Abstract
We have determined that I-mfa, an inhibitor of several basic helix-loop-helix (bHLH) proteins, and XIC, a Xenopus ortholog of human I-mf domain-containing protein that shares a highly conserved cysteine-rich C-terminal domain with I-mfa, inhibit the activity and DNA binding of the HMG box transcription factor XTcf3. Ectopic expression of I-mfa or XIC in early Xenopus embryos inhibited dorsal axis specification, the expression of the Tcf3/β-catenin-regulated genes siamois and Xnr3, and the ability of β-catenin to activate reporter constructs driven by Lef/Tcf binding sites. I-mfa domain proteins can regulate both the Wnt signaling pathway and a subset of bHLH proteins, possibly coordinating the activities of these two critical developmental pathways.
I-mfa directly inhibits the activity of Myf5 and related myogenic basic helix-loop-helix (bHLH) proteins by preventing nuclear localization and DNA binding (8). These activities are dependent upon the cysteine-rich C-terminal domain specific to I-mfa. During mouse embryogenesis, I-mfa mRNA is broadly expressed in both embryonic and extraembryonic tissues, including the presomitic mesoderm, the dermomyotome, and the sclerotome, but not in the myotome (23). This has led to the hypothesis that I-mfa inhibited the activity of the low levels of Myf5 expressed in the presomitic mesoderm until developmental signals, possibly including sonic hedgehog (SHH) and Wnt-1, relieved the I-mfa inhibition of Myf5 and permitted myogenic differentiation in the myotome (8). Targeted disruption of I-mfa in strain C57BL/6 mice results in lethality around embryonic day 9.5 (23). Phenotypic and molecular analysis has revealed that I-mfa inhibits the activity of bHLH protein MASH2, preventing normal differentiation of trophoblast giant cells. In the 129Sv mouse strain, homozygous disruption of I-mfa results in abnormal skeletal development and reduced expression of scleraxis, a bHLH protein implicated in chondrogenic differentiation (9). These results provide genetic and molecular support for the role of I-mfa as a negative regulator of a subset of bHLH transcription factors and implicate I-mfa as a necessary component of aspects of normal somite development.
Recently, human I-mfa domain-containing protein (HIC) has been identified as a protein that has a cysteine-rich C-terminal domain with a high degree of homology to the C-terminal domain of I-mfa (77% identity and 81% similarity) (40). HIC has been shown to be able to regulate Tat- and Tax-mediated expression of viral promoters differentially, stimulating the expression from the human T-cell leukemia virus type I (HTLV-I) long-terminal repeat and suppressing expression from the human immunodeficiency virus type 1 (HIV-1) long-terminal repeat. The I-mfa domain was necessary for the regulatory activity of HIC, just as it was necessary for the ability of I-mfa to regulate the activity of bHLH proteins. In this regard, HIC and I-mf represent two families of proteins that share a highly conserved cysteine-rich regulatory domain, termed the I-mfa domain. Given the conservation of this domain, it is likely that HIC and I-mfa might be capable of similar protein interactions, and, indeed, I-mfa also regulates HTLV-I transcription (40).
Tcf and Lef proteins are highly conserved members of the HMG domain family of architectural factors that bend DNA and facilitate assembly of nucleoprotein complexes to regulate transcription (12, 13). Originally characterized as factors which bind the enhancer of T-cell-specific genes (42), they have recently been identified as mediators of the Wnt/wingless signaling pathways through interaction with transcriptional coactivators β-catenin and armadillo (2, 16). Activation of the target genes in these pathways controls numerous cell fate and differentiation events, including axis specification in Xenopus laevis (29), somite patterning or paraxial mesoderm development in chickens and mice, and gastrulation (7, 31, 34) and axis formation in mice (14, 17), as well as colorectal epithelial stem cell maintenance and cancer (20, 21). In this report, we show that I-mfa and XIC, the Xenopus ortholog of HIC, can repress the Wnt signaling pathway and directly interact with XTcf3 to prevent DNA binding and transcriptional regulation by XTcf3. In both I-mfa and XIC, the cysteine-rich I-mfa domain is both necessary and sufficient for the interaction with XTcf3. The ability of the I-mfa domain to interact with both bHLH proteins and Tcf3 suggests that this class of proteins has the potential to play a role in integrating the signals from multiple regulatory pathways.
MATERIALS AND METHODS
Embryos and injection of synthetic RNA.
The culture of embryos and the synthesis and injection of capped mRNA were performed as described previously (37). For perturbation of the endogenous dorsal axis or inhibition of siamois and Xnr3 expression, injections were made subequatorially into the two dorsal blastomeres of four-cell embryos. In ectopic axis experiments, injections were into the equatorial region of two ventral blastomeres of four-cell embryos. For all other experiments, injections were into the animal pole at the two-cell stage. Control injections were performed with indicated amounts of β-galactosidase mRNA.
Western blotting.
Embryos were injected with myc-tagged β-catenin or myc-tagged pt β-catenin RNA into two ventral cells at the four-cell stage. At the early gastrula stage the embryos were collected and lysed in buffer containing 10 mM HEPES (pH 7.5), 150 mM NaCl, 2 mM EDTA, and 1% NP-40 and protease inhibitors. Proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and by immunoblotting with the anti-myc epitope antibody, 9E10.
Library screening.
A Xenopus oocyte lambda gt10 library (35) was screened using the I-mfa-specific domain of XIC obtained using degenerate PCR primers based on the sequences for mouse and human I-mfa.
RT-PCR.
RNA was isolated from 5 to 10 embryos following lysis in a solution containing 3 M LiCl and 6 M urea and treated for 30 min with RNase-free DNase. One to two micrograms of RNA was used for the reverse transcription (RT) reaction, and one-fifth of the RT product was a template for the PCR. PCR conditions for XIC, siamois, and Xnr3 included 35 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 1 min. Ef1α signals were detected using 20 cycles. All RT-PCR experiments included controls without RT and without a template. Primers were as follows: XIC, 5′ CTG AAT TCA CAC TGT GTA ACA TTG TAG TGG 3′ and 5′ GAT CTA GAT CCA AAC AGT CTG AAG ATT CAC 3′, ef1α (38), siamois (5), and Xnr3 (44).
Coimmunoprecipitations and MBP pull-down assays.
Coimmunoprecipitations were performed essentially as described (8). Embryos were injected twice in the animal pole with 3 ng of hemagglutinin (HA)-tagged XTcf3 and 3 ng of myc-tagged mouse I-mfa or I-mfb and collected at Nieuwkoop Faber (NF) stage 10.5 for lysis. Samples were lysed in a solution containing 100 mM Tris (pH 7.4), 150 mM NaCl, 2 mM EDTA, and 0.5% NP-40 and protease inhibitors and cleared by microcentrifugation for 20 min before adding polyclonal anti-mouse I-mf antisera. Proteins were collected using protein-A Dynabeads (Dynal), eluted with Laemmli SDS sample buffer, and examined by PAGE and Western blotting. Western blots were processed using 12CA5 anti-HA antibodies and then visualized using Amersham ECL reagents. In vitro pull-down assays using maltose-binding protein (MBP) fusions and XTcf3, β-catenin and XTcf3hmgG4A, and myogenin or MyoD synthesized in a reticulocyte lysate system (Promega) and labeled with [35S]methionine were performed as previously described (8). The MBP fusion proteins contained full-length I-mfa, amino acids 163 through 246 of I-mfa (I-mfa-specific C-terminal domain), full-length I-mfc, or amino acids 164 through 251 of I-mfb (I-mfb-specific C-terminal domain). Tcf3 HMGG4A (J. R. Miller, unpublished data) contained amino acid residues 259 through 415 fused to the GaI4 activation domain of pGAD424 (Clontech) and subcloned into pCS2+.
Gel mobility shift assays.
The DNA probe for Tcf3 binding was a 30-bp oligonucleotide encompassing the S1 Tcf3/Lef consensus binding site from the Xenopus siamois promoter (4), and the probe for the myogenic and E proteins contained an E-box consensus binding site (8). The probes were labeled to the same specific activity. XTcf3, β-catenin, myogenin, and the E12 and E47 proteins were synthesized in a wheat germ or reticulocyte lysate (Promega). The purified MBP–I-mf fusion proteins are described above. Proteins were combined in a mixture containing 20 mM HEPES (pH 8.0), 3 mM MgCl2, 1 mM dithiothreitol, and 1 mM EDTA; NaCl to a concentration of 18 mM was provided by the MBP protein preparation or an equivalent volume of phosphate-buffered saline–10% glycerol. Following incubation at 30°C for 15 min, 10 ng of end-labeled probe was added, the mixture was incubated for 10 min at room temperature, and samples were electrophoresed at 200 V for 4 h at 4°C.
RESULTS
In the mouse and in humans, three mRNA isoforms are expressed from the I-mf locus: I-mfa, I-mfb, and I-mfc. They share a common N terminal region and vary in the C terminus as a result of alternate splicing (8). Originally, we planned to assess the ability of I-mfa to inhibit myogenesis and the activity of XMyoD in Xenopus. Therefore, we injected mRNA for I-mf into the equatorial region of two-cell Xenopus embryos and evaluated the expression of myosin heavy chain, a protein expressed in skeletal muscle cells. I-mfa inhibited myosin heavy chain expression and myogenesis, whereas I-mfb and I-mfc isoforms did not (data not shown). Interpretation of myogenic inhibition by I-mfa was complicated by the unexpected lack of anterior and axial structures in many of the injected embryos, a phenotype that is characteristic of inhibition of dorsal axis development. This suggested that ectopic I-mfa was inhibiting the formation of the dorsal axis.
To further characterize the effect of I-mfa on axis specification, I-mfa mRNA was injected in the marginal zone of the two dorsal blastomeres of four-cell embryos. Injected embryos were cultured to the tailbud stage and scored according to the dorsoanterior index (DAI), a quantitative scale ranging from fully ventralized (a score of 0) through normal (a score of 5) to fully dorsalized (a score of 10) (18). I-mfa injection resulted in ventralization of the endogenous axis in a dose-dependent manner, whereas I-mfb and and I-mfc (I-mf isoforms that lack the I-mfa domain) did not alter normal axis development (Table 1). We concluded that I-mfa had an unexpected effect on axis formation and sought to determine whether a homolog of murine I-mfa was expressed during Xenopus development.
TABLE 1.
Inhibition of endogenous dorsal axis formation in Xenopus embryosa
| mRNA injected | Amt injected (ng) | No. examined | Avg DAI |
|---|---|---|---|
| I-mfa | 5 | 17 | 1.5 |
| I-mfa | 1 | 17 | 2.2 |
| I-mfa | 0.5 | 25 | 3.2 |
| I-mfa 163–246 | 5 | 32 | 1.2 |
| I-mfb or I-mfc | 5 | 63 | 5 |
| β-gal control | 5 | 30 | 5 |
| XIC | 5 | 32 | 1.1 |
| XIC | 1 | 27 | 3.0 |
| XIC | 0.5 | 25 | 3.8 |
Embryos were injected at the four-cell stage into the marginal zone of two dorsal blastomeres with the indicated mRNAs. Embryos were cultured to the tailbud stage and scored individually according to the DAI of Kao and Elinson (18). The average DAI score for each type of injection is shown.
Using a degenerate primer strategy based on comparison of the human and mouse I-mfa-specific sequence, we cloned XIC, a Xenopus cDNA that is the apparent ortholog of HIC (Fig. 1A). Compared to mouse I-mfa, XIC is 82% similar to I-mfa in the C-terminal I-mfa-specific region and 43% similar to the region common to all I-mf transcripts. XIC has a higher homology to the HIC protein, being 78% similar overall and 97% similar in the C-terminal region.
FIG. 1.
Sequence and RNA expression pattern of XIC. (A) Amino acid sequence comparison of mouse I-mfa, HIC, and XIC is shown. The conserved carboxy-terminal regions are boxed. (B) RNA from whole Xenopus embryos or the dorsal and ventral halves at specific developmental stages was isolated and reverse transcribed for amplification using primers specific for the specific domain of XIC cDNA. Expression of eflα was monitored as an internal control. There were no XIC PCR signals from control samples prepared without RT.
A developmental time course of XIC expression by RT-PCR demonstrated that XIC is present as maternal mRNA and expression persists throughout embryogenesis (Fig. 1B). During the late blastula and early gastrula stages of development there was no dorsal/ventral asymmetry in XIC RNA expression as determined by RT-PCR; in situ hybridization also revealed no spatially restricted expression (Fig. 1B; data not shown). Ubiquitous expression at the RNA level is characteristic of all currently known axis-determining factors, and the presence of the XIC mRNA in pre-mid-blastula-transition embryos is consistent with a possible role in axis specification.
Injection of XIC in the marginal zone of the two dorsal blastomeres resulted in a degree of ventralization that was similar to that obtained with injection of mouse I-mfa (Table 1). Both mouse I-mfa and XIC share the carboxy-terminal domain specific to the mouse I-mfa isoform, implicating this region as necessary for alteration of axis specification. Injecting mRNA that encoded only the I-mfa-specific domain (amino acids 163 through 246 of mouse I-mfa) resulted in ventralization of the embryo to the same degree as that caused by the full I-mfa protein (Table 1), demonstrating that the action of this carboxy-terminal region is sufficient to alter axis specification. Deletion of the I-mfa-specific domain from either protein eliminated its effect on axis formation. From these experiments we concluded that XIC has an effect on axis formation that is indistinguishable from that of I-mfa. Because this raises the possibility that XIC normally functions in axis development, we investigated the mechanism of action of both I-mfa and XIC.
The molecular steps in the axis specification of Xenopus have been intensively studied (6, 27, 29). Described briefly, signaling through a Wnt-like cascade reduces the function of GSK-3, protecting β-catenin from phosphorylation-dependent degradation. The stabilized β-catenin interacts with the Tcf/Lef transcription factor to initiate siamois expression, which is sufficient for dorsal axis specification. To ascertain the step at which I-mfa perturbs the pathway, we used this well-characterized system to test the ability of I-mfa to block axis duplication by specific components of the Wnt pathway. Injection of mRNA for Xwnt-8, a dominant negative GSK-3 (dnGSK-3) (33), β-catenin (46), or siamois into the marginal zone of the two ventral blastomeres of four-cell embryos resulted in the formation of a second axis containing eyes and cement gland (Table 2). Coinjection of I-mfa RNA completely blocked the specification of a second axis by Xwnt-8, dnGSK-3, and β-catenin, but it did not affect formation of a second axis by siamois. These data indicate that I-mfa acts prior to the transcriptional activation of siamois, possibly at the level of Tcf3 or β-catenin. To assess whether I-mfa facilitates the degradation of β-catenin, we injected mRNA encoding a phosphorylation site mutant of β-catenin (pt β-catenin) (46) that is resistant to GSK-3-mediated degradation. I-mfa efficiently blocked second-axis specification by pt β-catenin (Table 2), and the stability of pt β-catenin in the presence of I-mfa was demonstrated by Western analysis (Fig. 2, lane 4). These data demonstrate that I-mfa blocks the activity of β-catenin-dependent transactivation in a manner that does not rely on the degradation of β-catenin. To examine the effect of the proteins containing the I-mfa-specific domain on XTcf3 function, we injected XTcf3HMGGal4AD (J. R. Miller, unpublished), which contains only the HMG-box DNA binding domain of Tcf3, fused to a Gal4 activation domain. Transactivation by this construct is independent of β-catenin protein. Overexpression of XTcf3HMGG4A on the future ventral side of embryos leads to production of ectopic dorsal axes. Coinjection of I-mfa or the I-mfa domain (amino acids 163 to 246) alone but not I-mfb efficiently repressed the development of ectopic axes (Table 2). We concluded that I-mfa acts within the Wnt pathway, at the level of XTcf3.
TABLE 2.
Inhibition of ectopic dorsal axis in Xenopus embryos
| mRNA injecteda | Duplicate axes (n)b | Single axis (n)c |
|---|---|---|
| Xwnt-8 + control | 16 | 3 |
| Xwnt-8 + I-mfa | 0 | 25 |
| dnGSK3 + control | 20 | 1 |
| dnGSK3 + I-mfa | 0 | 23 |
| β-catenin + control | 15 | 1 |
| β-catenin + I-mfa | 0 | 15 |
| pt β-catenin + control | 49 | 9 |
| pt β-catenin + I-mfa | 2 | 46 |
| siamois + control | 29 | 2 |
| siamois + I-mfa | 22 | 3 |
| XTcf3HMGG4A | 36 | 2 |
| XTcf3HMGG4A + I-mfa | 0 | 66 |
| XTcf3HMGG4A + I-mfb | 12 | 0 |
| XTcf3HMGG4A + 163–246 | 0 | 5 |
Injection of 20 pg of Xwnt-8, 2 pg of dnGSK3, 300 pg of β-catenin, 25 pg of pt-β-catenin, 20 pg of siamois mRNA, or 50 pg of XTcf3HMGG4A was performed alone or in combination with 5 ng of I-mfa or β-galactosidase mRNA into the marginal zone of two ventral blastomeres of four-cell embryos. Two nanograms of RNA for I-mf proteins was coinjected with Tcf3HMGG4A.
Ectopic axes, scored at the tailbud stage, contained eyes and cement glands and were generated by minimal amounts of the Xwnt-8, dnGSK-3, or β-catenin mRNA.
Coinjection of I-mfa resulted in embryos containing only a single axis; inhibition of the ectopic axes was complete.
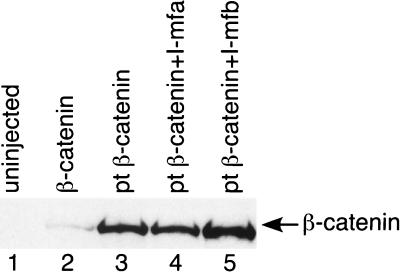
FIG. 2.
I-mfa inhibits dorsal axis in the presence of stable pt β-catenin. The two ventral cells of four-cell Xenopus embryos were injected with 60 pg of mRNA encoding myc-tagged β-catenin (lane 2) or myc-tagged pt β-catenin (lanes 3 through 5). I-mfa (2.5 ng; lane 4) or I-mfb (2.5 ng; lane 5) mRNA was coinjected with the mutant pt β-catenin mRNA in some embryos. Embryos were collected for analysis of proteins by Western blotting with antibody to the myc epitope. Lysate from the uninjected embryos is shown in lane 1.
Since I-mfa and XIC both block formation of the endogenous axis, we next examined whether they do so by blocking the expression of Wnt target genes that function in axis formation. Both siamois (4) and Xnr3 (26) are direct targets of the β-catenin/Tcf transcriptional activation complex that participate in specification of the dorsal axis in Xenopus embryos. To determine whether I-mfa prevents transcription of siamois and Xnr3, we monitored mRNA levels by RT-PCR in I-mfa- and control-injected embryos. Injection of I-mfa mRNA substantially reduced the abundance of both siamois and Xnr3 mRNA (Fig. 3A, lane 2). Coinjection of I-mfa mRNA with an excess of β-catenin mRNA partially rescued the expression of siamois and Xnr3 (lane 4). Consistent with ventralization of the dorsal axis, reduction of dorsal specific gene expression was dose dependent (lanes 6 through 8). As with other experiments, identical results were obtained when XIC mRNA was injected into embryos.
FIG. 3.
I-mfa is an inhibitor of dorsal marker gene expression and Tcf3/β-catenin-dependent promoter activation. (A) I-mfa reduces endogenous expression of siamois and Xnr3. Five nanograms of control mRNA (lane 1), I-mfa mRNA (lane 2), β-catenin (lane 3), or I-mfa plus 1.5 ng of β-catenin mRNA (lane 4) was injected subequatorially into the dorsal side of four-cell embryos. To determine a dose response 1 ng (lane 6), 2.5 ng (lane 7), or 5 ng (lane 8) of I-mfa mRNA was injected as described in Materials and Methods. At early gastrula stages of development, RNA was isolated from pools of eight embryos, and RT-PCR was performed using primer pairs for siamois and Xnr3 with ef1α as an internal control. Control reaction mixtures containing no RT showed no PCR signals. (B) I-mfa negatively regulates β-catenin-responsive siamois promoter activity. Two-cell embryos were injected into the animal pole with 300 pg of the siamois S013 (with Tcf/Lef sites) reporter construct and the indicated mRNAs—β-catenin (300 pg), I-mfa (5 ng), or I-mfb (5 ng). Results of a representative experiment are shown. (C) I-mfa inhibits β-catenin activation of a synthetic Lef/Tcf promoter fused to luciferase. Three hundred picograms of TOPFLASH or FOPFLASH reporter DNA was injected into the animal pole of two-cell embryos with 300 ng of β-catenin and 5 ng of I-mfa as indicated. Results of a representative experiment are shown. Embryos for all luciferase assays were collected at NF stage 10.25 in pools of five embryos and assayed in triplicate. Results were averaged and expressed as relative light units (RLU).
The regulatory sequences of the siamois and Xnr3 genes contain binding sites for Tcf/Lef (4, 26), through which β-catenin regulates transcription in a complex with Tcf/Lef. To determine whether I-mfa prevents expression of siamois and Xnr3 in a manner involving these Tcf/Lef binding sites, we injected reporter constructs containing the Tcf/Lef binding sites in the context of the siamois regulatory region into the animal pole of two-cell-stage embryos (Fig. 3B). The assay relies on endogenous XTcf3 and exogenous β-catenin. Activity from the siamois construct S013, containing an 800-nucleotide promoter fragment with three intact Tcf binding sites, was induced by β-catenin, as previously reported (4). I-mfa strongly repressed the β-catenin induction of reporter activity. In contrast, I-mfb had no significant effect on this or any other reporter used (Fig. 3B), indicating the necessity of the conserved carboxy-terminal region of I-mfa. We extended our analysis to the synthetic promoter TOPFLASH, which consists of multimerized Tcf/Lef binding sites (21). Again, I-mfa inhibited the β-catenin-mediated activation of this promoter, whereas mutation of the Tcf binding sites in the FOPFLASH mutant promoter (Fig. 3C) reduced the amount of both activation by β-catenin and suppression by I-mfa. We observed similar results using XIC mRNA. We concluded that proteins containing the I-mfa domain prevent the β-catenin/Tcf complex from positively regulating transcription.
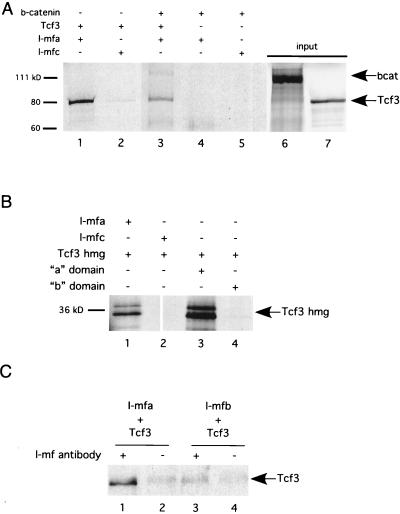
Because the observed inhibition of XTcf3-dependent transcriptional activity could be direct or indirect, we next examined whether I-mfa physically associates with XTcf3. In vitro-translated, 35S-labeled XTcf3 protein was incubated with purified MBP–I-mf fusions and the complexes were collected on amylose resin. The MBPs fused with I-mfc and a sample with no I-mf protein (not shown) were used as controls. As shown in Fig. 4A, XTcf3 interacts specifically with I-mfa (lane 1) but not with I-mfc (lane 2). β-Catenin is also pulled down with I-mfa and XTcf3 (lane 3) but not in the absence of XTcf3 (lane 4), indicating that β-catenin does not directly bind I-mfa. Our inability to detect direct interaction between I-mfa and β-catenin suggests that it joins the complex through its interaction with the N terminus of Tcf3. Further characterizations that mapped the regions required for the I-mfa and XTcf3 interaction were obtained using XTcf3HMGG4A as a template for protein used in the MBP–I-mfa pull-down assay.
FIG. 4.
I-mfa and XTcf3 interact directly in vitro and in vivo. (A) In vitro association between XTcf3 proteins and I-mf proteins. Full-length XTcf3 protein interacts with MBP–I-mfa (lane 1) but not with MBP–I-mfc (lane 2). β-Catenin (bcat) protein participates in a multiprotein complex with I-mfa, but only if XTcf3 is included (lane 3). β-Catenin does not interact with I-mfa alone (lane 4) or with I-mfc (lane 5). Lanes 6 and 7 contain aliquots of input proteins that represent 15% of the amounts in experimental reaction mixtures. (B) The DNA binding domain of XTcf3, expressed in the XTcf3HMGG4A protein, associates with full-length MBP–I-mfa (lane 1) and the C-terminal domain of I-mfa (lane 3) but not with full-length MPP–I-mfc (lane 2) or the C-terminal domain of I-mfb (lane 4). (C) XTcf3 coimmunoprecipitates with I-mfa but not I-mfb from Xenopus embryos. Embryos were injected with 3 ng of HA-tagged XTcf3 RNA, in combination with 3 ng of either myc-tagged I-mfa (lanes 1 and 2) or myc-tagged I-mfb RNA (lanes 3 and 4). Following immunoprecipitation using anti-I-mf antibodies (lanes 1 and 3), proteins were examined by Western blotting with anti-HA antibodies. Lanes 2 and 4 were from mock immunoprecipitation reaction mixtures containing no anti-I-mf antibodies. Background signals in lanes 2, 3, and 4 may be due to nonspecific binding of XTcf3 to the beads, as they are not dependent on the presence of antibody. +, included; −, not included.
Consistent with the results of in vivo functional analysis, the XTcf3 protein that contains only the DNA binding domain XTcf3HMGG4A physically interacts with full-length I-mfa (Fig. 4B, lane 1) as well as the I-mfa-specific domain alone (lane 3), but it does not interact with full-length I-mfc (lane 2) or the C-terminal region of I-mfb (lane 4). XTcf3 also interacts specifically with I-mfa in vivo, as demonstrated by results of coimmunoprecipitation from injected Xenopus embryos. We injected the animal pole of two-cell Xenopus embryos with mRNA encoding HA-tagged XTcf3 and either myc-tagged mouse I-mfa or myc-tagged I-mfb. Injected embryos were lysed, and the I-mf proteins were immunoprecipitated with an antibody that recognizes mouse I-mf isoforms, but not XIC, followed by Western blot detection of the HA-tagged XTcf3. XTcf3 coprecipitated with I-mfa (Fig. 4C, lane 1) but not I-mfb (lane 3), indicating that the I-mfa domain mediates complex formation, as was observed in the in vitro assays. We concluded that both in vivo and in vitro, I-mfa and XIC interact directly with XTcf3.
I-mfa is known to inhibit the activity of myogenic bHLH transcription factors by two mechanisms: (i) I-mfa masks the nuclear localization signal of the myogenic bHLH protein, resulting in cytoplasmic accumulation; and (ii) interaction of bHLH proteins with I-mfa also prevents DNA binding independently of nuclear localization (8). To determine whether I-mfa interferes with the binding of XTcf3 to DNA, we performed gel mobility shift assays using a single XTcf3 binding site from the siamois gene promoter as a probe (4). In vitro-translated XTcf3 protein bound to the probe and formed a low-mobility complex (Fig. 5A, lane 2). Addition of purified I-mfa fusion protein (MBP with the I-mfa C-terminal domain, residues 163 through 246) interfered with the assembly of the complex in a dose-dependent manner (Fig. 5A, lanes 3, 4, and 5). The unique carboxy-terminal domain (residues 164 through 251) of I-mfb and full-length I-mfc (Fig. 5A, lanes 6 and 7) did not affect the XTcf3-DNA complex formation even when included at the highest concentration that was tested for the I-mfa domain. I-mfa alone did not bind to the probe (data not shown). When I-mfa was added after the assembly of Tcf3-DNA complexes, it did not efficiently disrupt the preassembled complex (Fig. 5B). These data indicate that the I-mfa domain blocks the transcriptional activity of β-catenin by interacting with the DNA binding domain of Tcf3, thus preventing the Tcf3-β-catenin complex from binding DNA. This mechanism is consistent with the nuclear as well as cytoplasmic localization of I-mfa (8). Immunofluorescent colocalization of I-mfa, XTcf3, and β-catenin expressed in cultured fibroblasts and in Xenopus embryos demonstrated that I-mfa colocalized with β-catenin and Tcf3 in the nucleus and that I-mfa did not prevent the nuclear localization of Tcf3 or β-catenin (data not shown).
FIG. 5.
Inhibition of XTcf3 binding to DNA by the I-mfa domain. (A) In vitro-translated XTcf3 protein formed a complex with DNA in an electrophoretic mobility shift assay (lane 2) that was diminished by the addition of increasing amounts of purified I-mfa domain-containing protein (lanes 3, 4, and 5). Purified I-mfb (lane 6) and I-mfc (lane 7) did not reduce the amount of shifted probe. 1× corresponds to 0.2 μg of purified MBP fusion protein. A control reaction containing only the probe is shown in lane 1. Asterisks on right-hand side of panel A indicate nonspecific bands. (B) I-mfa reduces binding of Tcf3 to DNA less efficiently when added after assembly of the DNA-protein complex. Purified I-mfa domain-containing protein was added either before (lanes 2 through 5) or after (lane 6) in vitro-translated XTcf3 protein was combined with probe DNA. Lanes 5 and 6 show reactions containing equivalent amounts of I-mfa, and the sample shown in lane 1 contained only control lysate and probe.
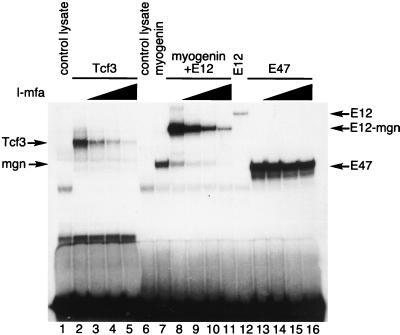
As a test of specificity, we compared the ability of I-mfa to inhibit DNA binding of Tcf3 with its ability to inhibit binding of a previously characterized I-mfa-sensitive protein complex (a myogenin-E12 heterodimer) and an I-mfa-insensitive complex (an E47 homodimer). As in the previous experiment, Tcf3 formed a low-mobility complex with a probe containing its binding site, and increasing amounts of I-mfa diminished formation of that complex (Fig. 6, lanes 2 through 5). An E12-myogenin heterodimer similarly formed a low-mobility complex with a probe containing its E-box binding site, and increasing amounts of I-mfa diminished the formation of this complex (lanes 8 through 11). In contrast, I-mfa did not substantially inhibit the binding of an E47 homodimer to the same E-box probe (lanes 13 through 16).
FIG. 6.
I-mfa inhibits binding by Tcf3 and myogenin-E12 heterodimers, but not by E47 homodimers. In vitro-translated XTcf3 (lanes 2 through 5) and heterodimers of myogenin and E12 protein (lanes 8 through 11) formed complexes with probes containing their respective DNA binding sites that were reduced by increasing amounts (0.2 μg [lanes 3, 9, and 14], 1 μg [lanes 4, 10, and 15], and 4 μg [lanes 5, 11, and 16]) of I-mfa domain-containing protein. The band shift generated by E47 homodimers (lanes 13 through 16) binding to the same DNA probe as the myogenin-E12 heterodimer was not affected by the same amounts of I-mfa protein.
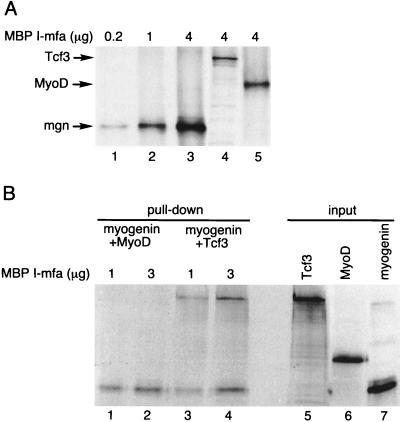
We next sought to determine whether I-mfa bound to Tcf3 with approximately the same affinity that it bound to the myogenic bHLH proteins. Previous studies had demonstrated that I-mfa interacted with myogenin and Myf5 with a higher affinity than with MyoD, and we therefore compared the relative abilities of myogenin, MyoD, and Tcf3 to bind to I-mfa. We used an amount of MBP–I-mfa that was limiting relative to the amount of in vitro-translated myogenin protein, as demonstrated by the increased amounts of protein pulled down by increasing amounts of MBP–I-mfa (Fig. 7A, lanes 1 through 3). This amount of I-mfa also pulled down Tcf3 and MyoD (lanes 4 and 5, respectively). Based on this titration, we used intermediate amounts of MBP–I-mfa with saturating amounts of myogenin and added equivalent amounts of MyoD or Tcf3. Roughly equal amounts of myogenin and Tcf3 bound to the limiting amounts of I-mfa when both were added together (Fig. 7B, lanes 3 and 4), indicating relatively equal affinities of myogenin and Tcf3 for I-mfa. In contrast, the presence of myogenin precluded MyoD from binding to I-mfa (Fig. 7B, lanes 1 and 2), despite the fact that MyoD would bind I-mfa in the absence of myogenin (Fig. 7A, lane 5), which is consistent with the previously demonstrated higher affinity of I-mfa for myogenin than for MyoD.
FIG. 7.
I-mfa has similar binding affinity for XTcf3 and for myogenin. (A) In a pull-down assay using a constant amount of input in vitro-translated protein, increasing amounts of MBP–I-mfa (0.2 μg, 1 μg, and 4 μg) recovered increasing amounts of myogenin (lanes 1 through 3), demonstrating that the I-mfa was limiting relative to the amount of myogenin. In vitro-translated XTcf3 and MyoD were also recovered by the MBP–I-mfa (lanes 4 and 5). (B) In a competitive pull-down assay, limiting amounts of I-mfa protein were incubated with relatively equal amounts (as determined by phosphorimager quantitation and adjustment for the number of methionine residues in each protein) of myogenin together with either MyoD or XTcf3. Relatively equal amounts of myogenin and XTcf3 (lanes 3 and 4) were pulled down by the limiting amount of I-mfa, whereas myogenin showed preferential binding compared to MyoD (lanes 1 and 2). Lanes 5 through 7 show representative aliquots of the in vitro-translated input proteins.
DISCUSSION
We have demonstrated that two I-mfa domain-containing proteins, I-mfa and XIC, can regulate the transcriptional activity of a β-catenin-Tcf3 complex. Mechanistically, the I-mfa domain directly interacts with XTcf3 and prevents its binding to DNA, but the I-mfa does not disrupt the formation of a complex between Tcf3 and β-catenin. As a consequence, the I-mfa domain proteins inhibit the ability of β-catenin to mediate transcription through Tcf3/Lef binding sites in promoters responsive to an activated Wnt pathway. In vivo this results in the inhibition of dorsal axis formation when I-mfa RNA or XIC RNA is injected into early Xenopus embryos. Since XIC is expressed ubiquitously as both a maternal and zygotic transcript in Xenopus embryos, it has the potential both to regulate the axis determination activity of XTcf3 and to modulate the proposed functions of XTcf3 or Lef at later stages of development, such as neural patterning (25) or mesoderm development (15).
The knowledge that I-mfa can interact with Tcf/Lef proteins and modulate the Wnt signaling pathway allows reconsideration of the phenotype of the I-mfa knockout mouse (23). In the 129/Sv mouse strain, I-mfa knockout resulted in delayed neural tube closure and decreased expression of Pax-1 and scleraxis in the ventral sclerotome and suppressed the outgrowth of the cartilagenous rib primordia, resulting in skeletal-patterning defects. Although some or all of these features could reflect dysregulated bHLH activity, it is interesting that dorsoventral patterning of the somite is regulated by a combination of Wnt and SHH signals (7, 10, 39). Wnt signaling appears critical for the formation of the paraxial mesoderm and dorsal somite development, whereas SHH acts to ventralize the somite (7, 24, 45). It is likely that the Wnt signaling is mediated at least in part through Tcf-1 and Lef-1, since the double knockout of both Tcf-1 and Lef-1 has a phenotype similar to the Wnt3a knockout (11). Therefore, the relatively high level of I-mfa expression in the ventral somite and sclerotome (8, 23) could act to limit the dorsalizing activity of Wnt signaling. In this regard, overexpression of Wnt-1 inhibits Pax-1 expression and suppresses chondrogenesis (7), phenotypes seen in the I-mfa-null mouse. In contrast to the 129/Sv mouse strain, deletion of the I-mf gene in the C57BL/6 mouse strain results in placental failure that is secondary to a decreased number of trophoblast giant cells. In this case, we demonstrated that I-mfa binds the bHLH protein Mash2 and prevents its transcriptional activity in a manner analogous to the inhibition of the myogenic bHLH proteins by I-mfa. Disruption of Wnt-2 causes placentation defects that are associated with ectopic giant cells in the placenta (28). Therefore, Wnt-2 suppresses the ectopic giant cells, whereas I-mfa promotes the formation of giant cells. Since Tcf-1 is expressed in the derivatives of the trophectoderm (32) and the combined Tcf-1/Lef-1 double-mutant mouse does not form a placenta (11), it is plausible to suggest that some of the placentation defects in the I-mfa-null mice might be secondary to a loss of inhibition of the Wnt signaling pathway.
Multiple roles for mammalian Tcf/Lef proteins during early development are consistent with their complex overlapping expression patterns (22, 32). Tcf-1 and Lef-1 have wide, coincident expression patterns until embryonic day 13.5 when they are expressed in common regions such as tooth buds and the thymus as well as unique regions such as trophectoderm-derived cells and thoracic prevertebrae for Tcf-1 and tail prevertebrae and brain for Lef-1. Both are expressed exclusively in lymphoid cells of adult animals. The phenotype of double knockout mice for Tcf-1 and Lef-1 correlates the activity of these proteins with Wnt signal transduction and reveals redundant roles that affect paraxial mesoderm differentiation as well as development of placenta and limb buds. Tcf4 expression overlaps with that of Lef-1 in the embryonic brain, but it is uniquely expressed in the intestinal epithelium. Mice lacking the Tcf4 gene are normal with the exception that they fail to maintain crypt stem cells of the small intestine (20). Tcf3 is expressed for a short period during gastrulation (22). The widespread expression pattern and knockout phenotype of I-mfa (23) are consistent with a potential role in regulation of Tcf/Lef activities, particularly in skeletal, placental, and mesoderm development. That additional effects of the knockout were not observed may be due to overlapping expression of a murine ortholog of HIC. While we currently know nothing about HIC RNA levels during mammalian development, the expression of this RNA in adult lymphoid tissue and the small intestine encourages investigation of its role in Tcf-1- or Tcf3-mediated thymocyte (41) or colorectal epithelial differentation and carcinoma (20). It is interesting that HIC suppression of transcription from the HIV-1 long-terminal repeat in T cells may be explained by the presence of a functionally significant (19) Tcf-1 binding site in the retroviral enhancer. Factors which negatively regulate HIV transcription could contribute to the development of cell populations that contain latent virus. A clearer understanding of the importance for I-mfa domain proteins, particularly with regard to Wnt signaling through Tcf/Lef proteins during early development, will necessitate a description of the embryonic expression patterns of other family members such as HIC and possibly the generation of null mutations of both genes.
The canonical Wnt signal transduction cascade is subject to multiple levels of negative control. Most negative regulation of the Wnt signaling pathway occurs through control of the degradation of β-catenin by factors including adenomatous polyposis coli and axin (1, 30). In addition, members of the Sox protein family have been shown to interact with the same armadillo repeat region of β-catenin as do Tcf and Lef, thereby repressing its signaling activity (47). Corepressors of Tcf3 also play a role in regulation of Wnt pathway activity. Specifically, Groucho (36) and CtBP (3) are corepressors of XTcf3 that affect Xenopus development. In contrast, I-mfa does not promote the degradation of β-catenin or prevent the translocation of β-catenin to the nucleus (L. Snider, unpublished observations). Instead, I-mfa prevents the binding of the β-catenin-Tcf3 complex to DNA by masking the Tcf3HMGDNA binding domain. It is interesting that I-mfa interacts with an alpha-helical region of the myogenic bHLH proteins to mask their DNA binding and nuclear localization signals and that the Tcf3 HMG domain also has an alpha-helical structure (43). In summary, we have demonstrated that I-mfa domain proteins that are known to interact with a subset of bHLH proteins also interact with the HMG transcription factor Tcf3 and repress Wnt signal transduction. Because members of these protein families participate in multiple differentiation pathways, the I-mfa domain has the potential to coordinate their regulation.
ACKNOWLEDGMENTS
L.S. dedicates this paper to the memory of Hal Weintraub.
We are grateful to Andrew Lassar and David Kimelman for their comments on this manuscript and to former and current members of the Weintraub and Tapscott labs for their support and discussions. David Kimelman and Jurgen Behrens provided plasmids, Doug Melton the Xenopus oocyte library, and Don Bergstrom invaluable assistance with production of the figures.
R.T.M. is an Investigator and J.R.M. is an Associate of the Howard Hughes Medical Institute. This work was supported by NIAMS AR45113 to S.J.T.
REFERENCES
- 1.Behrens J, Jerchow B-A, Wurtele M, Grimm J, Asbrand C, Wirtz R, Kuhl M, Wedlich D, Birchmeier W. Functional interaction of an axin homolog, conductin, with β-catenin, APC, and GSK3B. Science. 1998;280:596–599. doi: 10.1126/science.280.5363.596. [DOI] [PubMed] [Google Scholar]
- 2.Behrens J, von Kies J P, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Functional interaction of β-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 3.Brannon M, Brown J D, Bates R, Kimelman D, Moon R T. XCtBP is a XTcf-3 co-repressor with roles throughout Xenopus development. Development. 1999;126:3159–3170. doi: 10.1242/dev.126.14.3159. [DOI] [PubMed] [Google Scholar]
- 4.Brannon M, Gomperts M, Sumoy L, Moon R T, Kimelman D. A β-catenin/XTcf-3 complex binds to the siamois promoter to regulate dorsal axis specification in Xenopus. Genes Dev. 1997;11:2359–2370. doi: 10.1101/gad.11.18.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brannon M, Kimelman D. Activation of siamois by the Wnt pathway. Dev Biol. 1996;180:344–347. doi: 10.1006/dbio.1996.0306. [DOI] [PubMed] [Google Scholar]
- 6.Cadigan K M, Nusse R. Wnt signaling: a common theme in animal development. Genes Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- 7.Capdevila J, Tabin C, Johnson R L. Control of dorsoventral somite patterning by Wnt-1 and β-catenin. Dev Biol. 1998;193:182–194. doi: 10.1006/dbio.1997.8806. [DOI] [PubMed] [Google Scholar]
- 8.Chen A C-M, Kraut N, Groudine M, Weintraub H. I-mf, a novel myogenic repressor, interacts with members of the MyoD family. Cell. 1996;86:731–741. doi: 10.1016/s0092-8674(00)80148-8. [DOI] [PubMed] [Google Scholar]
- 9.Cserjesi P, Brown D, Ligon K L, Lyons G E, Copeland N G, Gilbert D J, Jenkins N A, Olson E N. Scleraxis; a basic helix-loop-helix protein that prefigures skeletal formation during mouse embryogenesis. Development. 1995;121:1099–1110. doi: 10.1242/dev.121.4.1099. [DOI] [PubMed] [Google Scholar]
- 10.Fan C-M, Lee C S, Tessier-Lavigne M. A role for Wnt proteins in induction of dermomyotome. Dev Biol. 1997;191:160–165. doi: 10.1006/dbio.1997.8713. [DOI] [PubMed] [Google Scholar]
- 11.Galceran J, Farinas I, Depew M J, Clevers H, Grosschedl R. Wnt3a−/−-like phenotype and limb deficiency in Lef1−/−Tcf1−/− mice. Genes Dev. 1999;13:709–717. doi: 10.1101/gad.13.6.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giese K, Cox J, Grosschedl R. The HMG domain of lymphoid enhancer factor 1 bends DNA and facilitates assembly of functional nucleoprotein structures. Cell. 1992;69:185–195. doi: 10.1016/0092-8674(92)90129-z. [DOI] [PubMed] [Google Scholar]
- 13.Grosschedl R, Giese K, Pagel J. HMG domain proteins: architectural elements in the assembly of nucleoprotein structures. Trends Genet. 1994;10:94–100. doi: 10.1016/0168-9525(94)90232-1. [DOI] [PubMed] [Google Scholar]
- 14.Haegel H, Larue L, Ohsugi M, Fedorov L, Herrenknecht K, Kemler R. Lack of β-catenin affects mouse development at gastrulation. Development. 1995;121:3529–3537. doi: 10.1242/dev.121.11.3529. [DOI] [PubMed] [Google Scholar]
- 15.Hoppler S, Brown J D, Moon R T. Expression of a dominant-negative Wnt blocks induction of MyoD in Xenopus embryos. Genes Dev. 1996;10:2805–2817. doi: 10.1101/gad.10.21.2805. [DOI] [PubMed] [Google Scholar]
- 16.Huber O, Korn R, McLaughlin J, Ohsugi M, Hermann B G, Kemler R. Nuclear localization of β-catenin by interaction with transcription factor LEF-1. Mech Dev. 1996;59:3–10. doi: 10.1016/0925-4773(96)00597-7. [DOI] [PubMed] [Google Scholar]
- 17.Huelsken J, Vogel R, Brinkmann V, Erdmann B, Birchmeier C, Birchmeier W. Requirement for beta-catenin in anterior-posterior axis formation in mice. J Cell Biol. 2000;148:567–568. doi: 10.1083/jcb.148.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kao K R, Elinson R P. The entire mesodermal mantle behaves as Spemann's organizer in dorsoanterior enhanced Xenopus laevis embryos. Dev Biol. 1988;127:64–77. doi: 10.1016/0012-1606(88)90189-3. [DOI] [PubMed] [Google Scholar]
- 19.Kim J Y H, Gonzalez-Scarano F, Zeichner S L, Alwine J C. Replication of type 1 human immunodeficiency viruses containing linker substitution mutations in the −201 to −130 region of the long terminal repeat. J Virol. 1993;67:1658–1662. doi: 10.1128/jvi.67.3.1658-1662.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korineck V, Barker N, Moerer P, van Donselaar E, Huls G, Peters P J, Clevers H. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat Genet. 1998;19:379–383. doi: 10.1038/1270. [DOI] [PubMed] [Google Scholar]
- 21.Korineck V, Barker N, Morin P, van Wichen D, de Weger R, Kinzler K W, Vogelstein B, Clevers H. Constitutive transcriptional activation by a β-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 22.Korineck V, Barker N, Willert K, Molenaar M, Roose J, Wagenaar G, Markman M, Lamers W, Destree O, Clevers H. Two members of the Tcf family implicated in Wnt/β-catenin signaling during embryogenesis in the mouse. Mol Cell Biol. 1998;18:1248–1256. doi: 10.1128/mcb.18.3.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kraut N, Snider L, Chen C-M A, Tapscott S J, Groudine M. Requirement of the mouse i-mfa gene for placental development and skeletal patterning. EMBO J. 1998;17:6276–6288. doi: 10.1093/emboj/17.21.6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marcelle C, Stark M R, Bonner-Fraser M. Coordinate actions of BMPs, Wnts, Shh and noggin mediate patterning of the dorsal somite. Development. 1997;124:3955–3963. doi: 10.1242/dev.124.20.3955. [DOI] [PubMed] [Google Scholar]
- 25.McGrew L L, Takemaru K-I, Bates R, Moon R T. Direct regulation of the Xenopus engrailed-2 promoter by the Wnt signaling pathway, and a molecular screen for Wnt-responsive genes, confirm a role for Wnt signaling during neural patterning in Xenopus. Mech Dev. 1999;87:21–32. doi: 10.1016/s0925-4773(99)00136-7. [DOI] [PubMed] [Google Scholar]
- 26.McKendry R, Hsu S-C, Harland R M, Grosschedl R. LEF-1/TCF proteins mediate Wnt-inducible transcription from the Xenopus nodal-related 3 promoter. Dev Biol. 1997;192:420–431. doi: 10.1006/dbio.1997.8797. [DOI] [PubMed] [Google Scholar]
- 27.Miller J, Moon R T. Signal transduction through β-catenin and specification of cell fate during embryogenesis. Genes Dev. 1996;10:2527–2539. doi: 10.1101/gad.10.20.2527. [DOI] [PubMed] [Google Scholar]
- 28.Monkley S J, Delaney S J, Pennisi D J, Christiansen J H, Wainwright B. Targeted disruption of the Wnt2 gene results in placentation defects. Development. 1996;122:3343–3353. doi: 10.1242/dev.122.11.3343. [DOI] [PubMed] [Google Scholar]
- 29.Moon R T, Kimelman D. From cortical rotation to organizer gene expression: toward a molecular explanation of axis specification in Xenopus. BioEssays. 1998;20:536–545. doi: 10.1002/(SICI)1521-1878(199807)20:7<536::AID-BIES4>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 30.Munemitsu S, Albert I, Souza B, Rubinfeld B, Polakis P. Regulation of intracellular β-catenin levels by the adenomatous polyposis coli (APC) tumor-suppressor protein. Proc Natl Acad Sci USA. 1995;92:3046–3050. doi: 10.1073/pnas.92.7.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Munsterberg A E, Kitajewski J, Bumcrot D A, McMahon A P, Lassar A B. Combinatorial signaling by sonic hedgehog and Wnt family members induces myogenic bHLH gene expression in the somite. Genes Dev. 1995;9:2911–2922. doi: 10.1101/gad.9.23.2911. [DOI] [PubMed] [Google Scholar]
- 32.Oosterwegel M, van de Wetering M, Timmerman J, Kruisbeek A, Destree O, Meijlink F, Clevers H. Differential expression of the HMG box factors TCF-1 and LEF-1 during murine embryogenesis. Development. 1993;118:439–448. doi: 10.1242/dev.118.2.439. [DOI] [PubMed] [Google Scholar]
- 33.Pierce S B, Kimelman D. Regulation of Spemann organizer formation by the intracellular kinase Xgsk-3. Development. 1995;121:755–765. doi: 10.1242/dev.121.3.755. [DOI] [PubMed] [Google Scholar]
- 34.Ranganayakulu G, Schulz R A, Olson E N. Wingless signaling induces nautilus expression in the ventral mesoderm of the Drosophila embryo. Dev Biol. 1996;176:143–148. doi: 10.1006/dbio.1996.9987. [DOI] [PubMed] [Google Scholar]
- 35.Rebagliati M R, Weeks D L, Harvey R P, Melton D A. Identification and cloning of localized maternal RNAs from Xenopus eggs. Cell. 1985;42:769–777. doi: 10.1016/0092-8674(85)90273-9. [DOI] [PubMed] [Google Scholar]
- 36.Roose J, Molenaar M, Peterson J, Hurenkamp J, Brantjes H, Moerer P, van de Wetering M, Destree O, Clevers H. The Xenopus Wnt effector Xtcf-3 interacts with Groucho-related trascriptional repressors. Nature. 1998;395:608–612. doi: 10.1038/26989. [DOI] [PubMed] [Google Scholar]
- 37.Rupp R A W, Snider L, Weintraub H. Xenopus embryos regulate the nuclear localization of XMyoD. Genes Dev. 1994;8:1311–1323. doi: 10.1101/gad.8.11.1311. [DOI] [PubMed] [Google Scholar]
- 38.Rupp R A W, Weintraub H. Ubiquitous MyoD transcription at the midblastula transition precedes induction-dependent MyoD expression in presumptive mesoderm of X. laevis. Cell. 1991;65:927–937. doi: 10.1016/0092-8674(91)90545-a. [DOI] [PubMed] [Google Scholar]
- 39.Tajbakhsh S, Borello U, Vivarelli E, Kelly R, Papkoff J, Duprez D, Buckingham M, Cossu G. Differential activation of Myf5 and MyoD by different Wnts in explants of mouse paraxial mesoderm and the later activation of myogenesis in the absence of Myf5. Development. 1998;125:4155–4162. doi: 10.1242/dev.125.21.4155. [DOI] [PubMed] [Google Scholar]
- 40.Thebault S, Gachon F, Lemasson I, Devaux C, Mesnard J-M. Molecular cloning of a novel human I-mfa domain-containing protein that differentially regulates human T-cell leukemia virus type I and HIV-1 expression. J Biol Chem. 2000;275:4848–4857. doi: 10.1074/jbc.275.7.4848. [DOI] [PubMed] [Google Scholar]
- 41.Verbeek S, Izon D, Hofhuis F, Robanus-Maandag E, te Riele H, van de Wetering M, Oosterwegel M, Wilson A, MacDonald H R, Clevers H. An HMG-box-containing T-cell factor required for thymocyte differentiation. Nature. 1995;374:70–74. doi: 10.1038/374070a0. [DOI] [PubMed] [Google Scholar]
- 42.Waterman M, Jones K. Purification of TCF-1α, a T-cell-specific transcription factor that activates the T cell receptor Cα gene enhancer in a context-dependent manner. New Biol. 1990;2:621–636. [PubMed] [Google Scholar]
- 43.Waterman M L, Fischer W H, Jones K A. A thymus-specific member of the HMG protein family regulates the human T cell receptor Cα enhancer. Genes Dev. 1991;5:656–669. doi: 10.1101/gad.5.4.656. [DOI] [PubMed] [Google Scholar]
- 44.Yang-Snyder J, Miller J R, Brown J D, Lai C J, Moon R T. A frizzled homolog functions in a vertebrate Wnt signaling pathway. Curr Biol. 1996;6:1302–1306. doi: 10.1016/s0960-9822(02)70716-1. [DOI] [PubMed] [Google Scholar]
- 45.Yoshikawa Y, Fujimori T, McMahon A P, Takada S. Evidence that absence of Wnt-3a signaling promotes neuralization instead of paraxial mesoderm development in the mouse. Dev Biol. 1997;183:243–249. doi: 10.1006/dbio.1997.8502. [DOI] [PubMed] [Google Scholar]
- 46.Yost C, Torres M, Miller J, Huang R, E, Kimelman D, Moon R T. The axis-inducing activity, stability, and subcellular distribution of β-catenin is regulated in Xenopus embryos by glycogen synthase kinase 3. Genes Dev. 1996;10:1443–1454. doi: 10.1101/gad.10.12.1443. [DOI] [PubMed] [Google Scholar]
- 47.Zorn A, Barish G D, Wiliams B O, Lavender P, Klymkowsky M W, Varmus H E. Regulation of Wnt signaling by Sox proteins: XSox17a/b and XSox3 physically interact with β-catenin. Mol Cell. 1999;4:487–498. doi: 10.1016/s1097-2765(00)80200-2. [DOI] [PubMed] [Google Scholar]