Abstract
Blood vessels are ubiquitously distributed within all tissues of the body and perform diverse functions. Thus, derivation of mature vascular endothelial cells, which line blood vessel lumens, from human pluripotent stem cells is crucial for a multitude of tissue engineering and regeneration applications. In vivo, primordial endothelial cells are derived from the mesodermal lineage and are specified toward specific subtypes, including arterial, venous, capillary, hemogenic and lymphatic. Hemogenic endothelial cells are of particular interest because, during development, they give rise to hematopoietic stem and progenitor cells, which then generate all blood lineages throughout life. Thus, creating a system to generate hemogenic endothelial cells in vitro would provide an opportunity to study endothelial-to-hematopoietic transition, and may lead to ex vivo production of human blood products and reduced reliance on human donors. While several protocols exist for the derivation of progenitor and primordial endothelial cells, generation of well-characterized hemogenic endothelial cells from human stem cells has not been described. Here, a method for the derivation of hemogenic endothelial cells from human embryonic stem cells in approximately one week is presented: a differentiation protocol with primitive streak cells formed in response to GSK3β inhibitor (CHIR99021), then mesoderm lineage induction mediated by bFGF, followed by primordial endothelial cell development promoted by BMP4 and VEGF-A, and finally hemogenic endothelial cell specification induced by retinoic acid. This protocol yields a well-defined population of hemogenic endothelial cells that can be used to further understand their molecular regulation and endothelial-to-hematopoietic transition, which has the potential to be applied to downstream therapeutic applications.
Keywords: Pluripotent human stem cells, human endothelial cells, hemogenic endothelial cells, arterial endothelial cells, cell culture protocol, directed differentiation
SUMMARY:
Presented here is a simple protocol for the directed differentiation of hemogenic endothelial cells from human pluripotent stem cells in approximately one week.
INTRODUCTION:
Endothelial cells (ECs) are a heterogeneous population of cells that perform multiple functions throughout the human body and in engineered tissues. In addition to supporting and regulating other cell types (i.e., cardiomyocytes1, osteoblastic cells2), these functions include forming a selective barrier between blood and tissues and assisting in tissue formation3. Differentiation of mature ECs during normal development requires diverse signaling pathways. Primordial ECs are derived from mesoderm progenitors, and are then specified toward mature arterial, venous, capillary and lymphatic phenotypes4. Additionally, a small subset of ECs in the extraembryonic yolk sac and embryonic Aorta-Gonad-Mesonephros (AGM) region are also specified to become hemogenic ECs, which give rise to hematopoietic stem and progenitor cells (HSPCs) that migrate to the fetal liver and fetal bone marrow, where they remain postnatally and generate all blood cell types throughout life4. The diverse range of EC phenotypes is essential for all tissue development and maintenance.
Thus, ECs and their derivatives are critical components of studies aimed at modeling, and elucidating mechanisms of, human development and/or disease, as well as regenerative medicine and tissue engineering applications5–8. However, the main limitation for these types of studies is the lack of availability of primary human ECs in the quantity required. It has been estimated that a minimum of 3 × 108 ECs would be required for the majority of therapeutic applications6. To solve this problem, the use of human embryonic stem cells (hESCs) and human induced pluripotent stem cells (hiPSCs) has been proposed due to their diverse lineage potential and their ability to generate large numbers of progeny6,9.
Indeed, the usefulness of cells derived from hESCs or hiPSCs has been demonstrated in multiple studies focused on disease modeling and drug screening10–12. Organ-on-a-Chip (OOC) technology has been used to more faithfully recapitulate the physiology of the human body by integrating cells and tissues into three-dimensional scaffolds. Furthermore, connection of multiple individual OOCs (a so-called body- or human-on-a-chip, BOC/HOC) can be accomplished via microfluidics to allow for crosstalk between the organs of interest13–15. Supporting tissues, such as the vasculature, are critical components of OOCs and BOC/HOCs; incorporating vasculature allows for the transport of nutrients, oxygen, and paracrine factors throughout the tissues, thereby promoting the requisite tissue-specific microenvironment3,12. Thus, methods for deriving mature human ECs, such as arterial, venous, lymphatic, and hemogenic ECs, are crucial to advancing these tissue engineering approaches.
Multiple protocols have been published detailing steps for the derivation of human primordial or progenitor ECs from hESCs or hiPSCs5,16–26. Many of these protocols rely on embryoid body (EB) formation or co-culture of ESCs/iPSCs with a murine feeder layer of stromal cells. These strategies tend to be difficult and time consuming, with low EC yields and/or contamination of human ECs with murine cells. Protocols that rely strictly on 2D culture without the use of stromal cells often require long inductions, utilize complex combinations of growth factors and/or inhibitors for induction, have extended expansion periods following cell separation, or a combination of these factors. Advancing knowledge of signaling pathways and factors involved in the derivation of mature EC types in vivo provides the groundwork for a straight-forward and robust in vitro differentiation protocol.
Previously, key roles for Notch and Retinoic Acid (RA) signaling pathways in the specification of murine arterial and hemogenic ECs, respectively, during development were identified. The Notch signaling pathway plays multiple roles in the specification and maintenance of the arterial EC phenotype. Work using the murine retinal vascularization model identified a pathway in which fluid shear stress induces a Notch-Cx37-p27 signaling axis, promoting G1 cell cycle arrest, which enables arterial EC specification27. Cell cycle states have been hypothesized to play a role in cell fate decisions by providing distinct windows of opportunity in which cells are receptive to certain signals that can induce gene expression and phenotypic changes28. This Notch-mediated G1 arrest enabled the expression of genes enriched in arterial ECs, including ephrinB2, Cx40, DLL4, Notch1 and Notch 4 (reviewed in 29, 30). It has also been shown that hemogenic EC specification is promoted in vivo via RA signaling31,32. Additional studies identified that, downstream of RA signaling, expression of c-Kit and Notch upregulate p27, which enables hemogenic specification in the murine yolk sac and AGM33. Murine hemogenic ECs can be minimally identified by expression of both endothelial (i.e., CD31, KDR) and hematopoietic (i.e., c-Kit, CD34) markers4. Finally, hemogenic ECs undergo an endothelial-to-hematopoietic transition (EHT) to form HSPCs, which can give rise to all blood cell types4, 34, 35.
Recent work tested whether this same signaling hierarchy can promote human hemogenic EC specification. To do so, a serum- and feeder-free 2D culture protocol to derive hemogenic ECs from hESCs was developed, and these hemogenic ECs were characterized on a single cell level as CD31+ KDR+ c-Kit+ CD34+ VE-Cadherin− CD45−. This study also took advantage of the Fluorescent Ubiquitination Cell Cycle Indicator (FUCCI) reporter, which identifies different cell cycle states, by using H9-hESCs that express the FUCCI reporter construct (H9-FUCCI-hESC)36. In studies with these cells, it was demonstrated that RA promotes early G1 cell cycle arrest in ECs, and early G1 state enables hemogenic specification in vitro37. Herein, a detailed protocol for the differentiation of these human hemogenic endothelial cells and assays confirming their identity are provided. This straight-forward method provides a useful means of generating this specialized subset of ECs for future studies of mechanisms of human blood cell development.
PROTOCOL:
1. Reagents and reagent preparation
Note: A list of reagents is provided in Table of Materials.
Materials.
| Name | Company | Catalog Number | Comments |
|---|---|---|---|
| 15 cm dishes | Corning | 430599 | tissue culture treated |
| 35 mm dishes | Corning | 430165 | tissue culture treated |
| 6-well plates | Corning | 3516 | tissue culture treated |
| Antimicrobial reagent Brand Name: Normocin | Invitrogen | ant-nr-1 | |
| bFGF | R&D systems | 233-FB-025 | use at 50 ng/mL |
| BMP4 | BioLegend | 595202 | use at 25 ng/mL |
| Bovine Serum Albumin (BSA) | Fisher Scientific | BP1600–1 | |
| Cell Detatchment Solution Brand Name: Accutase | Stemcell Technologies | 7920 | |
| Dimethyl Sulfoxide (DMSO) | Sigma Aldrich | D2650–100mL | |
| Dispase | Stemcell Technologies | 7913 | |
| DLL4 | R&D systems | 1506-D4/CF | recombinant human; use at 10 μg/mL |
| DMEM:F12 | Gibco | 11320–033 | |
| Dulbecco’s Phosphate Buffered Saline (PBS) | Gibco | 14190144 | |
| Endothelial cell growth medium Brand Name: EGM-2 Endothelial Cell Growth Medium-2 BulletKit (EGM-2) | Lonza | CC-3162 | |
| FACS tubes | Corning | 352235 | polystyrene round bottom with filter cap |
| Fetal Bovine Serum (FBS) | Gemini Bio | 100–106 | |
| Fibronectin | ThermoFisher Scientific | 33016015 | use at 4 μg/cm2 |
| GSK3i/CHIR99021 | Stemgent | 04–0004–02 | 10 mM stock; use at 5 μM |
| Hanks Balanced Salt Solution (HBSS) | Gibco | 14175–095 | |
| Hydrochloric Acid (HCl) | Fisher Scientific | A144S-500 | |
| Matrix protein Brand Name: Matrigel | Corning | 356230 | Growth factor reduced. Refer to the Certificate of Analysis for the lot to determine the protein (Matrigel) concentration. This concentration is required to calculate the volume of Matrigel that contains 1 mg of protein. |
| Methylcellulose-based medium Brand Name: MethoCult H4435 Enriched | Stemcell Technologies | 4435 | |
| Pluripotent stem cell differentiation medium Brand Name: STEMdiff APEL 2 | Stemcell Technologies | 5270 | |
| Pluripotent stem cells: H1, H9, H9-FUCCI | WiCell | WA09 (H9), WA01 (H1) | human; H9-FUCCI were obtained from Dr. Ludovic Vallier’s lab at Cambridge Stem Cell Institute |
| Protein-Free Hybridoma Medium (PFMH) | Gibco | 12040077 | |
| Retinoic Acid | Sigma Aldrich | R2625–50mg | use at 0.5 μM |
| Reverse transcription master mix Brand Name: iScript Reverse Transcription Supermix | BioRad | 1708840 | |
| RNA extraction kit Brand Name: RNeasy Mini Kit | Qiagen | 74104 | |
| Sodium Hydroxide (NaOH) | Fisher Scientific | SS255–1 | |
| Stem cell growth medium Brand Name: mTeSR1 | Stemcell Technologies | 85850 | |
| SYBR Green master mix Brand Name: iTaq Universal SYBR Green Master Mix | BioRad | 1725121 | |
| Trypsin-EDTA | Gibco | 25299956 | 0.25% |
| VEGF165 (VEGF-A) | PeproTech | 100–20 | use at 50 ng/mL |
| α-CD31-FITC | BioLegend | 303104 | 2 μg/mL* |
| α-CD45-APC/Cy7 | BioLegend | 304014 | 2 μg/mL* |
| α-Flk-1-PE/Cy7 | BioLegend | 359911 | 2 μg/mL* |
| α-c-Kit-APC | BioLegend | 313206 | 2 μg/mL* |
| α-VE-Cadherin-PE | BioLegend | 348506 | 2 μg/mL* |
| α-CD34-Pacific Blue | BioLegend | 343512 | 2 μg/mL* |
Antibody fluorescent conjugates should be optimized based on the cell sorter used. Presented here are the final concentrations utilized in this study.
1.1.
Obtain human pluripotent stem cell lines: H1-hESC, H9-Fucci-hESC.
NOTE: The generation of hemogenic ECs may be more efficient in the H1 cell line.
1.2.
Prepare matrix protein stocks: Aliquot matrix protein into pre-chilled 1.5 mL tubes (on ice) so that each tube contains 1 mg of matrix protein. 1 mg of matrix protein is enough to coat all wells of two 6-well plates (12 wells total). Store the aliquots at −20 °C until use.
NOTE: Perform all steps involving matrix protein on ice or at 4 °C. Thaw the frozen stock vial of matrix protein on ice at 4 °C overnight. Once thawed, swirl the vial to ensure the contents are mixed. Prechill 1.5 mL microcentrifuge tubes for at least 1 h at −20 °C and transfer to ice immediately prior to aliquoting.
1.3.
Prepare matrix protein-coated plates: Thaw matrix protein aliquots on ice at 4 °C. Add 12.5 mL of ice-cold DMEM:F12 to a prechilled conical tube on ice. Utilizing pre-chilled pipette tips, transfer one aliquot (1 mg) of matrix protein to the conical tube and mix well by pipetting up and down. Aliquot 1 mL of the diluted matrix protein to each well of pre-chilled 6-well plates (on ice) using a pre-chilled serologic pipette. Swirl and rock the plates so that the entire well is coated evenly. Incubate the plates for a minimum of 30 min at room temperature to allow the matrix protein to solidify. Wrap the plates with parafilm and store at 4 °C until use. Use matrix protein-coated plates within 2 weeks of preparation.
NOTE: Use matrix protein-coated 6-well plates for routine passaging of cells and differentiation to primordial and hemogenic endothelial cells. Perform all the steps on ice. Pre-chill 6-well plates, pipette tips, serologic pipettes, and conical tubes prior to use for at least 1 h at −20 °C. Transfer these items to ice when ready to use.
1.4.
Prepare the base differentiation medium by adding 5 mL of PFHM to 100 mL of pluripotent stem cell differentiation medium. Store at 4 °C.
1.5.
Prepare 0.1% BSA-PBS by dissolving 0.1 g BSA in 100 mL PBS. Filter sterilize by passing through a 0.22 μm filter and store at 4 °C.
1.6.
Prepare 5 mM HCl by diluting stock HCl (12 M) 1:2400 with water. Adjust the pH to 3.0 utilizing NaOH. Filter sterilize by passing through a 0.22 μm filter and store at 4 °C.
1.7.
Prepare bFGF, BMP4, VEGF-A, Retinoic Acid (RA), and DLL4 stocks.
1.7.1.
bFGF: Reconstitute the lyophilized powder to 100 μg/mL in 0.1% BSA-PBS. Aliquot and store at −20 °C. Use bFGF stocks within 3 months of preparation. Thaw aliquots immediately prior to use.
1.7.2.
BMP4: Reconstitute the lyophilized powder to 1 mg/mL in 5mM HCl, pH 3.0. Further dilute to 50 μg/mL in 0.1% BSA-PBS. Aliquot and store at −20 °C. Use BMP4 stocks within 3 months of preparation. Thaw aliquots immediately prior to use.
1.7.3.
VEGF-A: Reconstitute the lyophilized powder to 1 mg/mL in dH2O. Further dilute to 100 μg/mL in 0.1% BSA-PBS. Aliquot and store at −20 °C. Use VEGF-A stocks within 3 months of preparation. Thaw aliquots immediately prior to use.
1.7.4.
RA: Reconstitute the lyophilized powder to 100 mM in DMSO. Aliquot and store at −80 °C. Use RA stocks within 1 month of preparation. Thaw aliquots immediately prior to use.
NOTE: Protect RA stocks from light.
1.7.5.
DLL4: Reconstitute the lyophilized powder to 1 mg/mL in PBS. Aliquot and store at −20 °C. Use DLL4 stocks within 12 months of preparation. Thaw aliquots immediately prior to use.
1.8.
Prepare the endothelial cell differentiation medium immediately prior to use by diluting VEGF-A and BMP4 in base differentiation medium (Step 1.4) so that the final concentrations are 25 ng/mL and 50 ng/mL, respectively.
1.9.
Prepare the working RA immediately prior to use by diluting 100 mM stock 1:1000 in DMSO to 100 μM.
NOTE: Protect working RA from light.
1.10.
Prepare the hemogenic endothelial cell differentiation medium immediately prior to use by diluting VEGF-A, BMP4, and working RA in base differentiation medium (Step 1.4) so that the final concentrations are 25 ng/mL, 50 ng/mL, and 0.5 μM, respectively.
1.11.
Prepare the antibody staining buffer by making HBSS containing 10% FBS and supplement with 1:500 diluted antimicrobial reagent. Sterile filter and use immediately.
1.12.
Prepare the cell sorting buffer by making HBSS containing 1% FBS and supplement with 1:500 diluted antimicrobial reagent. Sterile filter and use immediately.
1.13.
Prepare 1 mg/mL fibronectin stocks by adding 5 mL of sterile water to 5 mg lyophilized fibronectin. Aliquot and store at −20 °C. Use fibronectin stocks prior to the expiry date on the lyophilized product label. Thaw aliquots immediately prior to use.
1.14.
Prepare the fibronectin-coated 35 mm dishes. Dilute fibronectin stocks (1 mg/mL) to 4 μg/mL in sterile water. Add 1 mL of this fibronectin coating solution to each dish and incubate at 37 °C for 30 min to 1 h. Use dishes immediately following coating.
1.15.
Create 3- or 4-mL aliquots of methylcellulose-based medium following the manufacturer’s instructions. Store aliquots at −20 °C until use. Use methylcellulose-based medium aliquots prior to the expiry date indicated on the stock product label. Thaw aliquots immediately prior to use.
1.16.
Prepare the DLL4 coating solution by diluting recombinant human DLL4 stock to a final concentration of 10 μg/mL in PBS.
1.17.
Prepare and store the endothelial cell growth medium according to manufacturer’s instructions.
1.18.
Prepare the flow cytometry analysis buffer by making PBS containing 0.1% BSA. Sterile filter and store at 4 °C until use.
2. Cell culture and passaging of hESCs
2.1.
Allow the matrix protein-coated plates, stem cell growth medium, and DMEM:F12 to warm to room temperature.
2.2.
Grow the hESC cell lines in stem cell growth medium (2 mL/well) on matrix protein-coated 6-well plates in a 37 °C, 5% CO2 incubator.
2.3.
Check the cells daily and remove differentiated cells as necessary by gently scraping them off the plate using a p200 pipette tip.
NOTE: Differentiated cells will appear on the periphery of colonies. Refer to the stem cell growth medium product manual for examples of differentiated cells in culture.
2.4.
Passage the cells once they reach 70–80% confluency. To passage cells:
NOTE: Cells should be split prior to reaching 70–80% confluency if increased differentiation occurs.
2.4.1.
Remove the medium above the cells and gently wash with 1 mL DMEM:F12 per well.
2.4.2.
Add 1 mL DMEM:F12 per well.
2.4.3.
Add 160 μL/mL Dispase to per well and incubate the cells at 37 °C in a 5% CO2 incubator for 45 min.
2.4.4.
Following Dispase incubation, add an additional 1 mL DMEM:F12 per well and gently pipette to lift the cells.
NOTE: Avoid dissociating the cells into a single-cell suspension. Passage the cells as small clumps.
2.4.5.
Transfer the cells to a conical tube containing 12 mL DMEM:F12 and allow the cells to settle by gravity (~5–10 min).
2.4.6.
Remove the supernatant and gently resuspend the pellet in 0.5 mL of stem cell growth medium per each well of lifted cells to obtain small clumps of cells.
NOTE: Avoid dissociating the cells into a single-cell suspension. Passage the cells as small clumps.
2.4.7.
Aspirate the matrix protein coating solution from the wells of the prepared matrix protein-coated plate and add 1.5 mL stem cell growth medium per well.
2.4.8.
Add the desired volume of resuspended cells (step 2.4.6) to each well of the prepared matrix protein-coated plate.
2.4.9.
Incubate the plate in a 37 °C, 5% CO2 incubator. Change the medium every 24 h to fresh stem cell growth medium.
3. Differentiation of hESCs to primordial endothelial cells
3.1.
Day −1: Culture and passage the cells as described above in section 2. Seed the cells (step 2.4.6) in small clumps (~50 μm) at a density of approximately 2 clumps per square centimeter38.
NOTE: Evaluate the seed density and refine empirically if necessary.
3.2.
Day 0: 24 h after seeding the cells, aspirate the medium from each well and gently wash the cells with 1 mL DMEM:F12 per well. Add 1 mL base differentiation medium containing 5 μM GSK3i (CHIR99021, added fresh) to each well and incubate for 24 h (37 °C, 5% CO2).
NOTE: For all wash steps, slowly add the indicated wash media to the plate by pipetting against the wall of the plate well. Gently swirl the plate so that the entire surface of the well is covered by wash media. Slightly tilt the plate so that the wash media pools at the six o-clock position and carefully aspirate the wash media.
3.3.
Day 1: Aspirate the medium from each well and gently wash the cells with 1 mL DMEM:F12 per well. Add 1 mL base differentiation medium containing 50 ng/mL bFGF (added fresh, 1:2000 from frozen stock) to each well and incubate 24 h (37 °C, 5% CO2).
3.4.
Day 2: Aspirate the medium from each well and gently wash the cells with 1 mL DMEM:F12 per well. Add 1 mL endothelial cell differentiation medium to each well and incubate 24 h (37 °C, 5% CO2).
3.5.
Day 3: Replace the medium above the cells with 1 mL freshly prepared endothelial cell differentiation medium per each well and incubate 24 h (37 °C, 5% CO2).
3.6.
Day 4: Replace the medium above the cells with 1 mL freshly prepared endothelial cell differentiation medium per each well and incubate 24 h (37 °C, 5% CO2).
3.7.
Day 5: FACS purify the cells to assess EC phenotype (sections 4–5) OR keep in culture and differentiate towards hemogenic endothelial cells (section 6).
4. FACS Purification of Primordial Endothelial Cells
4.1.
Aspirate the medium above the cells and gently wash once with 1 mL DMEM:F12 per well.
4.2.
Add 1 mL cell detachment solution per well and incubate the cells for 12 min in a 37 °C, 5% CO2 incubator, or until cells have dissociated.
4.3.
Transfer the dissociated cells to a conical tube containing 12 mL DMEM:F12 and pellet by centrifugation for 5 min, 1000 × g.
4.4.
Remove the supernatant and resuspend the pellet in 12 mL DMEM:F12 to wash.
4.5.
Pellet the cells by centrifugation for 5 min, 1000 × g.
4.6.
Remove the supernatant and resuspend the cell pellet in ice-cold antibody staining buffer and count the cells. Adjust the concentration using ice-cold antibody staining buffer to 1 × 105 cells/mL.
4.7.
Divide the cells evenly into microcentrifuge tubes on ice, each containing a minimum of 600 μL cells at 1 × 105 cells/mL, for antibody staining.
NOTE: Four tubes of cells are required for staining of primordial ECs: unstained control, CD31 single antibody control, CD45 single antibody control, and sample containing both CD31 and CD45 antibodies. Information about antibodies is provided in Table of Materials.
4.8.
Add antibodies to the tubes containing cells, as appropriate, and incubate on ice and protected from light for 30 min.
4.8.1.
Unstained control: Do not add any antibody.
4.8.2.
CD31 single antibody control: Add only CD31 antibody.
4.8.3.
CD45 single antibody control: Add only CD45 antibody.
4.8.4.
Sample: Add both CD31 and CD45 antibodies.
NOTE: The final antibody concentration in the sample, as well as the fluorescent conjugates, should be optimized based on the specific antibodies and cell sorter used.
4.9.
Pellet the cells by centrifugation in a 4 °C tabletop microcentrifuge for 5 min at 1000 × g.
4.10.
Remove the supernatant and resuspend the cell pellets in 600 μL ice-cold sorting buffer.
4.11.
Strain the samples through the mesh filter caps of 5 mL FACS tubes and store the cells on ice, protected from light, for immediate FACS.
4.12.
Obtain primordial endothelial cells (CD31+ CD45−, by performing the following steps:
4.12.1.
Identify the CD45-negative cell population and gate (CD45−).
4.12.2.
Within (CD45−), identify the CD31-positive (CD31+) cell population and sort cells into 6 mL ice-cold cell sorting buffer. Use these cells for downstream applications (see section 5).
5. Assay to Confirm Primordial Endothelial Cell Phenotype
5.1.
Coat 3 wells of a 6-well plate with 1 mL/well DLL4 coating solution for 30 min in a 37 °C, 5% CO2 incubator. As a control, mock-coat the other 3 wells of the plate with 1 mL/well PBS.
5.2.
Aspirate the DLL4 coating solution and PBS, and plate 25,000 sorted primordial endothelial cells (see Step 4.12) in 2 mL endothelial cell growth medium per well.
5.3.
Incubate the cells for 24 h in a 37 °C, 5% CO2 incubator.
5.4.
Aspirate the medium above the cells and wash once with 2 mL/well PBS. Analyze the cells by qPCR or flow cytometry (for cells expressing the FUCCI construct):
5.4.1.
To analyze the cells by qPCR, perform the following steps:
5.4.1.1.
Aspirate the liquid above the cells and isolate the RNA in the cells using an RNA extraction kit according to the manufacturer’s protocol.
5.4.1.2.
Perform reverse transcription reactions with a reverse transcription master mix according to the manufacturer’s protocol.
5.4.1.3.
Perform qPCR reactions with a SYBR Green master mix according to the manufacturer’s protocol.
NOTE: Primers utilized (EFNB2, GJA5, GJA4, NR2F2, EPHB4, HEY2) are listed in Table 1.
Table 1:
qPCR primer information
| Name | Forward | Reverse |
|---|---|---|
| EFNB2 | TATGCAGAACTGCGATTTCCAA | TGGGTATAGTACCAGTCCTTGTC |
| EPHB4 | CGCACCTACGAAGTGTGTGA | GTCCGCATCGCTCTCATAGTA |
| GJA5 | CCGTGGTAGGCAAGGTCTG | ATCACACCGGAAATCAGCCTG |
| GJA4 | ACACCCACCCTGGTCTAC | CACTGGCGACATAGGTGCC |
| HEY2 | GCCCGCCCTTGTCAGTATC | CCAGGGTCGGTAAGGTTTATTG |
| NR2F2 | GGACCACATACGGATCTTCCAA | ACATCAGACAGACCACAGGCAT |
5.4.2.
To analyze cells expressing the FUCCI construct by flow cytometry, perform the following steps:
5.4.2.1.
Aspirate the liquid above the cells and add 500 μL/well 0.25% trypsin-EDTA.
5.4.2.2.
Incubate the cells at 37 °C in a 5% CO2 incubator until lifted, ~5 min.
5.4.2.3.
Transfer the cells to a microcentrifuge tube and pellet the cells in a 4 °C microcentrifuge for 5 min at 1000 × g.
5.4.2.4.
Aspirate the supernatant and resuspend the pellet in 500 μL ice-cold flow cytometry analysis buffer.
5.4.2.5.
Strain the samples through the mesh filter cap of a 5 mL FACS tube and store the cells on ice, protected from light, for immediate flow cytometry analysis.
5.4.2.6.
Analyze the percentage of cells plated on DLL4 vs. PBS control in early G1 (no color), late G1 (mCherry+/mVenus−), G1/S (mCherry+/mVenus+), and S/G2/M (mCherry−/mVenus+).
6. Differentiation of hESCs to Hemogenic Endothelial Cells
NOTE: Differentiate cells to day 4 primordial ECs, as described above in sections 3.1–3.6.
6.1.
Day 5: Aspirate the medium above the cells and gently wash the cells with 1 mL DMEM:F12 per well. Add 1 mL freshly prepared hemogenic endothelial cell differentiation medium per each well and incubate 24 h (37 °C, 5% CO2).
6.2.
Day 6: Replace the medium above the cells with 1 mL freshly prepared hemogenic endothelial cell differentiation medium per each well and incubate 24 h (37 °C, 5% CO2).
6.3.
Day 7: Replace the medium above the cells with 1 mL freshly prepared hemogenic endothelial cell differentiation medium per each well and incubate 24 h (37 °C, 5% CO2).
6.4.
Day 8: FACS isolate hemogenic endothelial cells (see section 7).
7. FACS-Isolation of Hemogenic Endothelial Cells
7.1.
Lift and wash the cells as described above in sections 4.1–4.6.
7.2.
Divide the cells evenly into microcentrifuge tubes on ice, each containing a minimum of 600 μL cells at 1 × 105 cells/mL, for antibody staining.
NOTE: Eight tubes of cells are required for antibody staining of hemogenic ECs: unstained control, CD31 single antibody control, CD45 single antibody control, KDR single antibody control, c-Kit single antibody control, CD34 single antibody control, VE-cadherin single antibody control, and sample containing all six antibodies.
NOTE: Antibody information is provided in Table of Materials.
7.3.
Add antibodies to the tubes containing cells, as appropriate, and incubate on ice and protected from light for 30 min.
7.3.1.
Unstained control: Do not add antibody.
7.3.2.
CD31 single antibody control: Add only CD31 antibody.
7.3.3.
CD45 single antibody control: Add only CD45 antibody.
7.3.4.
KDR single antibody control: Add only KDR antibody.
7.3.5.
c-Kit single antibody control: Add only c-Kit.
7.3.6.
CD34 single antibody control: Add only CD34 antibody.
7.3.7.
VE-cadherin single antibody control: Add only VE-cadherin antibody.
7.3.8.
Sample: Add CD31, CD45, KDR, c-Kit, CD34, and VE-cadherin antibodies.
NOTE: The final antibody concentration in the sample, as well as the fluorescent conjugates, should be optimized based on the specific antibodies and cell sorter used.
7.4.
Pellet the cells by centrifugation in a 4°C microcentrifuge for 5 min at 1000 × g.
7.5.
Remove the supernatant and resuspend the pellets in 600 μL ice-cold sorting buffer.
7.6.
Strain the samples through the mesh filter cap of 5 mL FACS tubes and store cells on ice, protected from light, for immediate FACS.
7.7.
To obtain hemogenic endothelial cells (CD31+ KDR+ c-Kit+ CD34+ VE-Cadherin− CD45−), perform the following steps:
7.7.1.
Identify the CD45− cell population and gate (CD45−).
7.7.2.
Within (CD45−), identify the CD31+ cell population and gate (CD31+).
7.7.3.
Within (CD31+), identify the VE-Cadherin− cell population and gate (CDH5−).
7.7.4.
Within (CDH5−), identify the c-Kit+ cell population and gate (KIT+).
7.7.5.
Within (KIT+), identify the CD34+ cell population and gate (CD34+)
7.7.6.
Within (CD34+), identify and sort the KDR+ cells into 6 mL ice-cold cell sorting buffer. Use these cells in downstream applications (See section 8).
8. Colony Forming Unit Assay
8.1.
Thaw aliquots of methylcellulose-based medium following manufacturer’s instructions.
NOTE: One 3mL aliquot is sufficient for two samples and one 4mL aliquot is sufficient for three samples.
8.2.
Count the sorted hemogenic endothelial cells obtained in Step 7.12.6.
8.3.
Following the manufacturer’s instructions, calculate the volume of sorted cells to add to each methylcellulose-based medium aliquot so that each sample will contain a minimum of 1000 hemogenic endothelial cells.
NOTE: The number of cells can be adjusted as necessary.
8.4.
Add the calculated volume of cells to the methylcellulose-based medium aliquot and vortex thoroughly. Allow the methylcellulose-based medium cultures to sit at room temperature for 10 min, or until the bubbles dissipate.
8.5.
Aspirate the fibronectin coating solution from the prepared 35mm dishes. Following the manufacturer’s instructions, dispense 1 mL of methylcellulose-based medium culture per 35 mm dish. Gently rotate the dish to evenly distribute the culture.
8.6.
Place these 35 mm dishes, as well as two additional 35 mm dishes filled with sterile water, within a 15 cm tissue culture dish.
8.7.
Incubate the cultures in a 37 °C, 5% CO2 incubator, and observe the cultures on days 8 and 14:
8.7.1.
On day 8, count CFU-E and BFU-E colonies.
8.7.2.
On day 14, count CFU-GM and CFU-GEMM colonies.
NOTE: Refer to the methylcellulose-based medium manual for morphological identification of colony types.
REPRESENTATIVE RESULTS:
A schematic outlining the specification of primordial ECs and hemogenic ECs from hESCs, and a representative image of cells 24 h after plating are shown in Figure 1. Following specification, primordial ECs and hemogenic ECs are FACS purified on days 5 and 8, respectively. Primordial ECs are defined as CD31+ CD45− and hemogenic ECs are defined as CD31+ KDR+ c-Kit+ CD34+ VE-Cadherin− CD45−. A representative flow cytometric gating strategy for primordial ECs and hemogenic EC purification is shown in Figure 2. Cells are initially gated based on negative expression of CD45 and positive expression of CD31 to obtain purified primordial ECs (Figure 2A). To obtain purified hemogenic ECs, cells are initially gated as in Figure 2A and are then further purified based on positive or negative expression of (in order) VE-Cadherin (CDH5), c-Kit (KIT), CD34 and KDR (Figure 2B).
Figure 1: Protocol for the specification of primordial and hemogenic ECs.
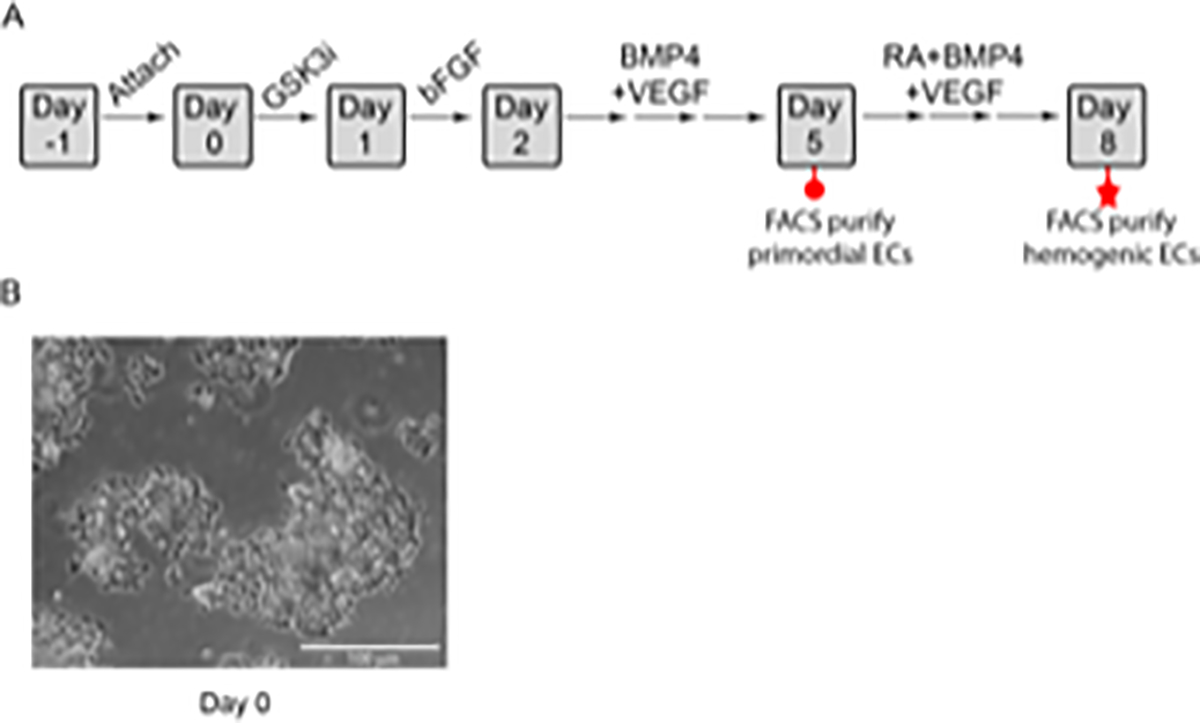
A) Schematic diagram of the differentiation protocol. Embryonic stem cells are plated on Day −1 on matrix protein-coated plates and are allowed to attach overnight. The cells are then treated on Days 0 and 1 with GSK3i inhibitor (CHIR99021) and bFGF, respectively, to induce primitive streak and mesoderm specification, respectively. Beginning on Day 2, the cells are treated with a combination of BMP4 and VEGF-A to promote primordial EC development. Primordial ECs (red circle) are FACS purified on Day 5. Alternatively, to generate hemogenic ECs, the medium above the primordial ECs is exchanged on Day 5 to fresh hemogenic differentiation medium containing BMP4, VEGF-A, and RA. This medium is replaced daily until Day 8, when hemogenic ECs (red star) are FACS purified. B) Colonies on day 0 of differentiation, scale bar= 100 μm. Panel A has been modified from Qiu et al.37 with permission from Elsevier.
Figure 2: FACS analysis of hemogenic ECs derived from hESCs.
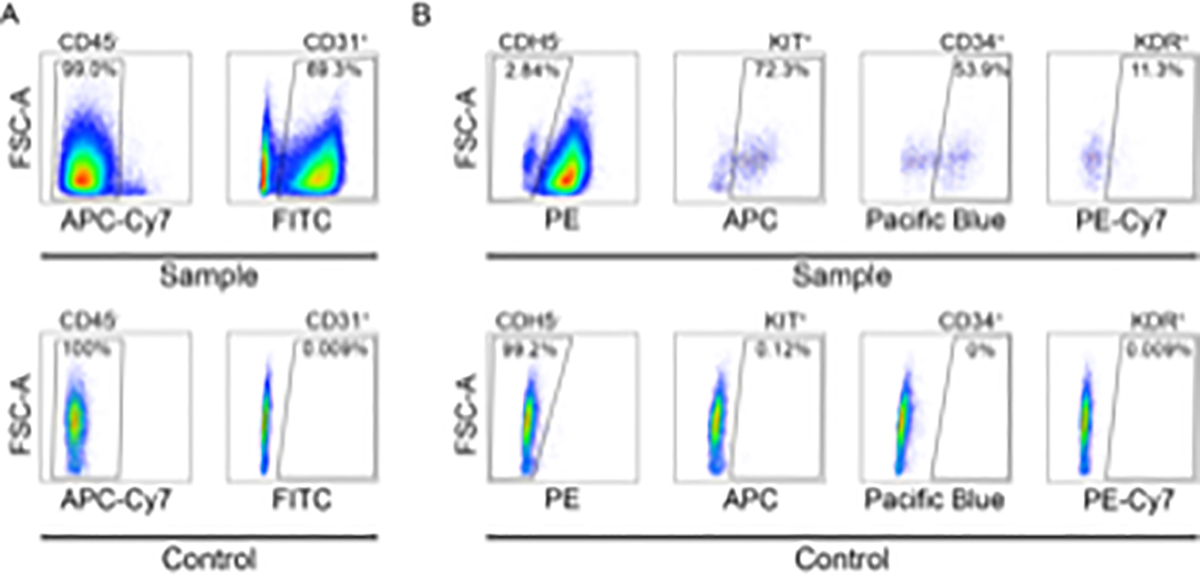
Representative flow cytometric gating strategy for the purification of A) primordial ECs and B) hemogenic ECs. Note that since hemogenic ECs are derived from primordial ECs, the flow cytometry gating strategy for CD45 and CD31 is identical for both cell populations. Shown in the top row of each panel (sample) are cells that were differentiated to hemogenic ECs as described in protocol section 6 and stained with antibodies as described in protocol section 7.3.8. Shown in the bottom row of each panel (control) are unstained cells that were differentiated for 8 days without RA treatment.
To assess the potential of H9-Fucci-hES-derived primordial CD31+ CD45− endothelial cells, isolated via FACS at day 5 of differentiation (protocol section 4), to give rise to endothelial subtypes, the purified cells are seeded onto plates coated with either the Notch ligand DLL4 to induce arterial specification, or PBS (control), and incubated for 24 h in a 37 °C, 5% CO2 incubator. The cells are then lysed with RNA lysis buffer, the RNA extracted, and reverse transcribed to cDNA, and qPCR is performed to compare gene expression levels in the DLL4-treated vs. control cells. As expected, endothelial cells grown on DLL4 have increased expression of the Notch-responsive gene HEY2, as well as the arterial-associated genes EFNB2, GJA5, GJA4. Additionally, these cells also have decreased expression of the venous transcription factor NR2F2 (Figure 3A). Alternatively, to determine the effect of DLL4 treatment on cell cycle state, the FACS purified CD31+ CD45− cells are incubated on plates coated with either DLL4 or PBS for 24 h, lifted, and analyzed based on expression of hCdt1(30/120)-mCherry (late G1) and hGem(1/110)-mVenus (S/G2/M). Consistent with findings that Notch signaling promotes late G1 cell cycle arrest27, a greater percentage of primordial ECs are arrested in late G1 after growth in the presence of DLL4, compared to control cells (Figure 3B,C).
Figure 3: DLL4-induction of H9-Fucci CD31+ CD45− primordial endothelial cells results in late G1 arrest and increased arterial gene expression.
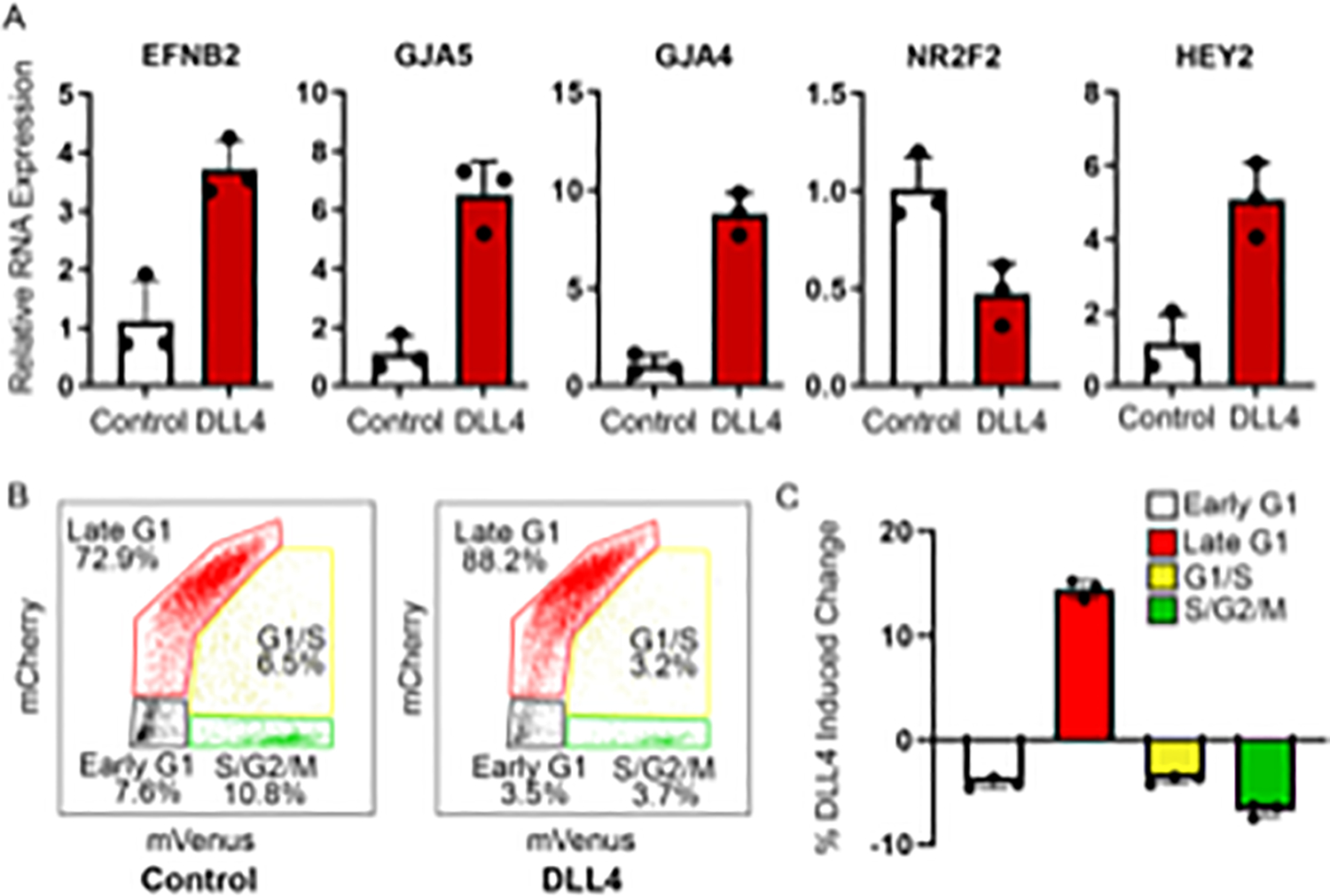
A) DLL4 treatment of CD31+ CD45− H9-hESC- derived primordial endothelial cells expressing the Fucci construct purified at day 5 of differentiation results in increased expression of arterial genes (i–iii) and the Notch responsive gene Hey2 (v), which is accompanied by a concomitant decrease in the expression of the venous gene NR2F2 (iv). B) Representative FACS plots showing the cell cycle state distribution of 5,000 H9-Fucci-derived CD31+ CD45− grown on PBS (control) or DLL4 for 24 h. C) DLL4 induction results in a 15% increase in cells in late G1 phase compared to control. Data are the averages of triplicate samples from the same experiment shown in panel B. Error bars indicate standard deviation.
To verify the hematopoietic potential of hemogenic endothelial cells, the CD31+ KDR+ c-Kit+ CD34+ VE-Cadherin− CD45− endothelial cells isolated through FACS (Methods Section 7) are seeded in a methylcellulose-based medium formulated for growth of hematopoietic progenitor cells in colony-forming unit (CFU) assays and are allowed to grow for 14 days. CFU-E erythroid colonies and blast-forming unit (BFU)-E erythroid colonies are counted on day 8 (Figure 4A,B), and CFU-GM granulocyte/macrophage and GFU-GEMM (granulocyte, erythroid, macrophage, and megakaryocyte) multipotent hematopoietic progenitor colonies are counted on day 14 (Figure 4C and D). Per 1000 hemogenic ECs plated, approximately 20 CFU are generated (Figure 4F). Cells with endothelial cell morphology can also be seen in the cultures (Figure 4E); these are the hemogenic endothelial cells that give rise to multi-lineage hematopoietic progenitors on a single cell level37.
Figure 4: Analysis of hematopoietic potential of hemogenic ECs derived from hESCs.
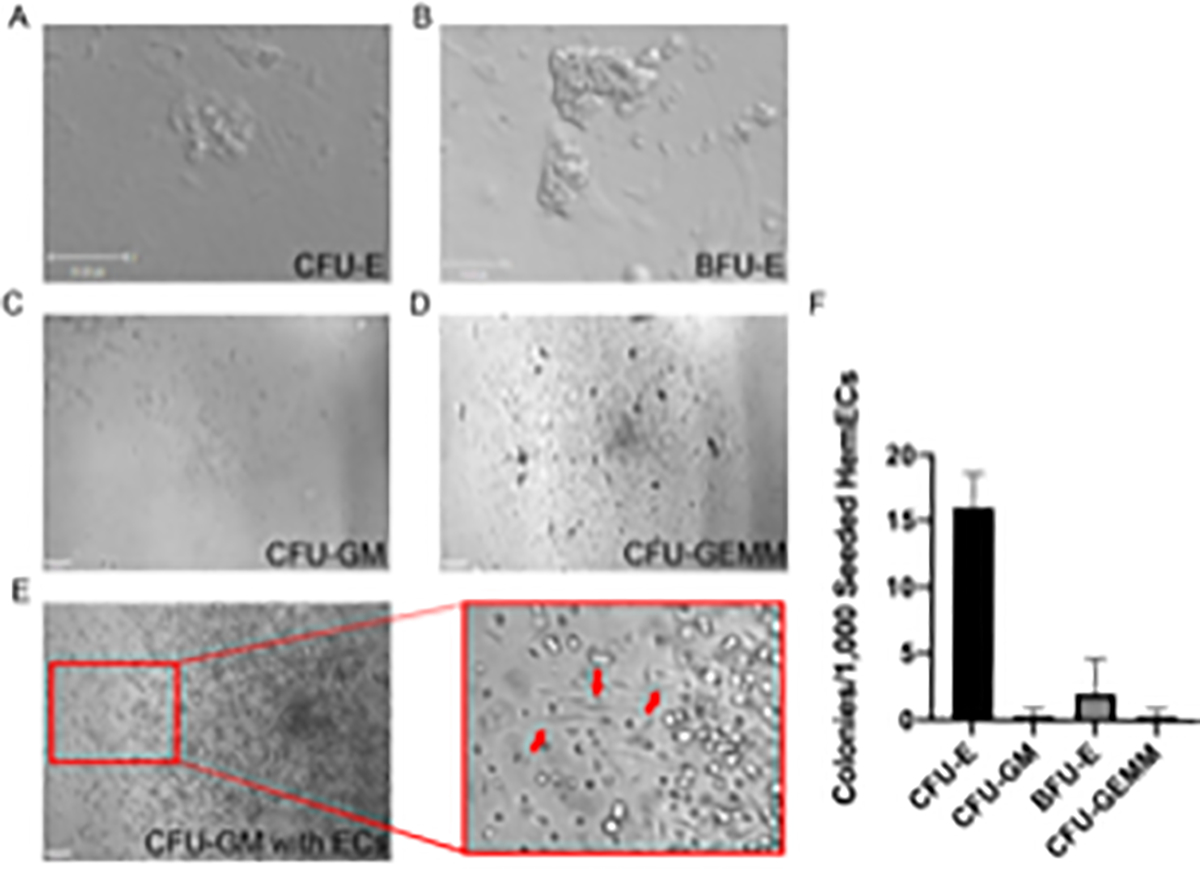
Representative images showing morphology of H1-hESC derived A) CFU-E erythroid colony (scale bar= 35 μm), B) BFU-E erythroid colony (scale bar= 75 μm), C) CFU-GM granulocyte/macrophage colony, D) CFU-GEMM multipotent hematopoietic progenitor colony, and E) CFU-GM granulocyte/macrophage colony with underlying endothelial cells (ECs) (red arrows). F) number and distribution of CFUs formed per 1000 plated hemogenic endothelial cells. Scale bar = 100 μm in C–E. Additional images of CFUs differentiated using this protocol can be found in Qiu et al.37. Panel F has been modified from Qiu et al.37 with permission from Elsevier.
DISCUSSION:
Herein, the steps for producing hemogenic endothelial cells from human embryonic stem cells in approximately 1 week using a murine feeder- and serum-free 2D culture system (Figure 1) are outlined. This protocol expands on a method described by Sriram et al. (2015) to obtain primordial ECs38. The primordial nature and specification potential of the CD31+ CD45− ECs is demonstrated by culturing these cells on DLL4-coated plates and observing gene expression changes consistent with arterial specification (Figure 3). Additionally, the gain of arterial identity is associated with late G1 cell cycle arrest (Figure 3), which is consistent with previous studies27. After culturing primordial ECs for an additional 3 days in the presence of 0.5 μM RA, 25 ng/mL BMP4, and 50 ng/mL VEGF-A, it was possible to generate and FACS-isolate hemogenic ECs (Figure 2) that are capable of giving rise to CFU-erythroid, BFU-erythroid, CFU-granulocyte/macrophage, and CFU-granulocyte, erythrocyte, macrophage, and megakaryocyte colonies (Figure 4). Using this method in a recently published study, gene expression changes over an 8-day time period consistent with loss of pluripotency, primitive streak and mesoderm induction, acquisition of endothelial cell identity, and finally hematopoietic identity were observed37. Furthermore, RA treatment induced early G1 cell cycle arrest to enable hemogenic EC specification37.
Recently, Ohta et al. (2019) described a protocol for the differentiation of hemogenic ECs from hPSCs39. However, the protocol described above offers significant advantages: 1) this method does not require the formation of spheroids, 2) this protocol utilizes a standard 37°C, 5% CO2 incubator rather than a hypoxic incubator, eliminating the need for dedicated specialty equipment, and 3) this protocol utilizes only one medium (pluripotent stem cell differentiation medium supplemented with PFHM), a cost-saving advantage, whereas the Ohta protocol requires mediums for induction. Another recently published study by Galat et al. (2017) described a protocol in which CHIR99021 induction was utilized to generate a population of CD34+ hemogenic endothelial cells40. These cells also expressed CD31 and were capable of giving rise to endothelial cells when cultured under monolayer conditions or cells expressing myeloid and lymphoid markers after co-culturing with OP9 or OP9-DLL4 cells, respectively, in the presence of additional cytokines. The requirement for additional co-culture could lead to potential contamination of desired cell populations with murine cells. Additionally, although Ohta et al. and Galat et al. utilized a hemogenic induction period that was shorter than the one described here (4 days and 5 days, respectively vs. 8 days), both defined hemogenic ECs as CD34+, whereas this protocol utilized a more stringent definition: CD31+ KDR+ c-Kit+ CD34+ VE-Cadherin− CD45−. While CD34 is recognized as a marker of hematopoietic cells, it is also expressed by other non-hematopoietic cell types, such as mesenchymal stromal cells and endothelial cells41. The definition of hemogenic ECs in this protocol (CD31+ KDR+ c-Kit+ CD34+ VE-Cadherin− CD45−) is therefore more rigorous and represents a more defined population.
One limitation to the use of hESCs or hiPSCs in therapeutic applications is the large number of cells required, and standard 2D derivation methods are primarily restricted to small-scale differentiations. Utilizing hiPSC lines, Olmer et al. (2018) demonstrated the feasibility of scaling up production of functional CD31+ ECs that expressed both arterial (DLL4) and venous (EPHB4) cell markers utilizing either suspension culture or a stirred-tank bioreactor6. Importantly, they showed that they were able to obtain 1.18 × 107 CD31+ ECs that co-express CD34 and KDR from a single flask containing 20 mL suspension culture. In order to obtain the requisite 3 × 108 ECs necessary for the majority of therapeutic applications, just over two 500 mL flasks would be required6. Future experiments should explore the application of scaling techniques to the protocol presented here for large-scale production of hemogenic ECs.
ACKNOWLEDGMENTS:
This work was partially supported by NIH grants HL128064 and U2EB017103. Further support was provided by CT Innovations 15-RMB-YALE-04 grant.
Footnotes
DISCLOSURES:
None
REFERENCES:
- 1.Giacomelli E et al. Three-dimensional cardiac microtissues composed of cardiomyocytes and endothelial cells co-differentiated from human pluripotent stem cells. Development. 144 (6), 1008–1017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu J, Wu Z, Xue Z, Li H, Liu J PHBV/bioglass composite scaffolds with co-cultures of endothelial cells and bone marrow stromal cells improve vascularization and osteogenesis for bone tissue engineering. RSC Advances. 7 (36), 22197–22207 (2017). [Google Scholar]
- 3.Lee H, Chung M, Jeon NL Microvasculature: An essential component for organ-on-chip systems. MRS Bulletin. 39 (1), 51–59 (2014). [Google Scholar]
- 4.Gritz E, Hirschi KK Specification and function of hemogenic endothelium during embryogenesis. Cellular and Molecular Life Sciences. 73 (8), 1547–1567 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams IM, Wu JC Generation of endothelial cells from human pluripotent stem cells: Methods, considerations, and applications. Arteriosclerosis, Thrombosis, and Vascular Biology. 39 (7), 1317–1329 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olmer R et al. Differentiation of human pluripotent stem cells into functional endothelial cells in scalable suspension culture. Stem Cell Reports. 10 (5), 1657–1672 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cossu G et al. Lancet Commission: Stem cells and regenerative medicine. The Lancet. 391 (10123), 883–910 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Fox IJ et al. Use of differentiated pluripotent stem cells in replacement therapy for treating disease. Science. 345 (6199), 1247391–1247391 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahla RS Stem cells applications in regenerative medicine and disease therapeutics. International Journal of Cell Biology. 2016, 1–24 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ebert AD, Liang P, Wu JC Induced pluripotent stem cells as a disease modeling and drug screening platform: Journal of Cardiovascular Pharmacology. 60 (4), 408–416 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rowe RG, Daley GQ Induced pluripotent stem cells in disease modelling and drug discovery. Nature Reviews Genetics. 20 (7), 377–388 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu C, Oikonomopoulos A, Sayed N, Wu JC Modeling human diseases with induced pluripotent stem cells: from 2D to 3D and beyond. Development. 145 (5), dev156166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wnorowski A, Yang H, Wu JC Progress, obstacles, and limitations in the use of stem cells in organ-on-a-chip models. Advanced Drug Delivery Reviews. 140, 3–11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramme AP et al. Autologous induced pluripotent stem cell-derived four-organ-chip. Future Science OA. 5 (8), 1–12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ronaldson-Bouchard K, Vunjak-Novakovic G Organs-on-a-Chip: A fast track for engineered human tissues in drug development. Cell Stem Cell. 22 (3), 310–324 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kane NM et al. Derivation of endothelial cells from human embryonic stem cells by directed differentiation: analysis of microrna and angiogenesis in vitro and in vivo. Arteriosclerosis, Thrombosis, and Vascular Biology. 30 (7), 1389–1397 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Costa M et al. Derivation of endothelial cells from human embryonic stem cells in fully defined medium enables identification of lysophosphatidic acid and platelet activating factor as regulators of eNOS localization. Stem Cell Research. 10 (1), 103–117 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Nguyen MTX et al. Differentiation of human embryonic stem cells to endothelial progenitor cells on laminins in defined and xeno-free systems. Stem Cell Reports. 7 (4), 802–816 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ikuno T et al. Efficient and robust differentiation of endothelial cells from human induced pluripotent stem cells via lineage control with VEGF and cyclic AMP. PLOS ONE. 12 (3), e0173271 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aoki H et al. Efficient differentiation and purification of human induced pluripotent stem cell-derived endothelial progenitor cells and expansion with the use of inhibitors of ROCK, TGF-β, and GSK3β. Heliyon. 6 (3), e03493 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lian X et al. Efficient differentiation of human pluripotent stem cells to endothelial progenitors via small-molecule activation of Wnt signaling. Stem Cell Reports. 3 (5), 804–816 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orlova VV et al. Generation, expansion and functional analysis of endothelial cells and pericytes derived from human pluripotent stem cells. Nature Protocols. 9 (6), 1514–1531 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Kusuma S, Gerecht S Derivation of endothelial cells and pericytes from human pluripotent stem cells. Human Embryonic Stem Cell Protocols. Edited by Turksen K, 213–222, Humana Press. New York, NY: (2016). [DOI] [PubMed] [Google Scholar]
- 24.Bao X, Lian X, Palecek SP Directed endothelial progenitor differentiation from human pluripotent stem cells via wnt activation under defined conditions. Wnt Signaling: Methods and Protocols. Edited by Barrett Q, Lum L, 183–196., Humana Press. New York, NY: (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu M, He J, Zhang C, Xu J, Wang Y Strategies for derivation of endothelial lineages from human stem cells. Stem Cell Research & Therapy. 10 (1), 200 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arora S, Yim EKF, Toh Y-C Environmental specification of pluripotent stem cell derived endothelial cells toward arterial and venous subtypes. Frontiers in Bioengineering and Biotechnology. 7, 143 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fang JS et al. Shear-induced Notch-Cx37-p27 axis arrests endothelial cell cycle to enable arterial specification. Nature Communications. 8 (1), 2149 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dalton S Linking the cell cycle to cell fate decisions. Trends in Cell Biology. 25 (10), 592–600 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolf K, Hu H, Isaji T, Dardik A Molecular identity of arteries, veins, and lymphatics. Journal of Vascular Surgery. 69 (1), 253–262 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rocha SF, Adams RH Molecular differentiation and specialization of vascular beds. Angiogenesis. 12 (2), 139–147 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Goldie LC, Lucitti JL, Dickinson ME, Hirschi KK Cell signaling directing the formation and function of hemogenic endothelium during murine embryogenesis. Blood. 112 (8), 3194–3204 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chanda B, Ditadi A, Iscove NN, Keller G Retinoic acid signaling is essential for embryonic hematopoietic stem cell development. Cell. 155 (1), 215–227 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Marcelo KL et al. Hemogenic endothelial cell specification requires c-kit, notch signaling, and p27-mediated cell-cycle control. Developmental Cell. 27 (5), 504–515 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dejana E, Hirschi KK, Simons M The molecular basis of endothelial cell plasticity. Nature Communications. 8 (1), 14361 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ottersbach K Endothelial-to-haematopoietic transition: an update on the process of making blood. Biochemical Society Transactions. 47 (2), 591–601 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pauklin S, Vallier L The cell-cycle state of stem cells determines cell fate propensity. Cell. 155 (1), 135–147 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qiu J, Nordling S, Vasavada HH, Butcher EC, Hirschi KK Retinoic acid promotes endothelial cell cycle early g1 state to enable human hemogenic endothelial cell specification. Cell Reports. 33 (9) 108465 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sriram G, Tan JY, Islam I, Rufaihah AJ, Cao T Efficient differentiation of human embryonic stem cells to arterial and venous endothelial cells under feeder- and serum-free conditions. Stem Cell Research & Therapy. 6 (1), 261 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohta R, Sugimura R, Niwa A, Saito MK Hemogenic endothelium differentiation from human pluripotent stem cells in a feeder- and xeno-free defined condition. Journal of Visualized Experiments: JoVE. (148), 59823 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Galat Y, Dambaeva S, Elcheva I, Khanolkar A, Beaman K, Iannaccone PM, Galat V Cytokine-free directed differentiation of human pluripotent stem cells efficiently produces hemogenic endothelium with lymphoid potential. Stem Cell Research & Therapy. 8 (1), 67 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sidney LE, Branch MJ, Dunphy SE, Dua HS, Hopkinson A Concise review: Evidence for CD34 as a common marker for diverse progenitors. Stem Cells (Dayton, Ohio). 32 (6), 1380–1389 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]


