Abstract
Escherichia coli is a common inhabitant of the human microbiota, and isolates exhibit probiotic, commensal, and pathogenic roles in the host. E. coli members often use diverse small molecule chemistry to regulate intra-bacterial, inter-microbial, and host-bacterial interactions. While E. coli is considered to be a well-studied model organism in biology, much of its chemical arsenal has only more recently been defined and much remains to be explored. Here, we describe chemical signaling systems in E. coli in context with the broader field of metabolism at the host-bacteria interface and its role in disease modulation.
Introduction
Escherichia coli strains constitute a minute fraction of the overall healthy human microbiome (≤0.1% abundance)1. However, they are highly prevalent throughout the human population (>90%), they are often the first bacterial species to colonize the gastrointestinal tract of infants, and they can establish lifelong colonization in adults2. The underlying molecular mechanisms by which E. coli maintain this strong association are not well characterized. E. coli members typically serve commensal roles, producing the essential vitamin K2 and B-complex vitamins, maintaining an anaerobic environment for other commensal inhabitants, and excluding pathogenic competitors, among other roles. However, the species also encompasses a variety of virulent pathotypes falling under two primary categories, IPEC (intestinal pathogenic E. coli) and ExPEC (extraintestinal pathogenic E. coli), that frequently cause enteric/diarrheal disease, urinary tract infections (UTIs), and sepsis/meningitis, amongst others3. While many of the virulence systems in individual E. coli pathotypes have been characterized, the detailed molecular mechanisms by which E. coli strains regulate diverse microbe-microbe, environment-microbe, and host-microbe interactions in the host remain a major area of research. Indeed, modern metabolomic studies of human-derived microbial communities show that the vast majority of detectable molecular features (~98%) remain uncharacterized, including those from E. coli4. Some of these unknown molecular features will undoubtedly serve as crucial signaling molecules and await discovery. Here, we review characterized E. coli small molecules and our current understanding of their roles at the host-microbe interface.
Quorum sensing molecules
Quorum sensing (QS) is a bacterial cell-extrinsic cell-to-cell communication strategy that couples population density to collective genetic behaviors using small-molecule signals called autoinducers. The production and detection of autoinducers allow a bacterial population to coordinate behaviors such as bioluminescence, biofilm formation, virulence factor expression, antimicrobial production, sporulation and DNA-uptake5. E. coli utilize several QS systems, most of which are shared with other bacterial species. Questions remain as to whether additional autoinducers exist and how they work within this larger QS network. In this section, we discuss established autoinducer responses in E. coli.
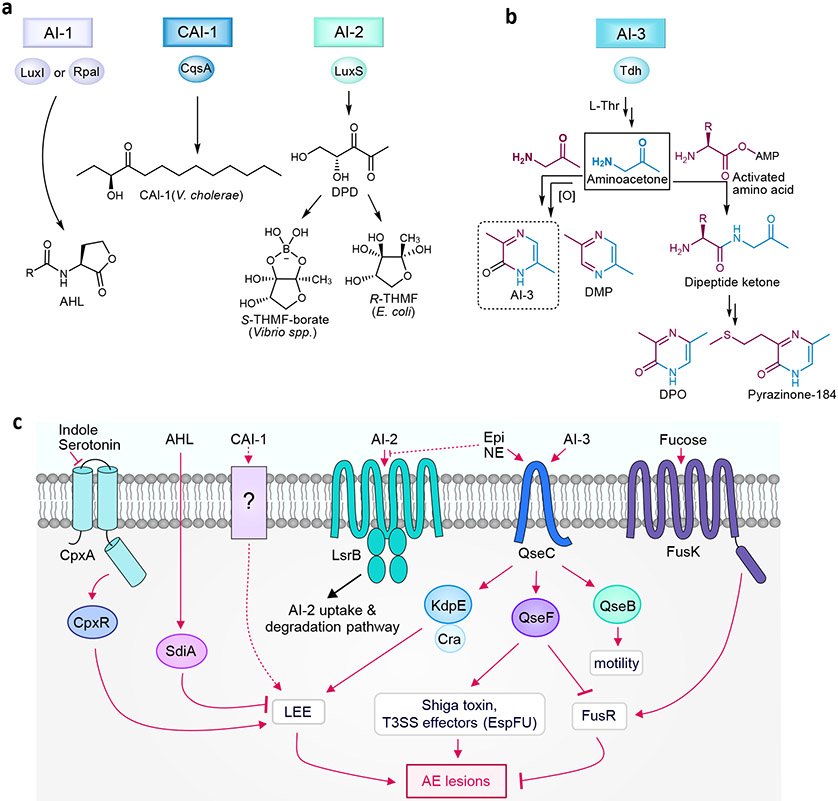
Acyl-homoserine lactones (AHLs), also known as autoinducer-1 (AI-1), are a common class of autoinducers among Gram-negative bacteria with hundreds of bacterial species possessing the AHL synthase, a LuxI homolog. They consist of a homoserine-lactone core and an N-acyl moiety, such as fatty acids with differing chain lengths (4-18 carbons) or other modifications (Fig 1a). While E. coli does not encode the AHL synthase, it does encode the AI-1 receptor SdiA (a LuxR homolog) permitting inter-bacterial gene regulation in response to AHLs released by other bacteria (i.e., a LuxR solo)6. E. coli AHL responses include the regulation of glutamate metabolism and biofilm related genes6,7. Alternative ligands for SdiA have been identified in E. coli, including 1-octanoyl-rac-glycerol, promoting basal SdiA activity8. Consequently, the general regulatory spectrum of LuxR receptors extend beyond that of AHLs.
Figure 1∣. Gram-negative quorum sensing synthases, structures, and E. coli receptors.
a∣ Synthases and structures of autoinducer-1 (N-acyl homoserine lactone (AHL)), Vibrio cholerae autoinducer-1 (CAI-1), and autoinducer-2 (AI-2). b∣ Biosynthesis of autoinducer-3 (AI-3), 2,5-dimethylpyrazine (DMP), 3,5-dimethyl-pyrazin-2-one (DPO), and analogs. c∣ Quorum sensing receptors and signaling pathways in E. coli. Epi, epinephrine. NE, norepinephrine.
Enteropathogenic E. coli (EPEC) has recently been demonstrated to respond to the Vibrio cholerae QS signal (S)-3-hydroxytridecan-4-one, which is known as cholera autoinducer-1 (CAI-1)9. CAI-1 was found to enhance EPEC virulence, though neither an E. coli CAI-1 synthase nor receptor has been identified. Of note, CAI-1 did not upregulate virulence in the closely related enterohemorrhagic E. coli (EHEC) pathotype, suggesting an evolutionary basis for EPEC CAI-1 detection due to its common niche with V. cholerae, both of which cause diarrheal disease through infection in the small intestine. These studies highlight that inter-bacterial chemical communication in E. coli can be pathotype specific.
The second class of autoinducer (AI-2) consists of an interconvertible set of small molecules formed by the spontaneous cyclization of 4,5-dihydroxy-2,3-pentanedione (DPD) (Fig 1a). DPD is a reactive degradation product of S-adenosylhomocysteine produced by the broadly conserved bacterial enzyme S-ribosylhomocysteine lyase (LuxS)10. While only two forms – a furanosyl borate diester and tetrahydroxy furan – have been structurally elucidated, AI-2 is used as a collective term for DPD-derived interconvertible signaling molecules5. Its broad production and structural variability make AI-2 well-suited for inter-species communication5,11. In E. coli, production of AI-2 serves as a chemoattractant, promoting swimming motility and auto-aggregation. Auto-aggregation, in turn, enhances AI-2 signaling, bacterial stress resistance, and biofilm formation, phenotypes often associated with virulence12,13.
E. coli harbors a pathway for AI-2 uptake and degradation, which is encoded by the lsrACDBFGE operon (lsr stands for LuxS regulated)11. AI-2 accumulates extracellularly during exponential growth and then is imported upon entry into stationary phase by the LsrACDB ABC transporter. Imported AI-2 undergoes phosphorylation by LsrK which is then thought to bind to the LsrR repressor, leading to derepression of the lsr operon. This creates a positive feedback loop, whereby uptake and phosphorylation of AI-2 enhances the expression of the Lsr transporter, resulting in increased uptake and rapid depletion of extracellular AI-2. Recently, LsrK activity was found to be regulated by the phosphoenolpyruvate (PEP)–dependent sugar phosphotransferase system (PTS) protein HPr, linking the AI-2 QS system to cell metabolism14. It is postulated that by internalizing AI-2 from the environment, lsr-expressing bacteria can interfere with the AI-2 mediated communication of other members within their niche.
Along with indirectly affecting gut health by manipulating microbiome populations, AI-2 has been demonstrated to directly induce host cell responses. Exposure of mammalian epithelial cells to AI-2 initiates the rapid production of cytokine interleukin 8 (IL-8)15. IL-8 is an important mediator of the innate immune response, acting as a potent neutrophil chemoattractant. Reciprocally, host epithelial cells have been shown to produce their own AI-2 mimic thought to dysregulate bacterial colonization. Bacterial exposure and the disruption of tight junctions promote the release of this AI-2 mimic, which activates the bacterial AI-2 receptors LuxP and LsrB16. The structure of the AI-2 mimic and its importance for inter-kingdom signaling is not yet fully understood.
The third E. coli QS system, AI-3, was initially reported as a virulence regulator of the foodborne pathogen EHEC, a causative agent of acute human gastroenteritis and hemorrhagic colitis17. EHEC virulence stems from a chromosomal pathogenicity island, termed the locus of enterocyte effacement (LEE), that encodes a type III secretion system (T3SS) and effector proteins used to create attaching and effacing lesions within the gastrointestinal tract18. Several signaling molecules have been implicated in LEE regulation, including AI-3 and the host-derived adrenergic signals, epinephrine and norepinephrine17,19. It is thought that epinephrine and norepinephrine pass through the host intestinal epithelium, cueing EHEC to the site of colonization and infection. These three signals modulate LEE expression through EHEC’s QseBC two-component signal transduction pathway20 (Fig 1c). Additionally, indole and serotonin have been shown to regulate LEE through the CpxAR two-component system and will be discussed further in the next section.
AI-3 consists of a family of pyrazinone analogs, with all but one formed from the spontaneous coupling of threonine dehydrogenase (Tdh)-derived aminoacetone with an aminoacyl-AMP derived from “abortive” tRNA synthetase reactions21 (Fig 1b). The one exception, 3,6-dimethylpyrazin-2-one, is produced by the dimerization of aminoacetone. All of the pyrazinones require an oxidation step, which coincides with pathogenic E. coli’s microaerobic niche environment along the epithelium and the location of LEE expression22. Disruption of the epithelium by EHEC-induced lesions results in increased environmental oxygen, further driving EHEC colonization and amplifying virulence expression23. Of the pyrazinones characterized from cell culture, only 3,6-dimethylpyrazin-2-one exhibited an identical LEE activity spectrum as that previously reported for uncharacterized AI-3 and could be detected in mouse fecal samples17. Thus, 3,6-dimethylpyrazin-2-one is suggested to be the primary form of AI-3. The other pyrazinones derived from the abortive tRNA synthetase reactions were upregulated in response to ribosome inhibitor stress in cell culture, providing a molecular mechanism for how cellular stress signaling could be integrated with QS responses. While only select members of this group regulated LEE expression, they were produced in diverse Gram-negative and Gram-positive organisms. Thus, the fecal detection of select analogs likely derive from community-level production rather than E. coli alone. Interestingly, the V. cholerae signal, 3,5-dimethylpyrazin-2-one (DPO), an alanine analog, was discovered as a minor constituent amongst the characterized pyrazinones in E. coli21. In V. cholerae, DPO represses genes required for biofilm formation and toxin production24. These data suggest that tdh-derived aminoacetone regulation may be a more general signaling mechanism, as aminoacetone is incorporated in several signaling molecules including AI-3, DPO, and the ant pheromone, 2,5-dimethylpyrazine (DMP)25.
Unlike the E. coli receptors QseC and QseF, which respond to host epinephrine and norepinephrine, mammalian androgenetic receptors do not reciprocally respond to AI-3 pyrazinones. However, data suggest that AI-3 and its analogs can act as microbe- or pathogen-associated molecular patterns (PAMPs) to modulate host immune responses. For example, a methionine derived analog increased IL-8 secretion at 100 nM and higher in human THP-1 cells differentiated into their macrophage cell type21. The host sensory system(s) used to recognize these PAMPs remain an open area of investigation.
Aromatic amino acid-derived metabolites
The gut microbiome converts tryptophan into indole containing catabolites that serve as intra-species, inter-species, and inter-kingdom signaling molecules, capable of modulating bacterial and host physiology. These tryptophan metabolites play central roles in mitigating intestinal inflammation and conferring protection against diseases such as inflammatory bowel diseases (IBDs). Determining the structures, producers, and fate of these tryptophan related metabolites are important to understanding their underlying role in human health and their potential use as therapeutics or nutraceuticals.
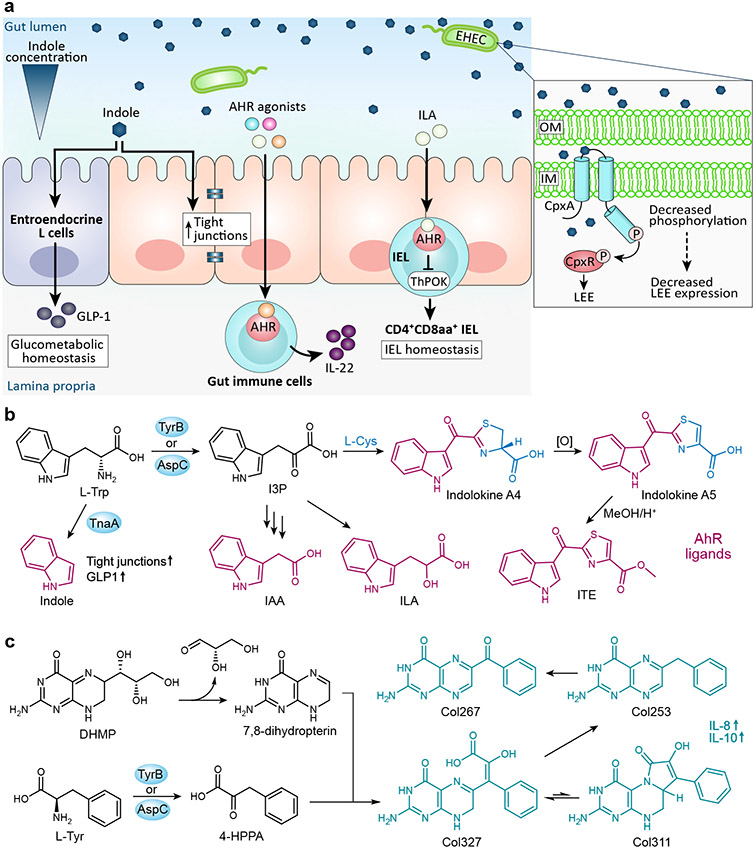
In E. coli, indole is formed through catalytic cleavage of L-tryptophan by the pyridoxal phosphate (PLP)-dependent enzyme tryptophanase (TnaA) and is sensed by the membrane-bound histidine kinase sensors CpxA and BaeS and a transcriptional activator for acid resistance, GadX26. Beyond this, indole’s import and export mechanisms and molecular targets remain largely obscure27. A diverse range of metabolic and physiological effects have been associated with indole signaling in E. coli. Indole is generally thought to affect responses to stressors such as antibiotics, pH changes, heat, and reactive oxygen species (ROS), as well as, influence biofilm formation, persister cell formation, and virulence27.
It has been suggested that E. coli sense an indole concentration gradient – from the lumen to the epithelial mucosa – to regulate the appropriate biogeographical gene machinery to aide in niche establishment28,29. EHEC is reported to use this indole sensory system to regulate the expression of LEE virulence genes, which favors expression near the epithelial surface29. High concentrations of indole within the lumen result in deregulation of LEE-encoded T3SS, saving EHEC the metabolic burden of unnecessary T3SS production. From an alternative perspective, nonpathogenic competitors may secrete indole to prevent the invasion of enteric pathogens along the epithelium by downregulating the expression of their virulence repertoire, dually protecting themselves and the host30. The indole containing neurotransmitter, serotonin, was also recently discovered to reduce EHEC virulence by downregulating LEE through the indole receptor, CpxA31. Expression of the mammalian serotonin selective reuptake transporter (SERT) decreases during inflammation and in principle could lead to elevated serotonin levels at the inflammatory site of infection to inhibit pathogens such as EHEC32.
Studies have revealed the importance of indole signaling on host physiology and homeostasis. Indole stimulates the production of tight junctions in intestinal epithelial cells (IECs), which promotes epithelial integrity and reduces inflammation33,34. Additionally, indole induces the secretion of glucagon-like peptide-1 (GLP-1) from enteroendocrine L cells35. GLP-1 plays a critical role in glucometabolic homeostasis by enhancing insulin secretion, delaying gastric emptying, and decreasing appetite36,37. Outside the gastrointestinal tract, indole treatment of hepatic cells was discovered to reduce proteins of the NF-κB pathway and down-regulate NLRP3 inflammasome expression, thus alleviating liver inflammation in mice38.
Research defining tryptophan catabolite pathways have been mainly focused on a few major producing strains, leaving many microbiome producers unidentified. A couple of papers have reported the detection of several of these catabolites (indole-3-lactic acid (ILA), indole-3-acetic acid (IAA), and indole-3-pyruvate (I3P)) in E. coli cultures, though information on how E. coli form, transport, and sense these catabolites are incomplete39,40. ILA can be found in a diverse range of microorganisms including bacteria, fungi, other human microbes, and plants. It is a prominent aryl hydrocarbon receptor (AhR) regulator, which is a human ligand-activated transcription factor that modulates essential biological processes involved in tissue homeostasis as well as the development of pathological conditions like autoimmune, neoplastic, metabolic and degenerative diseases41. Via AhR, ILA maintains intraepithelial lymphocyte (IEL) homeostasis. Deficiencies in either AhR or its ligands impair crucial IEL functions, such as epithelial cell turnover, the ability to control microbial load and composition, and the regulation of immunopathology41,42. In a mouse study, ILA from the gut commensal Lactobacillus reuteri activated an AhR-dependent reprogramming of intraepithelial CD4+ T helper cells into the recently discovered immunoregulatory cells, CD4+CD8αα+ double-positive IELs (DP IELs)43. Germ free mice lack DP IELs, highlighting the importance microbial metabolites have on host cell differentiation. ILA has also been inversely linked to hypertension and autoimmunity caused by high salt consumption44. High salt intake in mice depleted the ILA producer, Lactobacillus murinus, resulting in an increased T helper (TH)17 cell response, an autoimmune response associated with hypertension. Further, ILA inhibited polarization of TH17 cells in vitro. It was previously established that dietary salt promotes TH17 cell differentiation, which plays a role in the pathogenesis of many autoimmune diseases, including IBD, psoriasis, rheumatoid arthritis, and multiple sclerosis45,46. Thus, these new findings suggest that in addition to direct induction, salt indirectly induces TH17 cell response via disruption of the microbiota. Further insights into this molecular link between salt consumption, the microbiota, and the immune system will be critical for future research into modulating autoimmune diseases.
E. coli also produce IAA, another widely generated AhR agonist. AhR activity is thought to affect the severity of IBD, as AhR activation enhances regulatory T cells in humanized mice to prevent colitis and AhR downregulation is observed in the intestinal tissue of IBD patients37,47,48. IAA levels are reduced in IBD patients, implicating IAA and other microbially-produced tryptophan catabolites as important regulators of IBD49. In murine macrophage and hepatocyte cultures, IAA attenuated fatty acid- and LPS-induced pro-inflammatory cytokine responses via AhR50. It was further observed that mice fed high-fat diets experienced reduced IAA, thus connecting the microbiota and reduced IAA to liver inflammatory response and diet. Together, these findings suggest that along with modifying diet, supplementary ILA and IAA could potentially become a useful avenue for combating inflammatory conditions. However, AhR exhibits promiscuous ligand selectivity and responses, and the wider functional roles of microbiota-derived AhR (ant)agonists remain a subject of current investigation51,52.
Tryptophan catabolites can also be derivatized and incorporated into more complex bioactive compounds. For example, E. coli produce a family of redox stress induced metabolites termed indolokines that are dependent on the conversion of tryptophan into indole-3-pyruvic acid (I3P) by the aromatic amino transferases AspC/TyrB (Fig 2b)53. I3P is then likely converted into 3-indoleglyoxylic acid (or an activated form) prior to spontaneous coupling with L-Cys or the glutathione-associated metabolite, L-Cys-Gly. Interestingly, four of the members have been previously reported as phytochemicals associated with plant pathogen defense, suggesting conservation in cellular stress signaling between plants and E. coli. These molecules are closely related to the potent synthetic AhR agonist 2-(1′H-indole-3′-carbonyl)-thiazole-4-carboxylic acid methyl ester (ITE).
Figure 2∣. Aromatic amino acid-containing bacterial small molecules and their proposed mechanisms of action.
a∣ Here we highlight a few of the signaling pathways induced by E. coli tryptophan catabolites. Indole stimulates tight junctions and glucagon-like peptide 1 (GLP-1), respectively, affecting intestinal barrier integrity and glycometabolism homeostasis. An indole concentration gradient in the human gut influences the virulence gene expression in EHEC, with the expression of the LEE pathogenicity island decreasing in the presence of indole. Several tryptophan catabolites act as aryl hydrocarbon receptor (AhR) ligands to influence host immune response. Indole-3-lactic acid (ILA) stimulates the development of CD4+CD8αα+ intraepithelial lymphocytes. b∣ Tryptophan catabolites produced by E. coli that also serve as AhR ligands include indole, indole acetic acid (IAA), and ILA. E. coli also produce the AhR ligands indolokine A4 and indolokine A5, which are formed from indole-3-pyruvic acid (I3P) and L-cysteine, and resemble the potent AhR agonist 2-(1′H-indole-3′-carbonyl)-thiazole-4-carboxylic acid methyl ester (ITE). c∣ The tryptophan derivative 3-(4-hydroxyphenyl)pyruvate (4-HPPA) reacts with 7,8 dihydropterin, a degradation product of the folate/monapterin pathway, to form a molecular family termed the colipterins.
E. coli also produces another recently characterized family of AspC/TyrB dependent metabolites termed colipterins (Fig 2c)54. These metabolites consist of a pterin core derived from a degradation product of the monapterin pathway, 7,8 dihydropterin, that is largely functionalized with an AspC/TyrB-dependent phenylpyruvate precursor. Major members of the colipterin family were observed to activate anti-inflammatory interleukin-10 (IL-10) and improve colitis symptoms in a colitis mouse model. It was further found that treatment with the antifolate inhibitor sulfamethoxazole enhanced colipterin production in E. coli through the monapterin pathway. These studies supported a new model, in which antibiotics could indirectly regulate immune system function through the activation of bacterial stress responses.
RiPPs
Microbes have developed a variety of strategies to compete with other species for resources and niche colonization, including the secretion of ribosomally-encoded and posttranslationally-modified peptides (RiPPS). RiPPs encompass a large family of ribosomal precursor peptides that have undergone diverse posttranslational modifications, including proteolytic cleavage, cyclization, glycosylation, phosphorylation, alkylation, and redox modification55. The biosynthetic genes for RiPPS are usually clustered together at a single gene locus and encode for the precursor peptide, modification enzymes, and proteins involved in immunity and export. Metagenomic studies have revealed that biosynthetic gene clusters encoding modified RiPPs are widely distributed among the human microbiota56.
E. coli strains produce about eleven low molecular weight (<10 kDa) RiPPs termed microcins. Microcins (Mcc) are a subclass of bacteriocins; a class of Gram-negative and Gram-positive antimicrobial RiPPs defined by their ability to kill or inhibit the growth of closely related bacterial strains57. These include the oxazole/thiazole-containing MccB17, the nucleotide carrying MccC7/C51, the lasso peptide MccJ25, and the siderophore conjugated MccM and MccH47. Microcins are typically hydrophobic, often showing high stability to heat, extreme pH, and proteases. They target susceptible bacteria through a variety of mechanisms, such as inhibiting DNA gyrase (MccB17) and aspartyl tRNA synthetase (MccC), blocking the secondary RNA polymerase channel and cytochromes required for cellular respiration (MccJ25), among others58. As such, they act as mediators of inter- and intra-species competition and modulate pathogenicity. For example, MccM and MccH47 enable the probiotic bacterium E. coli Nissle 1917 to limit the expansion of closely related Enterobacteriaceae, such as commensal E. coli, adherent-invasive E coli (AIEC), and the related pathogen Salmonella enterica, during intestinal inflammation by targeting specific siderophore receptors59.
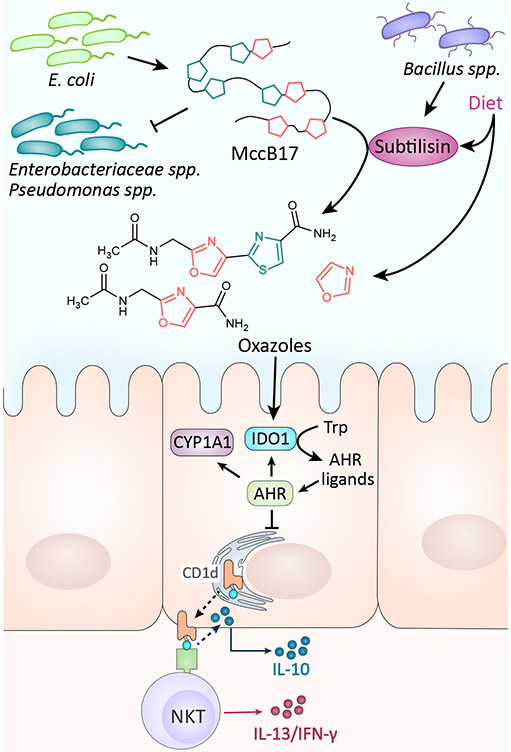
Bacteriocins have also been implicated in host-microbe interactions, including anticancer interactions. For instance, E. coli’s microcin J25 exhibits pro-apoptotic activity against breast cancer cells60. A recent study has also illustrated an immunoregulatory role for the oxazole/thiazole containing microcin, MccB17. While microcins are generally thought to possess low toxicity, oxazole/thiazole containing fragments from the proteolysis of MccB17 by subtilisin were shown to induce colonic inflammation by suppressing IEC IL-10 production in a CD1d-dependent manner through activation of the aryl hydrocarbon receptor (AhR) pathway61 (Fig 3). This surprising finding challenges other recent reports of AhR activity in IECs which demonstrate AhR-dependent inhibition of proinflammatory pathways through increased IL-10 production62 and provides a mechanistic explanation for the oxazolone-induced colitis previously observed in animal models63. Future research is needed to disentangle the multi-faceted small molecule-AhR interactions and their disparate context-dependent roles in pro-inflammatory and/or anti-inflammatory activities.
Figure 3∣. Microcin B17 (Mcc17) biological mechanisms.
MccB17 is produced by E. coli strains to inhibit the growth of closely related bacteria by targeting their DNA gyrase. MccB17 also can be cleaved by the protease subtilisin to form small oxazole containing fragments. Oxazoles derived from diet and microbes activate indoleamine 2,3-dioxygenase (IDO1) which induces the production of AhR ligands in intestinal epithelial cells (IECs). AhR activation in IECs leads to invariant natural killer T (iNKT) cell-mediated intestinal inflammation61.
Siderophores
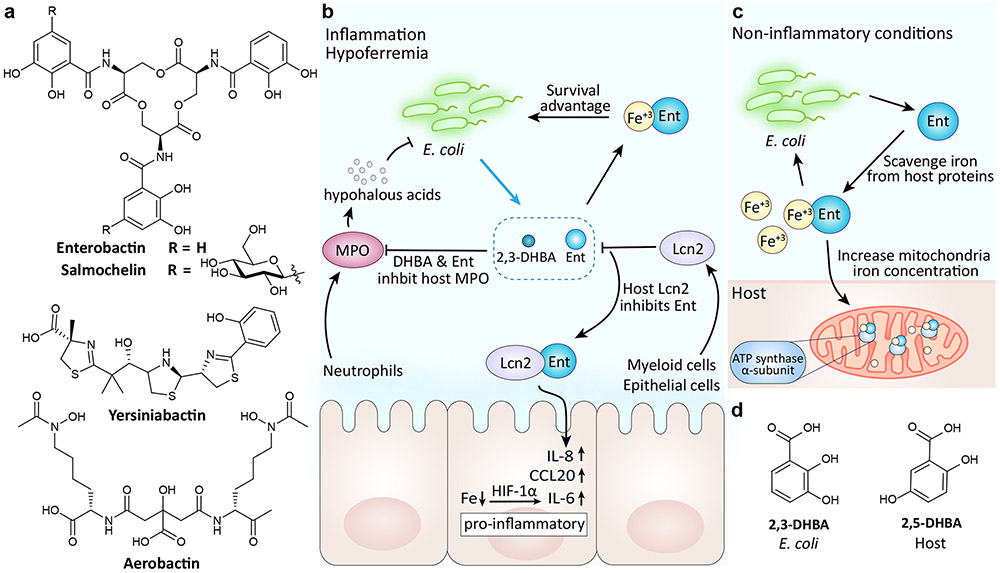
Like most organisms, E. coli require iron for growth and other biological processes. However, in a mammalian host environment, this essential nutrient is limited. One limiting factor is the insolubility of extracellular iron III in neutral pH environments (i.e., serum)64. The other is bacterial competition with the host’s iron storage mechanisms, which provide protection against bacterial pathogens via nutritional immunity. To survive, bacteria have evolved a number of ways to evade iron nutritional immunity, including the use of siderophores. Siderophores are low molecular weight, high-affinity iron chelating metabolites that are synthesized and secreted by bacteria (and fungi) to scavenge iron from host chelators (i.e., transferrin and lactoferrin). Many pathogens sense low iron levels in the host (i.e., hypoferremia of inflammation) as a cue to initiate virulence64,65. Four distinct siderophores have been reported from E. coli strains: the catecholates enterobactin and salmochelin, the hydroxamate aerobactin, and the mixed-ligand siderophore yersiniabactin (Fig 4a).
Figure 4∣. Siderophore structures and their interactions within the human gut.
a∣ Structures of E. coli siderophores. b∣ During inflammation, environmental iron levels decrease. In turn, enterobactin secretion is increased to maintain E. coli’s iron load. Enterobactin and its precursor 2,3-dihydroxybenzoic acid (DHBA) also promote E. coli survival by inhibiting the bactericidal enzyme myeloperoxidase (MPO). As a countermeasure, the host protein lipocalin 2 (Lcn2) binds enterobactin. The Lcn2-enterobactin complex prevents the pirating of iron and induces a host immune response. c∣ As part of a mutualistic relationship between healthy gut bacteria and the host, enterobactin promotes mitochondrial iron uptake by binding the to host ATP synthase α subunit72. d∣ Structure of bacterial 2,3-DHBA and its human analog 2,5-DHBA.
Enterobactin (also called enterochelin) has the highest affinity toward iron than any other known siderophore (Kd 10−49 M)66 and is produced by both commensal and pathogenic strains of E. coli, as well as in other Enterobacteriaceae members. Like all other siderophores, enterobactin is secreted and therefore can act as a public good that can be sensed and utilized by both producers and some non-producers with appropriate uptake mechanisms67,68. For instance, pathogens such as Pseudomonas aeruginosa and Bordetella pertussis do not produce enterobactin but have evolved the appropriate receptors to initiate xenosiderophore uptake allowing them to access iron without bearing the metabolic cost of siderophore production67,69. For the bacterial respiratory pathogens Bordetella pertussis and Bordetella bronchiseptica, enterobactin is incorporated into one of many iron-source-sensing circuits termed ferrimone sensing. This allows the pathogen to exploit different iron sources during infection69.
In addition to iron acquisition, enterobactin has also been shown to manipulate host processes by interacting directly with host proteins (Fig 4b). Hypoferremia in the inflamed gut stimulates enterobactin production in E. coli. Enterobactin along with its precursor 2,3-dihydroxybenzoic acid (DHBA) inhibit the bactericidal human enzyme myeloperoxidase (MPO), thus affording E. coli a survival advantage through both iron acquisition and protection against oxidative stress70. Enterobactin and DHBA mediated inhibition is counter regulated by the host siderophore binding protein, lipocalin 2 (Lcn2). Enterobactin-bound Lcn2 regulates the host innate immune response by inducing the secretion of IL-8 in the mucosal epithelia.
There have been mixed reports as to whether enterobactin is required for E. coli virulence. Mutant studies have shown enterobactin to have a negligible role in ExPEC virulence, unlike the siderophores aerobactin and salmochelin71. This has been attributed to the host’s ability to inhibit enterobactin through Lcn2, which cannot bind the non-catecholate siderophores, aerobactin and yersiniabactin, or the glycosylated catecholate siderophore, salmochelin. While enterobactin is often presumed to negatively affect the host, the high prevalence of enterobactin-producing commensal bacteria and its uncertain role in virulence has put into question whether alternative beneficial enterobactin-regulated host pathways exist. Recently it was discovered that enterobactin binds to the mitochondrial ATP synthase α-subunit in the nematode Caenorhabditis elegans, in turn promoting C. elegans growth through mitochondrial iron-uptake (Fig 4c)72. Conservation of this mechanism was further observed in mammalian HEK293T cells. This mutualistic relationship may explain how host animals tolerate abundant enterobactin concentrations from non-infectious gut microbiota members and points to a larger role for enterobactin in host-microbe interactions.
Colibactin
Some strains of E. coli harbor an ~54 kb genomic island (clb or pks) that encodes for the polyketide synthase-nonribosomal peptide synthetase (PKS-NRPS) hybrid genotoxin termed colibactin73. Exposure of mammalian cells to colibactin-producing (clb+) E. coli induces mammalian cell genotoxicity, including DNA double stranded breaks, cell cycle arrest, and genomic instability phenotypes73,74. The pathway in the AIEC pathotype has been shown to promote carcinogenesis under inflammatory conditions in multiple mouse models, and clinical data indicate an increased prevalence of the pathway in inflammatory bowel disease (IBD) and colorectal cancer (CRC) patients relative to healthy human subjects75-77. Because colibactin has been the subject of several recent reviews78-80, we briefly highlight recent discoveries and discuss some areas that require further investigation.
While the clb pathway produces a collection of metabolites with variable activities, the primary genotoxic mode of action of colibactin has been attributed to DNA alkylation and interstrand crosslinking. This crosslinking event has been demonstrated in mammalian cells transiently infected by colibactin-producing enterobacteria, activating host DNA damage responses such as the DNA crosslink repair machinery of the Fanconi anemia pathway, γH2AX (a downstream marker of DNA double strand breaks), and megalocytosis (cellular enlargement)73,81 (Figure 5a). Consistent with these observations, colibactin has been characterized as a heterodimer containing two electrophilic spirocyclopropane warhead motifs that create interstrand crosslinks by alkylating the N3-position of adenine nucleobases82,83 (Figure 5a). Recently, it was demonstrated that prolonged exposure of wild-type human organoids to clb+ E. coli promotes the formation of two unique mutational signatures whose presence are enriched in human CRC tumors84,85. These mutational signatures in cancer patients were consistent with colibactin adenine alkylation events.
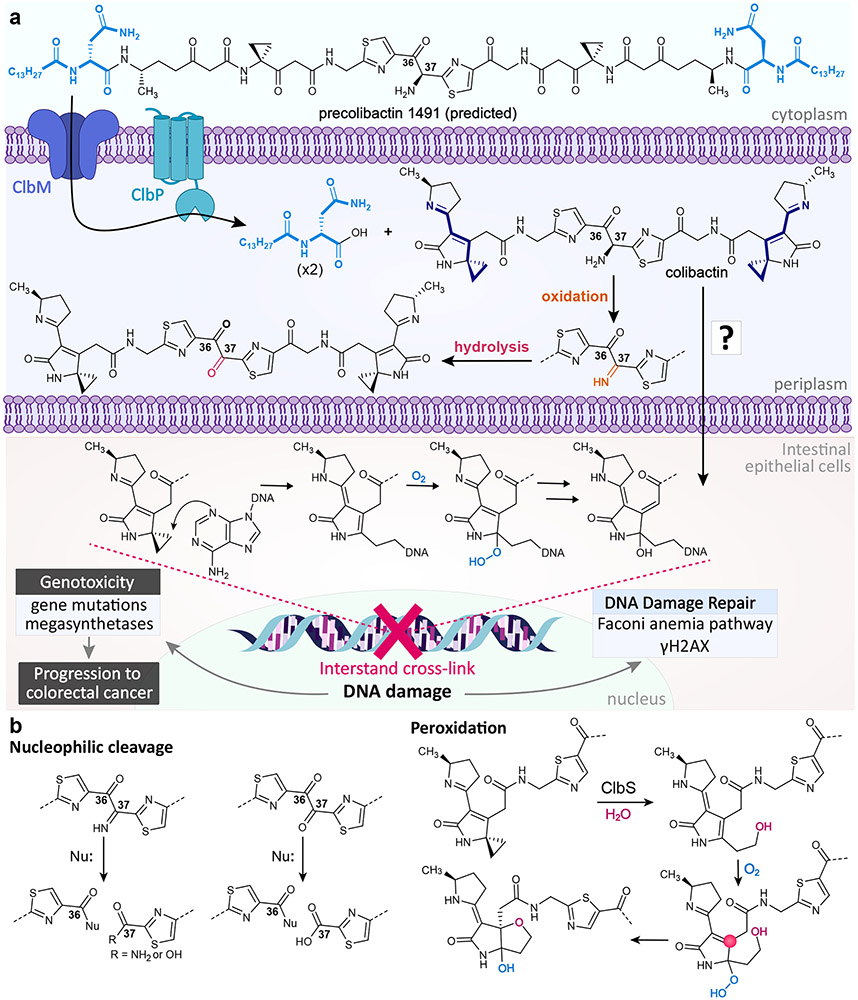
Figure 5∣. Proposed biosynthesis and biology of colibactin.
a∣ Precolibactin 1491 is transported into the periplasm and cleaved to form a mature colibactin. The α-amino-ketone bithiazole spacer is subject to autoxidation at the C37 position in vitro, though it is unclear to what extent oxidation would occur in vivo. Colibactin is transported into host epithelial cells through an unknown mechanism, where its electrophilic spirocyclopropane moieties cross-link DNA. b∣ In the absence of ClbP, precolibactin 1491 undergoes oxidation and macrocyclization to form precolibactin 1489, a prediction based on tandem MS, isotopic labeling, genome editing studies, and smaller characterized macrocycles. c∣ Colibactin has been observed to undergo reactions other than DNA cross-linking, such as nucleophilic cleavage of the oxidized α-amino-ketone bithiazole spacer and peroxidation. In the peroxidation mechanism, the C4 position (pink circle) can act as an electrophilic site, although it can also undergo stable tautomerization.
While considerable work has gone into elucidating the structure of colibactin and its biological role in human health, some important mechanistic questions that remain to be addressed include the following. 1) The α-amino-ketone bithiazole spacer is subject to autoxidation and nucleophilic cleavage (Figure 5b). This activity underlies the challenges in isolating mature colibactin for detailed chemical biological analysis and explains many of the degradation products observed in cell culture. While the intestinal tract is largely anaerobic, oxygen becomes available at the epithelial surface. The extent and biological relevance of oxidation that occurs during colibactin trafficking to the host cell nucleus remains unknown. Thus, the role of the α-amino-ketone, which likely participates in the competitive colibactin inter-bacterial interactions in the largely anaerobic gut86 has not been addressed due to its instability. 2) As noted, once the amino-ketone is oxidized, its diketone motif is subject to nucleophilic attack, providing an unexplored electrophilic site. Additionally, the cyclopropane ring-opened product is highly subject to peroxidation and presents an electrophile at the C4 of the lactam ring (Figure 5b), which was identified during investigations of the colibactin resistance protein ClbS87. The roles of the organic peroxide (a ROS), the diketone electrophile, and the C4 electrophile, which is subject to stable tautomerization83, remain unknown. We speculate that these electrophilic sites could participate in additional crosslinks with other molecules (e.g., histones). 3) Colibactin activity requires bacteria-human cell contact, even when colibactin is produced in standard laboratory cloning strains73. Outer membrane vesicle delivery and/or colibactin instability may contribute to this phenotype88, but the detailed mechanisms of contact dependency and cellular trafficking remain undefined. 4) A clbA mutant that abrogates colibactin and siderophore biosynthesis89 in the probiotic E. coli Nissle 1917 no longer provides a protective benefit in a colitis mouse model86. How molecules from the colibactin pathway participate in this phenotype, if at all, still remains an open question. 5) Finally, with the knowledge that colibactin is causal of mutational signatures in cancer patients84,85, how can mechanistic information be exploited to reduce colorectal cancer burden? Both general and targeted approaches could be considered. For example, sodium tungstate is being explored in clinical trials90 where treatment leads to a general reduction in Enterobacteriaceae like E. coli and reduces colorectal cancer burden in animal models91. However, sodium tungstate itself can be toxic. Inhibitors of colibactin peptidase ClbP reduce DNA damage and block tumorigenesis in animal models, although the compounds required prophylactic treatment at millimolar levels92. Exogenous supplementation of colibactin resistance protein ClbS protects human cells from genotoxic action in a human-bacteria transient infection model,81,88 although this would similarly require a prophylactic strategy. Lastly, the prodrug activation mechanism inherent to colibactin could be exploited for Trojan horse delivery of prodrug antibiotics.
Cyclic dinucleotides
Cyclic dinucleotides (cdNs) are versatile and ubiquitous second messengers found in both bacteria and humans. They are synthesized by dinucleotide cyclases and nucleotidyltransferases (NTases), and degraded by phosphodiesterase enzymes (PDEs). The most well-studied cdN to date is 3’-5’/3’-5’ cyclic diguanylate (c-di-GMP), a prevalent Gram-negative and Gram-positive signaling molecule that has been implicated in a wide range of molecular processes. In E. coli, this includes the regulation of motility, morphogenesis, biofilm formation, and virulence93. Along with acting as an E. coli signal, c-di-GMP is recognized by the human receptor STING (stimulator of interferon genes) and acts as a PAMP to trigger an innate immune response. C-di-GMP regulation in E. coli has been discussed in several reviews93,94. Here we will focus on more recently identified cdNs and their roles at the host-bacteria interface.
One of these recently identified cdNs is 3’-5’/3’-5’ cyclic GMP-AMP (3’,3’-cGAMP). 3’,3’-cGAMP is synthesized by DncV (dinucleotide cyclase in Vibrio cholerae), named for the bacterium from which it was discovered95. DncV belongs to a family of cdN NTases termed cGAS/DncV-like nucleotidyltransferases (CD-NTases)96. cGAS refers to the metazoan homolog, cyclic GMP-AMP synthase (cGAS), which synthesizes the mammalian signal 2’-5’/3’-5’ cyclic GMP-AMP (2’,3’-cGAMP). Recently, 3’,3’-cGAMP signaling has been implicated in regulating anti-phage resistance mechanisms97. In the original V. cholerae O1 El Tor Biotype system, cGAMP activates the phospholipase CapV, resulting in membrane degradation and growth inhibition. The operon containing the capV-dncV gene pair also appears in the genome of ETEC strain TW11681. The presence of the operon was shown to confer resistance to a variety of phages by initiating altruistic suicide through membrane degradation prior to phage-mediated killing, thus preventing phage progeny from spreading amongst a bacterial population (Fig 6a). This abortive infection mechanism is termed CBASS for cyclic-oligonucleotide-based anti-phage signaling. In the mammalian cGAS-STING pathway, cGAS produces the endogenous STING ligand 2’,3’-cGAMP in response to viral infection to initiate antiviral STING responses (Fig 6b). This new finding suggests that the mammalian cGAS-STING antiviral pathway is evolutionary linked to microbial antiphage systems and raises the question to whether additional links exist between human innate immunity and bacterial defense systems.
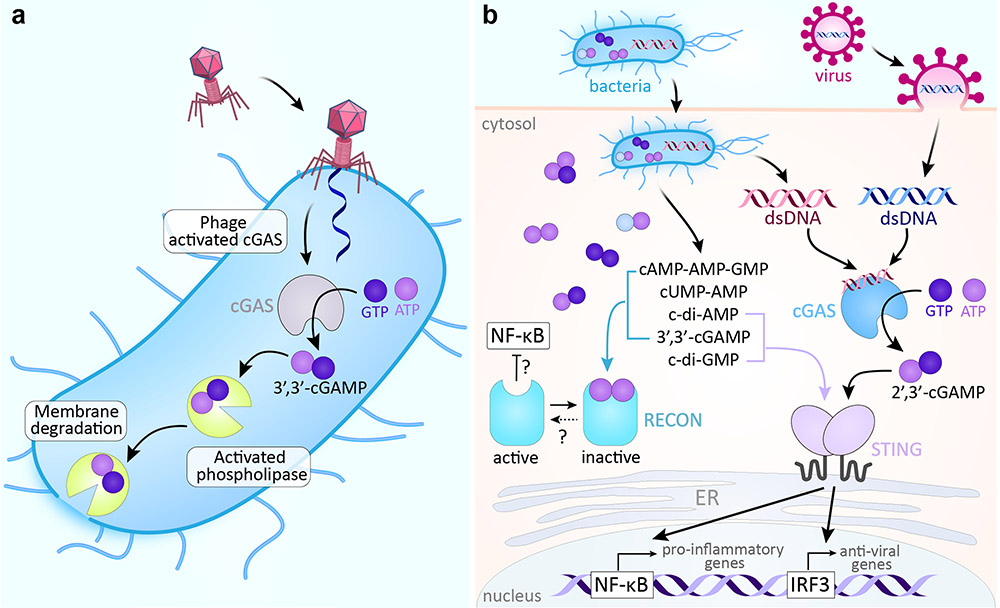
Figure 6∣. Cyclic nucleotide signaling defense pathways in bacteria and mammalian cells.
a∣ In CBASS, phage infection triggers the synthesis of 3’,3’-cGAMP, which then activates phospholipase. The phospholipase degrades the bacterial membrane, leading to altruistic suicide prior to the completion of phage reproduction and thus preventing the further spread of infection97. b∣ In the mammalian cGAS–STING pathway, cGAS senses the DNA of bacterial and viral invaders and synthesizes 2’,3’-cGAMP, which in turn binds and activates STING. STING activates transcription factors needed for downstream transcription of innate immune response genes. The bacterial cdNs, c-di-GMP and 3’,3’-cGAMP, also directly activate STING. Several bacterial signals bind to RECON, inhibiting its undefined oxidoreductase activity and leading to increased activation of the proinflammatory transcription factor NF-κB. CBASS, cyclic-oligonucleotide-based anti-phage signaling system; cGAMP, cyclic GMP-AMP; cGAS, cyclic GMP-AMP synthase; STING, stimulator of interferon genes; cdN, cyclic dinucletotide; RECON, reductase controlling NF-κB.
Recent investigations into DncV homologues have also resulted in the identification of an E. coli purine-pyrimidine CD-NTase, CdnE, found to synthesize cyclic 3’-5’/3’-5’ UMP-AMP (cUMP-AMP)96. Similar to bacterial cGAMP, cUMP-AMP activates a cognate phospholipase, CapE, though the physiological relevance of this system to E. coli has yet to be defined. Unlike c-di-GMP, 3’,3’-cGAMP, and 2’,3’-cGAMP, cUMP-AMP is not detected by STING; however, it is sensed by a separate cdN pattern recognition receptor in mammals known as RECON (reductase controlling NF-κB, Fig. 6b)96. RECON has been reported to antagonize STING by reducing cdN bioavailability and to regulate antibacterial inflammation through a currently undefined immunometabolic mechanism in cell culture studies98. Thus, with the discovery of new cdNs like cUMP-AMP comes many exciting questions regarding their biological roles in bacteria and their immunological roles in the host.
Conclusion
While commensal E. coli provide several benefits to their human host, pathogenic E. coli account for well over 200,000 infections per year in the United States alone99. In one hand, the substantial genomic variation among E. coli strains complicates our understanding of E. coli’s molecular roles at the host-bacteria interface,100 but on the other hand, this variation defines their mechanisms of host engagement (i.e., pathotype specificity). Many of the underlying biological mechanisms are governed by small molecule signaling. However, the majority of detectable small molecule metabolites in E. coli strains remains uncharacterized. Thus, characterization of new E. coli metabolites in a functional context continues to be a promising area of biomedical research. E. coli as a model provides a solid foundation for these efforts with a wealth of chemical signaling information already established. The chemical diversity of these molecules derives from a range of biosynthetic strategies, including dedicated biosynthetic gene clusters of secondary metabolism, dysregulated primary metabolic pathways, and spontaneous chemical reactions of cellular precursors. These molecular strategies extend well beyond that of E. coli. From a chemical biology perspective, identifying the structures, abundance, biological activity, and prevalence of uncharacterized small molecules across microbes (and host cells) are critical first steps toward understanding how the microbiota modulates host physiology at the molecular level.
ACKNOWLEDGMENTS.
Our work on E. coli metabolism at the host-microbe interface is supported by the National Institutes of Health (R01CA215553), the Burroughs Wellcome Fund (1016720), the Camille & Henry Dreyfus Foundation (TC-17-011), the Yale Comprehensive Cancer Center (18-001952), and Yale University.
Footnotes
ETHICS DECLARATION. The authors declare no competing interests.
References:
- 1.Huttenhower C et al. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Secher T, Brehin C & Oswald E Early settlers: Which E. coli strains do you not want at birth? Am. J. Physiol. - Gastrointest. Liver Physiol 311, G123–G129 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Kaper JB, Nataro JP & Mobley HLT Pathogenic Escherichia coli. Nature 2, 123–140 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Ricardo R, Dorrestein PC & Quinn RA Illuminating the dark matter in metabolomics. PNAS 112, 12549–12550 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Papenfort K & Bassler BL Quorum sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol 14, 576–588 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Houdt R, Aertsen A, Moons P, Vanoirbeek K & Michiels CW N-acyl-L-homoserine lactone signal interception by Escherichia coli. FEMS Microbiol. Lett 256, 83–89 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Lee J, Maeda T, Hong SH & Wood TK Reconfiguring the quorum-sensing regulator sdiA of escherichia coli to control biofilm formation via indole and N-acylhomoserine lactones. Appl. Environ. Microbiol 75, 1703–1716 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nguyen Y et al. Structural and Mechanistic Roles of Novel Chemical Ligands on the SdiA Quorum-Sensing Transcription Regulator. MBio 6, e02429–14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorelik O et al. Vibrio cholerae autoinducer-1 enhances the virulence of enteropathogenic Escherichia coli. Sci. Rep 9, 1–13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schauder S, Shokat K, Surette MG & Bassler BL The LuxS family of bacterial autoinducers: Biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol 41, 463–476 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Pereira CS, Thompson JA & Xavier KB AI-2-mediated signalling in bacteria. FEMS Microbiol. Rev 37, 156–181 (2013). [DOI] [PubMed] [Google Scholar]
- 12.González Barrios AF et al. Autoinducer 2 Controls Biofilm Formation in Escherichia coli through a Novel Motility Quorum-Sensing Regulator ( MqsR , B3022 ). J. Bacteriol 188, 305–316 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laganenka L, Colin R & Sourjik V Chemotaxis towards autoinducer 2 mediates autoaggregation in Escherichia coli. Nat. Commun 7, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ha JH et al. Evidence of link between quorum sensing and sugar metabolism in Escherichia coli revealed via cocrystal structures of LsrK and HPr. Sci. Adv 4, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zargar A et al. Bacterial Secretions of Nonpathogenic Escherichia coli Elicit Inflammatory Pathways : a Closer Investigation of Interkingdom. MBio 6, 1–10 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ismail AS, Valastyan JS & Bassler BL A Host-Produced Autoinducer-2 Mimic Activates Bacterial Quorum Sensing. Cell Host Microbe 19, 470–480 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sperandio V, Torres AG, Jarvis B, Nataro JP & Kaper JB Bacteria-host communication: The language of hormones. Proc. Natl. Acad. Sci 100, 8951–8956 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furniss RCD & Clements A Regulation of the locus of enterocyte effacement in attaching and effacing pathogens. J. Bacteriol 200, 1–12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bearson BL & Bearson SMD The role of the QseC quorum-sensing sensor kinase in colonization and norepinephrine-enhanced motility of Salmonella enterica serovar Typhimurium. Microb. Pathog 44, 271–278 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Njoroge J & Sperandio V Enterohemorrhagic escherichia coli virulence regulation by two bacterial adrenergic kinases, QseC and QseE. Infect. Immun 80, 688–703 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim CS et al. Characterization of Autoinducer-3 Structure and Biosynthesis in E. coli . ACS Cent. Sci (2020) doi: 10.1021/acscentsci.9b01076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carlson-Banning KM & Sperandio V Enterohemorrhagic Escherichia coli outwits hosts through sensing small molecules. Curr. Opin. Microbiol 41, 83–88 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lopez CA et al. Virulence factors enhance Citrobacter rodentium expansion through aerobic respiration. Science 353, 1249–1254 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Papenfort K et al. A Vibrio cholerae autoinducer-receptor pair that controls biofilm formation. Nat. Chem. Biol 13, 551–557 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Silva-Junior EA et al. Pyrazines from bacteria and ants: Convergent chemistry within an ecological niche. Sci. Rep 8, 4–10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee JH & Lee J Indole as an intercellular signal in microbial communities. FEMS Microbiol. Rev 34, 426–444 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Zarkan A, Liu J, Matuszewska M, Gaimster H & Summers DK Local and Universal Action: The Paradoxes of Indole Signalling in Bacteria. Trends Microbiol. xx, 1–12 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Bansal T et al. Differential Effects of Epinephrine, Norepinephrine, and Indole on Escherichia coli O157:H7 Chemotaxis, Colonization, and Gene Expression. Infect. Immun 75, 4597–4607 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar A & Sperandio V Indole Signaling at the Host-Microbiota-Pathogen Interface. BMC Microbiol. 10, e01031–19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kohli N et al. The microbiota metabolite indole inhibits Salmonella virulence: Involvement of the PhoPQ two-component system. PLoS One 13, 1–19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar A et al. The Serotonin Neurotransmitter Modulates Virulence of Enteric Pathogens. Cell Host Microbe 28, 41–52.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Esmaili A et al. Enteropathogenic Escherichia coli Infection Inhibits Intestinal Serotonin Transporter Function and Expression. Gastroenterology 137, 2074–2083 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bansal T, Alaniz RC, Wood TK & Jayaraman A The bacterial signal indole increases epithelial-cell tight-junction resistance and attenuates indicators of inflammation. Proc. Natl. Acad. Sci. United States 107, 228–233 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shimada Y et al. Commensal bacteria-dependent indole production enhances epithelial barrier function in the colon. PLoS One 8, 1–10 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chimerel C et al. Bacterial Metabolite Indole Modulates Incretin Secretion from Intestinal Enteroendocrine L Cells. Cell Rep. 9, 1202–1208 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holst JJ The physiology of glucagon-like peptide 1. Physiol. Rev 87, 1409–1439 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Roager HM & Licht TR Microbial tryptophan catabolites in health and disease. Nat. Commun 9, 1–10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beaumont M et al. The gut microbiota metabolite indole alleviates liver inflammation in mice. FASEB J. 32, 6681–6693 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith EA & Macfarlane GT Enumeration of human colonie bacteria producing phenolic and indolic compounds : Effects of pH, carbohydrate availability and retention time on dissimilatory aromatic amino acid metabolism. J. Appl. Bacteriol 81, 288–302 (1996). [DOI] [PubMed] [Google Scholar]
- 40.Liu Y, Hou Y, Wang G, Zheng X & Hao H Gut Microbial Metabolites of Aromatic Amino Acids as Signals in Host–Microbe Interplay. Trends Endocrinol. Metab 1–17 (2020) doi: 10.1016/j.tem.2020.02.012. [DOI] [PubMed] [Google Scholar]
- 41.Rothhammer V & Quintana FJ The aryl hydrocarbon receptor: an environmental sensor integrating immune responses in health and disease. Nat. Rev. Immunol 19, 184–197 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Li Y et al. Exogenous stimuli maintain intraepithelial lymphocytes via aryl hydrocarbon receptor activation. Cell 147, 629–640 (2011). [DOI] [PubMed] [Google Scholar]
- 43.Cervantes-Barragan L et al. Lactobacillus reuteri induces gut intraepithelial CD4+CD8αα+ T cells. Science 357, 806–810 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilck N et al. Salt-responsive gut commensal modulates TH17 axis and disease. Nature 551, 585–589 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu C et al. Induction of pathogenic TH 17 cells by inducible salt-sensing kinase SGK1. Nature 496, 513–517 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang J, Sundrud MS, Skepner J & Yamagata T Targeting Th17 cells in autoimmune diseases. Trends Pharmacol. Sci 35, 493–500 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Goettel JA et al. AHR Activation Is Protective against Colitis Driven by T Cells in Humanized Mice. Cell Rep. 17, 1318–1329 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Monteleone I et al. Aryl hydrocarbon receptor-induced signals up-regulate IL-22 production and inhibit inflammation in the gastrointestinal tract. Gastroenterology 141, 237–248.e1 (2011). [DOI] [PubMed] [Google Scholar]
- 49.Lamas B et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat. Med 22, 598–605 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krishnan S et al. Gut Microbiota-Derived Tryptophan Metabolites Modulate Inflammatory Response in Hepatocytes and Macrophages. Cell Rep. 23, 1099–1111 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Giani Tagliabue S, Faber SC, Motta S, Denison MS & Bonati L Modeling the binding of diverse ligands within the Ah receptor ligand binding domain. Sci. Rep 9, 1–14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Denison MS & Faber SC And now for something completely different: Diversity in ligand-dependent activation of Ah receptor responses. Curr. Opin. Toxicol 2, 124–131 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim CS et al. Cellular Stress Upregulates Indole Signaling Metabolites in Escherichia coli. Cell Chem. Biol 1–10 (2020) doi: 10.1016/j.chembiol.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Park HB et al. Sulfamethoxazole drug stress upregulates antioxidant immunomodulatory metabolites in Escherichia coli. Nature Microbiology vol. 5 1319–1329 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arnison PG et al. Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Nat. Prod. Rep 30, 108–160 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hetrick KJ & van der Donk WA Ribosomally synthesized and post-translationally modified peptide natural product discovery in the genomic era. Curr. Opin. Chem. Biol 38, 36–44 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Duquesne S, Destoumieux-Garzón D, Peduzzi J & Rebuffat S Microcins, gene-encoded antibacterial peptides from enterobacteria. Nat. Prod. Rep 24, 708–734 (2007). [DOI] [PubMed] [Google Scholar]
- 58.Baquero F, Lanza VF, Baquero MR, del Campo R & Bravo-Vázquez DA Microcins in Enterobacteriaceae: Peptide Antimicrobials in the Eco-Active Intestinal Chemosphere. Front. Microbiol 10, 1–25 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sassone-Corsi M et al. Microcins mediate competition among Enterobacteriaceae in the inflamed gut. Nature 540, 280–283 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Soudy R, Etayash H, Bahadorani K, Lavasanifar A & Kaur K Breast Cancer Targeting Peptide Binds Keratin 1: A New Molecular Marker for Targeted Drug Delivery to Breast Cancer. Mol. Pharm 14, 593–604 (2017). [DOI] [PubMed] [Google Scholar]
- 61.Iyer SS et al. Dietary and Microbial Oxazoles Induce Intestinal Inflammation by Modulating Aryl Hydrocarbon Receptor Responses. Cell 173, 1123–1134.e11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lanis JM et al. Tryptophan metabolite activation of the aryl hydrocarbon receptor regulates IL-10 receptor expression on intestinal epithelia. Mucosal Immunol. 10, 1133–1144 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Heller F, Fuss IJ, Nieuwenhuis EE, Blumberg RS & Strober W Oxazolone Colitis, a Th2 Colitis Model Resembling Ulcerative Colitis, Is Mediated by IL-13-Producing NK-T Cells. Immunity 17, 629–638 (2002). [DOI] [PubMed] [Google Scholar]
- 64.Skaar EP The battle for iron between bacterial pathogens and their vertebrate hosts. PLoS Pathog. 6, 1–2 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Beare PA, For RJ, Martin LW & Lamont IL Siderophore-mediated cell signalling in Pseudomonas aeruginosa: Divergent pathways regulate virulence factor production and siderophore receptor synthesis. Mol. Microbiol 47, 195–207 (2003). [DOI] [PubMed] [Google Scholar]
- 66.Loomis LD & Raymond KN Solution Equilibria of Enterobactin and Metal-Enterobactin Complexes. Inorg. Chem 30, 906–911 (1991). [Google Scholar]
- 67.Scholz RL & Greenberg EP Sociality in Escherichia coli: Enterochelin is a private good at low cell density and can be shared at high cell density. J. Bacteriol 197, 2122–2128 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kramer J, Özkaya Ö & Kümmerli R Bacterial siderophores in community and host interactions. Nat. Rev. Microbiol (2019) doi: 10.1038/s41579-019-0284-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brickman TJ & Armstrong SK Temporal signaling and differential expression of Bordetella iron transport systems: The role of ferrimones and positive regulators. BioMetals 22, 33–41 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Singh V et al. Interplay between enterobactin, myeloperoxidase and lipocalin 2 regulates E. Coli survival in the inflamed gut. Nat. Commun 6, 1–11 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Garénaux A, Caza M & Dozois CM The Ins and Outs of siderophore mediated iron uptake by extra-intestinal pathogenic Escherichia coli. Vet. Microbiol 153, 89–98 (2011). [DOI] [PubMed] [Google Scholar]
- 72.Qi B & Han M Microbial Siderophore Enterobactin Promotes Mitochondrial Iron Uptake and Development of the Host via Interaction with ATP Synthase. Cell 175, 571–582.e11 (2018). [DOI] [PubMed] [Google Scholar]
- 73.Nougayrède J-P et al. Escherichia coli Induces DNA Double-Strand Breaks in Eukaryotic Cells. Science 313, 848–852 (2006). [DOI] [PubMed] [Google Scholar]
- 74.Cuevas-Ramos G et al. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc. Natl. Acad. Sci. U. S. A 107, 11537–11542 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Arthur JC et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338, 120–123 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Buc E et al. High Prevalence of Mucosa-Associated E. coli Producing Cyclomodulin and Genotoxin in Colon Cancer. PLoS One 8, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Arthur JC et al. Microbial genomic analysis reveals the essential role of inflammation in bacteria-induced colorectal cancer. Nat. Commun 5, 1–11 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Taieb F, Petit C, Nougayrède J-P & Oswald E The Enterobacterial Genotoxins: Cytolethal Distending Toxin and Colibactin. EcoSal Plus 7, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Faïs T, Delmas J, Barnich N, Bonnet R & Dalmasso G Colibactin: More than a new bacterial toxin. Toxins (Basel). 10, 16–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wernke KM et al. Structure and bioactivity of colibactin. Bioorganic Med. Chem. Lett 30, 127280 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bossuet-Greif N et al. The colibactin genotoxin generates DNA interstrand cross-links in infected cells. MBio 9, 1–15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xue M et al. Structure elucidation of colibactin and its DNA cross-links. Science 365, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wilson MR et al. The human gut bacterial genotoxin colibactin alkylates DNA. Science 363, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pleguezuelos-manzano C et al. Mutational signature in colorectal cancer caused by genotoxic pks + E . coli. Nature 580, 269–273 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dziubańska-Kusibab PJ et al. Colibactin DNA-damage signature indicates mutational impact in colorectal cancer. Nat. Med. 26, (2020). [DOI] [PubMed] [Google Scholar]
- 86.Massip C et al. Deciphering the interplay between the genotoxic and probiotic activities of Escherichia coli Nissle 1917. PLoS Pathog. 15, e1008029 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tripathi P et al. ClbS Is a Cyclopropane Hydrolase That Confers Colibactin Resistance. J. Am. Chem. Soc 139, 17719–17722 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shine EE et al. Model Colibactins Exhibit Human Cell Genotoxicity in the Absence of Host Bacteria. ACS Chem. Biol 13, 3286–3293 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Martin P, Tronnet S, Garcie C & Oswald E Interplay between siderophores and colibactin genotoxin in Escherichia coli. IUBMB Life 69, 435–441 (2017). [DOI] [PubMed] [Google Scholar]
- 90.Francisco Nualart RB Preclinical and Clinical Studies for Sodium Tungstate: Application in Humans. J. Clin. Cell. Immunol 06, 1–12 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhu W et al. Precision editing of the gut microbiota ameliorates colitis. Nature 553, 208–211 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cougnoux A et al. Small-molecule inhibitors prevent the genotoxic and protumoural effects induced by colibactin-producing bacteria. Gut 65, 278–285 (2016). [DOI] [PubMed] [Google Scholar]
- 93.Povolotsky TL & Hengge R ‘Life-style’ control networks in Escherichia coli: Signaling by the second messenger c-di-GMP. J. Biotechnol 160, 10–16 (2012). [DOI] [PubMed] [Google Scholar]
- 94.Jenal U, Reinders A & Lori C Cyclic di-GMP: Second messenger extraordinaire. Nat. Rev. Microbiol 15, 271–284 (2017). [DOI] [PubMed] [Google Scholar]
- 95.Davies BW, Bogard RW, Young TS & Mekalanos JJ Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell 149, 358–370 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Whiteley AT et al. Bacterial cGAS-like enzymes synthesize diverse nucleotide signals. Nature 567, 194–199 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cohen D et al. Cyclic GMP–AMP signalling protects bacteria against viral infection. Nature 574, 691–695 (2019). [DOI] [PubMed] [Google Scholar]
- 98.McFarland AP et al. Sensing of Bacterial Cyclic Dinucleotides by the Oxidoreductase RECON Promotes NF-κB Activation and Shapes a Proinflammatory Antibacterial State. Immunity 46, 433–445 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Scallan E et al. Foodborne illness acquired in the United States-Major pathogens. Emerg. Infect. Dis 17, 7–15 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Martinson JNV et al. Rethinking gut microbiome residency and the Enterobacteriaceae in healthy human adults. ISME J. 13, 2306–2318 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]