Abstract
Pulsed-field gel electrophoresis (PFGE) and coagulase gene restriction profile (CRP) analysis techniques were used to analyze 71 Staphylococcus aureus isolates recovered from nine food-borne disease outbreaks. Twenty-two PFGE profiles and 11 CRPs were identified, with discrimination indices of 0.86 and 0.72, respectively. In addition, the variable regions of the coagulase genes of 39 isolates were sequenced and showed extensive identity, indicating that this is not an efficient alternative for the molecular typing of S. aureus.
Staphylococcus aureus is one of the major causative agents of food poisoning. In Taiwan, S. aureus was responsible for 30% of cases of bacterial food-borne outbreaks between 1986 and 1995 (17). Staphylococcal food poisoning is characterized by nausea, vomiting, abdominal pain, and diarrhea and has an incubation period of 30 min to 8 h after ingestion of the contaminated food (8). Enterotoxins produced by the bacteria are believed to be wholly responsible for the symptoms of food poisoning (3); therefore, only enterotoxigenic strains of S. aureus are thought to be able to cause food poisoning. To date, nine S. aureus enterotoxins, designated SEA, SEB, SEC, SED, SEE, SEG, SEH, SEI, and SEJ, have been identified (4, 16, 22, 26). The first five of these (SEA through SEE [SEA-E]) can be detected with commercially available antisera. Tests with such antisera are routinely performed with staphylococcal isolates in the laboratories of the Taiwanese health department in order to confirm the source of a food-borne outbreak. However, while these tests can identify SEA-E-producing isolates, they do not address the possibility that non-SEA-E-producing isolates may be the cause of a food-poisoning outbreak. Another problem of S. aureus identification is that, because the scale of food-borne outbreaks in which S. aureus could be involved is frequently small, only very limited numbers of isolates are usually recovered.
Molecular typing of staphylococcal isolates can provide useful clonality information for confirmation of a staphylococcal food-borne outbreak. Numerous such methods for S. aureus typing have been described (5, 6, 7, 12, 19, 23, 25, 27). Among these methods, pulsed-field gel electrophoresis (PFGE) has been demonstrated to have advantages in discriminatory power, typeability, and reproducibility and has been taken as the “gold standard” for the typing of S. aureus (2, 18, 20), even though it is labor-intensive and time-consuming. Compared to PFGE, coagulase gene restriction profile (CRP) analysis, a PCR-based method, is easy to perform and has high levels of specimen typeability and reproducibility, and it has been used successfully for the typing of a large number of methicillin-resistant S. aureus isolates (9, 13). In this study, we compared PFGE and CRP analysis for the characterization of staphylococcal isolates recovered from nine food-borne outbreaks. The relationship between the staphylococcal isolates and the food-borne outbreaks is also discussed.
MATERIALS AND METHODS
Bacterial strains.
Bacterial isolates were recovered from rectal swabs of patients and from nasal and hand swabs of suspected food handlers from nine food-borne disease outbreaks in central Taiwan between 1995 and 1997. The specimens were streaked onto Baird-Parker agar plates (Merck Taiwan Ltd., Taichung City, Taiwan), and the plates were incubated at 35°C for 24 h. Two or three colonies were picked and subcultured onto nutrient agar plates (Eiken Chemical Co., Tokyo, Japan). The bacteria were tested with staphylase agglutination testing kits (Oxoid Unipath, Hampshire, England), and the bacteria that tested positive were considered to be S. aureus. A total of 71 isolates were obtained. The isolates were immediately tested for the presence of enterotoxins (see below) and were then kept in 15% glycerol at −70°C until use.
Detection of toxins by RPLA.
Staphylococcal isolates were screened for the expression of SEA, SEB, SEC, SED, and SEE by reverse passive latex agglutination (RPLA) according to the manufacturer's instructions (Denka Seiken, Co., Tokyo, Japan).
Detection of toxin genes by PCR.
Isolates were examined for the presence of sea, seb, sec, sed, and see by PCR, according to the work of Johnson and colleagues (11).
CRP analysis.
Amplification of the repeated region of the coagulase gene by PCR was performed as described by Goh and colleagues (7), except that a new forward primer, primer COAG-5 (5′-GGTATTCGTGAATACAACGATGGAA-3′), located 40 bp upstream from COAG-2, was used in the reaction with primer COAG-3 (5′-AAAGAAAACCACTCACATCA-3′). Restriction profiles were determined by digesting the amplified fragment with AluI (Promega Corp., Madison, Wis.) and separating the DNA fragments in 2.5% agarose gels.
DNA sequencing and data analysis.
The PCR-amplified DNA was sequenced (done by MB Mission Biotech Corp., Taipei, Taiwan) with COAG-5 as the primer. About 700 bp of each sequence was determined in a single gel run, and 560 bp was used for sequence analysis. The GAP program of the GCG package (version 9) was used to align sequences in order to determine the maximum identity.
PFGE.
Preparation and SmaI restriction treatment of the genomic DNA for PFGE analysis were performed by a rapid preparation procedure (15). DNA fragments were separated in 0.9% agarose gels at 13°C with a Rotaphor type V apparatus (Biometra, Göttingen, Germany) in 0.5× TBE buffer (0.045 M Tris-borate, 0.1 mM EDTA [pH 8.3]). Electrophoresis was performed at a fixed voltage (190 V), at a 120° fixed angle, and with pulse time intervals from 8 to 14 s for 10 h and then from 19 to 28 s for 12 h. A ladder of bacteriophage λ DNA concatemers (New England BioLabs, Inc., Beverly, Mass.) was included to estimate the sizes of the DNA fragments. The PFGE pattern was interpreted in accordance with a previously published guideline (24).
Statistics.
Discriminatory power was evaluated by use of the discrimination index, as described by Hunter (10).
Epidemiological data.
Epidemiological data relating to the food-borne disease outbreaks being investigated were obtained from standardized case report forms filled in by the county public health authority. The reports included the dates, times, and locations of suspected food ingestion, the suspected foods, the persons involved in eating and poisoning, and basic information about each patient's sex, age, date and time of onset, residency, symptoms, and medical treatment.
RESULTS
Production of enterotoxins.
A total of 71 S. aureus isolates were screened for the expression of enterotoxin. First, isolates recovered from the specimens were immediately subjected to screening by RPLA. A total of 17 isolates were identified in this process: 15 isolates from outbreaks 1, 2, and 3 and 2 isolates from outbreak 7, which produced SEC, SED, SEA, and SEA in the four outbreaks, respectively (Table 1). Second, isolates revived from stocks stored at −70°C prior to further molecular characterization were also screened. In the second test, a total of 29 isolates were detected. In addition to the 17 isolates detected in the first screening, an additional 12 isolates from six outbreaks were identified. Ten of these 12 isolates produced SEC, while the other 2 isolates were found to produce SEA and SEB, respectively.
TABLE 1.
Phenotypes and genotypes of S. aureus isolates from the food-borne disease outbreaks
| Outbreak | Date, location | Incubation period (median) | Isolate codea | Toxin productionb
|
Toxin genec | PFGE profiled | CRP | Sequence (no. of bp differences)e | |
|---|---|---|---|---|---|---|---|---|---|
| First RPLA | Second RPLA | ||||||||
| 1 | 18 March 1995, Nantou County | 1 h–1 h and 10 min (1 h) | 398 | SEC | SEC | sec | 3 | 3 | Seq3 (14) |
| 400 | SEC | SEC | sec | 4 | 4 | Seq4 | |||
| 404 | SEC | SEC | sec | 4 | 4 | ||||
| 405 | SEC | SEC | sec | 4 | 4 | ||||
| 2 | 8 June 1995, Nantou County | 4 h and 30 min–9 h and 30 min (6 h) | 191 | SED | SED | sed | 1 | 1 | Seq1 |
| 199 | SED | SED | sed | 1 | 1 | ||||
| 206∗ | SED | SED | sed | 1 | 1 | Seq1 | |||
| 193 | SED | SED | sed | 2 | 2 | Seq2 | |||
| 200 | SED | SED | sed | 2 | 2 | ||||
| 3 | 12 September 1995, Taichung County | 1 h and 20 min–2 h and 40 min (2 h and 30 min) | 620 | SEA | SEA | sea | 6 | 3 | Seq3 |
| 621 | SEA | SEA | sea | 6 | 3 | ||||
| 622 | SEA | SEA | sea | 6 | 3 | ||||
| 623 | SEA | SEA | sea | 6 | 3 | ||||
| 625 | SEA | SEA | sea | 6 | 3 | ||||
| 627∗ | SEA | SEA | sea | 6 | 3 | Seq3 | |||
| 4 | 24 October 1995, Miauoli County | 1 h–2 h (2 h) | 934 | — | — | — | 5 | 5 | Seq5 |
| 936 | — | — | — | 5 | 5 | Seq5 | |||
| 940 | — | — | — | 5 | 5 | Seq5 | |||
| 941 | — | — | — | 13 | 5 | Seq5 | |||
| 947 | — | — | — | 8 | 6 | Seq6 | |||
| 946 | — | SEC | sec | 7a | 8 | Seq8 | |||
| 953-1∗ | — | — | — | 10a | 5 | Seq5 | |||
| 953-2∗ | — | — | — | 10b | 5 | Seq5 | |||
| 953-3∗ | — | — | — | 9 | 7 | Seq7 | |||
| 5 | 5 January 1996, Taichung County | 30 min–20 h (5 h and 30 min) | 1120 | — | — | — | 11a | 5 | Seq5 |
| 1121 | — | — | — | 11a | 5 | Seq5 | |||
| 1126 | — | — | — | 11b | 5 | Seq5 | |||
| 1130 | — | — | — | 11b | 5 | Seq5 | |||
| 1131 | — | SEC | sec | 7a | 8 | Seq8 | |||
| 6 | 12 January 1996, Taichung County | 1 h–20 h and 30 min (5 h and 30 min) | 1172 | — | — | — | 11a | 5 | Seq5 |
| 1173 | — | — | — | 11a | 5 | Seq5 | |||
| 1174 | — | — | — | 11a | 5 | Seq5 (4) | |||
| 1176 | — | — | — | 11a | 5 | ||||
| 1180 | — | — | — | 11a | 5 | ||||
| 1181 | — | — | — | 11a | 5 | ||||
| 1183 | — | — | — | 11a | 5 | ||||
| 1184 | — | — | — | 11a | 5 | ||||
| 1185 | — | — | — | 11a | 5 | ||||
| 1187 | — | — | — | 11a | 5 | ||||
| 1189 | — | — | — | 11a | 5 | ||||
| 1190 | — | — | — | 11a | 5 | ||||
| 1191 | — | — | — | 11a | 5 | ||||
| 1194 | — | — | — | 11a | 5 | ||||
| 1195 | — | — | — | 11a | 5 | ||||
| 1199 | — | — | — | 11a | 5 | ||||
| 1200 | — | — | — | 11a | 5 | ||||
| 1202 | — | — | — | 11a | 5 | ||||
| 1203 | — | — | — | 11a | 5 | ||||
| 1197 | — | — | — | 12 | 5 | Seq5 | |||
| 1179 | — | SEC | sec | 7a | 8 | Seq8 | |||
| 1186 | — | SEC | sec | 7a | 8 | ||||
| 7 | 8 January 1997, Taichung County | 1 h–5 h (2 h and 30 min) | 4807 | SEA | SEA | sea | 14a | 3 | Seq3 |
| 4811 | SEA | SEA | sea | 14a | 3 | ||||
| 4813 | — | SEC | sec | 7b | 8 | Seq8 | |||
| 8 | 12 March 1997, Taichung County | 4 h–17 h (13 h) | 5126 | — | — | — | 11a | 5 | Seq5 (2) |
| 5166 | — | — | — | 11a | 5 | Seq5 | |||
| 5180 | — | — | — | 11a | 5 | Seq5 (2) | |||
| 5190 | — | — | — | 6 | 3 | ||||
| 5174 | — | — | — | 14b | 3 | Seq3 (1) | |||
| 5178 | — | SEC | sec | 7a | 8 | ||||
| 5199 | — | — | — | 15 | 9 | Seq9 | |||
| 9 | 17 March 1997, Taichung County | 1 h–25 h and 45 min (8 h and 30 min) | 5235 | — | — | — | 17 | 7 | Seq7 |
| 5224 | — | — | — | 17 | 7 | ||||
| 5239 | — | — | — | 11c | 5 | Seq5 | |||
| 5231-1 | — | SEB | seb | 11c | 5 | ||||
| 5231-2 | — | SEC | sec | 7a | 8 | Seq8 | |||
| 5242-1 | — | SEA | sea | 6 | 3 | ||||
| 5242-2 | — | SEC | sec | 7a | 8 | Seq8 | |||
| 5233 | — | SEC | sec | 7a | 8 | ||||
| 5222 | — | SEC | sec | 16 | 10 | Seq10 | |||
| 5220 | — | — | — | 6 | 11 | Seq11 | |||
| Discrimination index | 0.86 | 0.72 | |||||||
∗, isolates recovered from food handlers.
—, no SEA, SEB, SEC, SED, or SEE was detected by the RPLA test.
—, no sea, seb, sec, sed, or see gene was detected.
The major PFGE types are designated by numerals as patterns that differed by at least four DNA fragments; minor PFGE subtypes are designated by letter suffixes as patterns that differed by at most three DNA fragments.
Each sequence type differs from the others by less than 97% identity over 560 bp.
Toxin genes.
The types of toxin genes carried by isolates as detected by PCR were concordant with the types of expressed toxins as determined in the second RPLA test (Table 1). Of the 71 isolates, 29 carried one of the toxin genes (sea, seb, sec, sed, or sed), and 42 isolates did not carry sea, seb, sec, sed, or see.
Genotypes.
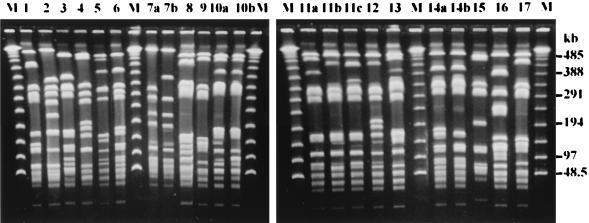
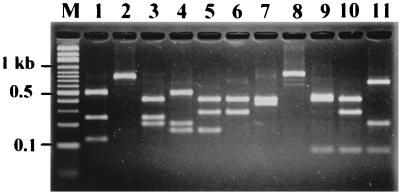
Screening by PFGE and CRP analysis grouped the 71 isolates into 22 PFGE types and 11 CRP types (Table 1). The PFGE profiles and CRPs are shown in Fig. 1 and 2, respectively. The typeabilities for both techniques were 100%. PFGE produced better discrimination than CRP analysis, with discrimination indices of 0.86 and 0.72, respectively. PFGE further divided CRP-3 into four PFGE types (types 3, 6, 14a, and 14b), CRP-5 into eight PFGE types (types 5, 10a, 10b, 11a, 11b, 11c, 12, and 13), CRP-7 into two PFGE types (types 9 and 17), and CRP-8 into two PFGE types (types 7a and 7b). In contrast, only PFGE type 6 could be separated into two CRPs (CRP-3 and CRP-11). In individual outbreaks, both methods showed the same discriminatory ability for isolates from outbreaks 1, 2, 3, and 7, but PFGE showed a higher discriminatory ability than CRP analysis for isolates from outbreaks 4, 5, 6, 8, and 9 (Table 1).
FIG. 1.
PFGE profiles of S. aureus isolates. Chromosomal DNA was digested with SmaI, and the fragments were fractionated on a 0.9% agarose gel. The numbers above the lanes represent the assigned PFGE types and subtypes (1 to 17). Lanes M, bacteriophage lambda molecular weight markers.
FIG. 2.
CRPs of S. aureus isolates. PCR amplicons of the 3′ region of the coagulase gene were digested with AluI and were then fractionated on a 2.5% agarose gel. The numbers above the lanes represent different CRPs (1 to 11). Lane M, molecular weight marker.
Variable-region sequences of coagulase genes.
To determine whether sequencing of the variable segment of the coagulase gene can be used as an alternative method for molecular typing of S. aureus isolates, the 3′ variable regions of the coagulase genes of 39 isolates, representing all CRP and PFGE types, were sequenced. Sequences of 560 bp with the same starting base pair were used for analysis and comparison. Eleven unique sequences, designated Seq1 through Seq11, were identified (Table 1). Among the sequences, 63 to 97% identities were found (data not shown). With only a few exceptions, sequences of more than 560 bp from isolates with the same CRP were identical. The most divergence was found in the sequence for the CRP-3 and PFGE type 3 isolate, which showed a 14-bp difference in Seq3. The rest showed mismatches of only 1 to 4 bp.
DISCUSSION
PFGE, because of its great discriminatory power and high degree of specimen typeability, is accepted as the gold standard for the molecular typing of S. aureus isolates. It has successfully been used to study the epidemiology of S. aureus nosocomial infection and methicillin resistance (12, 25). In this study, PFGE also exhibited superiority as a technique for the identification of S. aureus isolates collected from food-borne disease outbreaks. The discrimination index (0.86) for PFGE was found to be much higher than that for CRP analysis (0.72) (Table 1). Nevertheless, PFGE is time-consuming and labor-intensive and can be performed only in reference laboratories with skillful technicians. Due to these drawbacks, PFGE is not an ideal typing method for health departments undertaking routine analysis of large numbers of S. aureus isolates in the investigation of food-borne disease outbreaks in which S. aureus is the suspected causative agent.
In contrast, PCR-based CRP analysis is fast and easy to manipulate. As the data in this study show, CRP analysis exhibits the same discriminatory power as PFGE for SEA-E-producing isolates. However, CRP analysis is much less discriminatory than PFGE in the case of non-SEA-E-producing isolates, and its results could therefore lead to the misjudging of the causes of some outbreaks (for example, outbreaks 4 and 5 in this study [Table 1]). Thus, while CRP analysis has been used successfully in several epidemiological studies of S. aureus (9, 13), one study has indicated that it is insufficient as a sole method for the typing of staphylococcal isolates (21). Nevertheless, by combining CRP analysis with other fast PCR-based analytical techniques such as protein A gene typing (6) or inter-IS256-PCR typing (5), this methodology may be able to meet the needs of health departments. In any event, the development of a fast and highly discriminatory typing method for S. aureus remains an important focus.
It has been hypothesized that the determination of the nucleotide sequence of the highly variable repeated region of the coagulase gene could be an alternative method for the typing of S. aureus isolates (21). However, our results from the sequencing of 39 of the 71 isolates show that the variable region of the coagulase gene is much more conserved than expected. Even though isolates were recovered from different disease outbreaks, most isolates with the same CRP differed by only 1 to 4 bp over a 560-bp sequence. This indicates that sequencing of the variable segment of the coagulase gene may not be an efficient method for the typing of S. aureus isolates.
In our experience, most staphylococcal food-borne disease outbreaks occur on small scales. As such, in most cases only a few staphylococcal isolates are obtained, leading to difficulty in identifying the causative agent. Since toxigenic strains of S. aureus are quite prevalent in the general population (1), it is easy to misdiagnose the cause of a food-borne disease outbreak as being S. aureus on the basis of routine enterotoxin analysis of limited isolates. Characterization of staphylococcal isolates recovered from patients and food handlers by various techniques, in particular, molecular typing methods, can provide useful information in investigations of the relatedness of bacterial isolates and disease outbreaks. In outbreaks 1, 2, 3, and 7, only limited numbers of isolates were recovered. Yet, using PFGE and CRP analysis, we traced the contamination sources of two outbreaks (outbreaks 2 and 3) to food handlers and obtained sufficient evidence to identify enterotoxigenic isolates as the causes of the outbreaks.
One of the criteria for confirmation of staphylococcal food poisoning is whether the recovered isolates are enterotoxigenic. As only SEA-E antisera are available for testing, staphylococcal isolates are routinely tested for production of SEA-E only in the laboratories of the public health department in Taiwan. Consequently, non-SEA-E-producing S. aureus strains are not considered etiologic organisms for food poisoning. Our typing results indicate that a non-SEA-E-producing strain (CRP-5, PFGE type 11a) may be strongly implicated in outbreak 6 and possibly in a further two outbreaks. Staphylococcal enterotoxins belong to a big superantigen family (14). In addition to SEA-E, four new enterotoxins (SEG, SEH, SEI, and SEJ) have recently been characterized (16, 22, 26). Some non-SEA-E-producing strains presented in our study appear to be implicated in food poisoning and are likely to belong to one of these new groups of enterotoxin producers.
The identification of a new enterotoxin requires complicated biochemical procedures and bioassays, such as emetic responses in rhesus monkeys and stimulation of murine T-cell proliferation (16), which are not easily conducted in general laboratories. Molecular typing provides a way of linking non-SEA-E-producing strains of S. aureus with food-poisoning outbreaks and helps to select potential candidates which may be new enterotoxin producers.
In conclusion, molecular typing of S. aureus isolates provides useful information in the investigation of food-borne disease outbreaks. The results of this study demonstrate that PFGE is a more precise method for molecular typing of S. aureus isolates than CRP analysis. However, to meet the requirements of health departments undertaking routine typing of large numbers of S. aureus isolates, a faster and more discriminatory method of molecular typing is needed. Sequencing of variable regions of the coagulase gene in S. aureus isolates would not appear to be an efficient alternative, but other methods combined with CRP analysis may achieve this aim.
ACKNOWLEDGMENTS
This work was supported by grant DOH87-TD-1095 from the Department of Health of Taiwan.
We thank S. Y. Li, of the Vaccine Research & Production Center of the Center for Disease Control, for critical review of this manuscript.
REFERENCES
- 1.al Bustan M A, Udo E E, Chugh T D. Nasal carriage of enterotoxin-producing Staphylococcus aureus among restaurant workers in Kuwait City. Epidemiol Infect. 1996;116:319–322. doi: 10.1017/s0950268800052638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bannerman T L, Hancock G A, Tenover F C, Miller J M. Pulsed-field gel electrophoresis as a replacement for bacteriophage typing of Staphylococcus aureus. J Clin Microbiol. 1995;33:551–555. doi: 10.1128/jcm.33.3.551-555.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergdoll M S. Staphylococcus aureus. In: Doyle M P, editor. Bacterial foodborne pathogens. New York, N.Y: Marcel Dekker, Inc.; 1989. pp. 464–523. [Google Scholar]
- 4.Betley M J, Borst D W, Regassa L B. Staphylococcal enterotoxins, toxic shock syndrome toxin and streptococcal pyrogenic exotoxins: a comparative study of their molecular biology. Chem Immunol. 1992;55:1–35. [PubMed] [Google Scholar]
- 5.Deplano A, Vaneechoutte M, Verschraegen G, Struelens M J. Typing of Staphylococcus aureus and Staphylococcus epidermidis strains by PCR analysis of inter-IS256 spacer length polymorphisms. J Clin Microbiol. 1997;35:2580–2587. doi: 10.1128/jcm.35.10.2580-2587.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frenay H M, Theelen J P, Schouls L M, Vandenbroucke-Grauls C M, Verhoef J, van Leeuwen W J, Mooi F R. Discrimination of epidemic and nonepidemic methicillin-resistant Staphylococcus aureus strains on the basis of protein A gene polymorphism. J Clin Microbiol. 1994;32:846–847. doi: 10.1128/jcm.32.3.846-847.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goh S H, Byrne S K, Zhang J L, Chow A W. Molecular typing of Staphylococcus aureus on the basis of coagulase gene polymorphisms. J Clin Microbiol. 1992;30:1642–1645. doi: 10.1128/jcm.30.7.1642-1645.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holmberg S D, Blake P A. Staphylococcal food poisoning in the United States. New facts and old misconceptions. JAMA. 1984;251:487–489. [PubMed] [Google Scholar]
- 9.Hookey J V, Richardson J F, Cookson B D. Molecular typing of Staphylococcus aureus based on PCR restriction fragment length polymorphism and DNA sequence analysis of the coagulase gene. J Clin Microbiol. 1998;36:1083–1089. doi: 10.1128/jcm.36.4.1083-1089.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hunter P R. Reproducibility and indices of discriminatory power of microbial typing methods. J Clin Microbiol. 1990;28:1903–1905. doi: 10.1128/jcm.28.9.1903-1905.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson W M, Tyler S D, Ewan E P, Ashton F E, Pollard D R, Rozee K R. Detection of genes for enterotoxins, exfoliative toxins, and toxic shock syndrome toxin 1 in Staphylococcus aureus by the polymerase chain reaction. J Clin Microbiol. 1991;29:426–430. doi: 10.1128/jcm.29.3.426-430.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumari D N, Keer V, Hawkey P M, Parnell P, Joseph N, Richardson J F, Cookson B. Comparison and application of ribosome spacer DNA amplicon polymorphisms and pulsed-field gel electrophoresis for differentiation of methicillin-resistant Staphylococcus aureus strains. J Clin Microbiol. 1997;35:881–885. doi: 10.1128/jcm.35.4.881-885.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lawrence C, Cosseron M, Mimoz O, Brun-Buisson C, Costa Y, Samii K, Duval J, Leclercq R. Use of the coagulase gene typing method for detection of carriers of methicillin-resistant Staphylococcus aureus. J Antimicrob Chemother. 1996;37:687–696. doi: 10.1093/jac/37.4.687. [DOI] [PubMed] [Google Scholar]
- 14.Marrack P, Kappler J. The staphylococcal enterotoxins and their relatives. Science. 1990;248:705–711. doi: 10.1126/science.2185544. [DOI] [PubMed] [Google Scholar]
- 15.Matushek M G, Bonten M J, Hayden M K. Rapid preparation of bacterial DNA for pulsed-field gel electrophoresis. J Clin Microbiol. 1996;34:2598–2600. doi: 10.1128/jcm.34.10.2598-2600.1996. . (Erratum, 35:536, 1997.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Munson S H, Tremaine M T, Betley M J, Welch R A. Identification and characterization of staphylococcal enterotoxin types G and I from Staphylococcus aureus. Infect Immun. 1998;66:3337–3348. doi: 10.1128/iai.66.7.3337-3348.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan T M, Wang T K, Lee C L, Chien S W, Horng C B. Food-borne disease outbreaks due to bacteria in Taiwan, 1986 to 1995. J Clin Microbiol. 1997;35:1260–1262. doi: 10.1128/jcm.35.5.1260-1262.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prevost G, Jaulhac B, Piemont Y. DNA fingerprinting by pulsed-field gel electrophoresis is more effective than ribotyping in distinguishing among methicillin-resistant Staphylococcus aureus isolates. J Clin Microbiol. 1992;30:967–973. doi: 10.1128/jcm.30.4.967-973.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richardson J F, Aparicio P, Marples R R, Cookson B D. Ribotyping of Staphylococcus aureus: an assessment using well-defined strains. Epidemiol Infect. 1994;112:93–101. doi: 10.1017/s0950268800057459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saulnier P, Bourneix C, Prevost G, Andremont A. Random amplified polymorphic DNA assay is less discriminant than pulsed-field gel electrophoresis for typing strains of methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 1993;31:982–985. doi: 10.1128/jcm.31.4.982-985.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwarzkopf A, Karch H. Genetic variation in Staphylococcus aureus coagulase genes: potential and limits for use as epidemiological marker. J Clin Microbiol. 1994;32:2407–2412. doi: 10.1128/jcm.32.10.2407-2412.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su Y C, Wong A C. Identification and purification of a new staphylococcal enterotoxin, H. Appl Environ Microbiol. 1995;61:1438–1443. doi: 10.1128/aem.61.4.1438-1443.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tambic A, Power E G, Talsania H, Anthony R M, French G L. Analysis of an outbreak of non-phage-typeable methicillin-resistant Staphylococcus aureus by using a randomly amplified polymorphic DNA assay. J Clin Microbiol. 1997;35:3092–3097. doi: 10.1128/jcm.35.12.3092-3097.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshida T, Kondo N, Hanifah Y A, Hiramatsu K. Combined use of ribotyping, PFGE typing and IS431 typing in the discrimination of nosocomial strains of methicillin-resistant Staphylococcus aureus. Microbiol Immunol. 1997;41:687–695. doi: 10.1111/j.1348-0421.1997.tb01912.x. [DOI] [PubMed] [Google Scholar]
- 26.Zhang S, Iandolo J J, Stewart G C. The enterotoxin D plasmid of Staphylococcus aureus encodes a second enterotoxin determinant (sej) FEMS Microbiol Lett. 1998;168:227–233. doi: 10.1111/j.1574-6968.1998.tb13278.x. [DOI] [PubMed] [Google Scholar]
- 27.Zuccarelli A J, Roy I, Harding G P, Couperus J J. Diversity and stability of restriction enzyme profiles of plasmid DNA from methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 1990;28:97–102. doi: 10.1128/jcm.28.1.97-102.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]