Abstract
Although substantial progress has been made with Ebola virus (EBOV) vaccine measures, the immune correlates of vaccine-mediated protection remain uncertain. Here, five mucosal vaccine vectors based on human and avian paramyxoviruses provided nonhuman primates with varying degrees of protection, despite expressing the same EBOV glycoprotein (GP) immunogen. Each vaccine produced antibody responses that differed in Fc-mediated functions and isotype composition, as well as in magnitude and coverage toward GP and its conformational and linear epitopes. Differences in the degree of protection and comprehensive characterization of the response afforded the opportunity to identify which features and functions were elevated in survivors and could therefore serve as vaccine correlates of protection. Pairwise network correlation analysis of 139 immune- and vaccine-related parameters was performed to demonstrate relationships with survival. Total GP-specific antibodies, as measured by biolayer interferometry, but not neutralizing IgG or IgA titers, correlated with survival. Fc-mediated functions and the amount of receptor binding domain antibodies were associated with improved survival outcomes, alluding to the protective mechanisms of these vaccines. Therefore, functional qualities of the antibody response, particularly Fc-mediated effects and GP specificity, rather than simply magnitude of the response, appear central to vaccine-induced protection against EBOV. The heterogeneity of the response profile between the vaccines indicates that each vaccine likely exhibits its own protective signature and the requirements for an efficacious EBOV vaccine are complex.
INTRODUCTION
Ebola virus (EBOV) causes a severe disease with a high case fatality rate of 25 to 90% (1). EBOV vaccine efficacy testing in humans is ethically impossible given the lethality of the virus. Testing during outbreaks of Ebola virus disease (EVD) presents logistical challenges due to their sporadic nature. In these situations, the U.S. Food and Drug Administration’s (FDA’s) “Animal Rule” grants approval of vaccines based on preclinical efficacy studies in relevant animal models that show likely clinical benefit to humans (2). Therefore, deciphering the immune responses to vaccination that correlate with protection is imperative to predict efficacy in humans.
The concept that all EBOV vaccines have a common correlate of protection is under debate (3). Protective correlates can depend on several variables including vaccine type, delivery route, and route of infection, making their definition complex and a single correlate to predict survival questionable. As with most vaccines, immunoglobulin G (IgG) titers are thought to be the best correlate of protection (4, 5). However, depending on the type of vaccine, cell-mediated responses may also be associated with survival (6–8). The unprecedented 2013–2016 EBOV epidemic in West Africa, followed by additional outbreaks in the Democratic Republic of Congo, sparked the emergency rollout of the most promising vaccine candidates into clinical trials up to phase 3 (9–14). Despite FDA (15) and European Commission (16) approvals and despite receipt of the vaccine by more than 303,000 people (17), the immune correlates of protection for the recombinant vesicular stomatitis virus–Zaire Ebola virus (rVSV-ZEBOV) or other EBOV vaccines are still under investigation. Establishing the signatures of vaccine-generated immunity remains crucial for vaccine design, assessment, and application.
Previously, we developed a panel of nine respiratory virus–vectored vaccines, based on human and avian paramyxoviruses (APMVs), expressing the glycoprotein (GP) of EBOV, and tested them in a guinea pig challenge model (18). The panel was narrowed down to the five vectors that conferred 100% protection and elicited high-magnitude responses against a broader spectrum of GP antigenic regions. In this study, nonhuman primates (NHPs) received two doses of the selected vaccine candidates via the combined intranasal and intratracheal (IN/IT) route and were subsequently exposed to a lethal dose of EBOV by intramuscular injection. The candidates conferred varying degrees of protection from death and disease, ranging from disease-free survival to only partial protection. We focused on characterizing the antibody response profile rather than T cell responses, because we have previously shown that vaccination through the respiratory tract elicits robust EBOV-specific T cell responses, mostly at the site of vaccination, which may be more relevant for protection against respiratory challenge (19). However, protection against challenge may be supported by an anamnestic T cell response. Through in-depth characterization of the humoral response, we found that even though all vaccine vectors express the same antigen, they differed in multiple aspects, including response coverage toward GP, neutralizing and linear epitopes, isotype distribution, the facilitation of innate immune effector cells, and the magnitude of total antibody bound to GP and its truncated structures. The correlates of protection appeared to be unique to the vaccine platform, composed of multiple and possibly complementary features that integrate to determine the outcome of infection.
We used a pairwise network analysis approach to integrate 139 vaccine response features measured in this study, including the properties of vaccine-induced antibodies beyond conventional IgG and neutralization titers, and identified which features best correlated with survival. We also identified features that correlated with the quality of survival that accounted for the degree of EVD observed in each NHP. We show that the total GP-binding antibody response, as determined by biolayer interferometry (BLI), but not by enzyme-linked immunosorbent assay (ELISA), is an influential factor in survival. Furthermore, Fc-mediated effector functions, as well as the response coverage toward the receptor binding domain (RBD) of GP, were strongly associated with a better outcome after EBOV challenge, alluding to potential mechanisms involved in survival. The relevance of the functional aspects of vaccine-induced antibodies, and not just their quantities, in survival has important implications on how we currently assess the efficacy of vaccines in humans.
RESULTS
Five needle-free mucosal EBOV vaccines expressing the same immunogen confer varying degrees of protection in NHPs
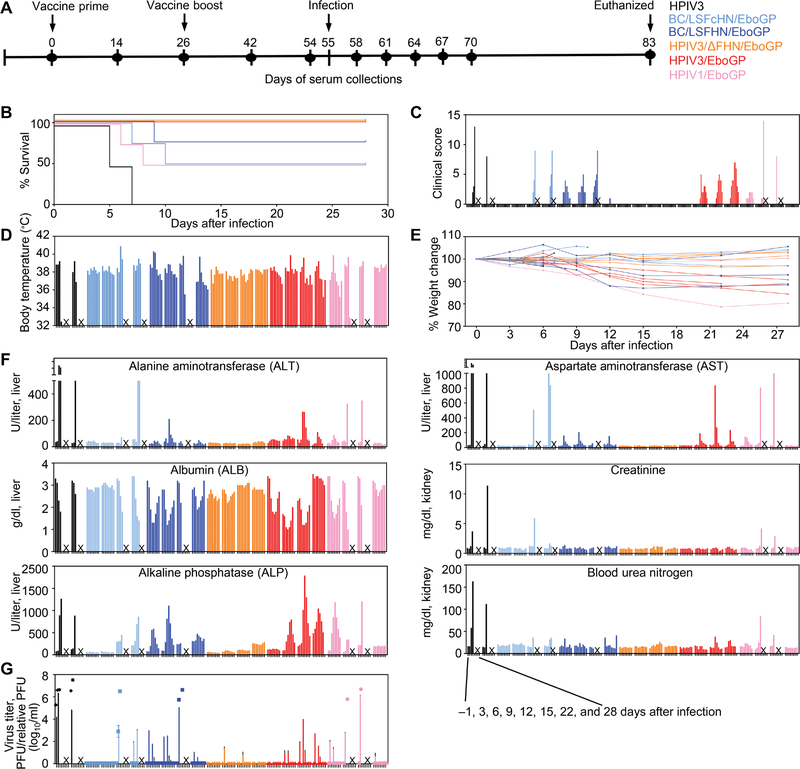
Our goal was to test the efficacy of five mucosal vaccines expressing GP against lethal EBOV (Fig. 1A). Three vaccines were derived from human paramyxoviruses, human parainfluenza virus (HPIV) 1 (HPIV1/EboGP), and HPIV3 with (HPIV3/EboGP) or without their fusion (F) and hemagglutinin-neuraminidase (HN) surface proteins (HPIV3/ΔFHN/EboGP) (fig. S1). Two vaccines were based on the APMV, Newcastle disease virus (NDV) strain Beaudette C (BC), which had the NDV LaSota (LS) strain’s F protein cleavage site (Fc) and HN protein (BC/LSFcHN/EboGP) or the F and HN proteins (BC/LSFHN/EboGP). Cynomolgus macaques (n = 4 per vaccine group) were prime vaccinated on day 0 and homologously boosted on day 26 via a combined IN/IT route (Fig. 1A). One group (n = 2) was vaccinated with HPIV3 for the study’s control. All vaccines replicated in the upper respiratory tract after each dose (fig. S2A), whereas replication in the lower respiratory tract was only detectable after the prime dose and not observed for HPIV1/EboGP recipients (fig. S2B). Typically, vector-specific immune responses to the first dose restrict the replication of the vaccine vector after a second dose (20). On day 55, the NHPs were infected with the targeted dose of 1000 plaque-forming units (PFUs) of EBOV and were monitored over 28 days for signs of EVD (Fig. 1, B to F). Survival was defined when the animal did not reach a moribund condition requiring euthanasia. The two control animals met end point criteria on days 5 and 7 (Fig. 1B). All animals from the HPIV3/ΔFHN/EboGP- and HPIV3/EboGP-vaccinated groups survived. However, although the HPIV3/ΔFHN/EboGP group displayed no signs of illness throughout the infection course, some animals from the HPIV3/EboGP group developed disease. Three of the four recipients of the BC/LSFHN/EboGP vaccine survived but were not free of disease. Two of the four NHPs from both the HPIV1/EboGP and BC/LSFcHN/EboGP groups survived, but one HPIV1/EboGP-vaccinated survivor exhibited EVD, whereas BC/LSFcHN/EboGP-vaccinated survivors did not. Vaccinated animals that met euthanasia criteria presented with severe signs of EVD, markers of impaired liver and kidney function (Fig. 1F), and viremia comparable to the control group detected by both plaque assay and the more sensitive quantitative reverse transcription polymerase chain reaction (qRT-PCR) (Fig. 1G). Vaccinated animals that survived infection but presented with clinical illness and elevated liver and kidney enzyme readings during the acute stage had transient viremia detectable only by qRT-PCR. Some vaccinated animals, free of EVD symptoms, had low viremia detectable only by qRT-PCR at a single sampling point, soon after infection. Thus, vaccines conferred NHPs with varying degrees of protection despite expressing the same GP immunogen.
Fig. 1. Survival, clinical scores, weight loss, temperature, markers of liver and kidney function, and viremia in vaccinated and infected NHPs.
(A) NHPs were vaccinated (n = 4 per group) via the IN/IT route on day 0 and boosted on day 26 with rBC/LSFcHN/EboGP (light blue), rBC/LSFHN/EboGP (dark blue), HPIV3/ΔFHN/EboGP (orange), HPIV3/EboGP (red), and HPIV1/EboGP (pink). A control group received the HPIV3 vector control (n = 2; black). (B to E) Animals were challenged with 1000 PFU of EBOV by the intramuscular route at day 55. All animals were monitored for changes in survival (B), clinical scores (C), body temperature (D), and percent weight change (E) for 28 days after the challenge. Each bar represents an individual NHP at the indicated time points. (F) Serum was collected from all animals at 3-day intervals over the course of infection and measured for blood markers of EVD. Each bar represents an individual NHP at the indicated time points. (G) Viremia in serum determined by plaque titration (PFU/ml; symbols) and in qRT-PCR (relative PFU/ml; bars). Each symbol or bar represents the mean of replicates for each NHP. Error bars represent ±SEM. “X” indicates nonsurviving animals.
Immune correlates of protection could not be defined with conventional immunologic assays
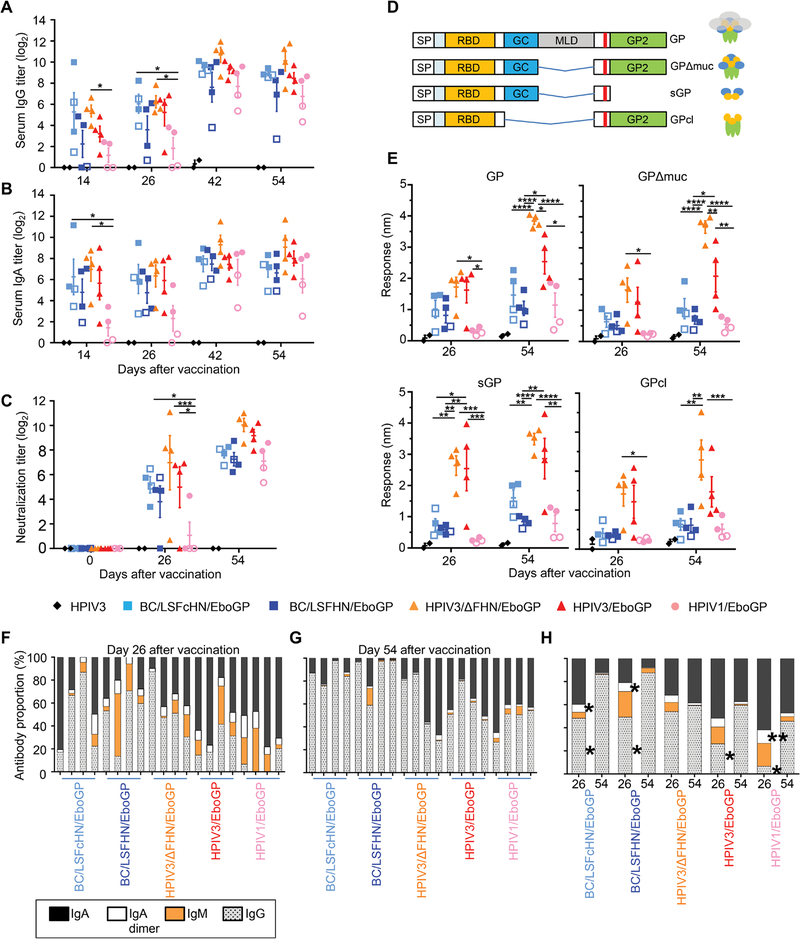
To determine whether the abundance of antibodies could explain the varying degrees of protection, we used both classical and BLI-based high-resolution serological assays. Most vaccinated NHP groups induced GP-specific IgG response 2 weeks after the prime dose based on serum ELISA results (Fig. 2A). Mean IgG titers increased after administration of the boost dose and were comparable between all vaccine groups, despite some vaccines conferring only partial protection. Vector-specific immunity after prime vaccination did not appear to impede the response to the booster. The serum GP-specific IgA titers reflected GP-specific IgG titers (Fig. 2B). However, the trends observed in serum were not mirrored by mucosal GP-specific IgA and IgG titers in the respiratory tract (fig. S3). The HPIV3/ΔFHN/EboGP vaccine did not produce higher GP-specific IgA and IgG titers compared to the other vaccines (Fig. 2, A and B). However, differences in IgA or IgG titers were significantly different between vaccinated NHPs grouped according to survivors versus nonsurvivors, irrespective of the vaccine they received (P ≤ 0.05; fig. S4, A and B). Vaccine groups did not demonstrate differences in IgM titers after the first or second vaccine dose (fig. S4C), nor were differences in IgM titers observed between survivors and nonsurvivors (fig. S4D). To determine whether serum GP-specific antibody binding titers correlate with the quality of survival, we assigned each animal a survival index defined by the criteria of clinically observed scores, liver and kidney disease markers, and viremia (table S1). All HPIV3/ΔFHN/EboGP recipients had the highest index scores, because they were completely protected. A moderate but significant positive correlation was observed between the quality of survival and serum GP-specific IgG (P = 0.0051) or IgA (P = 0.0032) ELISA titers (fig. S4, E and F). IgM titers did not correlate with the quality of survival (fig. S4G).
Fig. 2. Serum antibody responses in vaccinated NHP.
(A to C) Total GP-specific IgG (A) and IgA (B) in serum and serum EBOV neutralization titers (C) were measured in vaccinated groups before EBOV infection using ELISA and plaque reduction assays, respectively. Post-vaccination ELISA titers were corrected by subtraction of respective preimmune sera baseline values. Open symbols indicate nonsurviving animals. Bars denote group means ± SEM. (D) Schematic of the truncated GP forms evaluated by BLI. (E) Maximum response of total post-vaccination sera binding to the different truncated GP forms using BLI. (F and G) Proportion of the antibody isotypes in the total GP-specific antibody response for each NHP at days 26 (F) and 54 (G). IgA (black), IgA dimer (white), IgM (orange), and IgG (gray). Each bar represents an individual NHP. (H) Average isotype proportions for each vaccine group at days 26 and 54. Significance was measured by a two-way ANOVA with (A) to (C) and (E) Tukey’s correction or (H) Sidak’s correction for multiple comparisons. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001.
Most NHPs elicited detectable neutralizing titers after the prime dose, which increased after the boost dose (Fig. 2C). By day 54, neutralizing titers were comparable between HPIV3/ΔFHN/EboGP and HPIV3/EboGP groups, suggesting that this parameter cannot account for why the latter group exhibited EVD. The difference in neutralizing titers between individuals within each APMV group was minimal and again could not explain why some of these animals did not survive. The two HPIV1 vaccine recipients that did not survive infection, however, did exhibit somewhat lower neutralizing titers when compared to their surviving counterparts. The neutralizing titers in vaccinated survivors were not different to vaccinated nonsurvivors (fig. S4H). However, a moderate significant correlation was observed between neutralizing titers and the survival index (P = 0.0014; fig. S4I). Therefore, ELISA binding or neutralizing antibody titers should be cautiously used as protective correlates, because they may only be relevant on a per vaccine mode basis, given the modest correlations we observed with our survival index.
For higher-resolution analysis of antibody binding, BLI was performed using the Octet platform. The sensors of the instrument were coated with the full GP ectodomain or its truncated forms lacking the mucin-like domain (MLD) (GPΔmuc), secreted GP (sGP), and cathepsin-cleaved GP (GPcl) (Fig. 2D). Contrary to the ELISA data, BLI revealed pronounced differences between the vaccine groups when we examined total antibody binding to all GP forms (Fig. 2E). The sera from the HPIV3/ΔFHN/EboGP group displayed the highest binding response to all GP forms; antibody binding to GP, GPΔmuc, and GPcl exceeded that of the other groups, potentially associating antibodies specific for epitopes in full GP and epitopes exposed upon removal of the MLD or cathepsin cleavage with the absence of disease. We found that sera from vaccinated survivors have a higher capacity to bind all GP forms than sera from vaccinated nonsurvivors (fig. S5A). Moreover, the antibody binding response to all GP forms strongly correlated with the survival index (P < 0.0001, GP; P = 0.0002, GPΔmuc; P = 0.0002, sGP; P = 0.0007, GPcl; fig. S5B). HPIV3/ΔFHN/EboGP and HPIV3/EboGP groups exhibited disparate protection against EVD but did not differ in their sGP and GPcl antibody binding response (Fig. 2E). Therefore, disease-free survival may not be facilitated by these sGP- and GPcl-binding antibodies or may depend on the functionality of these antibodies rather than their abundance.
We quantified the IgG, IgA, and IgM isotype composition of serum binding to EBOV GP, relative to the total amount of bound antibodies, using an ELISA (Fig. 2, F to H). For each serum sample, the amount of a specific anti-GP isotype was calculated from an isotype control standard curve and divided by the total quantity of GP-specific isotypes and represented as a percentage. After the prime dose, most HPIV-based vaccine recipients had a higher portion of their GP-specific response composed of the IgA isotype compared to APMV-based vaccine recipients (Fig. 2F). After the boost dose, the composition of HPIV sera was more balanced between IgA and IgG isotypes, whereas the IgG isotype dominated in APMV sera (Fig. 2G). For HPIV3/ΔFHN/EboGP, the IgG and IgA proportions remained relatively constant after the boost dose, which conflicted with the congruent increases in IgG and decreases in IgA observed for all other groups (Fig. 2H). All groups, except HPIV3/ΔFHN/EboGP and HPIV3/EboGP, had significant reductions in the frequency of their IgA dimers after the boost dose (P ≤ 0.05; Fig. 2H). Although the mean absolute IgM ELISA titers for vaccine groups after the booster were not different from titers achieved after the prime dose (fig. S4C), IgM proportions relative to the IgG and IgA isotypes were much lower when compared to the prime dose (Fig. 2H). Thus, as IgA and IgG titers increased after the boost dose, a switch away from the IgM isotype did not occur. Therefore, IgM titers and the kinetics of the IgM response are unlikely to influence protection. Survivors demonstrated no differences in isotype frequencies when compared to nonsurvivors after each vaccine dose (fig. S6). Together, these data demonstrate that the total GP-specific antibody response determined by BLI provides greater clarity on the disparity between vaccine modes and between survivors versus nonsurvivors, rather than classical isotype-specific ELISAs or neutralization titers, and thus may be better-suited for predicting protection.
Survivors have a greater proportion of antibodies targeting the RBD and GP2 + GP1/2 base regions of GP
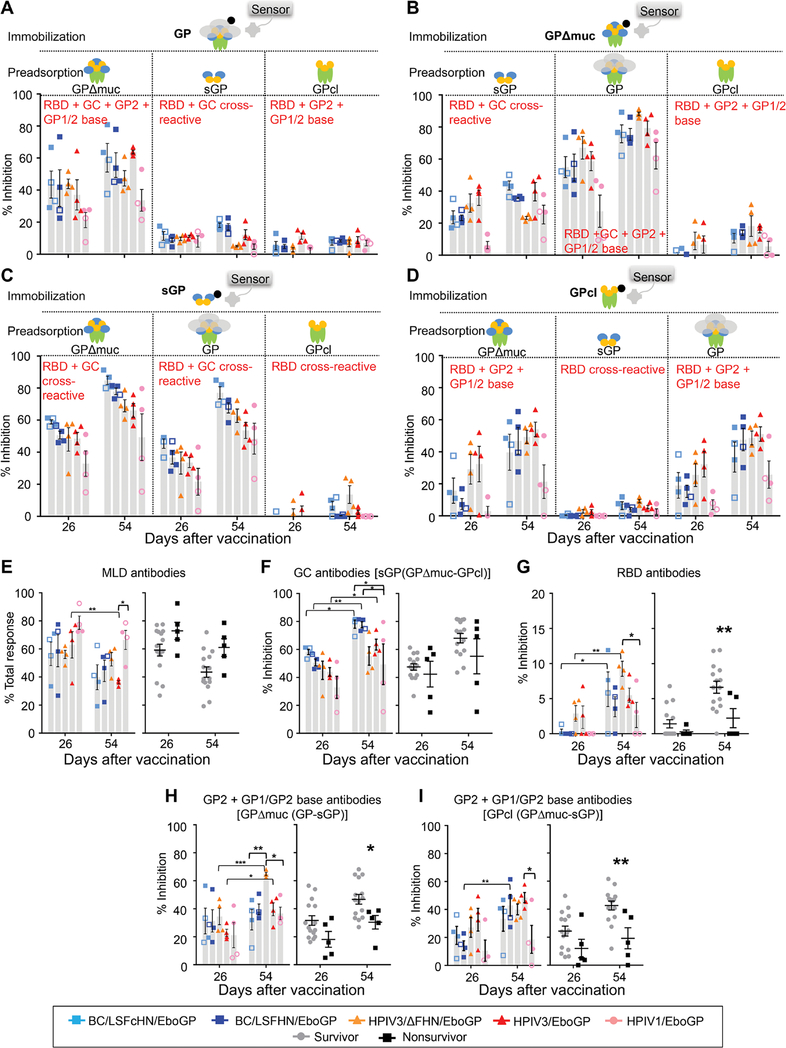
We sought to determine the proportion of the vaccine-induced response directed toward key regions on GP [MLD, glycan cap (GC), RBD, GP1/2 base, and GP2] and thereby identify the regions that elicit protective responses. GP region–specific responses were measured using BLI competition assays (Fig. 3, A to D). Sera from vaccinated animals were allowed to bind a GP protein immobilized on the BLI sensor after preadsorption treatment with a GP variant to remove antibodies targeting regions shared between the competing and immobilized GP. The proportion of MLD-specific antibodies was inferred from the percent of serum antibody binding not removed by GPΔmuc preadsorption (Fig. 3A). MLD antibodies decreased significantly for HPIV3/EboGP after the boost dose (P = 0.003) but remained unchanged or marginally decreased in other groups (Fig. 3E). The proportion of MLD antibodies in vaccinated survivors was not different to vaccinated nonsurvivors.
Fig. 3. Distribution of serum antibodies binding to EBOV GP antigenic domains.
(A to D) Results of BLI assays of sera binding to different immobilized biotinylated GP forms after sera preadsorption with various GP forms are shown. Binding inhibition to immobilized GP (A), GPΔmuc (B), sGP (C), and GPcl (D) after preadsorption of serum with GP forms indicated in each figure panel, expressed as a percentage of total binding response (in nanometers) obtained without sera preadsorption. (E) MLD domain antibodies according to vaccine groups or survivors (n = 15) versus nonsurvivors (n = 5). (F) GC antibodies in vaccine groups or survivors and nonsurvivors; binding to biotinylated sGP, inhibited by GPcl adsorption (targeted to the RBD), was subtracted from the binding removed by GPΔmuc adsorption (targeted to the GC and RBD). (G) RBD antibodies in vaccine groups or survivors versus nonsurvivors. (H and I) Proportion of GP2 + GP1/GP2 base antibodies for vaccine groups or survivors versus nonsurvivors, calculated by either subtracting percent binding inhibition to GPΔmuc caused by sGP from the percent inhibition caused by GP (H) or subtracting percent binding inhibition to GPcl caused by sGP from the percent inhibition caused by GPΔmuc (I). NHPs were organized according to groups (symbols, open symbols denote nonsurvivors; and gray bars represent mean values ± SEM) or survivors (gray circles) and nonsurvivors (black squares) ± SEM. (E to I) Significance measured by two-way ANOVA with post hoc tests for multiple comparisons between vaccine groups at a given time point (Tukey’s correction) or between time points and between survivors and nonsurvivors (Sidak’s correction). *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001.
RBD and GC antibodies that cross-react with sGP and GP could be quantified by measuring the binding of the sGP-preadsorbed serum to immobilized GP (Fig. 3A) or GPΔmuc (Fig. 3B). Although the binding pattern was similar using these two immobilized GP forms, measurements with immobilized GPΔmuc excludes the MLD antibody pool and therefore augments the proportion of GC and RBD antibodies. The APMV and HPIV1 responses directed to the sGP/GP cross-reactive epitopes in the GC and RBD increased after the boost dose but remained unchanged for the HPIV3 vectors (Fig. 3B). This trend was also observed when the GPcl-preadsorbed serum bound to immobilized GPΔmuc (Fig. 3B) but not GP (Fig. 3A). Therefore, the increases observed for the APMV and HPIV1 groups may be attributed to increases in RBD antibodies’ binding epitopes exposed by the removal of the MLD on GPΔmuc. RBD and GC, sGP/GP cross-reactive antibodies could also be measured by the binding of the GP- or GPΔmuc-preadsorbed serum to immobilized sGP (Fig. 3C). In this configuration, cross-reactive antibodies after the boost dose increased for all vaccine groups, including the HPIV3 vectors. GPΔmuc and GP have more surface area for interaction with serum antibodies compared to sGP and could explain the discrepancy between the two configurations used to measure sGP/GP cross-reactive antibodies in the HPIV3-vaccinated groups. Any increase in antibodies targeting these additional surfaces on immobilized GPΔmuc or GP after two doses may conceal the increase in sGP/GP cross-reactive antibodies so that amounts appeared constant. Conversely, for the APMV and HPIV1 groups, the ratio of sGP/GP cross-reactive antibodies to antibodies targeting the additional surfaces on GP may have increased.
GC antibodies that were cross-reactive with sGP and GP were calculated as previously described (18). APMV vaccine recipients had more GC antibodies compared to HPIV recipients (Fig. 3F). Over the vaccination course, GC antibodies increased in APMV [BC/LSFcHN/EboGP (P = 0.0377) and BC/LSFHN/EboGP (P = 0.0040)] and HPIV3/EboGP (P = 0.0421) vaccine groups but did not change for HPIV3/ΔFHN/EboGP and HPIV1 vaccine groups. GC antibody amounts did not appear to delineate survival given the minimal difference observed between survivors and nonsurvivors. GPΔmuc and sGP are structures where tertiary contact points involving the MLD are not possible and the GC is exposed. Sera from all vaccine groups preadsorbed by GPcl, which lacks the GC, were minimally impeded in their ability to bind immobilized GPΔmuc (Fig. 3B) and sGP (Fig. 3C). Therefore, GC antibodies likely comprise a large portion of the response induced by all vaccines, more so than the RBD, GP2, and GP1/GP2 base antibodies. RBD antibodies were measured from the percent of binding to immobilized GPcl, removed by sGP preadsorption (Fig. 3, D and G). RBD antibodies significantly increased in the HPIV3/ΔFHN/EboGP (P = 0.0081)–and BC/LSFcHN/EboGP (P = 0.0126)–vaccinated groups after the boost dose (Fig. 3G). Survivors also had higher amounts of RBD antibodies after the booster (P = 0.0066).
GP2 + GP1/GP2 base antibodies were deduced by subtracting binding inhibition to immobilized GPΔmuc caused by sGP adsorption from the inhibition caused by GP adsorption (Fig. 3H). The HPIV3/ΔFHN/EboGP group had more antibodies targeting GP2 + GP1/GP2 after the boost dose compared to the other vaccine groups (P = 0.0065, HPIV3/ΔFHN/EboGP versus BC/LSFcHN/EboGP; P = 0.0233, HPIV3/ΔFHN/EboGP versus HPIV1/EboGP). Alternately, GP2 + GP1/2 base antibodies could also be determined by subtracting the percent of binding inhibition to GPcl caused by GPΔmuc adsorption from the percent of inhibition caused by sGP adsorption (Fig. 3I). The GP2 + GP1/2 base antibody pool binding to GPcl (Fig. 3I) differs from the pool binding to GPΔmuc (Fig. 3H). Specifically, the HPIV3/ΔFHN/EboGP group had more of GP2 + GP1/2 base antibodies binding to GPΔmuc compared to other groups after the boost dose (Fig. 3H). Against GPcl, these antibodies bound comparably with other groups (Fig. 3I). HPIV3/ΔFHN/EboGP therefore likely has greater amounts of GP1/GP2 base antibodies targeting GPΔmuc, whose binding is disrupted against the proteolytically modified GPcl. Antibodies targeting these GP1/GP2 base sites lost after proteolytic cleavage also increased for HPIV3-derived vaccine groups and remained steady for APMV groups after the booster (Fig. 3H). Regardless of the binding configuration used, GP2 + GP1/GP2 base antibodies were consistently higher in survivors (P = 0.0389, immobilized GPΔmuc; P = 0.0042, immobilized GPcl; Fig. 3, H and I).
The changes in the composition of the antibody response specific to regions on GP were divergent between groups after two vaccine doses with the fully protective HPIV3/ΔFHN/EboGP vaccine response increasing in RBD (P = 0.0081; Fig. 3G) and GP2 + GP1/GP2 base (P = 0.0004; Fig. 3H) antibodies while maintaining steady GC antibody amounts (Fig. 3F). GC and MLD antibodies were the most abundant in the vaccine response, but only the RBD and GP2 + GP1/GP2 base antibodies were enriched in survivors (Fig. 3, G to I). Together, these data demonstrate that the RBD and GP2 + GP1/GP2 base are important antigenic regions on GP that contribute to the protection.
Vaccine vectors affect antigenic regions involved in EBOV neutralization
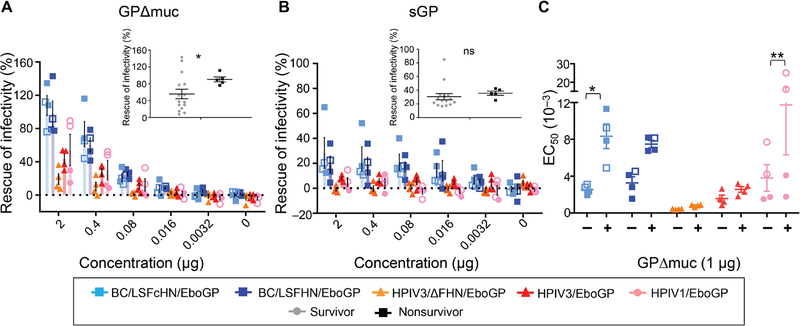
Having determined that RBD and GP2 + GP1/2 base regions were preferentially targeted by survivors and disparities in the response toward GP regions existed between vaccine groups, we next addressed whether the GP regions targeted by neutralizing antibodies were specific to each vaccine. The regions of GP responsible for eliciting neutralizing antibodies were determined by preadsorbing day 54 serum diluted at the concentration required to achieve at least 80% of neutralization, with increasing concentrations of GPΔmuc or sGP (Fig. 4, A and B). The ability of APMV-derived serum to neutralize virus was nearly abolished in the presence of GPΔmuc, indicating that non–MLD-targeting antibodies were heavily involved in neutralization (Fig. 4A). Conversely, in surviving HPIV-vaccinated animals, non–MLD-binding antibodies were not the main contributors toward neutralization, because infectivity was only partially restored in the presence of GPΔmuc. This difference between APMV- and HPIV-based vaccines was further confirmed by determining the effect of GPΔmuc on the concentration of serum required to reduce half the maximal infectivity (EC50), which revealed that EC50 values shifted considerably for APMV vaccines but not HPIV-vaccinated survivors (Fig. 4C). The neutralizing antibodies from the two HPIV1-vaccinated animals, which did not survive, mainly targeted non-MLD regions, whereas the group’s survivors targeted MLD. Contrary to this, there were no differences among the surviving and nonsurviving APMV recipients, indicating that neutralizing antibodies are not determinants of protection for the APMV vaccines. This again exemplifies that each vaccine has different protective mechanisms. Although the contribution of MLD antibodies toward neutralization was different between and within the vaccine groups, the neutralizing response of survivors was associated with greater binding to the MLD (inferred from the low rescue of infectivity percentage; P = 0.0146; Fig. 4A, inset graph). However, there was no correlation between the abundance of MLD neutralizing antibodies and the survival index (fig. S7).
Fig. 4. Epitope specificity of neutralizing antibodies depends on the vaccine vector.
(A and B) Neutralizing activity of day 54 immune sera preincubated with GPΔmuc (A) or sGP (B) expressed as percentages of activity of the same sera not preincubated with proteins. Sera from vaccinated groups, diluted to achieve 80% of their neutralization activities, were incubated with increasing concentrations of GPΔmuc or sGP. The dashed line indicates 0%. Inset graphs represent the differences between survivors (gray circles) and nonsurvivors (black squares) at 2 μg of GPΔmuc or sGP. Significance measured by unpaired t test with Welch’s correction. *P ≤ 0.05; ns, not significant. (C) Serum dilution that reduces infectivity by 50% (EC50) in the absence (−) or presence (+) of 1 μg GPΔmuc. Symbols indicate individual NHPs. Open symbols indicate nonsurvivors. Significance was measured by a two-way ANOVA with Sidak’s correction for multiple comparisons. *P ≤ 0.05 and **P ≤ 0.01.
Preadsorption of sera from APMV groups with sGP had a slight effect on virus infectivity, suggesting that sGP/GP cross-reactive antibodies targeting the RBD and GC weakly contributed to mechanical neutralization (Fig. 4B). sGP had no effect on the ability of serum from HPIV-vaccinated groups to neutralize virus. Moreover, the effect of sGP on the neutralizing ability of sera from survivors was similar to nonsurvivors (Fig. 4B, inset graph). APMV groups were not fully protected against EBOV infection, and their neutralizing antibody repertoire targeted non-MLD regions, some of which cross-react with epitopes on sGP, indicating that they are not the delineating characteristics of survival. In addition, non-MLD antibodies only marginally contributed to the neutralizing ability of the HPIV3/ΔFHN/EboGP-derived sera, and amounts were similar to the surviving animals in the HPIV1/EboGP and HPIV3/EboGP groups who exhibited disease symptoms. Although most of the HPIV-vaccinated survivors’ neutralizing capacity is harnessed from MLD-targeting antibodies, they do not appear to be associated with disease-free survival, pointing to the involvement of other mechanisms in complete protection. Together, these results demonstrate that the neutralizing antibody response elicited by vaccine vectors differ in the regions that they predominately target on GP with HPIV-based vaccines targeting the MLD and APMV-derived vaccines targeting the non-MLD regions. However, neutralizing titers per se do not predict protection and may be more suited as a vaccine-specific correlate rather than applied to all vaccine modes to predict survival.
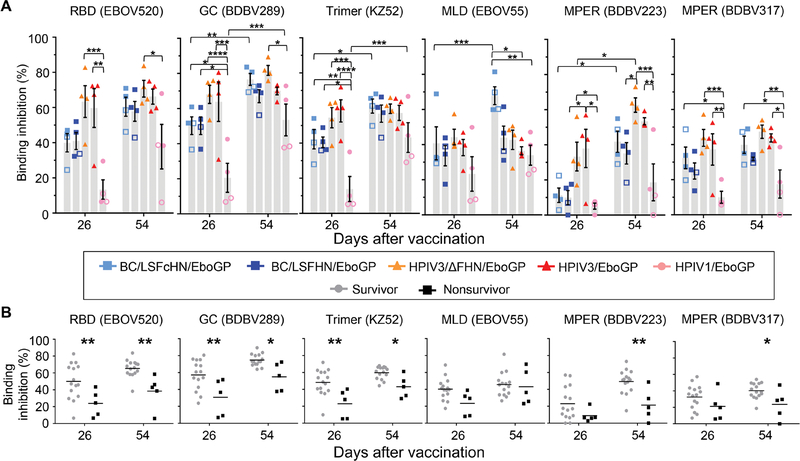
Survivors produce more antibodies toward known protective monoclonal antibody epitopes
We next evaluated the ability of antibodies induced by each vaccine to target conformational epitopes in GP to identify disparities between the vaccines, between survivors and nonsurvivors, and in the evolution of the protective response composition over two doses. Representative monoclonal antibodies (mAbs) isolated from human survivors with known epitopes in the antigenic regions of GP were selected to compete with serum for binding on the BLI platform. These mAbs bind the RBD [EBOV520 (21)], GC [BDBV289 (22)], MLD (EBOV55), the GP1/GP2 interface [KZ52 (23)], or the membrane proximal external region (MPER) [BDBV223 and BDBV317 (24)]. EBOV55 was the only mAb that lacked neutralizing capability and in vivo protection data. After the prime vaccine dose, recipients of HPIV3-derived vaccines generally had higher antibody binding response toward most of the epitopes compared to recipients of the HPIV1 and APMV vaccines (Fig. 5A). Differences between groups tapered after the boost dose, with the serum composition of antibodies targeting some epitopes increasing in both the APMV and HPIV1 recipients while remaining mostly unchanged in HPIV3-based recipients. However, competition between immune sera and the two MPER-specific antibodies differed from the other region-specific mAbs. Whereas epitope-specific antibody responses of both HPIV3/EboGP and HPIV3/ΔFHN/EboGP recipients were typically unaltered after the booster, antibodies targeting the BDBV223 epitope was the only example where binding increased significantly for the HPIV3/ΔFHN/EboGP group (P = 0.0226). Competition with BDBV317 for its MPER epitope saw no differences between the HPIV3 and APMV vaccine groups after the prime dose, with no further changes after the boost dose.
Fig. 5. Epitope diversity of immune sera characterized by competition assays.
(A) The results of a BLI-based competition assay whereby biotinylated GP was immobilized on a streptavidin sensor and saturated with preinfection sera are shown. MAbs were then applied to compete for a specific epitope. The level of inhibition is determined as a percentage of blocking activity compared to negative control sera against the tested mAb. Symbols indicate individual NHPs. Gray bars denote the average blocking of mAbs by sera from vaccinated groups ± SEM. Open symbols indicate nonsurvivors. (B) The percentage of mAb epitope binding inhibition for NHPs organized according to survivors (gray circles) and nonsurvivors (black squares) is shown. Significance measured by two-way ANOVA with (A) Tukey’s correction for multiple comparisons between vaccine groups and Sidak’s correction for multiple comparisons between (A) time points and between (B) survivors and nonsurvivors. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001.
The differential binding response to neutralizing epitopes observed between vaccine groups after prime vaccination coincided with survivors having greater antibody responses toward these epitopes compared to nonsurvivors (Fig. 5B). MPER-targeting antibodies, however, were the exception, because amounts after the prime dose were indistinguishable between survivors and nonsurvivors despite differences in amounts between the vaccine groups. After the booster, survivors had significantly more antibodies targeting all neutralizing epitopes compared to nonsurvivors (P ≤ 0.05), although negligible differences in response toward the majority of epitopes were observed between groups. The serum antibody responses specific for the EBOV55 epitope were not different between survivors and nonsurvivors.
Throughout the vaccination course, the serum antibody response targeting known mAb epitopes evolved separately for each vaccine, indicating divergent antibody response kinetics. However, the minimal differences between vaccine groups after the boost dose may indicate that two vaccine doses ensue similar neutralization responses contingent on the epitope. Although differences in binding activity were seen between survivors and nonsurvivors toward all neutralizing epitopes, the survival index strongly correlated with the RBD (P = 0.0003) and GC (P = 0.0002) epitopes and moderately correlated with MPER [BDBV223 (P = 0.0017) and BDBV317 (P = 0.0054)] and GP1/GP2 (P = 0.0362) interface epitopes (fig. S8). Therefore, a response directed to key epitopes, in this case, within the RBD and GC, may serve as vaccine-independent protective correlates, whereas a response toward other epitopes, for example, the MPER epitope for BDBV223, may be vaccine-specific correlates.
Survivors recognize the linear epitopes of known protective mAbs
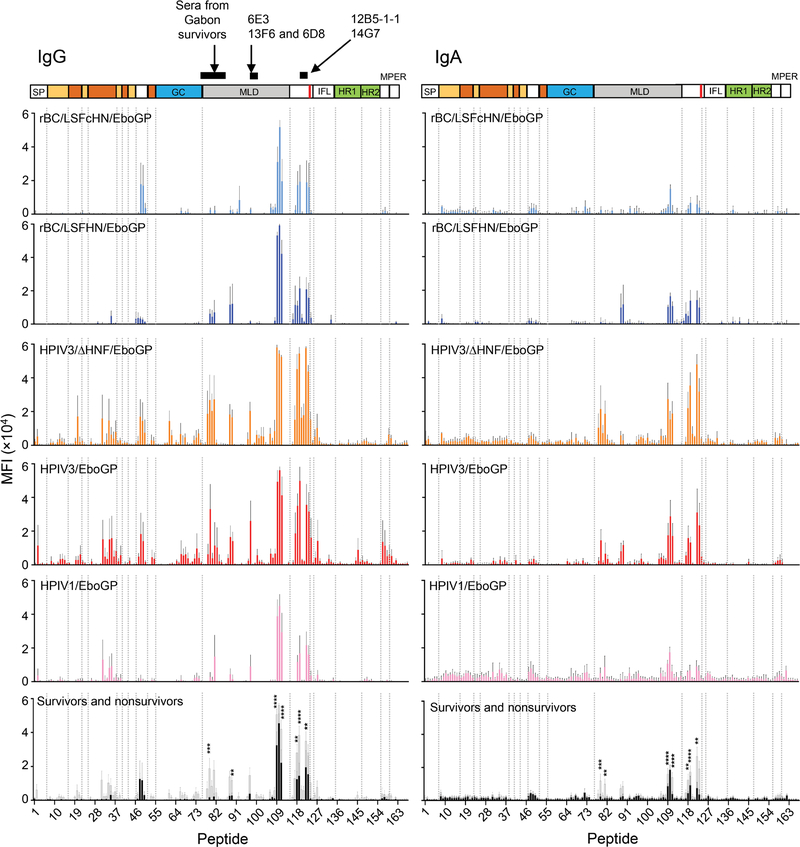
Given the disparities observed between vaccine groups and between survivors and nonsurvivors in their ability to recognize conformational epitopes, we considered whether this was also true for linear epitopes. We examined the ability of IgG and IgA antibodies collected after the boost dose to bind linear epitopes using a peptide array spotted with 15-mer peptides offset by four amino acids spanning the entire GP. There was a clear distinction between the vaccine vector group’s ability to recognize linear epitopes. HPIV3-based vaccine–derived IgG response bound more epitopes in the GC and MLD and at greater magnitudes when compared to the other vaccine groups (Fig. 6 and fig. S9A). HPIV3-based vaccine recipients also had considerably higher IgG magnitudes binding the GP1 C-terminal region, despite no differences in the response breadth between vaccine groups (fig. S9A). IgG from HPIV-based vaccine recipients, but not APMV-based vaccine recipients, recognized linear epitopes in the head region of GP1; HPIV-derived vaccines could generate a response to peptides 31 to 37, whereas detection of peptides 19 to 21 was unique to recipients of the HPIV3-based vaccines (Fig. 6).
Fig. 6. Binding of IgG and IgA from immune sera to peptides spanning full-length GP.
The mean fluorescence intensity (MFI) of day 54 immune sera binding specific, individual GP peptides according to vaccine groups or survivors versus nonsurvivors is plotted against each peptide. The vaccine constructs are indicated at the top left corner of each plot. Bars represent the average MFI ± SEM. The last panel shows the average MFI of survivors (gray bars) and nonsurvivors (black bars). Black lines above the GP schematic indicates previously identified epitopes for mAbs and polyclonal serum. Significance was measured by a two-way ANOVA with Sidak’s correction for all comparisons between survivors and nonsurvivors. **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001.
The binding profiles for IgA and IgG were only similar for the HPIV3-derived vaccine recipients in the MLD and GP1 C-terminal region (Fig. 6). IgA, however, did bind at a lower breadth and magnitude (fig. S9A). Outside these regions, IgA did not recognize the linear epitopes recognized by IgG (Fig. 6). Specific peptides in the MLD (peptides 108 to 111, amino acids 429 to 455) and in the C-terminal end of GP1, just before the furin cleavage site (peptides 117 to 119, amino acids 465 to 487 and peptides 122 to 123, amino acids 485 to 503), mapped according to Lee and Saphire (25), were detected by both the IgA and IgG from all vaccine groups. IgA binding was again lower, especially for the HPIV1- and APMV-based vaccine groups. The magnitude of IgA and IgG targeting these peptides were higher in survivors compared to nonsurvivors, suggesting that they contribute to improved survival, provided that a threshold is reached.
The IgG antibodies from survivors also targeted linear epitopes located at the start of the internal fusion loop (IFL; peptides 126 and 127), in the MLD (peptides 78 to 82, 88, 89, and 97) and GC (peptides 61, 66, 73, and 74), and the head region of GP1 (peptides 20, 31, 34, 35, and 37), which appeared to be absent or below detectable quantities in the nonsurvivors. Peptide 97, located in the MLD, contains epitope for murine mAb 6D8, a neutralizing component of the MB-003 therapeutic (26, 27), and highlights this epitope as a characteristic trait among survivors. In survivors, the IgG and IgA antibody footprints were only similar in the MLD and the GP1 C-terminus, albeit IgA antibodies bound at a lower magnitude and did not recognize peptide 97. The GC, GP1 head region, and IFL epitopes recognized by the IgG from survivors were not recognized by their IgA counterparts. Compared to nonsurvivors, survivors generally had a higher magnitude of antibodies binding to the C-terminus region of GP1 (P = 0.0009, IgG; P = 0.0103, IgA) and a greater breadth of IgA binding to the MLD (P = 0.0089; fig. S9B).
Several epitope footprints unique to the vaccinated survivors are also known epitopes for protective responses. Peptides that contained the epitopes for protective murine mAbs 12B5 (26, 28) and 14G7 (29, 30) and a region in the MLD recognized by sera from survivors of the outbreak in Gabon (31) were bound by the antibodies from vaccinated survivors (Fig. 6). Although the IgA or IgG linear epitope profiles are different between the vaccines, epitopes recognized by serum from our study’s survivors, particularly those historically identified as protective, may act as predictors of survival.
Fc-mediated effector functions correlate with improved survival
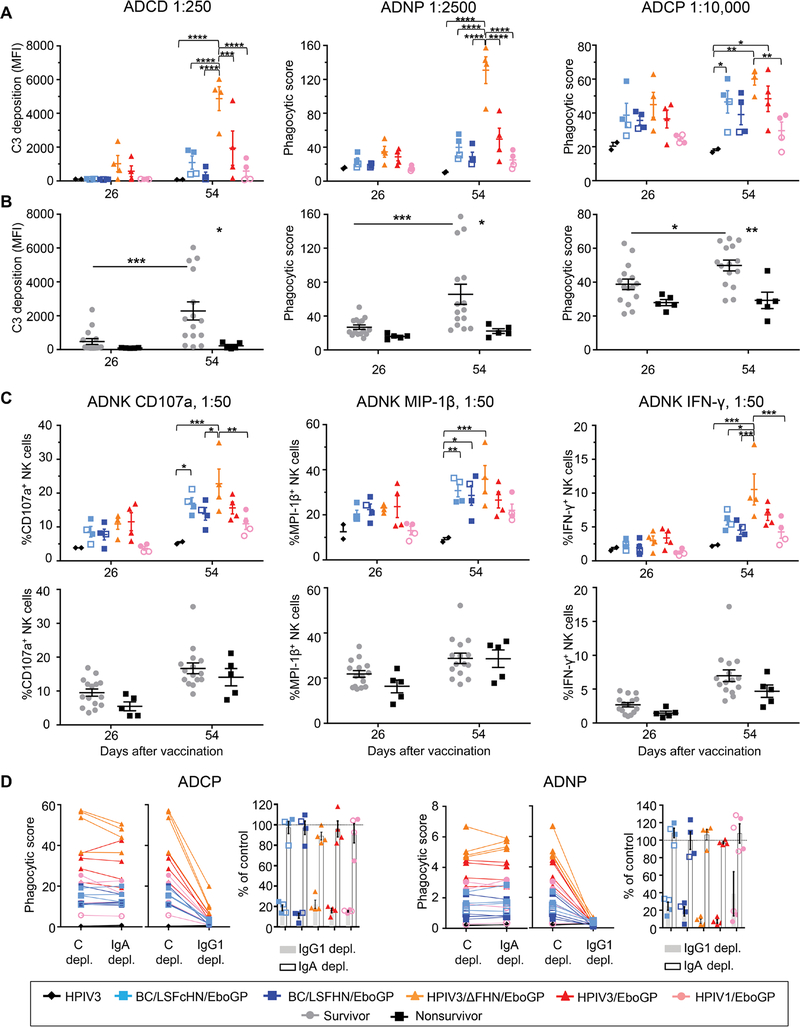
The role of antibody functions other than neutralization in vaccine-mediated protection was examined by the ability of antibody responses from vaccinated groups and survivors to activate Fc-mediated effector functions including antibody-dependent cellular phagocytosis mediated by monocytes (ADCP) and neutrophils (ADNP), antibody-dependent complement deposition (ADCD), and antibody-dependent activation of natural killer cells (ADNK) by analysis of the degranulation marker CD107a and activation markers macrophage inflammatory protein–1β (MIP-1β) and interferon-γ (IFN-γ) (Fig. 7, A to C). After two vaccine doses, the HPIV3/ΔFHN/EboGP group activated all arms of the tested Fc-mediated protective mechanisms to a greater capacity than other vaccines. The correlation between the survival index for NHPs and their ADCD (P < 0.0001) or ADNP (P < 0.0001) activities was strong but moderate for ADCP (P = 0.0019) and absent for ADNK (fig. S10). In survivors, all Fc mechanisms except ADNK significantly increased after the boost dose and were enriched compared to nonsurvivors (P ≤ 0.05; Fig. 7, B and C). To determine which antibody classes mediated the Fc effector functions that correlated with improved survival, we depleted IgG1 and IgA from samples collected after the boost dose and measured the impact on ADNP and ADCP activities. Regardless of the vaccine, IgG1 was responsible for most of these effector functions (Fig. 7D).
Fig. 7. Fc effector functions of the vaccine response.
(A and B) The ADCD, ADNP, and ADCP activity of immune sera at specified dilution from NHPs grouped according to vaccine (color symbol, NHP; open symbol, nonsurvivor) (A) or according to survivors (gray circles) and nonsurvivors (black squares) (B) at days 26 and 54 are shown. C3, complement component 3. (C) NK cell activation based on the production of CD107a, MIP-1β, and IFN-γ is shown using serum collected at days 26 and 54 at specified dilution from vaccinated groups or survivors and nonsurvivors. (D) ADCP and ADNP activity of day 54 samples depleted of IgA or IgG1 compared to the control depletion. Bar graph shows the percent of the functional activity in depleted samples as compared to the control (shown by the gray line at 100%). Significance was measured by two-way ANOVA with Tukey’s correction for multiple comparisons between vaccine groups or Sidak’s correction for all comparisons between survivors and nonsurvivors. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001.
Circulating monocytes and granulocytes, of which neutrophils are the most abundant, were measured by the hematological analysis over the infection course to determine whether different vaccines had varying effects on their frequencies. In human EBOV infections, perturbation of monocyte and neutrophil populations occurs, yet their numbers recover, coinciding with survival and control of virus (32–34). Monocyte numbers in vaccinated groups were marginally affected by infection (fig. S11A). At day 9 after infection, monocytes significantly declined in vaccinated nonsurvivors compared to survivors (P = 0.0454; fig. S11B), and higher monocyte numbers were associated with a better survival outcome (P = 0.0121; fig. S11C). Among the vaccine groups, granulocyte numbers were more stable in the HPIV3/ΔFHN/EboGP recipients, except for one with elevated numbers at day 6 after infection (fig. S11D). Vaccinated nonsurvivors generally experienced protracted granulocytosis compared to survivors (fig. S11E). Moreover, granulocytosis during acute infection was associated with a poor survival outcome (P = 0.0004, day 3; P = 0.0025, day 6; fig. S11F). Although antibody-driven phagocytosis was associated with the survival index (fig. S10), the frequencies of the Fc effector cells may also be influenced by different vaccines to affect the ability of antibodies to mediate effector functions.
In summary, ADCD, ADNP, and ADCP activities appear to promote survival, irrespective of the vaccine. Although there was no apparent involvement of NK cell activity in survival when we look at survivors, independent of the vaccine they received, we cannot exclude that the role of NK cells in protection may be vaccine specific.
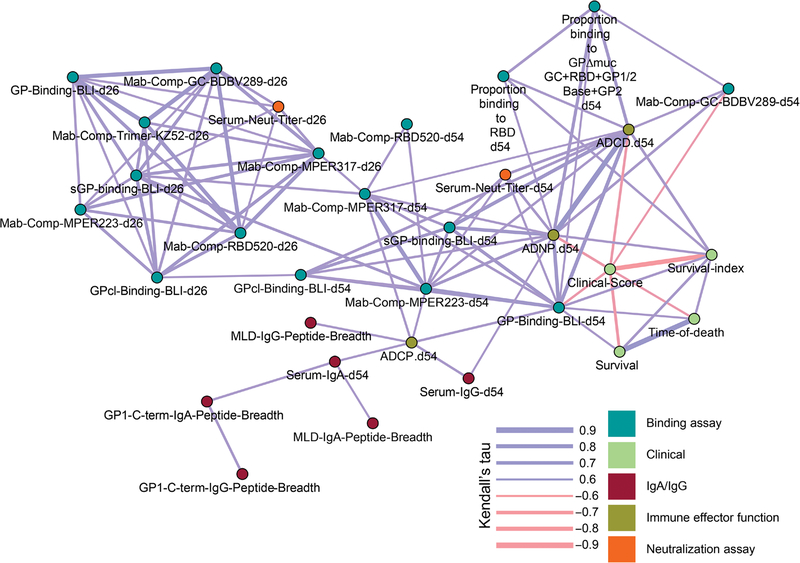
Pairwise correlation analysis identifies correlates of protection from 139 parameters of the vaccine response
Given that our vaccine panel produced varied response profiles and several vaccine response features were elevated in survivors or in some vaccine groups, we used univariate pairwise correlation analysis to integrate all data on response features from the vaccinated NHPs (n = 20) and distinguish which ones could best explain survival (Fig. 8). Because the outcome of severe EVD (survival versus death) can be stochastic, we also applied the survival index, a measure for disease severity based on viremia and clinical parameters we mentioned above, to our analysis. We constructed a correlation network to link relationships between 139 humoral response features and functions with survival and the survival index. No parameter measured after the prime dose correlated with survival. However, amounts of MPER and GPcl-specific antibodies after the prime dose correlated with amounts after the boost dose, which, in turn, was linked to features associated with survival or the survival index and may therefore remotely serve as early predictors of vaccine efficacy. The total antibody binding to GP after the booster that was analyzed by BLI, and not ELISA or neutralizing titers, correlated with survival (Τ = 0.611, P = 0.046) and the survival index (Τ = 0.646, P = 0.033). BLI-determined total antibody amounts also correlated with neutralizing titers (Τ = 0.6211, P = 0.039) and Fc-mediated effects (ADCD, Τ = 0.7789, P = 0.004; ADNP, Τ = 0.7684, P = 0.006; ADCP, Τ = 0.6421, P = 0.033), indicating that polyfunctionality, and not just magnitude, was important for survival. Significant correlations were identified between the survival index and ADCD (Τ = 0.668, P = 0.026) and ADNP (Τ = 0.634, P = 0.035) effector functions, and the proportion of the response binding the RBD (Τ = 0.634, P = 0.034) was measured after the boost dose. Higher measures for RBD antibodies and these effector functions may result in a better outcome of infection, including reduced disease severity and lower viremia.
Fig. 8. Pairwise correlation analysis identifies antibody features associated with improved survival.
Map of correlation network depicting pairs of parameters with statistically significant correlations. Line thickness indicates Kendall rank correlation coefficient values (tau), where thicker lines indicate higher correlation, and color represents direction of correlation (positive, blue; negative, red). Color nodes represent measured parameters grouped according to feature or function.
We also used the multivariate, Cox regression modeling approach to identify a set of vaccine-induced humoral response features that could best predict survival after EBOV infection (fig. S12). The variables selected as most predictive of survival were serum IgA titers, the response coverage of the GP2 + GP1/2 base region, neutralizing epitopes in the GP1/2 base and RBD, linear epitopes in the MLD, and their breadth in both the MLD and C-terminal regions of GP1, albeit they were not considered reliable, because the 95% confidence interval of the predictor coefficients included 0. Pairwise correlation analysis of all measured immune response features found that Fc-mediated functions and RBD antibodies correlated with improved survival outcome. These associations are suggestive of the immune mechanisms and GP regions involved in vaccine-mediated protection against EBOV.
DISCUSSION
EBOV vaccines have been instrumental in limiting recent outbreaks and have been given to more than 303,000 people, yet the correlates of protection have not been completely defined. In this study, we compared multiple respiratory EBOV vaccines to identify what humoral features constitute a protective vaccine-induced response against EBOV. Correlation network analysis evaluating qualitative and quantitative humoral parameters defined RBD-specific antibodies and Fc-mediated immune functions as contributing factors to improved survival. Previous studies have mechanistically determined the importance of antibodies (4), associated antibody features with IgG titers (35, 36), or focused on a limited number of classical serological parameters to draw associations with survival (37, 38). Here, we have determined the qualitative features associated with EBOV vaccine–mediated survival from a comprehensive assessment of the humoral response to multiple vaccines.
Of the five tested EBOV GP–expressing mucosal vaccines derived from human and avian paramyxoviral vectors, the HPIV3/ΔFHN/EboGP vaccine conferred NHPs with the best protection, leaving them free of disease with near-sterilizing immunity against EBOV. The varying degrees of protection afforded by our vaccines provided an opportunity to identify the correlates of vaccine-mediated protection. Binding and neutralizing titers, isotype composition, total sera binding to GP as determined by BLI, the proportion of binding and neutralizing antibodies against certain GP regions, linear epitope recognition patterns, and Fc-mediated effects were among some of the qualitative and quantitative response features analyzed. Conventional serological assays could not distinguish prominent differences between our vaccines; the antigen-specific IgG titers determined by ELISA and neutralization titers were similar, despite some vaccines conferring only partial protection. IgG titers achieved by the protective rVSV-ZEBOV vaccine have been used to partially infer the efficacy of the recently approved Ad26.ZEBOV/MVA-BN-Filo vaccine (14), but our results suggest that such comparisons should be cautiously used to measure efficacy. The differences between our vaccines become obvious when we examined the architecture of the antibody profile and the response kinetics. HPIV3 vectors elicited a greater BLI-determined total antibody response toward GP and its intermediate forms, a higher ratio of MLD to non-MLD neutralizing antibodies, and IgA and IgG targeting linear epitopes at a greater magnitude. Neutralizing antibodies composed of a higher ratio of MLD- to non–MLD-specific neutralizers and their magnitude may be important determinants of protection for the HPIV1 vaccine. APMV vaccines elicited proportionally more GC antibodies and had a lower ratio of MLD to non-MLD neutralizing antibodies. The response targeting certain GP regions and neutralizing epitopes diverged between HPIV3-derived and the other vaccines after two vaccine doses, indicating that the kinetics of the response, in addition to its evolving profile, is also dependent on the vaccine platform.
Although our vaccines are delivered via the respiratory tract, the distinct antibody response profiles that they elicit may be due to several factors, including differences in sequence, tropism, replication efficiency, and environmental cues at the replication site. HPIV1 was confined to the upper respiratory tract, and APMV and HPIV3 vaccines were detected in the upper and lower respiratory tract. Although both our APMV and HPIV1 vectored vaccines depend on a trypsin-like enzyme that is limited to the respiratory epithelial lining for the extracellular processing of their F protein (39), GP appeared to widen the tropism of APMV. Conversely, the HPIV3 F protein is intracellularly processed by the more widespread furin enzyme. Although APMV and HPIV3 vaccines were detected at the same sites, they likely have disparate capabilities to evade or subdue the host’s innate immune response (40, 41). Another consideration is a disproportionate dominance of additional antigenic epitopes in the vector-derived surface proteins in the B cell epitope hierarchy. These surface proteins are absent in HPIV3/ΔFHN/EboGP, enabling GP to act as a central immunogen. To compensate for any constraints on replication imposed by the host or milieu, increasing the doses of our partially protective vectors is likely to strengthen the antibody response to confer complete survival.
We created an extensive network integrating the relationships between 139 features of the vaccine response to define those that best associated with survival. No feature measured after the prime vaccine dose directly correlated with survival, indicating that efficacy prediction may not be possible after the first dose. After the boost dose, the BLI measure for total sera that bound to GP was the only feature to directly correlate with survival in the pairwise correlation analysis. This implies that the protective response may have evolved through affinity maturation, B cell class switching, and higher antibody titers. However, identification of correlates that reflect early or durable immunity may be time sensitive relative to the vaccination schedule or time of exposure after vaccination. Furthermore, a single dose of the best-performing candidate, HPIV3/ΔFHN/EboGP, likely confers complete protection, given its antibody response profile is divergent from other vaccines. Although survivors had higher IgG and IgA ELISA titers than nonsurvivors, the ELISA data were not as robust as the BLI readout for our network analysis to support a correlation with survival. BLI provides a better resolution of antibody binding that is not possible with an ELISA, measuring interactions with varying affinities between proteins in their conformationally accurate forms in real time. The choice of an immune feature or the stringency of the assay used for its characterization appears to be fundamental to reliably draw an association with survival.
A vaccine must impart more than survival and ideally will completely arrest virus replication and abate disease. The survival parameter alone is a rigid binary output that simplifies what is essentially a highly complex process. For this reason, we also used a survival index in the pairwise analysis that incorporates several parameters recapitulating the human disease and may therefore be the better tool for correlative measures. Several key antibody features, in addition to total sera bound to GP, correlated with the survival index across all vaccine groups, pointing to commonalities in mechanisms involved in survival. The response coverage toward RBD and GP2 + GP1/2 base was enriched in survivors of EBOV infection, and the presence of RBD antibodies associated strongly with an improved challenge outcome. Conversely, although the GC and MLD antibodies comprised most of the response for all vaccine types, their lack of association with the survival index points to a limited role in reducing EVD. Surviving NHP recipients of a virus-like particle–based vaccine induced antibodies that bind GP at low pH (37), pointing to the involvement of antibodies that bind the RBD and base domains of the proteolytic cleaved GP to prevent receptor recognition and fusion for subsequent entry. Rare mAbs targeting the RBD and GP2 regions that exhibit protective functions have been isolated from human survivors of EBOV infection and are emerging as therapeutic avenues against EBOV (21, 42). Although these regions are less immunogenic than the MLD or GC, they are worth pursuing for future targeted vaccine design.
The ADCD and ADNP effector arms of ADCC also strongly correlated with the survival index and may act as good predictors for vaccine efficacy, whereby elevated activities ensured reduced risk of EVD. Although aberrant neutrophil activity has been linked to the dysregulation of adaptive immunity and tissue damage during EVD (43), our findings indicate that neutrophils are likely to be required for vaccine-mediated immunity. Fc-mediated functions have been identified in the response of EVD survivors (44) and associated with protective mAbs isolated from survivors (45–47). In the human immunodeficiency virus field, ADCC is associated with vaccine-elicited protection from simian immunodeficiency virus infection in NHPs (48–50) and reduced the risk of infection or mortality for mother-to-child transmission (51, 52). Our demonstrated associations of ADNP, ADCD, and total GP-bound sera with the survival index also indicate that effector functions of the humoral response, and not just its magnitude, are important for survival. Vaccines that can elicit antibodies that orchestrate certain Fc effector functions and target specific GP regions at proportionally higher concentrations in relation to the other GP regions are likely to be more potent. Future studies will be required to determine whether the phenotype and distribution of Fc effector cells at sites relevant to vaccination or infection are influenced by different vaccines, which can ultimately affect the ability of antibodies to mediate the effector functions associated with survival.
In addition to the survival index, ADCD and ADNP correlated with other vaccine-elicited response features in the correlation network including neutralizing titers, antibodies targeting known neutralizing epitopes in the GC and MPER, and the proportion of the response targeting the RBD. This suggests that complementarity between several antibody functions directed at certain GP regions may promote survival. Although total neutralizing titers did not correlate with survival, we show that antibodies that recognize key neutralization-sensitive GP epitopes and domains are likely to be involved in protection. It is also feasible that the intensity of antibody Fc effector functions associated with survival could be influenced by where they bind on GP. GC-targeting mAbs isolated from human survivors of EBOV infection have been shown to induce more phagocytic activity compared to other GP epitopes and to have variable-neutralizing capabilities (46). Conversely, mAbs from human survivors of EBOV infection recognizing the MPER of GP2 were associated with strong neutralizing activity (45). However, Fc-mediated phagocytosis was enriched among neutralizing EBOV mAbs targeting these regions to promote protection (45, 53, 54). In EVD survivors, both neutralizing antibodies and polyfunctional IgG1 and IgA antibodies that induce Fc effector functions were natural constituents of humoral immunity (44). In the context of EBOV vaccine–induced immunity, studies have highlighted the importance of antibodies, specifically IgG titers (4, 38). We show vaccine-induced IgG1 as the major contributor to the Fc effector functions, which correlate with improved survival. However, unlike EVD survivors, the Fc effector functions of the circulating IgA elicited by our mucosal vaccines were limited, implying that their subclass and glycosylation state may be conducive to an effector response that maintains immune homeostasis (55). Our study suggests that vaccines that generate a humoral response with combined neutralizing activity and isotype-specific Fc effector functions that are associated with key vulnerable sites on GP will be more potent.
The GP binding profiles of mAbs isolated from human survivors of EBOV infection and the immune sera from rVSV-ZEBOV recipients were comparable with the majority of neutralizing antibodies mapped to the non-MLD regions of GP (46, 56). Here, we show that, although our vaccines elicit antibodies against known protective neutralizing epitopes, they differ in their neutralizing antibody coverage of GP regions. Contrasting with rVSV-ZEBOV recipients, we also show that a substantial proportion of the neutralizing antibodies generated by the HPIV-vaccinated survivors target the MLD. Unlike GP-based vaccines, the immune response to EBOV in human survivors reflects the broader antigenic content of the virus and involves other mechanisms such as T cell–mediated responses. Therefore, the correlates or mechanisms of protection identified in vaccine recipients are not interchangeable with those in survivors of EBOV infection and vary between the different vaccine platforms.
There are limitations to our study. Because of the inherent logistical factors of conducting large-scale NHP studies in an animal biosafety level (ABSL)–4 facility, our study was limited to a cohort of 20 vaccinated NHPs. A larger cohort would strengthen the predictive power of our multivariate survival modeling. Several features that were enriched in survivors were selected by the multivariate model as potential influencers of survival but were not reliable predictors. However, this indicates that more than one correlate is likely required to predict protection. Furthermore, the inclusion of variables from all our vaccines, most of which could be vaccine-specific correlates, may weaken the power of the multivariant analysis to select for protective correlates. However, our pairwise correlation network analysis on the vaccinated cohort identified features of the immune response that correlated with survival.
Immune parameters enriched only in vaccinated survivors could serve as universal protective correlates relevant to all EBOV vaccines. However, our vaccines had unique response profiles that were multi-faceted, despite expressing the same GP immunogen. Moreover, some response features were only enriched according to vaccine modes and not among all survivors, highlighting the likely existence of multiple vaccine-specific correlates of protection. The response heterogeneity that we observed indicates that protection may be achieved through synergy of several humoral features and functions, with specific Fc-mediated activities and RBD-targeting antibodies providing the important foundation for maximizing survival. Together with the ability to explain survival from BLI-measured total serum binding to GP, rather than from isotype-specific ELISA titers, we demonstrate here that full consideration of both the scope and role of the polyclonal response is required to unravel the complexity of protection. Using evolving technologies for comprehensive immune response characterization will enable clearer definitions on the requirements for a protective vaccine and fast-track the scrutiny of new candidates. It will also reveal how these features functionally coordinate to mediate the outcome of infection. Each EBOV vaccine mode likely adopts its own protective signature, and its correlates of protection may therefore be influenced accordingly.
MATERIALS AND METHODS
Study design
This study was originally designed to determine the immunogenicity and efficacy of five mucosal vaccines against EBOV in cynomolgus macaques (Macaca fascicularis). All animal protocols were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Texas Medical Branch (UTMB). Even distribution of NHPs into vaccine groups was based on gender and weights. Group sizes were informed by available data from previous EBOV vaccine immunogenicity or efficacy studies in NHPs (19, 57) and housing availability. No blinding was used throughout the study. All measurements were included in our analysis; no outliers were excluded. Primary data are included in data file S1.
Five groups of cynomolgus macaques (M. fascicularis; 3.5 to 6 kg, n = 4 per group) were vaccinated with the BC/LSFcHN/EboGP, BC/LSFHN/EboGP, HPIV3/ΔFHN/EboGP, HPIV3/EboGP, or HPIV1/EboGP vaccine via the combined IN/IT route (0.5 ml per nostril and 1 ml IT) with 2 × 108 PFU/ml on day 0. A control group (n = 2) received the HPIV3 empty-vectored virus at the same dose. On day 26, all animals were homologously boosted via the same route. Bronchoalveolar lavage samples and blood was collected just before each vaccination and EBOV challenge for serological studies, and nasal swabs and tracheal lavages were collected to measure vaccine vector shedding. On day 55, animals were inoculated with 1000 PFU of EBOV (Kikwit, 7U variant; GenBank KC242796.1) via the intramuscular route in BSL-4 biocontainment (Galveston National Laboratory, University of Texas Medical Branch). Over the infection course, NHPs were monitored for clinical symptoms of dyspnea, depression, recumbency, and rash or hemorrhage and scored accordingly. A score of 0 to 2 required no intervention, whereas a score of 9 required euthanasia as per the IACUC protocol. Peripheral blood markers of EVD, measured by hematologic (Beckman Coulter) and biochemistry analyzers (Vetscan), and viremia in serum, measured by both plaque assays and qRT-PCR, were also assessed (19). Surviving NHPs were euthanized on day 28 after infection.
Each NHP was assigned a survival index based on clinical scores, liver and kidney functions, and viremia detectable by both plaque assay and qRT-PCR. Animals recording clinical scores ≤2, normal liver and kidney functions, and no detectable viremia were assigned an index score of 100. Indices were adjusted if animals presented with clinical scores of 3 to 5 (deducted by 10) or >5 (deducted by 20), had abnormal liver and kidney functions (deducted by 20 for each function), and were positive for viremia by plaque assay (deducted by 20) and qRT-PCR (deducted by 10 or 20 if viremia was respectively detected at one sampling point only or detected at more than one sampling point).
Vaccine constructs expressing EBOV GP
The vaccine constructs were based on human and APMVs constructed to express the EBOV GP protein (isolate Mayinga; GenBank AAG40168.1) (fig. S1). Human paramyxoviral vectors were based on HPIV1 (HPIV1/EboGP) and HPIV3 (HPIV3/EboGP) viruses. The next-generation version of the HPIV3/EboGP vaccine construct lacked the F and HN surface proteins (HPIV3/ΔFHN/EboGP). APMV vaccines were based on the NDV strain BC, modified by substituting its Fc cleavage site and HN gene (BC/LSFcHN/EboGP) or both the F and HN genes (BC/LSFHN/EboGP) with that of the LS strain. The construction and growth of all vaccines were previously described (18). HPIV3 was used as an empty vector control.
Analysis of vector shedding
Nasal swabs and tracheal lavages were performed on days 2, 5, and 7 after the prime vaccine dose and on days 28, 30, and 33 after the boost dose. These samples were snap-frozen, and virus titers were later analyzed in triplicate by qRT-PCR. RNA was extracted from samples using the QIAamp Viral RNA Extraction Kit (QIAGEN) as per the manufacturer’s protocol. First-strand cDNA synthesis was performed with a sense strand, GP gene–specific primer, Superscript IV Reverse Transcriptase (Invitrogen), and 300 ng of RNA. Real-time RT-PCR was performed with the ABI 7900HT system, and primers and probes specific for the GP gene: forward, AGCTGGTGAATGGGCTGAAA; reverse, CTGGCGCTGCTGGTAGACA; probe, CTGCTACAATCTTGAAATC-FAM (Invitrogen). Absolute quantification was achieved using a standard curve generated by serial dilution of an 80–base pair Mayinga GP DNA amplicon (Integrated DNA Technologies).
Systemic and mucosal serology studies
EBOV GP–specific serum and mucosal IgG, IgA, and neutralizing antibody titers for each NHP were measured in duplicate as previously described (19). For IgG and IgA antibody detection, ELISA plates were respectively coated with 7 and 25 ng per well of GP protein [Integrated BioTherapeutics (IBT) Bioservices]. Serum IgG and IgA titers for post-vaccination sera determined by ELISA were corrected for baseline by subtracting preimmune sera values.
Isotyping ELISAs were performed similar to the ELISA mentioned above with some modifications. Briefly, 96-well plates were coated with 25 ng per well of EBOV GP (IBT) and blocked with 5% milk powder or SuperBlock (Pierce). Serum was either serially diluted in 5% milk powder or 2% bovine serum albumin. Biotinylated isotype–specific mAbs obtained from the National Institutes of Health (NIH) NHP Reagent Resource (IgG and IgA dimer) or Mabtech (anti-IgA and anti-IgM) were applied to the plates for 1 hour. Bound complexes were detected with streptavidin–horseradish peroxidase (Pierce). The optical density values for the IgM assay readout were also used to calculate titers. Baseline correction for post-vaccination sera was also performed by subtraction of preimmune sera values. A standard curve of known concentrations of purified NHP isotype antibodies (IgG, monomeric and dimeric IgA, NIH NHP Reagent Resource, and IgM; Rockland) was run alongside samples to determine the amount of GP-specific isotype antibody present in serum. Briefly, isotypes serially diluted in phosphate-buffered saline were coated onto the 96-well plate and, after a blocking step, were detected with biotinylated isotype-specific antibodies as described above. The concentration for each GP-specific isotype in an NHP sample was extrapolated from the standard curve and expressed as a proportion of the total concentration of all isotypes measured in the sample.
BLI binding and competition assays
A FortéBio Octet Red96 instrument was used to measure sera binding to EBOV GP and its intermediate forms (provided by E. O. Saphire). All assays were performed with agitation at 1000 rpm, at 28°C in black 96-well plates. All samples were diluted in 1× Kinetics buffer (FortéBio) with a final volume of 200 μl per well. Biotinylated EBOV GP, GPΔmuc, sGP, and GPcl (2 μg/ml) were immobilized onto streptavidin sensors for 300 s to capture between ~1 and 1.5 nm, with variability within a row of eight sensors not exceeding 0.1 nm. Biosensor tips were then equilibrated for 300 s in 1× Kinetics buffer before binding measurements. Sera were diluted 1:40, and binding was assessed for 600 s, followed by dissociation for 600 s in 1× Kinetics buffer. Parallel corrections for baseline drift were made by subtracting measurements recorded with GP-loaded sensors in the absence of sera.
For sera preadsorption studies, sensors were treated with biocytin for 120 s after immobilization of a GP form. Sera depleted with excess amounts of GP forms (2.5 μg of GP, GPΔmuc, or sGP and 0.5 μg of GPcl) were allowed to bind to sensors as described above. To determine nonspecific binding responses, binding of sera from HPIV3-vaccinated animals to GP variant–loaded probes was monitored and set as the background. We calculated the percent inhibition of binding to an immobilized GP after serum adsorption relative to the binding observed without preadsorption using the following formula: % inhibition = [100 − (binding of serum preadsorbed with GP form (nm)/binding of serum without preadsorption (nm))] × 100. The percent inhibition values, derived from one immobilized GP variant as the common denominator, were used to calculate the relative proportions of serum binding to a specific GP domain.
For site-specific antigenicity assessment, biotinylated GP-loaded sensors (captured at ~0.5 nm) were incubated with serum diluted 1:10 in 1× Kinetics buffer for 900 s to generate a saturating signal against the competing mAb. Probes were then washed twice for 60 s before the reactivity of competing mAbs (25 nM BDBV223, EBOV55, and KZ52 and 300 nM BDBV289, EBOV520, and BDBV317) toward GP was assessed for 600 s. The binding inhibition to GP was calculated as a percentage of the blocking activity of sera from vaccinated animals compared to the negative control sera against the tested mAb. Data analysis and curve fitting were carried out using Octet software, version 7.0.
Reversing neutralizing activity in the presence of GP forms
Competition neutralization assays were performed as previously described (18). Briefly, day 54 sera diluted to concentrations that neutralized at least 60 to 80% of EBOV were incubated in duplicate with increasing concentrations of sGP or GPΔmuc and later exposed to recombinant EBOV expressing green fluorescent protein (GFP) in a neutralization assay. Restoration of infectivity was determined as a percentage of the plaques formed in the presence of serum incubated with the competing GP forms compared to the serum without the GP forms. Serum was also titrated in the presence or absence of 1 μg of GPΔmuc to determine its effect on the concentration of serum required to reduce half the maximal EBOV infectivity (EC50).
Peptide array
Multiwell array slides spotted with 15-mer peptides offset by four amino acids spanning the entire EBOV GP (Mayinga Zaire 1976; GenBank: NP 066246) were manufactured by JPT Innovative Peptide Solutions (Germany). Serum was diluted at 1:200 (IgG array) or 1:100 (IgA array) in blocking buffer (RepliTope Peptide Microarray Incubation Kit, JPT Innovative Peptide Solutions) and incubated for 1 hour. Bound complexes were detected with biotinylated anti-IgG (1:10,000) or anti-IgA (1:5000), followed by streptavidin-Cy5 (0.1 μg/ml). Array slides were dried and scanned with the GenePix Array Scanner at 635 nm and analyzed using GenePix Pro 6 (Molecular Devices). The sera from all NHPs within a group were analyzed, and the mean fluorescence intensity (MFI) for each peptide was averaged and corrected for baseline by subtracting the corresponding prevaccination MFI. The breadth of a response was determined by calculating the number of positive peptides per GP region for each NHP. In a string of positive peptides, the number of binding sites was determined by the number of amino acids shared between the first and last peptide of the string; peptides that shared five or more amino acids were defined as a single binding site, and peptides with four or less common amino acids were recognized as two sites.
Serum depletion
Day 54 plasma was diluted 1:10 and depleted using CaptureSelect IgG1 and IgA resins or a control resin as previously described (44). Briefly, two rounds of 1-hour incubations with resin and diluted plasma were performed, followed by the collection of depleted samples through a spin column. Depletion was confirmed by isotype-specific ELISA as described above.
ADNP and ADCP
Recombinant EBOV GP was biotinylated and coupled to 1-μm fluorescein isothiocyanate (FITC)+ NeutrAvidin beads (Life Technologies). Sera from vaccinated NHPs, diluted as specified in the figures or 5 μg/ml of the positive control EBOV GP–specific humanized murine IgG1 c13C6 mAb were incubated with GP-coated beads for 2 hours at 37°C. Freshly isolated neutrophils (5.0 × 104 cells per well) from human donor peripheral blood (collected by the Ragon Institute or the Massachusetts General Hospital Blood bank from healthy human volunteers with signed informed consent) were added, and plates were incubated for 1 hour at 37°C. Neutrophil cells were stained at 1:100 with CD66b (Pacific Blue; clone G10F5; BioLegend), CD3 (Alexa Fluor 700; clone UCHT1; BD Biosciences), and CD14 (APC-Cy7; clone MφP9; BD Biosciences). Neutrophils were defined as positive for a high side scatter area (SSC-Ahigh), CD66b+, CD3−, and CD14−. ADCP was measured as previously described (58) using a human monocyte cell line (THP-1 cells), whose overall profile and expression of Fcγ receptors are comparable to primary human monocytes (58, 59). Briefly, THP-1 cells (2.0 × 104 cells per well) were incubated overnight at 37°C with the GP-coated FITC bead serum mixtures in duplicate. Cells were fixed with 4% paraformaldehyde, and a minimum of 30,000 (ADNP) or 10,000 (ADCP) events were recorded and analyzed using a BD LSRII flow cytometer and FlowJo, version 10. The phagocytic scores were determined using the following formula: [(percentage of FITC+ cells) × (MFI of the FITC+ cells)]/10,000.
ADNP and ADCP assays with IgA- and IgG1-depleted samples were performed by incubating plasma with a CellTrace Far Red–stained cell line stably expressing GP on its surface [EBOV GPkik-293FS EGFP CCR5-SNAP cells (21), 1.0 × 105 cells per well] for 30 min at room temperature. Neutrophils (2.0 × 105 cells per well) or donor peripheral blood mononuclear cells (2.0 × 105 cells per well; Gulf Coast Regional Blood Center) were then added to the samples and incubated for 2 hours at 37°C. Neutrophils and monocytes were stained with LIVE/DEAD Fixable Aqua (Life Technologies), CD66b [phycoerythrin (PE); clone G10F5, BioLegend], CD3 (PE-Cy7; clone UCHT1, BioLegend), and CD14 (BV421; clone MφP9, BD Biosciences) before analysis using a BD FACSCanto flow cytometer. The phagocytic scores were determined using the following formula: {(percentage of Far Red+ cells) × [geometric mean fluorescence intensity (gMFI) of the Far Red+ cells]}/10,000. The scores presented for each NHP are averaged from two donor blood samples or duplicate THP-1 samples.
Antibody-dependent complement deposition
Biotinylated EBOV GP was coupled to 1-μm red fluorescent NeutrAvidin beads (Life Technologies). Diluted serum was incubated with GP-coated beads for 2 hours at 37°C, followed by the addition of reconstituted guinea pig complement (Cedarlane Labs) diluted in gelatin veronal buffer containing magnesium and calcium (Boston BioProducts). C3 deposition onto beads was measured by flow cytometry using an anti–guinea pig C3 antibody conjugated to FITC (1:100; MP Biomedicals). The average complement deposition MFI for duplicate samples is presented for each NHP.
Antibody-dependent activation of NK cells
The NK cell activation assay was performed as previously described (45). Briefly, NK cells isolated from the peripheral blood from three human donors by negative selection per the manufacturer’s instructions (STEMCELL Technologies) were added at 5 × 104 cells per well to EBOV GP (IBT Bioservices) protein absorbed on plates with diluted serum, brefeldin A (Sigma-Aldrich), GolgiStop (Life Technologies), and anti-CD107a antibody (1:40; PE-Cy5; clone H4A3, BD Biosciences) and incubated for 5 hours. Cells were surface-stained with anti-CD3 (1:100; Alexa Fluor 700; clone UCHT1, BD Biosciences), anti-CD16 [1:100, allophycocyanin (APC)–Cy7; clone 3G8, BD Biosciences], and anti-CD56 (1:100; PE-Cy7; clone B159, BD Biosciences) and intracellularly stained for IFN-γ (1:50; APC; clone B27, BD Biosciences) and MIP-1β (1:50; PE; clone D21–1351, BD Biosciences) after a Fix Perm treatment (Invitrogen) per the manufacturer’s instructions. Cells were analyzed by flow cytometry using a BD LSRII flow cytometer. The EBOV GP–specific humanized murine IgG1 c13C6 (IBT Bioservices) was used at 5 μg/ml as a positive control, and cynomolgus macaque sera were used as a negative control (dilution: 1:50). Background staining for MIP-1b and IFN-γ was confirmed to be within the acceptable range for the assay (45).
Statistical analysis
Statistical comparisons between groups were made using unpaired t tests or analysis of variance (ANOVA) with post hoc Tukey or Sidak tests (Prism version 9, GraphPad Software) as specified in the figure legends. Significance is denoted by *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, or ****P ≤ 0.0001. Correlations between the survival index and an antibody parameter for vaccinated NHPs (n = 20) were determined by two-sided Spearmanʼs rank tests.
For the correlation network analysis of 139 parameters measured in all vaccinated NHPs (n = 20) (data file S1), we used corr. test function in R to compute pairwise Kendall rank correlation coefficients, using the Benjamini-Hochberg procedure to adjust for multiple hypothesis testing (data file S2). The network was rendered using Cytoscape (60). The Cox regression model was computed using the Cyclops R package (61). The code is available at http://doi.org/10.5281/zenodo.4784498.
Supplementary Material
Acknowledgments:
We thank C. Klagis and the staff of Animal Resources Center at UTMB for performing vaccination and ABSL-2 procedures. We thank K. N. Agans, V. Borisevich, and D. J. Deer for NHP procedures and sampling in BSL-4. We thank K. S. Mohamed for assisting with experimental procedures.
Funding:
This work was supported by grants 1R01AI102887-01A1 (A.B.), U19 AI109711 (T.W.G., J.E.C. Jr., and A.B.), U19 AI142790 (E.O.S. and A.B.), and U19 AI135995 (K.G.A.) from the National Institute of Allergy and Infectious Diseases of the U.S. National Institutes of Health.
Competing interests:
J.E.C. Jr. has served as a consultant for Eli Lilly and Company, GlaxoSmithKline, and Luna Biologics; is a member of the Scientific Advisory Boards of CompuVax and Meissa Vaccines; and is Founder of IDBiologics. J.E.C. Jr. has received unrelated sponsored research agreements from IDBiologics and AstraZeneca. The other authors declare that they have no competing interests.
Footnotes
SUPPLEMENTARY MATERIALS
stm.sciencemag.org/cgi/content/full/13/602/eabg6128/DC1
Data files S1 and S2
View/request a protocol for this paper from Bio-protocol.
Data and materials availability:
All data associated with this study are present in the paper or the Supplementary Materials.
REFERENCES AND NOTES
- 1.WHO, Ebola virus disease (2020); www.who.int/news-room/fact-sheets/detail/ebola-virus-disease.
- 2.Electronic Code of Federal Regulations. e-CFR, Approval of new drugs when human efficacy studies are not ethical or feasible (2020); www.ecfr.gov/cgi-bin/text-idx?SID=07ae7117f4af9184631f0ba5ab8e9bec&mc=true&node=sp21.5.314.i&rgn=div6.
- 3.Meyer M, Malherbe DC, Bukreyev A, Can Ebola virus vaccines have universal immune correlates of protection? Trends Microbiol. 27, 8–16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marzi A, Engelmann F, Feldmann F, Haberthur K, Shupert WL, Brining D, Scott DP, Geisbert TW, Kawaoka Y, Katze MG, Feldmann H, Messaoudi I, Antibodies are necessary for rVSV/ZEBOV-GP-mediated protection against lethal Ebola virus challenge in nonhuman primates. Proc. Natl. Acad. Sci. U.S.A. 110, 1893–1898 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sullivan NJ, Martin JE, Graham BS, Nabel GJ, Correlates of protective immunity for Ebola vaccines: Implications for regulatory approval by the animal rule. Nat. Rev. Microbiol. 7, 393–400 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sullivan NJ, Hensley L, Asiedu C, Geisbert TW, Stanley D, Johnson J, Honko A, Olinger G, Bailey M, Geisbert JB, Reimann KA, Bao S, Rao S, Roederer M, Jahrling PB, Koup RA, Nabel GJ, CD8+ cellular immunity mediates rAd5 vaccine protection against Ebola virus infection of nonhuman primates. Nat. Med. 17, 1128–1131 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Olinger GG, Bailey MA, Dye JM, Bakken R, Kuehne A, Kondig J, Wilson J, Hogan RJ, Hart MK, Protective cytotoxic T-cell responses induced by venezuelan equine encephalitis virus replicons expressing Ebola virus proteins. J. Virol. 79, 14189–14196 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Winslow RL, Milligan ID, Voysey M, Luhn K, Shukarev G, Douoguih M, Snape MD, Immune responses to Novel adenovirus type 26 and modified vaccinia virus Ankara-vectored Ebola vaccines at 1 year. JAMA 317, 1075–1077 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Henao-Restrepo AM, Longini IM, Egger M, Dean NE, Edmunds WJ, Camacho A, Carroll MW, Doumbia M, Draguez B, Duraffour S, Enwere G, Grais R, Gunther S, Hossmann S, Konde MK, Kone S, Kuisma E, Levine MM, Mandal S, Norheim G, Riveros X, Soumah A, Trelle S, Vicari AS, Watson CH, Keita S, Kieny MP, Rottingen JA, Efficacy and effectiveness of an rVSV-vectored vaccine expressing Ebola surface glycoprotein: Interim results from the Guinea ring vaccination cluster-randomised trial. Lancet 386, 857–866 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Henao-Restrepo AM, Camacho A, Longini IM, Watson CH, Edmunds WJ, Egger M, Carroll MW, Dean NE, Diatta I, Doumbia M, Draguez B, Duraffour S, Enwere G, Grais R, Gunther S, Gsell PS, Hossmann S, Watle SV, Konde MK, Keita S, Kone S, Kuisma E, Levine MM, Mandal S, Mauget T, Norheim G, Riveros X, Soumah A, Trelle S, Vicari AS, Rottingen JA, Kieny MP, Efficacy and effectiveness of an rVSV-vectored vaccine in preventing Ebola virus disease: Final results from the Guinea ring vaccination, open-label, cluster-randomised trial (Ebola Ca Suffit!). Lancet 389, 505–518 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ewer K, Rampling T, Venkatraman N, Bowyer G, Wright D, Lambe T, Imoukhuede EB, Payne R, Fehling SK, Strecker T, Biedenkopf N, Krahling V, Tully CM, Edwards NJ, Bentley EM, Samuel D, Labbe G, Jin J, Gibani M, Minhinnick A, Wilkie M, Poulton I, Lella N, Roberts R, Hartnell F, Bliss C, Sierra-Davidson K, Powlson J, Berrie E, Tedder R, Roman F, De Ryck I, Nicosia A, Sullivan NJ, Stanley DA, Mbaya OT, Ledgerwood JE, Schwartz RM, Siani L, Colloca S, Folgori A, Di Marco S, Cortese R, Wright E, Becker S, Graham BS, Koup RA, Levine MM, Volkmann A, Chaplin P, Pollard AJ, Draper SJ, Ballou WR, Lawrie A, Gilbert SC, Hill AVS, A monovalent chimpanzee adenovirus Ebola vaccine boosted with MVA. N. Engl. J. Med. 374, 1635–1646 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Milligan ID, Gibani MM, Sewell R, Clutterbuck EA, Campbell D, Plested E, Nuthall E, Voysey M, Silva-Reyes L, McElrath MJ, De Rosa SC, Frahm N, Cohen KW, Shukarev G, Orzabal N, van Duijnhoven W, Truyers C, Bachmayer N, Splinter D, Samy N, Pau MG, Schuitemaker H, Luhn K, Callendret B, Van Hoof J, Douoguih M, Ewer K, Angus B, Pollard AJ, Snape MD, Safety and immunogenicity of novel adenovirus type 26- and modified vaccinia Ankara-vectored Ebola vaccines: A randomized clinical trial. JAMA 315, 1610–1623 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Ledgerwood JE, DeZure AD, Stanley DA, Coates EE, Novik L, Enama ME, Berkowitz NM, Hu Z, Joshi G, Ploquin A, Sitar S, Gordon IJ, Plummer SA, Holman LA, Hendel CS, Yamshchikov G, Roman F, Nicosia A, Colloca S, Cortese R, Bailer RT, Schwartz RM, Roederer M, Mascola JR, Koup RA, Sullivan NJ, Graham BS; VRC 207 Study Team, Chimpanzee adenovirus vector Ebola vaccine. N. Engl. J. Med. 376, 928–938 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Pollard AJ, Launay O, Lelievre JD, Lacabaratz C, Grande S, Goldstein N, Robinson C, Gaddah A, Bockstal V, Wiedemann A, Leyssen M, Luhn K, Richert L, Betard C, Gibani MM, Clutterbuck EA, Snape MD, Levy Y, Douoguih M, Thiebaut R; EBOVAC2 EBL2001 Study Group, Safety and immunogenicity of a two-dose heterologous Ad26. ZEBOV and MVA-BN-Filo Ebola vaccine regimen in adults in Europe (EBOVAC2): A randomised, observer-blind, participant-blind, placebo-controlled, phase 2 trial. Lancet Infect. Dis. 21, 493–506 (2020). [DOI] [PubMed] [Google Scholar]
- 15.FDA, First FDA-approved vaccine for the prevention of Ebola virus disease, marking a critical milestone in public health preparedness and response (2019); www.fda.gov/news-events/press-announcements/first-fda-approved-vaccine-prevention-ebola-virus-disease-marking-critical-milestone-public-health.
- 16.EMA, Human medicines European public assessment report (EPAR): Ervebo, Ebola Zaire vaccine (rVSVΔG-ZEBOV-GP, live), Hemorrhagic fever, Ebola, Date of authorization: 11/11/2019, Status: Authorized; www.ema.europa.eu/en/medicines/human/EPAR/ervebo.
- 17.WHO, Ebola virus disease—Democratic Republic of the Congo (2020); https://apps.who.int/iris/bitstream/handle/10665/332950/WER9527-eng-fre.pdf?ua=1&ua=1.
- 18.Meyer M, Yoshida A, Ramanathan P, Saphire EO, Collins PL, Crowe JE Jr., S. Samal, A. Bukreyev, Antibody repertoires to the same Ebola vaccine antigen are differentially affected by vaccine vectors. Cell Rep. 24, 1816–1829 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meyer M, Garron T, Lubaki NM, Mire CE, Fenton KA, Klages C, Olinger GG, Geisbert TW, Collins PL, Bukreyev A, Aerosolized Ebola vaccine protects primates and elicits lung-resident T cell responses. J. Clin. Invest. 125, 3241–3255 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bukreyev AA, Dinapoli JM, Yang L, Murphy BR, Collins PL, Mucosal parainfluenza virus-vectored vaccine against Ebola virus replicates in the respiratory tract of vector-immune monkeys and is immunogenic. Virology 399, 290–298 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilchuk P, Kuzmina N, Ilinykh PA, Huang K, Gunn BM, Bryan A, Davidson E, Doranz BJ, Turner HL, Fusco ML, Bramble MS, Hoff NA, Binshtein E, Kose N, Flyak AI, Flinko R, Orlandi C, Carnahan R, Parrish EH, Sevy AM, Bombardi RG, Singh PK, Mukadi P, Muyembe-Tamfum JJ, Ohi MD, Saphire EO, Lewis GK, Alter G, Ward AB, Rimoin AW, Bukreyev A, Crowe JE Jr., Multifunctional pan-ebolavirus antibody recognizes a site of broad vulnerability on the ebolavirus glycoprotein. Immunity 49, 363–374.e10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flyak AI, Shen X, Murin CD, Turner HL, David JA, Fusco ML, Lampley R, Kose N, Ilinykh PA, Kuzmina N, Branchizio A, King H, Brown L, Bryan C, Davidson E, Doranz BJ, Slaughter JC, Sapparapu G, Klages C, Ksiazek TG, Saphire EO, Ward AB, Bukreyev A, Crowe JE Jr., Cross-reactive and potent neutralizing antibody responses in human survivors of natural ebolavirus infection. Cell 164, 392–405 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee JE, Fusco ML, Hessell AJ, Oswald WB, Burton DR, Saphire EO, Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature 454, 177–182 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flyak AI, Kuzmina N, Murin CD, Bryan C, Davidson E, Gilchuk P, Gulka CP, Ilinykh PA, Shen X, Huang K, Ramanathan P, Turner H, Fusco ML, Lampley R, Kose N, King H, Sapparapu G, Doranz BJ, Ksiazek TG, Wright DW, Saphire EO, Ward AB, Bukreyev A, Crowe JE Jr., Broadly neutralizing antibodies from human survivors target a conserved site in the Ebola virus glycoprotein HR2-MPER region. Nat. Microbiol. 3, 670–677 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee JE, Saphire EO, Ebolavirus glycoprotein structure and mechanism of entry. Future Virol. 4, 621–635 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson JA, Hevey M, Bakken R, Guest S, Bray M, Schmaljohn AL, Hart MK, Epitopes involved in antibody-mediated protection from Ebola virus. Science 287, 1664–1666 (2000). [DOI] [PubMed] [Google Scholar]
- 27.Olinger GG Jr., Pettitt J, Kim D, Working C, Bohorov O, Bratcher B, Hiatt E, Hume SD, Johnson AK, Morton J, Pauly M, Whaley KJ, Lear CM, Biggins JE, Scully C, Hensley L, Zeitlin L, Delayed treatment of Ebola virus infection with plant-derived monoclonal antibodies provides protection in rhesus macaques. Proc. Natl. Acad. Sci. U.S.A. 109, 18030–18035 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitchell DAJ, Dupuy LC, Sanchez-Lockhart M, Palacios G, Back JW, Shimanovskaya K, Chaudhury S, Ripoll DR, Wallqvist A, Schmaljohn CS, Epitope mapping of Ebola virus dominant and subdominant glycoprotein epitopes facilitates construction of an epitope-based DNA vaccine able to focus the antibody response in mice. Hum. Vaccin. Immunother. 13, 2883–2893 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanchez-Lockhart M, Reyes DS, Gonzalez JC, Garcia KY, Villa EC, Pfeffer BP, Trefry JC, Kugelman JR, Pitt ML, Palacios GF, Qualitative profiling of the humoral immune response elicited by rVSV-ΔG-EBOV-GP using a systems serology assay, domain programmable arrays. Cell Rep. 24, 1050–1059.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Olal D, Kuehne AI, Bale S, Halfmann P, Hashiguchi T, Fusco ML, Lee JE, King LB, Kawaoka Y, Dye JM Jr., E. O. Saphire, Structure of an antibody in complex with its mucin domain linear epitope that is protective against Ebola virus. J. Virol. 86, 2809–2816 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Becquart P, Mahlakoiv T, Nkoghe D, Leroy EM, Identification of continuous human B-cell epitopes in the VP35, VP40, nucleoprotein and glycoprotein of Ebola virus. PLOS ONE 9, e96360 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ludtke A, Ruibal P, Becker-Ziaja B, Rottstegge M, Wozniak DM, Cabeza-Cabrerizo M, Thorenz A, Weller R, Kerber R, Idoyaga J, Magassouba N, Gabriel M, Gunther S, Oestereich L, Munoz-Fontela C, Ebola virus disease is characterized by poor activation and reduced levels of circulating CD16+ monocytes. J. Infect. Dis. 214, S275–S280 (2016). [DOI] [PubMed] [Google Scholar]
- 33.McElroy AK, Akondy RS, McLlwain DR, Chen H, Bjornson-Hooper Z, Mukherjee N, Mehta AK, Nolan G, Nichol ST, Spiropoulou CF, Immunologic timeline of Ebola virus disease and recovery in humans. JCI Insight 5, e137260 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hunt L, Gupta-Wright A, Simms V, Tamba F, Knott V, Tamba K, Heisenberg-Mansaray S, Tamba E, Sheriff A, Conteh S, Smith T, Tobin S, Brooks T, Houlihan C, Cummings R, Fletcher T, Clinical presentation, biochemical, and haematological parameters and their association with outcome in patients with Ebola virus disease: An observational cohort study. Lancet Infect. Dis. 15, 1292–1299 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Blaney JE, Marzi A, Willet M, Papaneri AB, Wirblich C, Feldmann F, Holbrook M, Jahrling P, Feldmann H, Schnell MJ, Antibody quality and protection from lethal Ebola virus challenge in nonhuman primates immunized with rabies virus based bivalent vaccine. PLOS Pathog. 9, e1003389 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rechtien A, Richert L, Lorenzo H, Martrus G, Hejblum B, Dahlke C, Kasonta R, Zinser M, Stubbe H, Matschl U, Lohse A, Krahling V, Eickmann M, Becker S, Consortium V, Thiebaut R, Altfeld M, Addo MM, Systems vaccinology identifies an early innate immune signature as a correlate of antibody responses to the Ebola vaccine rVSV-ZEBOV. Cell Rep. 20, 2251–2261 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Warfield KL, Howell KA, Vu H, Geisbert J, Wong G, Shulenin S, Sproule S, Holtsberg FW, Leung DW, Amarasinghe GK, Swenson DL, Bavari S, Kobinger GP, Geisbert TW, Aman MJ, Role of antibodies in protection against Ebola virus in nonhuman primates immunized with three vaccine platforms. J. Infect. Dis. 218, S553–S564 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wong G, Richardson JS, Pillet S, Patel A, Qiu X, Alimonti J, Hogan J, Zhang Y, Takada A, Feldmann H, Kobinger GP, Immune parameters correlate with protection against ebola virus infection in rodents and nonhuman primates. Sci. Transl. Med. 4, 158ra146 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Henrickson KJ, Parainfluenza viruses. Clin. Microbiol. Rev. 16, 242–264 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bukreyev A, Huang Z, Yang L, Elankumaran S, Claire MS, Murphy BR, Samal SK, Collins PL, Recombinant newcastle disease virus expressing a foreign viral antigen is attenuated and highly immunogenic in primates. J. Virol. 79, 13275–13284 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DiNapoli JM, Kotelkin A, Yang L, Elankumaran S, Murphy BR, Samal SK, Collins PL, Bukreyev A, Newcastle disease virus, a host range-restricted virus, as a vaccine vector for intranasal immunization against emerging pathogens. Proc. Natl. Acad. Sci. U.S.A. 104, 9788–9793 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.King LB, West BR, Moyer CL, Gilchuk P, Flyak A, Ilinykh PA, Bombardi R, Hui S, Huang K, Bukreyev A, Crowe JE Jr., Saphire EO, Cross-reactive neutralizing human survivor monoclonal antibody BDBV223 targets the ebolavirus stalk. Nat. Commun. 10, 1788 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eisfeld AJ, Halfmann PJ, Wendler JP, Kyle JE, Burnum-Johnson KE, Peralta Z, Maemura T, Walters KB, Watanabe T, Fukuyama S, Yamashita M, Jacobs JM, Kim YM, Casey CP, Stratton KG, Webb-Robertson BM, Gritsenko MA, Monroe ME, Weitz KK, Shukla AK, Tian M, Neumann G, Reed JL, van Bakel H, Metz TO, Smith RD, Waters KM, N’Jai A, Sahr F, Kawaoka Y, Multi-platform ‘omics analysis of human Ebola virus disease pathogenesis. Cell Host Microbe 22, 817–829 e818 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gunn BM, Roy V, Karim MM, Hartnett JN, Suscovich TJ, Goba A, Momoh M, Sandi JD, Kanneh L, Andersen KG, Shaffer JG, Schieffelin JS, Garry RF, Grant DS, Alter G, Survivors of Ebola virus disease develop polyfunctional antibody responses. J. Infect. Dis. 221, 156–161 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gunn BM, Yu WH, Karim MM, Brannan JM, Herbert AS, Wec AZ, Halfmann PJ, Fusco ML, Schendel SL, Gangavarapu K, Krause T, Qiu X, He S, Das J, Suscovich TJ, Lai J, Chandran K, Zeitlin L, Crowe JE Jr., D. Lauffenburger, Y. Kawaoka, G. P. Kobinger, K. G. Andersen, J. M. Dye, E. O. Saphire, G. Alter, A role for Fc function in therapeutic monoclonal antibody-mediated protection against Ebola virus. Cell Host Microbe 24, –221–233.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saphire EO, Schendel SL, Fusco ML, Gangavarapu K, Gunn BM, Wec AZ, Halfmann PJ, Brannan JM, Herbert AS, Qiu X, Wagh K, He S, Giorgi EE, Theiler J, Pommert KBJ, Krause TB, Turner HL, Murin CD, Pallesen J, Davidson E, Ahmed R, Aman MJ, Bukreyev A, Burton DR, Crowe JE Jr., Davis CW, Georgiou G, Krammer F, Kyratsous CA, Lai JR, Nykiforuk C, Pauly MH, Rijal P, Takada A, Townsend AR, Volchkov V, Walker LM, Wang CI, Zeitlin L, Doranz BJ, Ward AB, Korber B, Kobinger GP, Andersen KG, Kawaoka Y, Alter G, Chandran K, Dye JM, viral C hemorrhagic fever immunotherapeutic, systematic analysis of monoclonal antibodies against Ebola virus GP defines features that contribute to protection. Cell 174, 938–952. e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saphire EO, Schendel SL, Gunn BM, Milligan JC, Alter G, Antibody-mediated protection against Ebola virus. Nat. Immunol. 19, 1169–1178 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barouch DH, Alter G, Broge T, Linde C, Ackerman ME, Brown EP, Borducchi EN, Smith KM, Nkolola JP, Liu J, Shields J, Parenteau L, Whitney JB, Abbink P, Ng’ang’a DM, Seaman MS, Lavine CL, Perry JR, Li W, Colantonio AD, Lewis MG, Chen B, Wenschuh H, Reimer U, Piatak M, Lifson JD, Handley SA, Virgin HW, Koutsoukos M, Lorin C, Voss G, Weijtens M, Pau MG, Schuitemaker H, Protective efficacy of adenovirus/protein vaccines against SIV challenges in rhesus monkeys. Science 349, 320–324 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barouch DH, Stephenson KE, Borducchi EN, Smith K, Stanley K, McNally AG, Liu J, Abbink P, Maxfield LF, Seaman MS, Dugast AS, Alter G, Ferguson M, Li W, Earl PL, Moss B, Giorgi EE, Szinger JJ, Eller LA, Billings EA, Rao M, Tovanabutra S, Sanders-Buell E, Weijtens M, Pau MG, Schuitemaker H, Robb ML, Kim JH, Korber BT, Michael NL, Protective efficacy of a global HIV-1 mosaic vaccine against heterologous SHIV challenges in rhesus monkeys. Cell 155, 531–539 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ackerman ME, Das J, Pittala S, Broge T, Linde C, Suscovich TJ, Brown EP, Bradley T, Natarajan H, Lin S, Sassic JK, O’Keefe S, Mehta N, Goodman D, Sips M, Weiner JA, Tomaras GD, Haynes BF, Lauffenburger DA, Bailey-Kellogg C, Roederer M, Alter G, Route of immunization defines multiple mechanisms of vaccine-mediated protection against SIV. Nat. Med. 24, 1590–1598 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mabuka J, Nduati R, Odem-Davis K, Peterson D, Overbaugh J, HIV-specific antibodies capable of ADCC are common in breastmilk and are associated with reduced risk of transmission in women with high viral loads. PLOS Pathog. 8, e1002739 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Milligan C, Richardson BA, John-Stewart G, Nduati R, Overbaugh J, Passively acquired antibody-dependent cellular cytotoxicity (ADCC) activity in HIV-infected infants is associated with reduced mortality. Cell Host Microbe 17, 500–506 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ilinykh PA, Santos RI, Gunn BM, Kuzmina NA, Shen X, Huang K, Gilchuk P, Flyak AI, Younan P, Alter G, Crowe JE Jr., Bukreyev A, Asymmetric antiviral effects of ebolavirus antibodies targeting glycoprotein stem and glycan cap. PLOS Pathog. 14, e1007204 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bournazos S, DiLillo DJ, Goff AJ, Glass PJ, Ravetch JV, Differential requirements for FcγR engagement by protective antibodies against Ebola virus. Proc. Natl. Acad. Sci. U.S.A. 116, 20054–20062 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steffen U, Koeleman CA, Sokolova MV, Bang H, Kleyer A, Rech J, Unterweger H, Schicht M, Garreis F, Hahn J, Andes FT, Hartmann F, Hahn M, Mahajan A, Paulsen F, Hoffmann M, Lochnit G, Munoz LE, Wuhrer M, Falck D, Herrmann M, Schett G, IgA subclasses have different effector functions associated with distinct glycosylation profiles. Nat. Commun. 11, 120 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ehrhardt SA, Zehner M, Krahling V, Cohen-Dvashi H, Kreer C, Elad N, Gruell H, Ercanoglu MS, Schommers P, Gieselmann L, Eggeling R, Dahlke C, Wolf T, Pfeifer N, Addo MM, Diskin R, Becker S, Klein F, Polyclonal and convergent antibody response to Ebola virus vaccine rVSV-ZEBOV. Nat. Med. 25, 1589–1600 (2019). [DOI] [PubMed] [Google Scholar]
- 57.DiNapoli JM, Yang L, Samal SK, Murphy BR, Collins PL, Bukreyev A, Respiratory tract immunization of non-human primates with a Newcastle disease virus-vectored vaccine candidate against Ebola virus elicits a neutralizing antibody response. Vaccine 29, 17–25 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ackerman ME, Moldt B, Wyatt RT, Dugast AS, McAndrew E, Tsoukas S, Jost S, Berger CT, Sciaranghella G, Liu Q, Irvine DJ, Burton DR, Alter G, A robust, high-throughput assay to determine the phagocytic activity of clinical antibody samples. J. Immunol. Methods 366, 8–19 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kerntke C, Nimmerjahn F, Biburger M, There is (scientific) strength in numbers: A comprehensive quantitation of Fc gamma receptor numbers on human and murine peripheral blood leukocytes. Front. Immunol. 11, 118 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T, Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Suchard MA, Simpson SE, Zorych I, Ryan P, Madigan D, Massive parallelization of serial inference algorithms for a complex generalized linear model. ACM Trans. Model. Comput. Simul. 23, 1–17 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data associated with this study are present in the paper or the Supplementary Materials.