects via the enhancing Neuro imaging genetics through meta analysis (ENIGMA) consortium. Biological Psychiatry, 84(9), 644–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Swieten, J. , Koudstaal, P. , Visser, M. , Schouten, H. , & Van Gijn, J. (1988). Interobserver agreement for the assessment of handicap in stroke patients. Stroke, 19(5), 604–607. [DOI] [PubMed] [Google Scholar]
- Veer, I. , Waller, L. , Lett, T. , Erk, S. , & Walter, H. (2019). ENIGMA task‐based fMRI: A workgroup studying the genetic basis of task‐evoked brain activity. Paper presented at the Abstract presented at the 25th Organization of Human Brain Mapping Annual Meeting.
- Wang, Y. , Juliano, J. M. , Liew, S.‐L. , McKinney, A. M. , & Payabvash, S. (2019). Stroke atlas of the brain: Voxel‐wise density‐based clustering of infarct lesions topographic distribution. NeuroImage: Clinical, 24, 101981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, N. S. (2017). Restoring brain function after stroke—Bridging the gap between animals and humans. Nature Reviews Neurology, 13(4), 244–255. [DOI] [PubMed] [Google Scholar]
- Ween, J. E. , Alexander, M. P. , D'Esposito, M. , & Roberts, M. (1996). Factors predictive of stroke outcome in a rehabilitation setting. Neurology, 47(2), 388–392. [DOI] [PubMed] [Google Scholar]
- Whelan, C. D. , Altmann, A. , Botía, J. A. , Jahanshad, N. , Hibar, D. P. , Absil, J. , … Bartolini, E. (2018). Structural brain abnormalities in the common epilepsies assessed in a worldwide ENIGMA study. Brain, 141(2), 391–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf, S. L. , Catlin, P. A. , Ellis, M. , Archer, A. L. , Morgan, B. , Piacentino, A. (2001). Assessing Wolf motor function test as outcome measure for research in patients after stroke. Stroke, 32(7), 1635–1639. [DOI] [PubMed] [Google Scholar]
- World Health Organization . (2001). International classification of functioning, disability and health: ICF. Geneva: World Health Organization. [Google Scholar]
- Wu, O. , Cloonan, L. , Mocking, S. J. , Bouts, M. J. , Copen, W. A. , Cougo‐Pinto, P. T. , … Rosand, J. (2015). Role of acute lesion topography in initial ischemic stroke severity and long‐term functional outcomes. Stroke, 46(9), 2438–2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, O. , Winzeck, S. , Giese, A.‐K. , Hancock, B. L. , Etherton, M. R. , Bouts, M. J. , … Mocking, S. J. (2019). Big data approaches to phenotyping acute ischemic stroke using automated lesion segmentation of multi‐center magnetic resonance imaging data. Stroke, 50, 1734–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X. , Han, X. , Park, E. , Aylward, S. , Kwitt, R. , Niethammer, M. (2016). Registration of pathological images in Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics). Springer Verlag, 97–107. [Google Scholar]
- Zavaliangos‐Petropulu, A. , Bigjahan, B. , Borich, M. R. , Brown, T. R. , Buetefisch, C. M. , Byblow, W. D. , … Liew, S. L. (2019). Ipsilesional hippocampal volume is directly associated with motor performance in chronic stroke patients: An ENIGMA Stroke Recovery Analysis. Paper presented at the Society for Neuroscience, Chicago, IL.
- Zavaliangos‐Petropulu, A. , Nir, T. M. , Thomopoulos, S. I. , Reid, R. I. , Bernstein, M. A. , Borowski, B. , … Thompson, P. M. (2019). Diffusion MRI indices and their relation to cognitive impairment in brain aging: The updated multi‐protocol approach in ADNI3. Frontiers in Neuroinformatics, 13, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, R. J. , Hondori, H. , Khademi, M. , Cassidy, J. M. , Wu, K. M. , Yang, D. Z. , … McKenzie, A. (2018). Predicting gains with visuospatial training after stroke using an EEG measure of frontoparietal circuit function. Frontiers in Neurology, 9, 597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, A. H. , Moyer, D. C. , Nir, T. M. , Thompson, P. M. , & Jahanshad, N. (2018). Challenges and Opportunities in dMRI Data Harmonization. Paper presented at the International Conference on Medical Image Computing and Computer‐Assisted Intervention.
- Zigmond, A. S. , & Snaith, R. P. (1983). The hospital anxiety and depression scale. Acta Psychiatrica Scandinavica, 67(6), 361–370. [DOI] [PubMed] [Google Scholar]
- Zugman, A. , Harrewijn, A. , Cardinale, E. , Zwiebel, H. , Freitag, G. F. , Werwath, K. E. , … Hilbert, K. (2020). Mega‐analysis methods in ENIGMA: The experience of the generalized anxiety disorder working group. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


